Abstract
Abnormal phosphorylation contributes to the formation of neurofibrillary tangles in Alzheimer Disease (AD), but may play other signaling roles during AD pathogenesis. In this study, we employed immobilized metal affinity chromatography (IMAC) followed by liquid chromatography-tandem mass spectrometry to identify phosphopeptides from 8 individual AD and 8 age-matched control postmortem human brain tissues. Using this approach, we identified 5,569 phosphopeptides in frontal cortex across all 16 cases in which phosphopeptides represented 80 percent of all peptide spectral counts collected following IMAC enrichment. Marker selection identified 253 significantly altered phosphopeptides by precursor intensity, changed by at least 1.75 fold relative to controls, with an empirical false discovery rate below 7 percent. Approximately 21 percent of all significantly altered phosphopeptides in AD tissue were derived from tau. Of the other 142 proteins hyperphosphorylated in AD, membrane, synapse, cell junction, and alternatively spliced proteins were overrepresented. Of these, we validated differential phosphorylation of heat-shock protein-beta-1 (HSPB1) and crystallin-alpha-B (CRYAB) as hyperphosphorylated by western blotting. We further identified a network of phosphorylated kinases, which co-enriched with phosphorylated small heat shock proteins. This supports a hypothesis that a number of kinases are regulating and/or regulated by the small heat shock protein folding network.
Keywords: Immobilized metal-affinity chromatography (IMAC), mass spectrometry, neurodegeneration, proteostasis
1. Introduction
Protein phosphorylation is a central regulatory mechanism of protein function in the developing and mature central nervous system and underlies numerous cellular processes such as division, differentiation, alternative RNA splicing and cellular signaling [1]. Phosphorylation is one of the most prevalent post-translational modifications (PTMs), occurring on more than one-third of all cellular proteins [2] and there are approximately 500 kinases and 150 protein phosphatases that govern this dynamic PTM [3]. Abnormal protein phosphorylation of aggregate-prone proteins is observed in a number of neurodegenerative diseases including Alzheimer's Disease (AD). For example, AD is defined pathologically by the presence of detergent-insoluble extracellular amyloid-beta (Aβ) plaques and intracellular hyper-phosphorylated neurofibrillary tangles (NFTs) composed of the microtubule protein tau [4]. Abnormal tau phosphorylation is an early event in disease progression and strongly correlates with impairment of episodic memory and cognitive decline [5]. It is hypothesized that the increased phosphorylation of tau causes a conformational change in the protein triggering its dissociation from microtubules and inducing tangle formation, causing both functional deficits and neuronal toxicity [6]. Other structural proteins beyond tau, including neurofilaments [7], microtubule-associated protein 1B [8] and CRMP2 [9] are hyper-phosphorylated and co-aggregate with NFTs in AD brain tissue. Several kinases including GSK-3β, cyclin-dependent kinase 5 (CDK5), protein kinase C (PKC), microtubule-affinity regulating kinase (MARK), and rho-associated kinase 2 (ROCK2) have been directly implicated in the phosphorylation of these substrates in AD brain [10–14]. Moreover, reduced expression and activity of protein phosphatase 2A (PP2A) is also thought to contribute to enhanced phosphorylation of tau and other substrates in AD [15]. Thus, quantifying phosphorylated protein targets in AD brain tissue may reveal defects in kinase- or phosphatase-dependent signaling pathways involved in disease progression, as well as novel phosphorylation substrates.
Phosphopeptide enrichment strategies, including immobilized metal-affinity chromatography (IMAC) or titanium dioxide enrichment preceding liquid chromatography-tandem mass spectrometry (LC-MS/MS) have increased sensitivity for detection and quantification of phosphoproteins from complex mixtures, including human post-mortem brain tissue [16]. Alternatively, calcium phosphate precipitation (CPP) has also been employed as a simple and inexpensive approach to enrich phosphopeptides, previously used to identify 551 phosphopeptides on 185 proteins from AD brain tissue [17]. One drawback of CPP is the low yield of phosphopeptides, in which the number of phosphopeptides represent approximately 10 percent of all peptides analyzed [17]. Using FeCl3, we recently applied an IMAC-based peptide enrichment strategy followed by LC-MS/MS to identify differentially regulated phosphoproteins in detergent-soluble fractions from postmortem human brain tissue of a cohort of individuals with frontotemporal lobar degeneration (FTLD), compared to controls [18]. 786 phosphopeptides representing approximately 50 percent of the total peptides were identified. Quantification using total spectral counts revealed six proteins with significant changes in the FTLD phosphoproteome. NDRG2 and glial fibrillary acidic protein (GFAP) had an increased number of phosphospectra in FTLD, whereas microtubule associated protein 1A (MAP1A), reticulon 4, protein kinase C gamma, and heat shock protein 90 kDa alpha, had significantly fewer phosphospectra compared to control brain. Although this was a successful application of phosphoproteomics using post-mortem human brain tissue, this study was limited for three reasons. First, the study analyzed only the detergent-soluble fraction, limiting the identification of phosphopeptide differences to only proteins within this fraction. Second, analysis was conducted on pooled samples of either control or FTLD cases, which reduced biological variation [19], but also limited our statistical power and the ability to correlate phosphopeptide differences back to specific individual cases. The final limitation was that the quantification was based on the total number of peptide spectral counts to a given phosphoprotein, irrespective of signal for specific phosphorylation sites.
In the current study, we circumvented the above limitations by performing phosphoproteomic analysis using IMAC from whole brain extracts of eight control and eight pathologically confirmed AD cases matched as closely as possible for age and post-mortem interval. Using this approach, we observed on average an enrichment of phosphopeptides representing 80 percent of all MS/MS spectra across the 16 individual cases. Quantification was performed based on the extracted ion current measurements of identified phosphopeptides using in-house software [20–22]. Accurate peptide mass and retention time was used to derive signal intensity for every peptide across LC-MS/MS runs for each case. Pair-wise differential phosphopeptide analysis was performed using comparative marker selection software made available through the Broad Institute GenePattern service [23], revealing over 250 phosphopeptides significantly altered in AD cases compared to control cases. We confirmed some of the site specific changes on tau, heat-shock protein beta 1 (HSPB1) and crystallin alpha B (CRYAB) using phospho-specific antibodies by western blotting. We also noted increased phosphorylation on a number of substrates previously linked to AD, including neurofilaments [24] and adducin [17]. Finally, we highlight sites of increased phosphorylation on small heat shock proteins in AD that are highly correlated and potentially co-regulated with kinase phosphorylation events. Together, these data highlight the utility of label-free quantitative phosphoproteomics to define site-specific differences in phosphorylated targets and signaling events altered in AD.
2. Materials and Methods
2.1 Tissue Homogenization and Digestion
Frontal cortex brain samples were provided by the Emory Brain Bank. Control samples and AD cases were age and sex matched as closely as possible. Postmortem frontal cortex tissue from eight healthy control cases and eight pathologically confirmed AD cases were selected for comparison from the Emory Alzheimer's Disease Research Center (ADRC) brain bank. All AD cases were determined to be Braak stage V or VI and definite AD as classified by CERAD. Cases were matched as closely as possible for age of death, gender, and post mortem interval (Table 1). Each tissue piece was individually weighed (approximately 0.1 g) and homogenized (dounce homogenizer) in 500uL of urea lysis buffer (8M urea, 50mM Tris-HCl, pH 7.8), including both Protease Inhibitors (Roche) and the HALT (Pierce) phosphatase inhibitor cocktail, 0.6% (v/v). Samples were sonicated (Sonic Dismembrator, Fisher Scientific) 5 times for 5 s with 15 s intervals of rest at 30% amplitude to disrupt nucleic acids and then centrifuged at 22,800 r.c.f at 4°C for 5 minutes. Protein concentration was determined by the bicinchoninic acid (BCA) method, and then samples were frozen at −80°C. Protein samples (1 mg) were treated with 1 mM dithiothreitol (DTT) at 37°C for 30 minutes, followed by 5 mM iodoacetimide (IAA) at 37°C for 30 minutes, and digested with 1:200 (w/w) lysyl endopeptidase (Wako) at 37°C for 4 hours. Samples were diluted with 50 mM NH4HCO3 to a final concentration of 1.6 M urea and digested overnight with 1:50 (w/w) trypsin (Promega) at 37°C.
Table 1.
Individual case information and phosphopeptide enrichment
| Order | Case Number |
BRAAK Score |
Neocortical Neuritic Plaque Frequency |
PMI (hr)a |
Age at Onset |
Age at Death/Bx |
Duration | ApoE Genotype |
Race/ Sexb |
Frontal Cortex Neuritic Plaque Frequency |
Frontal Cortex Neurofibrillary Tangle Frequency |
Proteins | p(S/T/Y) Peptides |
p(S/T/Y) SCc |
Total SC |
% p(S/T/Y) SC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Control
| ||||||||||||||||
| CT1 | OS00-23 | 0 | Sparse | 11 | N/A | 68 | N/A | E3/3 | wf | None | None | 404 | 1234 | 1798 | 2332 | 77% |
| CT2 | OS00-06 | I | Sparse | 8 | N/A | 60 | N/A | E3/4 | bf | None | None | 570 | 1659 | 2491 | 3256 | 77% |
| CT3 | A86-46 | 0 | None | 6 | N/A | 65 | N/A | E3/3 | wm | None | None | 630 | 1884 | 2781 | 3349 | 83% |
| CT4 | OS03-299 | II | None | 6 | N/A | 69 | N/A | E3/3 | wm | None | None | 459 | 1433 | 2126 | 2420 | 88% |
| CT5 | E06-41 | II | None | 10 | N/A | 57 | N/A | E3/3 | wm | None | None | 641 | 1949 | 2869 | 3383 | 85% |
| CT6 | OS02-35 | I | None | 6 | N/A | 75 | N/A | E3/3 | wf | None | None | 500 | 1469 | 2408 | 2904 | 83% |
| CT7 | OS03-390 | II | None | 7 | N/A | 74 | N/A | E3/3 | wf | None | None | 434 | 1312 | 1872 | 3010 | 62% |
| CT8 | E05-130 | 0 | Sparse | 3 | N/A | 52 | N/A | E3/4 | wf | None | None | 535 | 1604 | 2406 | 2922 | 82% |
|
| ||||||||||||||||
|
AD
| ||||||||||||||||
| AD1 | E08-53 | VI | High | 8 | 70 | 78 | 10 | E3/3 | wf | High | High | 610 | 1902 | 2969 | 3516 | 84% |
| AD2 | OS00-32 | VI | High | 3.5 | 55 | 62 | 7 | E3/4 | wm | High | High | 652 | 2077 | 3200 | 3720 | 86% |
| AD3 | OS00-12 | V | High | 6 | 69 | 72 | 3 | E3/4 | wm | High | High | 608 | 1962 | 2790 | 3459 | 81% |
| AD4 | OS03-163 | V | High | 4.5 | 52 | 55 | 3 | E3/4 | wf | High | High | 354 | 1030 | 1617 | 2517 | 64% |
| AD5 | E06-155 | VI | High | 6.5 | 56 | 67 | 11 | E2/3 | wm | High | High | 297 | 988 | 1591 | 1881 | 85% |
| AD6 | E04-186 | VI | High | 7 | 59 | 72 | 13 | E3/4 | wf | High | High | 576 | 1809 | 2545 | 3395 | 75% |
| AD7 | E05-04 | VI | High | 4.5 | 52 | 64 | 12 | E3/4 | wf | High | High | 341 | 1116 | 1620 | 1906 | 85% |
| AD8 | E04-172 | V | High | 6 | 72 | 87 | 15 | E3/4 | wf | High | Moderate | 587 | 1773 | 2661 | 3282 | 81% |
Postmortem interval (PMI).
w (white/Caucasian), b (black/African American), a (Asian), h (Hispanic), N/A (not applicable); m (male), f (female).
SC, spectral counts.
2.2 Immobilized Metal-Affinity Chromatography (IMAC)
Digested peptides were acidified the next day to 0.4% TFA and desalted using a C18 Sep-Pak (Waters), vacuum dried, and resuspended in 500 μL binding buffer (80% ACN, 0.1% TFA). 1mL of Ni-NTA beads (Qiagen) was washed 3 times with HPLC grade water (Sigma), and treated with 1mL 40mM EDTA pH 8.1 for 1 hour at room temperature with constant vortexing at 70% intensity. Beads were then washed 3 more times in water, and treated with 1mL 100 mM FeCl3 for 1 hour at room temperature with constant vortexing at 70% intensity. Next, beads were washed once with 1:1:1 (v/v/v) ACN:methanol:water and 0.01 % acetic acid, followed by 3 washes in binding buffer before being split into 2 equal fractions. Each fraction of beads was incubated with peptides in binding buffer for 1 hour at room temperature with constant vortexing at 70% intensity, followed by 3 washes of binding buffer. Peptides were eluted in 100 μL elution buffer (1:1 ACN: 1:20 (v/v) ammonia water, pH 12) while being vortexed on and off for 1 minute. Beads were centrifuged at 22,800 r.c.f. for 1 minute, and then supernatant was transferred to an ACN-pretreated auto-sampler tube with 100 μL of 50% ACN, 5% formic acid and flash frozen in dry ice before being vacuum dried. Peptides were resuspended in 100 μL HPLC grade water, flash frozen, and vacuum dried twice more to ensure removal of ammonium salts, before being resuspended in loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% ACN) for LC-MS/MS analysis.
2.3 Peptide Analysis by LC-MS/MS
Extracted peptides were loaded onto a 20 cm nano-HPLC column (internal diameter 100 μm) packed with Reprosil-Pur 120 C18-AQ 1.9 μm beads (Dr. Maisch), eluted and sequenced (MS/MS) on an LTQ-Orbitrap mass spectrometer (Thermo Finnigan, San Jose, CA) using data-dependent acquisition. All MS/MS files were searched against the human database downloaded from the National Center for Biotechnology Information (RefSeq version 54) using the SEQUEST Sorcerer algorithm (version 3.11, SAGE-N). Searching parameters included mass tolerance of precursor ions (±20 ppm) and product ion (±0.5 m/z), semi-tryptic restriction, dynamic modifications for oxidized Met (+15.9949 Da), phosphorylation (+79.9663 Da), five maximal modification sites and a maximum of two missed cleavages. To evaluate false discovery rate (FDR) using a standard approach, all original protein sequences were reversed to generate a decoy database that was concatenated to the original database. To remove false positive matches, assigned peptides were grouped by a combination of trypticity (fully and partial) and precursor ion-charge state. Each group was first filtered by mass accuracy (10 ppm), and by dynamically increasing correlation coefficient (Xcorr) and ΔCn values to reduce protein FDR to less than 1 percent. If peptides were shared by multiple members of a protein family, the matched members were clustered into a single group in which each protein identified by a unique peptide represented a subgroup. The MS/MS spectra of matched phosphopeptides were also manually validated as described previously [17]. Phosphorylated Ser and Thr spectra were examined for signature phosphate neutral losses (−49 m/z for doubly charged, −32.7 m/z for triply charged, and −24.5 m/z for quadruply charged). In some cases, the product ions on MS/MS spectra did not provide sufficient information to distinguish the phosphorylation site(s) within the same peptide sequence. The higher the ΔCn, the more certain the phosphorylation site assignment [25]. Thus, when the two top matches of a spectrum were both phosphopeptides with the same sequence and the ΔCn was ≤0.125, the site of phosphorylation was considered ambiguous. Quantitative comparison of control and AD samples was performed using the software DQUAN as previously reported [20–22]. Briefly, ion intensities for identified peptides were extracted in MS survey scans of high-resolution (30,000), based on the isotopic ion selected for MS/MS sequencing. A number of features were derived, including precursor m/z, charge state, retention time, ion peak width, height, and noise level. The global noise level was derived by averaging signal intensity of all ions in the full MS scan after removing signal ion outliers that were at least two standard deviations away from the mean. The intensity of ions was presented by the peak height and normalized according to the noise intensity under the assumption that the noise level of MS scans reflects, at least partially, variable ionization efficiency. Peptide ion matching across LC-MS runs was allowed with a mass tolerance of 10 ppm. If a sequenced peptide could not be matched to a signal above general noise level, we estimated that the maximum ion current for undetected signal was equal to the local noise level (LNL) of ions within the 10 ppm window (LNL ≥ 0.5).
2.4 Comparative Marker Selection
Comparative marker selection was performed using the Broad Institute GenePattern online software service [23]. 5,569 peptide measurements (signal to noise) across 8 control and 8 AD samples were compared using the two-sided t test. 305 phosphopeptides were found to be significantly and consistently changed between AD and CT with p<0.05. We filtered this list, to keep 253 phosphopeptides for K-means clustering which had a minimum fold change of 1.75 (75 percent increase or decrease relative to the mean of the control population).
2.5 Empirical False Discovery Rate
A novel, more stringent means of determining false discovery using a null experiment has been previously described [22, 26]. Peptides for 7 of 8 control and 7 of 8 AD samples were analyzed by LC-MS/MS as a technical replicate (replicate 2; R2). 2,709 peptides were quantified across all 30 runs. Signal intensities for each of these peptides across the pair (R1 and R2 replicates) of 14 matched runs was analyzed as a null ratio R1/R2, then log2 transformed as a ratio of the average of 7 null AD measurements divided by the average of 7 null control measurements. These 2,709 log2-transformed null experiment ratios were plotted as a histogram, with the scale normalized to equal the maximum height of the histogram of the experimental values from R1. Both populations fit a Gaussian distribution, which enabled a calculation of the standard deviation (SD) of the null (0.286 log2 units) and experimental (0.304 log2 units) populations. A cutoff of 2.79 fold of the null SD (0.80 log2 units, or >1.74-fold change in AD/CT) was therefore consistent with a theoretical confidence interval of 99.5%. To empirically determine false discovery rate of the comparative marker selection described above, the null population of 14 paired measurement ratios (7 AD vs. 7 Control) was subjected to comparative marker selection. AD null vs. control null phosphopeptide markers with T test p<0.05 were counted and scaled by the multiplier 5,569/2,709 (2.06) to empirically determine the number of measurements in the experiment driven by technical variation that appear as significant markers in the null calculations. In total, 10 false positives (p<0.05) were discovered in the null population after phosphopeptide marker selection. In sharp contrast, in the experimental population (8 control vs. 8 AD) there were 305 markers with p<0.05 after marker selection. The 10 false positives in the null population imply a calculation for the false discovery rate, calculated as (10 × 2.06) / 305 (6.7%). Additional filtering on signal to noise, Xcorr or minimum fold change did not improve the false discovery rate, because the 10 false positives were changed more robustly than proteins falling off the marker list with additional filtering. To improve empirical FDR, it would thus be necessary to increase analytic power by increasing sample number.
2.6 Clustering Analysis
Following comparative marker selection, 253 peptides with p<0.05 and fold change ≥1.75 were clustered using a Spearman metric using Cytoscape 2.8.2 with DomainGraph release 3.0-enabled plugin clusterMaker v1.9 K-means cluster feature. Three clusters were generated with 35,000 iterations using as input log-transfromed ratio calculated as the single measurement (signal/noise) divided by 16 measurement ratio mean S:N. Clustering results were visualized with the cytoscape plugin or alternately, with Java TreeView 1.1.6r2. Multidimensional scaling (MDS) or principal component analysis (PCA) to determine distance between cases was performed in the WeiGhted Correlation Network Analysis (WGCNA) R package [27], followed by clustering using the hClust function on the distance matrix defined as 1 minus the adjacency matrix correlated using the Spearman correlation function with euclidian distance. All 5,569 peptides quantified were used for MDS clustering.
2.7 HSP27 Ser82 phosphopeptide correlation heat map
The WGCNA R netScreen function [27] was used to correlate the phosphopeptide abundance data set of 5,569 rows to abundances of HSP27 phospho-Ser82 across the 16 samples. Top identified correlated phosphopeptides were manually filtered to retain categories of interest, namely kinases, kinase effectors, and proteins involved in protein folding. The resulting heatmap ordered from highest correlation to lowest was visualized using Java TreeView 1.1.6r2.
2.8 Western blotting
Immunoblotting was performed according to standard procedures as reported previously [28]. Briefly, samples in Laemmli sample buffer were resolved by SDS-PAGE before an overnight transfer to Immobilon-P membranes (Millipore, Bedford, MA). Blots were blocked with Tris-buffered saline (TBS) plus blocking buffer (USB Corporation, Cleveland, OH) at room temperature for 30 min and probed with primary antibodies in TBS plus 0.1 % Tween-20 plus blocking buffer overnight at 4°C. The following day, blots were rinsed and incubated with secondary antibodies conjugated to fluorophores (Molecular Probes/Invitrogen) for one hour at room temperature. Images were captured using an Odyssey Image Station (LiCor, Lincoln, NE). Antibodies used: mouse monoclonal tau2 (Chemicon catalog MAB375; Millipore Corporation; Billerica, CA), pTau Thr231 (Chemicon catalog, MAB5450), pTau Ser404 (Invitrogen, 44758G), pSer59 Crystallin (Abcam, ab5577), pSer82 HSP27 (Cell Signaling, 2401S) and mouse anti-actin (Santa Cruz Biotech, sc-8432;) for a loading control.
3. Results
3.1 Label-free quantitative phosphopeptide analysis in post-mortem brain
We previously generated peptides for phosphoproteomic analysis of human post-mortem brain by precipitating proteins from detergent-soluble fractions using acetone prior to digestion [18]. In our current approach, individual AD and control case brain tissue was homogenized directly in urea buffer containing protease and phosphatase inhibitors. Notably, 8M urea buffer used for homogenization is directly compatible with LysC digestion, and urea denatured proteins are more likely to be completely digested at all available lysine residues by this approach, prior to subsequent dilution and digestion of total protein-derived peptides with trypsin.
Following in-solution digestion, phosphopeptides were enriched by an optimized IMAC protocol using FeCl3 [29] and analyzed in a single LC-MS/MS analysis for each control and AD case (n=16). After database searching and peptide filtering for uniqueness of each identified peptide, 5,569 mono-, di-, or tri-phosphorylated peptides belonging to 1,559 phosphoproteins remained. 4,185 unique phosphorylation sites were represented. The total number of phosphopeptide spectral counts represented approximately 80 percent of the total spectral counts collected, which was generally consistent across all cases sequenced (n=16) and showed no correlation with post-mortem interval (PMI) (Table 1). All phosphopeptide identifications and abundances are given in Supplemental Table 1. To empirically determine a false discovery rate, 7 of the 8 control and 7 of the 8 AD phosphopeptide samples were analyzed in technical replicate by LC-MS/MS. This enabled the construction of a null data set in which 14 replicate 1 (R1) values were each divided by replicate 2 (R2) values. Histograms of the log2 AD/Control for the R1 experiment and R1/R2 null are provided in Supplemental Figure 1. Both R1 experimental and null (R1/R2) values for individual AD and control measurements were compared for significant differences of phosphopeptide signal intensities using marker selection. In the experimental population (8 AD vs 8 Control), 305 phosphopeptides were found significantly altered (p<0.05). By contrast there were 10 false positives (p<0.05) arising from marker selection of AD to control null measurements, thus imputing a FDR of 6.7 percent. Calculations are provided in Supplemental Table 2.
3.2 Classification of individual cases based on phosphopeptide signal intensities
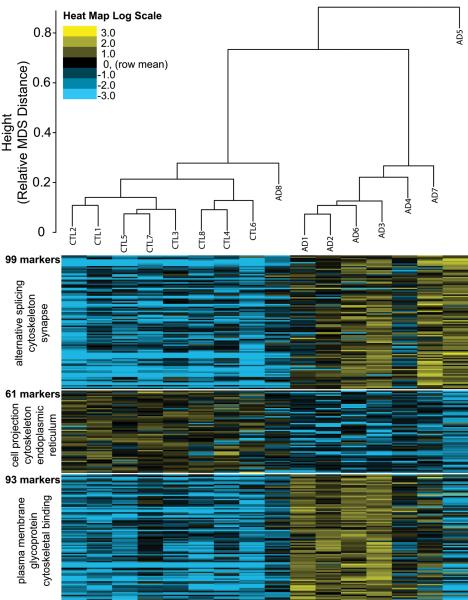
Of the 305 significantly altered phosphopeptides (6.7% FDR), 253 were found to be present at levels greater than 1.75 fold in AD relative to control or vice versa. All 5,569 phosphopeptide signatures defined a clustering distance between individual cases which clearly segregated controls from AD (Figure 1, top). The 253 phosphopeptide markers were sorted into 3 distinct groups by K-means clustering (Figure 1, bottom). Two of the groups had elevated levels of phosphopeptides in AD samples, whereas the remaining group had decreased levels in AD. Functional categories over-represented in each of the marker groups are given to the left, where cytoskeleton or cytoskeletal binding is a category of proteins identified by DAVID ontology analysis [30, 31] in each of the groups. Interestingly, phosphopeptides from endoplasmic reticulum were significantly reduced in AD brain homogenates compared to controls.
Figure 1. Clustering of individual cases based on phosphopeptide signal intensities.
Multidimensional scaling (MDS) was performed as described in methods and segregated samples into 2 general groups: controls with AD8, and all other AD cases. Heat mapping of the individual sample abundances for the 253 markers shows clear differentiation of makers into 3 clusters, as indicated. DAVID analysis indicating enrichment of functional groups of proteins compared to a background of all the proteins (identified via phosphopeptides) produced hits indicated to the left of each marker cluster.
3.3 Quantification of tau specific phosphopeptides in control and AD brain homogenates
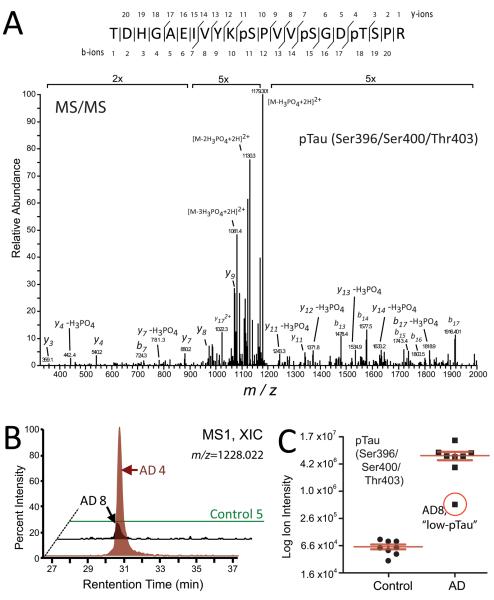
AD is defined pathologically by the presence of intracellular hyperphosphorylated NFTs composed of the microtubule protein tau. Consistently, approximately 21 percent (52 of 253) of the significantly altered phosphopeptides in AD tissue mapped to tau and all 52 of these tau phosphopeptides, on average, showed enriched signal intensities compared to control cases (Supplemental Table 1). Identification and quantification of a representative tau phosphopeptide corresponding to simultaneously occupied phosphosites Ser396, Ser400, and Thr403 on Tau is provided in Figure 2A. The raw extracted ion chromatogram for this peptide from a control and two AD cases, one with high phosphorylation on Tau (AD4) and one with a low level of phosphorylation (AD8), is shown in Figure 2B. Ion intensities for this tau phosphopeptide across all 16 cases are plotted on a log scale in Figure 2C, demonstrating that phosphorylation of this peptide is much more prevalent in AD cases, compared to controls.
Figure 2. Quantification of tau specific phosphopeptides in control and AD brain homogenates.
(A) Phospho-tau (pTau) peptides were identified by MS/MS and quantified by extracted ion chromatograms (XIC) of full precursor MS1 scans. (A) Representative MS/MS spectrum for the pTau peptide mapping to simultaneous modification sites on Ser396, Ser400 and Thr403. (B) A representative XIC [measured as the percentage intensity of the defined MS1 precursor (m/z 1228.002)] for the doubly charged Ser396/Ser400/Thr403 modified tau phosphopeptide was measured across one control and two AD cases. Peptide intensities were normalized to AD4 level (100%). (C) A “low-pTau” AD case 8 had 17-fold less Ser396/Ser400/Thr403 modified tau phosphopeptide signal intensity than the average of all other AD cases (n=7) measured (y-axis is log2 scale).
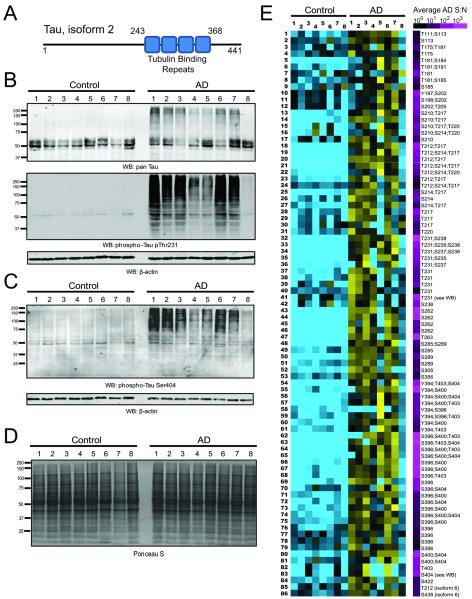
Tau isoform 2 is represented with its four tubulin binding repeat domains spanning residues 243 to 368 (Figure 3A). Western blotting for total tau compared to phospho-specific tau at residue Thr231 in whole homogenates indicates that high molecular weight tau polymerization above 60 kDa is a phenomenon that generally coincides with higher levels of phosphopeptide signal intensities harboring phosphorylated Thr231 (Figure 3B). This correlation was also observed for phosphorylated tau at residue Ser404 (Fig. 3C). Notably, there were no gross differences in protein integrity observed in any of the control and AD cases by Ponceau S staining of the transfer membrane prior to western blotting (Fig. 3D). All enriched tau phosphopeptides that were identified and quantified by LC-MS/MS across the 16 tissue samples are presented as a heat map in Figure 3E. This heat map indicates that AD samples have a significantly higher level of phosphorylation across most Tau phosphopeptides, with the notable exception of AD8, which is a low phospho-Tau containing sample regardless of the phosphopeptide measured. Interestingly, AD8 was classified as Braak stage 5, and described as having only moderate levels of NFTs in the prefrontal cortex after neuropathological examination (Table 1), whereas other AD cases analyzed by LC-MS/MS were classified with severe levels of NFTs. The reduced tau phosphorylation in AD8 may reconcile the clustering results presented in Figure 1. AD8 segregated more closely to control cases than other AD cases analyzed in this study. Peptide abundances for the 86 grossly hyperphosphorylated tau phosphopeptides shown are given in Supplemental Table 3 together with 26 additional Tau phosphopeptides that were not changed between control and AD samples.
Figure 3. AD-associated hyperphosphorylation events on Tau.
(A) The standard numbering of the amino acid residues of Tau phosphosites in this figure is given in the context of Tau isoform 2, which harbors four tubulin binding repeats. (B) Total or pan-Tau is visualized by western blot (top panel), followed by a phospho-Thr231 Tau specific blot (bottom panel); loading control is actin. (C) Phospho-Ser404 specific blotting across the 16 samples shows increased levels of high molecular weight phosphorylated Tau in AD. (D) No gross differences in protein integrity were observed in any of the control and AD cases by Ponceau S staining of the membrane following protein transfer prior to western blotting. (E) 86 phosphosite peptides across Tau were identified as differentially regulated (increased) in AD relative to control frontal cortex. Heat map has a blue-yellow log2 scale (ratio of measurement to average) from −3.0 to +3.0, with an additional scale to the right provided to indicate the average abundance in terms of AD sample signal-to-noise for each peptide. All peptide sequences are provided in Supplemental Table 3.
3.4 Small heat shock protein phosphorylation is correlated with a network of kinases
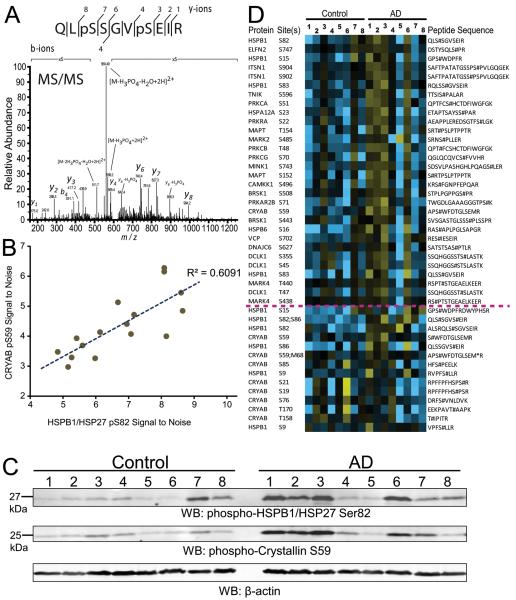
The heat shock protein (HSP) network plays a key role in protein folding and cytoskeletal protein assembly. Notably, small HSPs dynamically exist between polymeric and oligomeric states, which mediates their functional role in modulating protein aggregation in cells [32]. This process is regulated by phosphorylation downstream of stress and in the case of small HSP chaperones [33], this phosphorylation can enhance monomerization and chaperone activity [32, 34]. Therefore, we assessed whether the signal intensity for a functional phosphosite from small heat shock protein 27 (HSPB1), unambiguously identified in a peptide phosphorylated at residues Ser82 and Ser86 (Fig. 4A), was correlated with any other phosphopeptides in the control or AD phosphoproteome. As expected, there was strong correlation between Ser82/Ser86 HSPB1 phosphopeptide intensity and a Ser82 alone HSPB1 phosphopeptide. Moreover, strong correlation was also observed between Ser82 HSPB1 phosphopeptide intensity with a phosphopeptide corresponding to Ser59 phosphorylation on another small HSP, crystallin alpha B (CRYAB), which is known to be enriched in AD frontal cortex [35] (Fig. 4B). We confirmed the correlation observed at the phosphopeptide level between HSPB1 Ser82 and crystallin alpha B Ser59 phosphorylation by western blot (Fig. 4C). Notably, both these phosphorylation events are consistent with modulation of the dynamic oligomerization status of these small HSPs [33], and thereby, their function in regulating signaling in the cell. To ascertain other phosphopeptides correlating with phosphorylation of these two small HSP proteins, we utilized the Weighted Correlation Network Analysis (WGCNA) netScreen function [27]. This function correlated the phosphopeptide signal intensities of HSPB1 Ser82 phosphosite to the entire list of 5,569 phosphopeptides quantified across all 16 cases analyzed. This generated a heat map of highly correlated phosphopeptide signal intensities ranked from the highest to lowest correlation to HSPB1 Ser82 signal intensities (Fig. 4D). As expected, the CRYAB Ser59 phosphopeptide was highly correlated with HSPB1 Ser82 signal. However, a surprising number of additional phosphopeptides mapping to protein kinases also showed a strikingly similar pattern of signal intensities across cases. Although phosphopeptide signals varied case-to-case within the AD group, increased phosphorylation at HSP27 Ser82 strongly correlated with phosphopeptide intensities on a number of kinases (TNIK, PKA, PKC, MINK1, CAMKK1, MARK2 and 4, DCLK1), as well as phosphopeptides of a number of other chaperones involved in protein folding (HSPA12A, HSPB6, VCP, DNAJC6). The full list of phosphopeptides, including p-values and Z-scores, that strongly correlated with HSBP1 Ser82 phosphorylation is provided in Supplemental Table 4.
Figure 4. Co-enrichment of various phosphorylated protein kinases with small HSP (HSP27) Ser82 phosphorylation.
(A) MS/MS spectrum of doubly phosphorylated Ser82/Ser86 HSP27 peptide. (B) A positive correlation of HSP27 phospho-Ser82 abundance with that of phospho-Ser59 on crystallin α B was observed. (C) HSP27 Ser82 and Crystallin α B Ser59 phosphorylation are correlated by western blot. β-Actin was blotted as a loading control. (D) Highly correlated patterns of phosphopeptide signal intensities across the 16 samples revealed a co-enrichment of a network of kinases and small heat shock protein derived phosphopeptides. Heat map scale is log2 from −3.0 (blue) to +3.0 (yellow), with black indicating equivalence to the row average. Quantitation of all peptides in this figure with statistics is provided in Supplemental Table 4.
4. Discussion
In this study, we employed immobilized metal affinity chromatography (IMAC) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify phosphopeptides from eight individual AD and eight age-matched control postmortem human brain tissues. Using this approach, we identified 5,569 phosphopeptides in frontal cortex across 16 individuals in which phosphopeptides represented 80 percent of all peptide spectral counts collected following IMAC enrichment. Marker selection identified 253 significantly altered phosphopeptides by precursor ion intensity, changed by at least 1.75 fold relative to controls, with an empirical false discovery rate less than 7 percent. Approximately 21 percent of the significantly altered phosphopeptides in AD tissue mapped to tau and all showed enhanced signal intensities compared to controls. In addition to quantifying hyperphosphorylation on tau in AD, we also identified a broad array of phosphopeptides on other proteins including a surprising number that mapped to kinase-specific phosphosites, which showed a strong correlation to phosphopeptide signals found in small heat shock proteins (i.e., HSP27 and CRYAB).
It is important to note that global protein abundance differentially regulated at the transcript or protein level in a direction opposing a change in phosphopeptide abundance measured in this study would be expected to negate or “wash out” differences measured. Another drawback of examining whole brain tissue phosphorylation status is that phosphopeptides can be derived from any of the various brain cell types (glia, neurons, and even endothelium). The differences observed are the average across all the cell types. Thus, an increase in glial fibrillary acidic protein (GFAP) phosphopeptide intensity in disease can be indicative of gliosis or astrocyte proliferation in specific individuals. Also of note, AD cases 4 and 5 showed a lower overall level of phosphorylation by both western blot and/or in the overall IMAC enrichment efficiency. Two possible explanations are conceivable, the first being due to a potential biological difference such as a higher intrinsic phosphatase activity in the sampled brains. Alternatively, this could be due to technical variation beyond our control, such as subzonal sampling differences. The remainder of discussion focuses on specific phosphorylation events of potential relevance to AD identified in this study.
Stathmin-3 (STMN3) neuron specific microtubule destabilizing protein has a 5.5-fold AD-specific increase in signal intensity for the phosphopeptide with dual phosphorylation at Ser65 and Ser72 (Supplemental Table 1). As a negative control, Ser81 phosphopeptide did not change. Thus, it is possible that differential phosphorylation at Ser65 and Ser72, but not Ser81, may regulate STMN3 function directed at neuronal microtubules in AD.
The CAMK family kinase brain specific kinase 1 (BRSK1/SADK1), which phosphorylates Tau on residue Ser262 [36] and thereby regulates neuronal polarization, is differentially phosphorylated near its ubiquitin associated domain at residue Ser384. The Ser384 phosphopeptide on BRSK1 is 2.5-fold increased in AD relative to controls. In structural studies, it was shown that the ubiquitin associated domain of BRSK1 reorients relative to the catalytic domain during kinase activation following phosphorylation by an activating, upstream kinase [37] and this is thought to promote kinase activity. Tau Ser262 was indeed hyperphosphorylated in a majority of the 8 AD samples (Fig. 3C), and the fully tryptic Ser262 tau phosphopeptide was the most AD-enriched phosphopeptide detected in this study (580 fold increased in abundance). This result strongly implicates activation of BRSK1 in AD upstream of Tau Ser262 phosphorylation, potentially downstream of activating phosphorylation at Ser384 on BRSK1.
BAG6 is a chaperone required for tail-anchored membrane protein delivery to the endoplasmic reticulum (ER) membrane, and also promotes defective nascent polypeptide degradation by the proteasome. BAG6/BAT3 phosphorylation was detected at the novel sites Thr251, Thr260, and Thr265 with 3.5-fold increase in signal of the triply modified peptide spanning residues 243–266. Ten phosphopeptides representing other BAG6 phosphosites either between residues 100–117 or between 958–1075 were not enriched at all in AD, suggesting that the differentially modified sites spanning Thr251-Thr265 may have a role in regulating BAG6 function. Interestingly, among phosphoproteins with grossly reduced phosphopeptide levels in AD, ER proteins were overrepresented (Fig. 2).
Sequestosome-1/p62 is a protein implicated in tauopathies including AD, because it is found in neurofibrillary tangles, among other neurodegenerative aggregates [38]. Phosphorylation at Ser24 and Ser28 in the protein region arising from the alternatively spliced 5' coding exon (exon 3) was found to be increased more than 3-fold in separate phosphopeptides representing each phosphosite, and Ser24 phosphorylation has previously been seen to increase in response to proteasome inhibition [39]. p62 mediates TRAF6 K63-specific ubiquitin ligase interaction with effectors, so p62 differential phosphorylation may have a role in regulating the propensity for differential ubiquitin linkage accumulation seen in AD frontal cortex [40].
PTGES3/p23 HSP90 co-chaperone, which is a limiting component of HSP90/HSP70 chaperone complexes that prevents the formation of large insoluble protein aggregates [41], was previously reported to be significantly decreased in AD pyramidal neurons by immunohistochemistry [42]. Notably, this co-chaperone is differentially phosphorylated, with a 3-fold increase in Ser113 phosphorylation in AD phosphoproteome. The role of p23 phosphorylation in its co-chaperone activity, and in limiting protein aggregation specifically in AD, is unexplored.
In total, the above findings implicate significant signaling networks that differentially regulate the heat shock and protein misfolding response pathway in AD compared to healthy controls. Phosphosignaling through this network of quality control factors may prove to be important for age-dependent changes in surveillance and clearance of aggregate-prone proteins that ultimately disrupt proteostasis in AD.
Supplementary Material
The experimental population represents 5,569 phosphorylation site-containing peptides (Replicate 1; R1) where the average AD/CT ratio of phosphopeptide signal intensities give a log2-scaled population (red curve). To generate a null Gaussian distribution (blue curve) peptides for 7 of 8 control and 7 of 8 AD samples were analyzed by LC-MS/MS as a technical replicate (R2). Signal intensities for each of the phosphopeptides across the paired replicates (R1 and R2) of 14 matched LC-MS/MS runs were analyzed as a null ratio (R1/R2). These 2,709 log2-transformed null experiment ratios were plotted as a histogram, with the maximum scaled to equal the maximum height of the histogram of the experimental values from R1. Both populations fit a Gaussian distribution, which enabled a calculation of the standard deviation (SD) of the null (0.286 log2 units) and experimental (0.304 log2 units) populations. A cutoff of 2.79 fold of the null SD (0.80 log2 units, or >1.74-fold change in AD/CT) was therefore consistent with a theoretical confidence interval of 99.5% (green dashed lines). Empirical comparison of null and experimental marker counts obtained by comparative marker selection empirically determined false discovery to be 6.7 percent (see section 2.5).
Acknowledgments
We are appreciative of John Hanfelt (Emory School of Medicine, Department of Biostatistics) for comments and critical reading of this manuscript. NTS is supported by an Alzheimer Association New Investigator Research Grant (NIRG12-242297). This work was also supported by an NIH grants P50 (AG025688), P01 (AG014449), and mass spectrometry instrument time was subsidized by an Emory Neuroscience NINDS Core Facilities P30 grant (NS055077).
Footnotes
Conflict of Interest: The authors have no conflicts of interest to report.
6. References
- [1].Nestler EJ, Greengard P. Protein phosphorylation in the brain. Nature. 1983;305:583–588. doi: 10.1038/305583a0. [DOI] [PubMed] [Google Scholar]
- [2].Cohen P. The role of protein phosphorylation in human health and disease. European Journal of Biochemistry. 2001;268:5001–5010. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- [3].Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- [4].Querfurth HW, LaFerla FM. Alzheimer's Disease. New England Journal of Medicine. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- [5].Ghoshal N, Garcı, amp x, et al. Tau Conformational Changes Correspond to Impairments of Episodic Memory in Mild Cognitive Impairment and Alzheimer's Disease. Experimental Neurology. 2002;177:475–493. doi: 10.1006/exnr.2002.8014. [DOI] [PubMed] [Google Scholar]
- [6].Ballatore C, Lee VMY, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- [7].Sternberger NH, Sternberger LA, Ulrich J. Aberrant neurofilament phosphorylation in Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:4274–4276. doi: 10.1073/pnas.82.12.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ulloa L, Montejo de Garcini E, Gomez-Ramos P, Moran MA, Avila J. Microtubule-associated protein MAP1B showing a fetal phosphorylation pattern is present in sites of neurofibrillary degeneration in brains of Alzheimer's disease patients. Brain research. 1994;26:113–122. doi: 10.1016/0169-328x(94)90081-7. [DOI] [PubMed] [Google Scholar]
- [9].Cole AR, Noble W, Aalten L. v., Plattner F, et al. Collapsin response mediator protein-2 hyperphosphorylation is an early event in Alzheimer's disease progression. Journal of Neurochemistry. 2007;103:1132–1144. doi: 10.1111/j.1471-4159.2007.04829.x. [DOI] [PubMed] [Google Scholar]
- [10].Herskowitz JH, Feng Y, Mattheyses AL, Hales CM, et al. Pharmacologic inhibition of ROCK2 suppresses amyloid-beta production in an Alzheimer's disease mouse model. J Neurosci. 2013;33:19086–19098. doi: 10.1523/JNEUROSCI.2508-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hanger DP, Hughes K, Woodgett JR, Brion J-P, Anderton BH. Glycogen synthase kinase-3 induces Alzheimer's disease-like phosphorylation of tau: Generation of paired helical filament epitopes and neuronal localisation of the kinase. Neuroscience Letters. 1992;147:58–62. doi: 10.1016/0304-3940(92)90774-2. [DOI] [PubMed] [Google Scholar]
- [12].Noble W, Olm V, Takata K, Casey E, et al. Cdk5 Is a Key Factor in Tau Aggregation and Tangle Formation In Vivo. Neuron. 38:555–565. doi: 10.1016/s0896-6273(03)00259-9. [DOI] [PubMed] [Google Scholar]
- [13].Cole G, Dobkins KR, Hansen LA, Terry RD, Saitoh T. Decreased levels of protein kinase C in Alzheimer brain. Brain research. 1988;452:165–174. doi: 10.1016/0006-8993(88)90021-2. [DOI] [PubMed] [Google Scholar]
- [14].Chin JY, Knowles RB, Schneider A, Drewes G, et al. Microtubule-Affinity Regulating Kinase (MARK) Is Tightly Associated with Neurofibrillary Tangles in Alzheimer Brain: A Fluorescence Resonance Energy Transfer Study. Journal of Neuropathology & Experimental Neurology. 2000;59:966–971. doi: 10.1093/jnen/59.11.966. [DOI] [PubMed] [Google Scholar]
- [15].Gong CX, Singh TJ, Grundke-Iqbal I, Iqbal K. Phosphoprotein phosphatase activities in Alzheimer disease brain. J Neurochem. 1993;61:921–927. doi: 10.1111/j.1471-4159.1993.tb03603.x. [DOI] [PubMed] [Google Scholar]
- [16].Stasyk T, Huber LA. Mapping in vivo signal transduction defects by phosphoproteomics. Trends in Molecular Medicine. 2012;18:43–51. doi: 10.1016/j.molmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- [17].Xia Q, Cheng D, Duong DM, Gearing M, et al. Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J Proteome Res. 2008;7:2845–2851. doi: 10.1021/pr8000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Herskowitz JH, Seyfried NT, Duong DM, Xia Q, et al. Phosphoproteomic Analysis Reveals Site-Specific Changes in GFAP and NDRG2 Phosphorylation in Frontotemporal Lobar Degeneration. Journal of Proteome Research. 2010;9:6368–6379. doi: 10.1021/pr100666c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Diz AP, Truebano M, Skibinski DO. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis. 2009;30:2967–2975. doi: 10.1002/elps.200900210. [DOI] [PubMed] [Google Scholar]
- [20].Dammer EB, Duong DM, Diner I, Gearing M, et al. Neuron Enriched Nuclear Proteome Isolated from Human Brain. J Proteome Res. 2013;12:3193–3206. doi: 10.1021/pr400246t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gozal YM, Duong DM, Gearing M, Cheng D, et al. Proteomics analysis reveals novel components in the detergent-insoluble subproteome in Alzheimers disease. Journal of Proteome Research. 2009 doi: 10.1021/pr900474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Donovan LE, Dammer EB, Duong DM, Hanfelt JJ, et al. Exploring the Potential of the Platelet Membrane Proteome as a Source of Peripheral Biomarkers for Alzheimer's Disease. Alzheimers Res Ther. 2013;5:32. doi: 10.1186/alzrt186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gould J, Getz G, Monti S, Reich M, Mesirov JP. Comparative gene marker selection suite. Bioinformatics (Oxford, England) 2006;22:1924–1925. doi: 10.1093/bioinformatics/btl196. [DOI] [PubMed] [Google Scholar]
- [24].Rudrabhatla P, Grant P, Jaffe H, Strong MJ, Pant HC. Quantitative phosphoproteomic analysis of neuronal intermediate filament proteins (NF-M/H) in Alzheimer's disease by iTRAQ. The FASEB Journal. 2010;24:4396–4407. doi: 10.1096/fj.10-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- [26].Zhou JY, Hanfelt J, Peng J. Clinical proteomics in neurodegenerative diseases. Proteomics. Clinical applications. 2007;1:1342–1350. doi: 10.1002/prca.200700378. [DOI] [PubMed] [Google Scholar]
- [27].Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Seyfried NT, Gozal YM, Dammer EB, Xia Q, et al. Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyUb chains. Mol Cell Proteomics. 2010;9:705–718. doi: 10.1074/mcp.M800390-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ficarro SB, Adelmant G, Tomar MN, Zhang Y, et al. Magnetic bead processor for rapid evaluation and optimization of parameters for phosphopeptide enrichment. Anal Chem. 2009;81:4566–4575. doi: 10.1021/ac9004452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [31].Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Abisambra JF, Jinwal UK, Jones JR, Blair LJ, et al. Exploiting the diversity of the heat-shock protein family for primary and secondary tauopathy therapeutics. Current neuropharmacology. 2011;9:623–631. doi: 10.2174/157015911798376226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mehlen P, Arrigo A-P. The serum-induced phosphorylation of mammalian hsp27 correlates with changes in its intracellular localization and levels of oligomerization. European Journal of Biochemistry. 1994;221:327–334. doi: 10.1111/j.1432-1033.1994.tb18744.x. [DOI] [PubMed] [Google Scholar]
- [34].Ahmad MF, Raman B, Ramakrishna T, Rao Ch M. Effect of phosphorylation on alpha B-crystallin: differences in stability, subunit exchange and chaperone activity of homo and mixed oligomers of alpha B-crystallin and its phosphorylation-mimicking mutant. Journal of molecular biology. 2008;375:1040–1051. doi: 10.1016/j.jmb.2007.11.019. [DOI] [PubMed] [Google Scholar]
- [35].Shinohara H, Inaguma Y, Goto S, Inagaki T, Kato K. αB crystallin and HSP28 are enhanced in the cerebral cortex of patients with Alzheimer's disease. Journal of the Neurological Sciences. 1993;119:203–208. doi: 10.1016/0022-510x(93)90135-l. [DOI] [PubMed] [Google Scholar]
- [36].Kishi M, Pan YA, Crump JG, Sanes JR. Mammalian SAD kinases are required for neuronal polarization. Science. 2005;307:929–932. doi: 10.1126/science.1107403. [DOI] [PubMed] [Google Scholar]
- [37].Jaleel M, Villa F, Deak M, Toth R, et al. The ubiquitin-associated domain of AMPK-related kinases regulates conformation and LKB1-mediated phosphorylation and activation. Biochem J. 2006;394:545–555. doi: 10.1042/BJ20051844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Salminen A, Kaarniranta K, Haapasalo A, Hiltunen M, et al. Emerging role of p62/sequestosome-1 in the pathogenesis of Alzheimer's disease. Progress in Neurobiology. 2012;96:87–95. doi: 10.1016/j.pneurobio.2011.11.005. [DOI] [PubMed] [Google Scholar]
- [39].Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Molecular cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- [40].Dammer EB, Na CH, Xu P, Seyfried NT, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Morishima Y, Kanelakis KC, Murphy PJM, Lowe ER, et al. The Hsp90 Cochaperone p23 Is the Limiting Component of the Multiprotein Hsp90/Hsp70-based Chaperone System in Vivo Where It Acts to Stabilize the Client Protein·Hsp90 Complex. Journal of Biological Chemistry. 2003;278:48754–48763. doi: 10.1074/jbc.M309814200. [DOI] [PubMed] [Google Scholar]
- [42].Chaudhry UA, Dore S. Cytosolic prostaglandin E synthase: expression patterns in control and Alzheimer's disease brains. American journal of Alzheimer's disease and other dementias. 2009;24:46–51. doi: 10.1177/1533317508323655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The experimental population represents 5,569 phosphorylation site-containing peptides (Replicate 1; R1) where the average AD/CT ratio of phosphopeptide signal intensities give a log2-scaled population (red curve). To generate a null Gaussian distribution (blue curve) peptides for 7 of 8 control and 7 of 8 AD samples were analyzed by LC-MS/MS as a technical replicate (R2). Signal intensities for each of the phosphopeptides across the paired replicates (R1 and R2) of 14 matched LC-MS/MS runs were analyzed as a null ratio (R1/R2). These 2,709 log2-transformed null experiment ratios were plotted as a histogram, with the maximum scaled to equal the maximum height of the histogram of the experimental values from R1. Both populations fit a Gaussian distribution, which enabled a calculation of the standard deviation (SD) of the null (0.286 log2 units) and experimental (0.304 log2 units) populations. A cutoff of 2.79 fold of the null SD (0.80 log2 units, or >1.74-fold change in AD/CT) was therefore consistent with a theoretical confidence interval of 99.5% (green dashed lines). Empirical comparison of null and experimental marker counts obtained by comparative marker selection empirically determined false discovery to be 6.7 percent (see section 2.5).