Abstract
A diverse set of SUMO target proteins has been identified. Therefore, there is a growing interest in studying sumoylation and SUMO interactions in cells. When the sumoylation of a protein or a SUMO-interaction is suspected, a standard co-immunoprecipitation analysis using anti-SUMO and anti-target protein antibody is usually performed as a first step. However, the identification of endogenous sumoylated proteins is challenging because of the activity of isopeptidases, and often, only a small fraction of a target protein is sumoylated at a given time. Herein, we briefly summarize several important steps to ensure a successful co-immunoprecipitation analysis to detect possible sumoylation.
Keywords: SUMO, sumoylation, co-immunoprecipitation, isopeptidases, mass spectrometry, in vitro sumoylation
Introduction
Post-translational modification by Small Ubiquitin-like Modifiers (sumoylation) has been identified as an important regulatory event that is implicated in several cellular processes, seemingly based on the cell type [1–5]. Four SUMO paralogs have been identified: SUMO1, 2, 3 (often termed SUMO2/3 because of their 95% sequence identity) and 4. While SUMO1, 2, and 3 are abundantly expressed in different tissues, SUMO4 is restricted to the kidney, liver and lymph nodes [6–8]. For the initiation of sumoylation, SUMO is first proteolytically cleaved by a Sentrin-specific protease (SENP) to expose a diglycine motif at its C-terminus. The formation of an isopeptide bond between the C-terminal glycine of the SUMO and a lysine residue in the target protein is mediated by an enzymatic cascade that involves the actions of SUMO activating enzyme (E1, Uba2/Aos1), a SUMO-conjugating enzyme (E2, Ubc9), and a SUMO E3-ligase [9–13]. Sumoylation often occurs on a lysine residue within a consensus sequence: ψ-K-X-D/E, where ψ indicates a hydrophobic amino acid and X indicates any amino acid [14]. However, not all consensus sequences are sumoylated, and sumoylation often occurs outside of the consensus sequences [15]. Furthermore, there is a growing list of proteins that interact non-covalently with SUMO [16–20]. SUMO2/3 but not SUMO1 contain the consensus sequence and mixed SUMO chains with a terminal SUMO1 have been reported [14]. Sumoylation is reversed through the action of SENPs that cleave the isopeptide bond between SUMO and its substrate. A diverse set of SUMO target proteins has been identified, including factors that regulate transcription, replication, DNA repair, RNA metabolism, translation, and transport. [16– 21].
There is a growing interest in studying sumoylation and SUMO interaction in different fields of cell biology. When a post-translational modification of a protein or a SUMO-interaction is suspected, a standard co-immunoprecipitation (IP) analysis using anti-SUMO and anti-target protein antibodies is usually performed as a first step in many laboratories. However, the identification of endogenous sumoylated proteins is challenging. Although hundreds of proteins are modified by SUMO in different cell types, the SUMO moiety is easily lost from the sumoylated targets during cell lysis if the activity of isopeptidases (such as SENPs) is not inhibited. Furthermore, sometimes only a small fraction of a target protein (such as 5–10%) can be sumoylated at a given time, making identification difficult [22, 23]. Therefore, we briefly summarize herein several important steps to ensure a successful co-IP analysis with the use of commercially available anti-SUMO antibodies and the antibodies against the protein of interest to detect possible sumoylation. Omitting those steps during the IP or co-IP procedure will result in the inability to detect any specific signal.
1. Lysate concentration
As mentioned above, only a small fraction of a protein can be sumoylated at a given time; therefore, if the cell number is not a limiting factor, preparation of a highly concentrated protein lysate for co-IP studies is suggested. As a starting point, we suggest the use of 5×106 – 1×107 cells per each 300 µl of lysis buffer and approximately 600 µl of the lysate for each IP condition and each control. If the use of such a concentrated lysate results in the detection of a non-specific background in addition to the specific signal (including the background in the negative control), further experiments can be performed to reduce the protein concentration or/and increase the number of washes after the protein elution (see section 3) to eliminate the background but maintain the specific signal.
2. Lysis buffer composition and addition of an isopeptidase inhibitor
Many proteins are covalently and /or non-covalently modified by SUMO. It has therefore been suggested that the use of a denaturating lysis buffer containing, for example, a high percentage of SDS, would have the benefits of the immediate denaturation of isopeptidases and the elimination of non-covalent interactions with SUMO, thereby leaving only covalent SUMO binding in place [24–26]. The high percentage of SDS, however, would prevent sumoylated proteins from binding to anti-SUMO antibodies during the IP procedure, and as a result, SDS is usually either significantly diluted (e.g., 1:10) or removed from the lysis buffer by other means [24–26]. However, these manipulations can cause certain proteins to re-nature and non-covalent interactions to reform. In our studies of several SUMO targets, no significant difference has been found between the results of a co-IP analysis performed using denaturating and non-denaturating lysis buffers. In both cases, the conclusions concerning possible protein sumoylation were based on the presence of high-molecular weight protein conjugate(s) detected with both anti-SUMO and anti-target protein antibodies above the band corresponding to the non-modified form of the protein. It must be noted that using an SDS-containing buffer can still be favorable for the extraction of proteins that are difficult to extract with a regular buffer. If an SDS-containing buffer is used, SDS-removal columns (see below) can be employed so that the IP procedure can still be performed in small volumes; in our study, this approach worked better than diluting the lysate.
To produce the non-denatured lysate, approximately 5 × 106–107 cells should be re-suspended in 300 µl of whole cell extraction buffer and approximately 600 µl of the lysate should be used for each IP condition and each control. The buffer (for example, the Millipore Whole cell extraction kit, Life Technologies, 2910) should be supplemented with freshly prepared N-ethylmaleimide (NEM, an isopeptidase inhibitor that blocks the activities of SENPs [27]) at a final concentration of 15–20 mM. For example, a freshly prepared 20 mM NEM solution in Milli-Q water can be used to dilute the 5X concentrated lysis buffer. Isopeptidase inhibitors are not included in standard protein inhibitor cocktails but are critical for the prevention of protein desumoylation. As seen from Fig. 1A, when a lysis buffer is prepared without NEM, most of the high molecular weight (HMW) SUMO conjugates disappear, making their detection impossible. To ensure complete cell lysis, this cell suspension should be drawn through a 27-gauge needle 5 times and incubated on ice for 15 min. The lysate should be collected after high speed centrifugation at 4°C for 20 min.
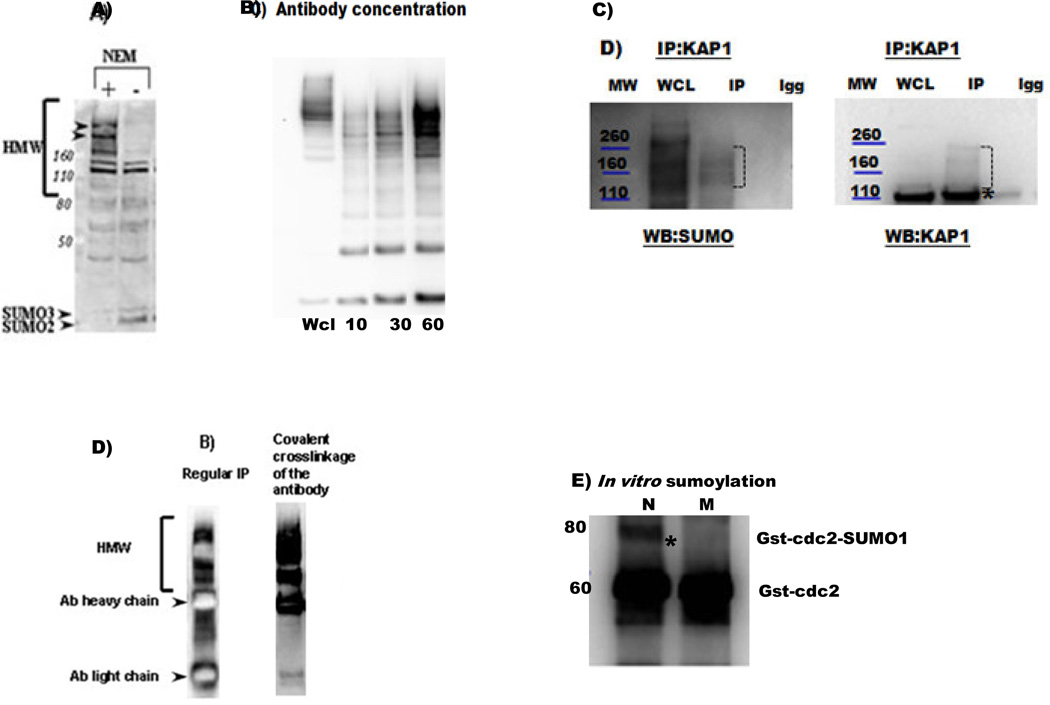
Figure 1.
A) A western blot analysis with anti-SUMO 2/3 antibody (ab3742) is shown following the preparation of a non-denaturative protein lysates (as described in section 2) with or without the addition of an isopeptidase inhibitor NEM. The absence of NEM results in a significant loss of high molecular weight (HMW) SUMO-conjugates (as shown, for example, for the two proteins indicated by arrowheads). The positions of free SUMO2 and SUMO3 and the migrating positions of MW markers are indicated.
B) Increasing the amount of anti-SUMO1 antibody (10, 30, 60 µl, 2, 6, 12 µg of antibody, respectively, (sc-5308 AC)) results in a better IP of sumoylated proteins; 1mg of protein in 600 µl of lysate was used. WCL-whole cell lysate.
C) Analysis of KAP1 sumoylation. Whole-cell lysate (WCL), negative control (Igg) and IP fractions are shown. The migrating positions of molecular weight (MW) markers are indicated. IP with anti- KAP1 antibody (ab 22553), followed by SUMO1(ab32058) and KAP1 western blot analyses identified high molecular weight SUMO-conjugates above a non-modified isoform of KAP1 (a band around 100 kDa, asterisk).
D) A cross-linkage of antibodies to the beads facilitates the elimination of the heavy and light chains in the eluate. An example of a SUMO2/3 IP analysis performed with and without anti-SUMO 2/3 antibody (ab3742) cross-linkage is shown.
E) To confirm the possible sumoylation of CDK1, an in vitro sumoylation reaction was performed with a recombinant GST- CDK1 protein, sumoylation enzymes (E1, E2), and either normal (N) SUMO or a mutant (M) SUMO incapable of forming an isopeptide bond (SUMOlink™ SUMO-1 kit from Active Motif (40120)). Western blot analysis with an anti-CDK1 antibody (Santa Cruz, sc-54) revealed the presence of a sumoylated CDK1 band above the non-modified CST-CDK1 when using the normal (N) but not mutant (M) SUMO isoform.
To produce denatured lysates, cell pellets should be re-suspended in a modified 2 × Laemmli buffer (150 mM Tris·HCl pH 7.2, 4% sodium dodecyl sulfate (SDS), 20% glycerol and 20 mM NEM; The re-activation of isopeptidases after the removal of SDS is possible, therefore, the inclusion of NEM in the denaturating buffer is still recommended). The solution should then be sonicated to reduce the viscosity of the sample and boiled at 100°C for 10 min. The lysates should then be collected after high speed centrifugation at room temperature for 15 min. Before the IP procedure, the denaturing lysate should be subjected to SDS removal using detergent removal spin columns (Thermo Scientific, (87778)), in accordance with the manufacturer’s instructions.
3. Antibody concentration required to immunoprecipitate most sumoylated proteins, covalent antibody cross-linkage, and additional immunoprecipitation procedure considerations
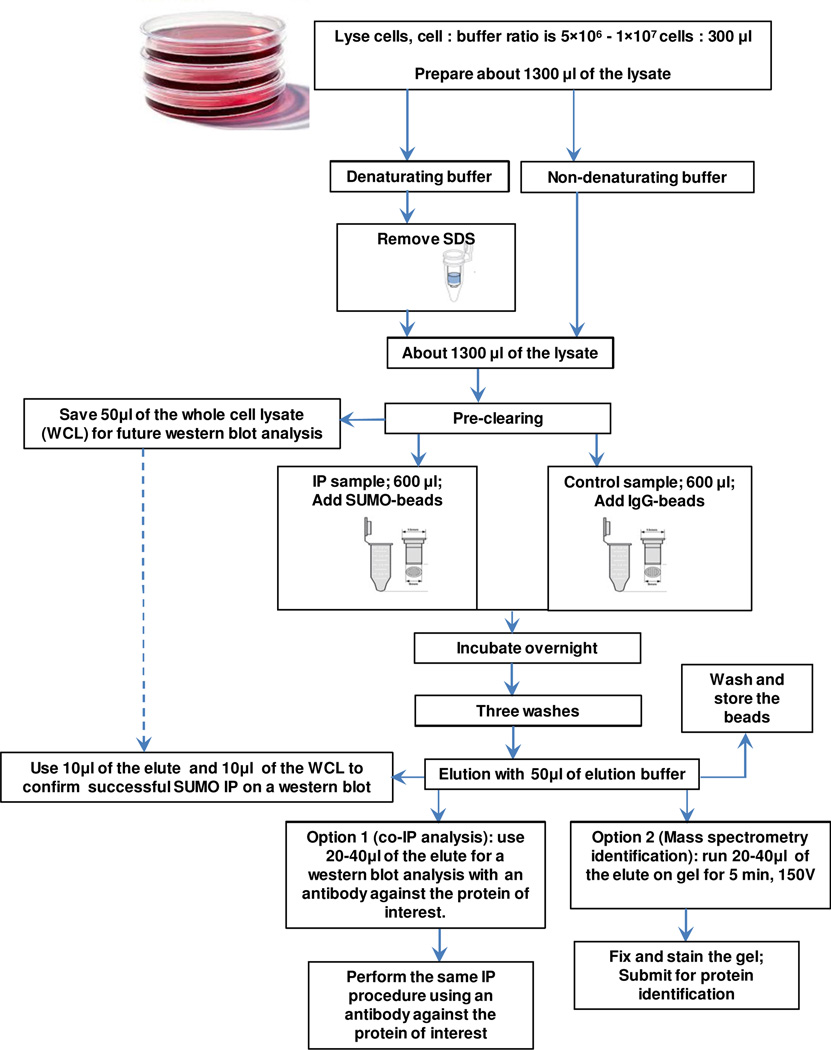
The IP procedure should follow a generally accepted protocol (a flow chat diagram of the procedure is depicted in Figure 2). Pre-clearing of the cell lysate by incubating the lysates with beads to decrease the non-specific binding is recommended. An aliquot of the whole cell lysate before IP should be kept aside for a later western blot analysis. We recommend the use of screw cap spin columns from Thermo Scientific (69705) for all IP steps to ensure successful washing steps without any bead loss.
Figure 2.
A flow chat diagram of co-IP analysis and optional identification of sumoylated proteins.
To precipitate all sumoylated proteins from a concentrated cell lysate, an unusually high amount of anti-SUMO antibody is required. As seen in Fig. 1B, increasing the amount of anti-SUMO antibody results in a better IP of sumoylated proteins. Preliminary experiments can be performed using a specific cell lysate and increasing concentrations of the anti-SUMO antibody to determine the optimal antibody/protein ratio for a particular cell line or tissue. As a starting point, we recommend the use of approximately 50 µl of SUMO antibody beads (Santa Cruz, sc-5308 AC), which contain 10 µg of the antibody per 500–1000 µg of protein in approximately 600 µl of the lysate buffer.
Overnight incubation with light agitation of the IP with anti-SUMO beads is recommended. IgG-cross-linked beads such as Mouse IgG–Agarose from Sigma (A0919) should be used as a negative control. After three washes with the lysis buffer, proteins should be incubated with approximately 50 µl of the elution buffer for 5 minutes at RT and then eluted. We recommend using a low-pH elution buffer, such as the one from Pierce (21004). An additional 50 µl of the elution buffer can be used as a column wash; however, the two eluate fractions should not be combined, and only the first fraction should be used for consecutive western blot analysis. The antibody beads can be re-used after rinsing with an elution buffer followed by washing with a lysis buffer and storing the beads in PBS supplemented with 1% (v/v) NaN3 (Fig. 2).The beads can be re-used several times (five or six) until they stop efficiently precipitating sumoylated proteins. Approximately 10 µl of the elution is generally sufficient to confirm a successful IP with anti-SUMO antibodies on a western blot. Anti-SUMO antibodies from Abcam (SUMO1, ab32058; SUMO2/3, ab3742) worked well in our group for western blotting, immunofluorescent, and immunohistochemistry analyses. A larger volume of the SUMO IP eluate (e.g., 20–30 µl, depending on the amount of sample buffer added and the wells of the gel) may be required for a western blot with an antibody against the target protein to detect its possible sumoylation. In addition, IP with the antibody against the target protein should be performed, preferably followed by two separate western blots (with anti-SUMO and and-anti target protein antibodies). For co-IP analysis of sumoylated proteins, we discourage stripping and re-probing the same membrane because some residual bands can interfere with the interpretation of the sumoylation results. Sumoylated isoform(s) should be detected with both SUMO and an anti-target protein antibody above the molecular weight corresponding to the non-sumoylated isoform of the protein (unless the sumoylated isoform is the predominant isoform, which is usually not the case). Many proteins interact non-covalently with sumoylated proteins, so it is possible to detect both covalent and/or non-covalent interactions with SUMO, which can be distinguished based on the molecular weight of the detected bands. Fig. 1C depicts a successful IP experiment with an anti-Kap1 antibody followed by a western blot with anti- SUMO1 and anti-KAP1 antibodies.
If the presence of both a heavy and light antibody chain interferes with the interpretation of the results for protein sumoylation or if all sumoylated proteins are identified by mass spectrometry after an IP with an anti-SUMO antibody, cross-linking of the antibodies to the bead support is recommended to avoid the presence of the antibody in the eluate. For SUMO IP, we recommend using the commercially available anti-SUMO antibody mentioned above, which is already conjugated to the bead support (Santa Cruz, sc-5308 AC). Alternatively, anti-SUMO and other antibodies of interest can be conjugated to the beads using an antibody conjugating kit from Thermo Scientific (26147). As seen in Figure 1D, a covalent cross-linkage of an anti-SUMO antibody eliminates the presence of the light and heavy antibody chains in the eluate, making both protein identification (see section 4) and a subsequent western blot analysis with an antibody of interest easier. It must be noted, however, that some antibodies can be modified by the covalent cross-linkage; thus, a successful IP procedure must be confirmed using a western blot analysis.
4. Mass spectrometric identification of sumoylated protein
If all sumoylated proteins must be identified, an IP using an anti-SUMO antibody beads should first be performed as described above, and the success of the IP procedure must be confirmed by a western blot analysis using SUMO antibodies and 10 µl of the eluate (Fig. 2). Subsequently, the maximum possible amount of the remaining eluate (e.g., 20–40 µl, depending on the amount of the sample buffer added and the wells of the gel) from both the IP and the negative control samples should be loaded on a gel, preferably spaced by 2 wells (to prevent any protein mixing or diffusion). The samples should be run at 150 V for approximately 5 min, and the gels should be fixed using a solution containing 50% (v/v) methanol and 7% (v/v) acetic acid at room temperature for 20 min followed by three washes with distilled water for 5 min each. The fixed gels should then be stained by incubation in GelCode® Blue Stain Reagent (Pierce, 24590) at room temperature for 1 hour. This step should be followed by washing with distilled water overnight to wash out excess staining solution. The gel can then be submitted to a mass spectrometry facility for identification of the sumoylated proteins (Fig. 2). For details of the procedure, see [21] The results from the IP and the negative control fractions should be carefully compared to identify specific results. For large-scale identification of sumoylated proteins, anti-SUMO antibodies can be produced in the laboratory using hybridoma cell lines, and the elution step can be performed using SUMO peptides to decrease non-specific binding [25].
5. Other methods to confirm sumoylation
Successful co-IP experiments strongly support the sumoylation of a protein. Other methods can be employed to support the observed results. In vitro sumoylation experiments can be performed using, for example, the SUMOlink™ SUMO-1 kit from Active Motif (40120). The reaction requires an in vitro synthesized protein and, in our experience, worked for some (maybe stably and/or heavily sumoylated) but not all proteins. It should be noted that sumoylation enzymes, which are added for the in vitro sumoylation reaction, are sumoylated themselves, resulting in numerous SUMO bands on the western blot. Therefore, conclusions regarding protein sumoylation can only be based on the western blot results with the antibody against the target protein. An example of the results from an in vitro sumoylation reaction using normal (N) and mutant (M), which is unable to form an isopeptide bond, isoforms of SUMO and in vitro translated GST-cdc2 is shown in Fig. 1E. For other in vitro methods to study possible protein sumoylation, such as the expression of both a wild type and a sumoylated deficient protein in cells, see [24, 28].
Acknowledgements
This study was supported by NIH; NICHD, Academic Research Enhancement Award 1R15HD067944-01A1 (MV, PI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ayaydin F, Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol Biol Cell. 2004;15(12):5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dasso M. Emerging roles of the SUMO pathway in mitosis. Cell Div. 2008;3:5. doi: 10.1186/1747-1028-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18(17):2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 4.Muller S, Ledl A, Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23(11):1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol Life Sci. 2007;64(23):3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohren KM, et al. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem. 2004;279(26):27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 7.Dohmen RJ. SUMO protein modification. Biochim Biophys Acta. 2004;1695(1–3):113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 8.Li M, et al. SUMO wrestling with type 1 diabetes. J Mol Med. 2005;83(7):504–513. doi: 10.1007/s00109-005-0645-5. [DOI] [PubMed] [Google Scholar]
- 9.Yang P, et al. Sumoylation modulates oxidative stress relevant to the viability and functionality of pancreatic beta cells. Am J Transl Res. 2014;6(4):353–360. [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh ET. SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem. 2009;284(13):8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 2007;17(8):370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32(6):286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J Cell Sci. 2009;122(Pt 23):4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276(16):12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 15.Blomster HA, et al. In vivo identification of sumoylation sites by a signature tag and cysteine-targeted affinity purification. J Biol Chem. 2010;285(25):19324–19329. doi: 10.1074/jbc.M110.106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chupreta S, et al. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol. 2005;25(10):4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8(6):550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin DY, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24(3):341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Song J, et al. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101(40):14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song J, et al. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280(48):40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 21.Vigodner M, et al. Localization and identification of sumoylated proteins in human sperm: excessive sumoylation is a marker of defective spermatozoa. Hum Reprod. 2013;28(1):210–223. doi: 10.1093/humrep/des317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu SY, Chiang CM. p53 sumoylation: mechanistic insights from reconstitution studies. Epigenetics. 2009;4(7):445–451. doi: 10.4161/epi.4.7.10030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker J, et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat Struct Mol Biol. 2013;20(4):525–531. doi: 10.1038/nsmb.2526. [DOI] [PubMed] [Google Scholar]
- 24.Tatham MH, et al. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4(9):1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- 25.Barysch SV, et al. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat Protoc. 2014;9(4):896–909. doi: 10.1038/nprot.2014.053. [DOI] [PubMed] [Google Scholar]
- 26.Sarge KD, Park-Sarge OK. Detection of proteins sumoylated in vivo and in vitro. Methods Mol Biol. 2009;590:265–277. doi: 10.1007/978-1-60327-378-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, et al. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J Biol Chem. 1999;274(44):31131–31134. doi: 10.1074/jbc.274.44.31131. [DOI] [PubMed] [Google Scholar]
- 28.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2(72):ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]