Highlights
-
•
Oncostatin M regulates nephronectin (Npnt) gene expression in a dose- and time dependent manner.
-
•
Nephronectin gene expression is regulated by JAK/STAT and MAPK pathways.
-
•
Down-regulation of Npnt influences inhibition of osteoblast differentiation by oncostatin M.
Abbreviations: Npnt, nephronectin; OSM, oncostatin M; JAK, janus kinase; STAT, signal transducer and activator of transcription; MAPK, mitogen-activated protein kinase; MAM, meprin, A5 protein, and receptor protein-tyrosine phosphatase μ; OSMR, OSM receptor; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; TGF-β, transforming growth factor-β; TNF-α, tumor necrosis factor-α; BMP-2, bone morphogenetic protein-2; MEF2, myocyte enhancer-binding factor 2A
Keywords: Nephronectin, Oncostatin M, JAK/STAT, MAPK
Abstract
Nephronectin (Npnt), also called POEM, is an extracellular matrix protein considered to play critical roles as an adhesion molecule in the development and functions of various tissues, such as the kidneys, liver, and bones. In the present study, we examined the molecular mechanism of Npnt gene expression and found that oncostatin M (OSM) strongly inhibited Npnt mRNA expression in MC3T3-E1 cells from a mouse osteoblastic cell line. OSM also induced a decrease in Npnt expression in both time- and dose-dependent manners via both the JAK/STAT and MAPK pathways. In addition, OSM-induced inhibition of osteoblast differentiation was recovered by over-expression of Npnt. These results suggest that OSM inhibits Npnt expression via the JAK/STAT and MAPK pathways, while down-regulation of Npnt by OSM influences inhibition of osteoblast differentiation.
1. Introduction
Nephronectin (Npnt), an extracellular matrix protein identified as a ligand that mediates the function of integrin α8β1 in developing kidneys, has 5 EGF-like domains, an Arg-Gly-Asp (RGD) cell binding motif, and MAM domain [1,2]. This protein also has critical roles in other tissues, such as kidney development [3], exacerbation of hepatitis [4], and promotion of osteoblast differentiation [5,6].
MC3T3-E1 cells are used for in vitro bone model of development systems, in which 2 time-dependent developmental sequences, proliferation and differentiation of osteoblasts are observed [6]. Kahai et al. found that when MC3T3-E1 cells were stably transfected with a small interfering RNA (siRNA) plasmid for inhibition of Npnt gene expression, osteoblast differentiation was inhibited, whereas over-expression of Npnt promoted osteoblast differentiation [6]. Other studies have also demonstrated that TGF-β inhibits Npnt expression in both time- and dose-dependent manners, while Npnt-induced osteoblast differentiation was shown to be inhibited by TGF-β in MC3T3-E1 cells [7,8]. Similarly, TNF-α inhibits Npnt expression in both time- and dose-dependent manners, and down-regulation of Npnt influences inhibition of osteoblast differentiation by TNF-α [7,9].
Oncostatin M (OSM) is a cytokine and member of the interleukin (IL)-6 subfamily that includes IL-6, leukemia inhibitory factor (LIF), ciliary neurotrophic factor (CNTF), IL-11, cardiotrophin-like cytokine (CLC), cardiotrophin-1 (CT-1), and novel neurotrophin-1/B cell stimulating factor-3 (NNT-1/BSF-3) [10,11]. OSM has unique and pleiotropic activities to regulate fetal liver hepatic cell differentiation, connective tissue framework balance, adipocyte differentiation, and proliferation of some types of cancer [12]. In mice, OSM binds the type II receptor, composed of the OSM receptor (OSMR) and gp130, a receptor subunit common among IL-6 subfamilies, and transmits signals via various routes including the JAK/STAT and MAPK pathways [13].
In the present study, we found that OSM regulates the expression of Npnt via the JAK/STAT and MAPK signaling pathways, while down-regulation of Npnt by OSM influences the inhibition of osteoblast differentiation in MC3T3-E1 cells, a mouse calvaria-derived osteoblast progenitor cell line.
2. Results
We first examined whether the expression of Npnt is regulated by cytokines belonging to the IL-6 subfamily, such as OSM, LIF, IL-6, and IL-11, using MC3T3-E1 cells. After treatment with 100 ng/ml of each cytokine separately for 24 h, OSM was found to sharply decrease the expression of Npnt mRNA in the cells (Fig. 1). We also examined the effects of OSM on Npnt gene expression using another cell line, C2C12 cells from a mouse myoblast cell line. OSM inhibited Npnt gene expression in C2C12 cells (Suppl. Fig. 1). Next, to more precisely investigate the influence of OSM, we treated MC3T3-E1 cells with a range of concentrations of OSM for various time periods. As shown in Fig. 2A, OSM significantly induced down-regulation of Npnt mRNA expression in a dose-dependent manner. Furthermore, when the cells were exposed to OSM for at least 6 h, a strong reduction of Npnt mRNA expression occurred in a time-dependent manner (Fig. 2B).
Fig. 1.
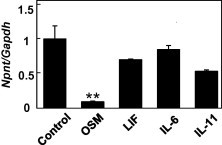
Reduction of Npnt mRNA expression by OSM. MC3T3-E1 cells were treated with 100 ng/ml of OSM, LIF, IL-6, or IL-11 for 24 h. Total cellular RNA was extracted, and mRNA levels for Npnt and Gapdh were examined by real-time PCR analysis. Results are shown as the mean ± SD from 3 samples as compared to the level at 0 ng/ml (without cytokine treatment). ∗∗P < 0.01, ∗P < 0.05, Student’s t test relative to the level without cytokine treatment.
Fig. 2.
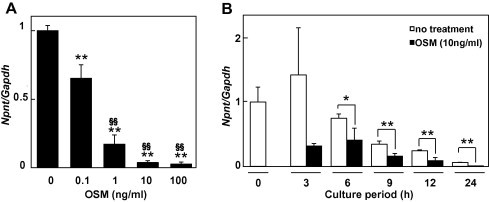
Dose- and time-dependent effects of OSM on Npnt mRNA expression. (A) Dose-dependent effects of OSM on Npnt mRNA expression. MC3T3-E1 cells were treated with 0, 0.1, 1, 10, or 100 ng/ml of OSM for 24 h. Results are shown as the mean ± SD from 3 samples as compared to the level with 0 ng/ml of OSM. ∗∗P < 0.01, ANOVA relative to the level with 0 ng/ml of OSM. §§P < 0.01, ANOVA relative to the level with 0.1 ng/ml of OSM. (B) Time course analysis of effects of OSM on Npnt mRNA expression. MC3T3-E1 cells were treated with 10 ng/ml of OSM for 3, 6, 9, 12, or 24 h. Total cellular RNA was extracted, and mRNA levels for Npnt and Gapdh were examined using real-time PCR analysis. Results are shown as the mean ± SD from 3 samples as compared to the level at 0 h. ∗∗P < 0.01, ∗P < 0.05, Student’s t test as compared to the level with 0 ng/ml of OSM at each time points.
Since the binding of cytokines to their specific cell surface receptor complexes is the start of cytokine signaling, we analyzed the presence of receptors that mediate OSM signaling using RT-PCR (Fig. 3A) and FACS (Fig. 3B) analyses. The RT-PCR shows that both the OSM receptor (OSMR) and gp130 are expressed by MC3T3-E1 cells, while OSMR is located on the cell surface.
Fig. 3.
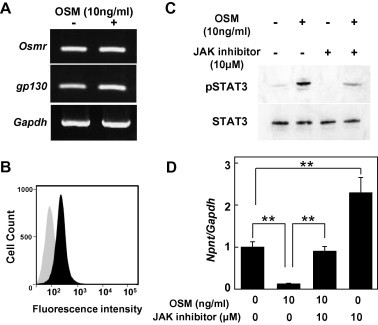
Analysis of OSM signaling molecules. (A) RT-PCR analysis for presence of OSM receptor (Osmr) and gp130 in MC3T3-E1 cells. With the present primers, the size of the PCR products for Osmr and gp130 were 132 and 100 bp, respectively. (B) MC3T3-E1 cells were subjected to flow cytometry analysis for the presence of the OSM receptor (OSMR) on the cell surface. The black area shows MC3T3-E1 cells incubated with biotinylated anti-OSMR and FITC streptavidin. The gray area shows MC3T3-E1 cells incubated with biotinylated anti-rat IgG2b κ isotype and FITC streptavidin. All cells were found positive for the OSM receptor. (C) OSM-induced phosphorylation of STAT3 blocked by JAK inhibitor. MC3T3-E1 cells were pretreated with 10 μM of the JAK inhibitor for 1 h, then treated with 10 ng/ml of OSM for 15 min. Total cellular proteins were extracted, and used to determine levels of phosphorylated and total STAT3 [23]. (D) Inhibition of down-regulation of OSM-induced Npnt expression by JAK inhibitor. MC3T3-E1 cells were treated with 10 ng/ml of OSM and 10 μM of the JAK inhibitor for 24 h. Total cellular RNA was extracted, and mRNA levels for Npnt and Gapdh were examined using real-time PCR analysis. Results are shown as the mean ± SD from 3 samples as compared to the level with 0 ng/ml of OSM and the JAK inhibitor. ∗∗P < 0.01, Student’s t test relative to the level with 0 ng/ml of the JAK inhibitor.
Following identification of the OSM receptor complex in MC3T3-E1 cells, we detected a signaling pathway utilized by OSM in MC3T3-E1 cells. Phosphorylated STAT3 was revealed by western blot analysis and that phosphorylation was reduced by exposure to the JAK inhibitor (10 μM) (Fig. 3C). Based on those results, the expression of Npnt in MC3T3-E1 cells treated with OSM and the JAK inhibitor was analyzed using real-time PCR. As shown in Fig. 3D, 10 μM of the JAK inhibitor blocked the inhibition induced by 10 ng/ml of OSM and restored Npnt mRNA expression. A previous study demonstrated that OSM stimulates MAPKs, such as ERK1/2, p38 MAPK, and JNK, in bovine and human chondrocytes [14]. To determine whether the MAPK pathway is involved in down-regulation of Npnt gene expression by OSM, MC3T3-E1 cells were treated with 10 μM of PD98059 [MAPK-ERK kinase (MEK) inhibitor], SB203580 (p38 MAPK inhibitor), or SP600125 (JNK inhibitor) following treatment with OSM at 10 ng/ml. Treatment with PD98059, SB203580, and SP600125 inhibited the down-regulation of Npnt gene expression by OSM (Fig. 4). These findings suggest that the mechanism of regulation of Npnt expression by OSM occurs via activation of MAPK pathway components and JAK/STAT.
Fig. 4.
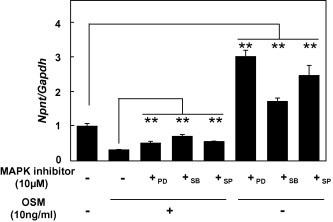
Inhibition of Npnt expression by OSM is regulated by ERK1/2. MC3T3-E1 cells were treated with 10 ng/ml of OSM and MAPK inhibitors (+PD; 10 μM PD98059,+SB; 10 μM SB203580, or +SP; 10 μM SP600125) for 24 h. Total cellular RNA was extracted, and mRNAs for Npnt and Gapdh were examined using real-time PCR analysis. Results are shown as the mean ± SD from 6 samples as compared to the level with 0 ng/ml of OSM and the MAPK inhibitors. ∗∗P < 0.01, Student’s t test relative to the level without MAPK inhibitors.
To elucidate the relationship between inhibition of osteoblast differentiation and reduction of Npnt gene expression by OSM, we established Npnt over-expressing MC3T3-E1 cells. MC3T3-E1 cells were transfected with an Npnt expression vector containing a neomycin resistant gene, then cultured in 3 mg/ml of medium containing G418 for 1 week. To examine whether over-expression of Npnt overcomes OSM-induced down-regulation of osteoblast differentiation with BMP-2 which is an inducer of bone genesis, MC3T3-E1 cells over-expressing Npnt were treated with OSM (Fig. 5). Our results showed that over-expression of Npnt reduced the level of OSM-induced inhibition of osteoblast differentiation with BMP-2.
Fig. 5.
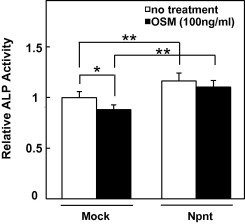
Over-expression of Npnt recovers OSM-induced inhibition of osteoblast differentiation. Npnt expressed vector or empty vector transfected MC3T3-E1 cells were treated with BMP-2 (500 ng/ml) and with/without OSM (■ OSM (100 ng/ml) or □ no treatment), then alkaline phosphatase (ALP) activity was determined after 2 days of treatment. Results are shown as the mean ± SD from 6 samples as compared to the level in empty vector transfected MC3T3-E1 cells with 0 ng/ml of OSM. ∗∗P < 0.01, ∗P < 0.05, Student’s t test relative to level in empty vector transfected MC3T3-E1 cells.
3. Discussion
The present novel findings demonstrated that OSM strongly inhibits Npnt gene expression via the JAK/STAT and MAPK signaling pathways, while OSM-induced inhibition of osteoblast differentiation is overcomed by Npnt over-expression (Fig. 6).
Fig. 6.

OSM strongly inhibits Npnt gene expression via the JAK/STAT and MAPK signaling pathways, while OSM-induced inhibition of osteoblast differentiation is recovered by Npnt over-expression in MC3T3-E1. RGD; motif of Arg-Gly-Asp tripeptide.
One of the physiological functions of Npnt is promotion of osteoblast differentiation. This Npnt-induced promotion of osteoblastogenesis is regulated by TGF-β [5]. Furthermore, TNF-α, a pro-inflammatory cytokine and inhibitory molecule of osteoblast differentiation, was also shown to regulate Npnt expression via NF-κB signaling [9]. OSM influences the differentiation and proliferation of osteoblasts [15], and Shih and Yen demonstrated that OSM significantly decreased cell proliferation and alkaline phosphatase activity, indicating the level of osteoblast differentiation, in a dose-dependent manner [16]. On the other hand, other reports noted that OSM induced alkaline phosphatase activity in MG63 cells obtained from a fibroblastic osteosarcoma, suggesting that its effect on osteoblast differentiation is dependent on the differentiation status of the osteoblastic cell line being tested [17–19]. Our results suggest that OSM-induced down-regulation of Npnt may be, at least in part, involved in regulation of osteoblast differentiation.
Previous studies demonstrated that OSM inhibits the activity of MEF2A, a transcription factor, through JAK/STAT signaling pathway in C2C12 cells [20], and the inhibition of MEF2A reduces the expression of Kruppel-like factor (Klf) 2 in COS-7 cells [21]. Sunadome et al. demonstrated that expression of Npnt is regulated by Klf2 via ERK5 activation in C2C12 cells during skeletal muscle differentiation [22]. In the Npnt promoter, there are eight copies of CACCC motifs which are Klf2 binding sites, and Npnt promoter activity is directly activated by Klf2. From those studies, OSM-induced down-regulation of Npnt can be explained by inhibition of Klf2 and MEF2 which are regulated by JAK/STAT signaling pathways, but further experiments are required to fully elucidate this mechanism.
4. Materials and methods
4.1. Cell cultures
MC3T3-E1 cells were maintained in MEMα with l-glutamine and phenol red medium (Wako Pure Chemical Industries, Ltd., Cat. No. 135-15175), supplemented with 10% fetal bovine serum (FBS) (Life Technologies, Cat. No. 10437) and 1% penicillin–streptomycin at 37 °C in a CO2 incubator (5% CO2, 95% air). To establish Npnt over-expressing cells, MC3T3-E1 cells were transfected with a Npnt-FLAG tagged vector or empty vector [2], then cultured in 10% FBS-MEMα containing 3 mg/ml G418 (Calbiochem-NovaBiochem Corp., Cat. No. A1720-1G). C2C12 cells were maintained in DMEM (Wako Pure Chemical Industries, Ltd., Cat. No. 044-29765) supplemented with 15% FBS and 1% penicillin–streptomycin at 37 °C in a CO2 incubator. For the experiments, cells were plated at 2 × 105 cells/well in 6-well plates (Thermo Scientific Inc., Cat. No. 140675).
4.2. Reagents
Mouse OSM (Cat. No. 495-MO-025), human LIF (Cat. No. 7734-LF-025), and mouse IL-11 (Cat. No. 418-ML-005), and human BMP-2 (Cat. No. 355-BEC-010) were purchased from R&D Systems, Inc., while human IL-6 (Cat. No. MAN0003501) came from Life Technologies and a JAK inhibitor (pyridone 6 [2-(1,1-dimethylethyl)-9-fluoro-3,6-dihydro-7H-benz[h]-imidaz[4,5-f]isoquinolin-7-one]) (Cat. No. 420099) from EMD Chemicals, Inc. PD98059 (Cat. No. P215-1 mg), SB203580 (Cat. No. S8307-1 mg), and SP600125 (Cat. No. S5567-10 mg) were purchased from SIGMA.
4.3. RT-PCR
Total RNA was extracted using TRIzol reagent (Life Technologies, Cat. No. 15596018), then reverse transcribed using SuperScript III (Life Technologies, Cat. No. 18080-044). PCR was performed with Taq polymerase (Promega, Cat. No. M7123) using the following specific PCR primers.
Glyceraldehyde 3-phosphate dehydrogenase (Gapdh):
5′-GAAGGTCGGTGTGAACGGATTTGGC-3′,
5′-CATGTAGGCCATGAGGTCCACCAC-3′.
Osm receptor (Osmr):
5′-ATCCAAAGGCTCCGCAGGAC-3′,
5′-GTAAGGTTGCAGGTCAAGGC-3′.
gp130:
5′-ACATCGTGTGGAAGACCAAC-3′,
5′-ACTCTGATTTCAAAGTGTAG-3′.
4.4. Quantitative real-time PCR
Quantitative real-time PCR was performed using a SYBR Green Fast PCR system (Applied Biosystems) with the following specific PCR primers.
Gapdh:
5′-AAATGGTGAAGGTCGGTGTG-3′,
5′-TGAAGGGGTCGTTGATGG-3′.
Nephronectin (Npnt):
5′-CACGAGTAATTACGGTTGACAACAG-3′,
5′-CTGCCGTGGAATGAACACAT-3′.
4.5. Western blotting
Protein samples were collected using Sample Buffer Solution with Reducing Reagent (6×) for SDS–PAGE (Nacalai Tesque, Cat. No. 09499-14) with a cell scraper (Corning Incorporated, Corning, Cat. No. 3010), then electrophoresed on a 7.5% SDS polyacrylamide gel, which was blotted onto a nitrocellulose membrane of the iBlot®2 Dry Blotting System (Life Technologies, Cat. No. IB21001). The nitrocellulose membrane was soaked in iBind™ solution (Life Technologies) for 24 h at 4 °C, after which the blots were rinsed with 10 μl of primary and secondary antibodies in iBind™ (Life Technologies, Cat. No. SLF1000). Antibodies for phospho-STAT3 (Cat. No. 9131S) and STAT3 (Cat. No. 9139) were purchased from Cell Signaling Technology, Inc. Anti-rabbit (Cat. No. NA934V) and anti-mouse (Cat. No. NA931VS) IgG horseradish peroxidase linked antibodies were purchased from GE Healthcare, and used as secondary antibodies to detect each of the primary antibodies. To visualize the locations of the antigenic bands, peroxidase reactions were developed using ECL Prime Western Blotting Detection Reagent (GE Healthcare).
4.6. Flow cytometry analysis
After performing cell dissociation with Cell Dissociation Solution (Sigma–Aldrich, Cat. No. C5914), MC3T3-E1 cells were incubated with FcR Blocking Reagent for mouse (MACS Miltenyi Biotec K.K., Cat. No. 120-003-855). Next, the cells were incubated with biotinylated rat IgG2b κ isotype as a control (Becton, Dickinson and Company, Cat No. 553987) or a biotinylated anti-mouse OSM receptor (OSMR) antibody (Medical & Biological Laboratories CO., Ltd., Cat. No. D059-3), followed by incubation with FITC streptavidin (Medical & Biological Laboratories CO., Ltd., Cat. No. 554060). Fluorescent immunostained samples were examined using FACSVerse Ver. 1.0 Suite 1.0.3 (Becton, Dickinson and Company), and data acquisition and analysis were performed using FACSuite™ (Becton, Dickinson and Company).
4.7. ALP (alkaline phosphatase) activity
ALP activity in the cell lysates was determined following incubation with the substrate p-nitrophenylphosphate (Wako Pure Chemical Industries, Ltd., Cat. No. 149–02342) in buffer (pH 10) containing 0.1 M 2-amino-2-methyl-1-propanol and 2 mM MgCl2.
4.8. Statistical analysis
All results are expressed as means ± SD. In Fig. 2A and Suppl. Fig. 1, statistical analysis was performed using one-way ANOVA. In other results, statistical analysis was performed using a two-tailed Student’s t test. A P value of P < 0.05 was considered statistically significant.
Conflict of interest
The authors have not conflicts of interest to declare in regard to this study.
Authors’ contributions
Conceived and designed the experiments: TK, AY, NM, RK. Performed the experiments: TK, AY, MT, DS, YS, KH, TE. Analyzed the data: TK, AY, MY, TI, TS, HI. Contributed reagents/materials/analysis tools: AY, NM. Wrote the manuscript: TK, AY, HI, RK.
Acknowledgments
We express our gratitude to Dr. Masato Yamamoto for the kind advice and encouragement. For one-way ANOVA, we thank Ms. Satomi Nimura for guidance on the use of statistics software produced by SSRI. This work was supported in part by the Project to Establish Strategic Research Center for Innovative Dentistry by the Ministry of Education, Culture, Sports, Science and Technology of Japan, and Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
Appendix A. Supplementary data
Dose-dependent effects of OSM on Npnt mRNA expressions in C2C12 cells. C2C12 cells were treated with 0, 1, 10, or 100 ng/ml of OSM for 24 h. Results are shown as the mean ± SD from 3 samples as compared to the level with 0 ng/ml of OSM. ∗∗P < 0.01, ANOVA relative to the level with 0 ng/ml of OSM.
References
- 1.Brandenberger R., Schmidt A., Linton J., Wang D., Backus C., Denda S., Muller U., Reichardt L.F. Identification and characterization of a novel extracellular matrix protein nephronectin that is associated with integrin alpha8beta1 in the embryonic kidney. J. Cell. Biol. 2001;154:447–458. doi: 10.1083/jcb.200103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morimura N., Tezuka Y., Watanabe N., Yasuda M., Miyatani S., Hozumi N., Tezuka Ki K. Molecular cloning of POEM: a novel adhesion molecule that interacts with alpha8beta1 integrin. J. Biol. Chem. 2001;276:42172–42181. doi: 10.1074/jbc.M103216200. [DOI] [PubMed] [Google Scholar]
- 3.Linton J.M., Martin G.R., Reichardt L.F. The ECM protein nephronectin promotes kidney development via integrin alpha8beta1-mediated stimulation of Gdnf expression. Development. 2007;134:2501–2509. doi: 10.1242/dev.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inagaki F.F. Nephronectin is upregulated in acute and chronic hepatitis and aggravates liver injury by recruiting CD4 positive cells. Biochem. Biophys. Res. Commun. 2013;430:751–756. doi: 10.1016/j.bbrc.2012.11.076. [DOI] [PubMed] [Google Scholar]
- 5.Kahai S., Lee S.C., Seth A., Yang B.B. Nephronectin promotes osteoblast differentiation via the epidermal growth factor-like repeats. FEBS Lett. 2010;584:233–238. doi: 10.1016/j.febslet.2009.11.077. [DOI] [PubMed] [Google Scholar]
- 6.Kahai S. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One. 2009;4:e7535. doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazono A. TGF-beta suppresses POEM expression through ERK1/2 and JNK in osteoblasts. FEBS Lett. 2007;581:5321–5326. doi: 10.1016/j.febslet.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Fang L., Kahai S., Yang W., He C., Seth A., Peng C., Yang B.B. Transforming growth factor-beta inhibits nephronectin-induced osteoblast differentiation. FEBS Lett. 2010;584:2877–2882. doi: 10.1016/j.febslet.2010.04.074. [DOI] [PubMed] [Google Scholar]
- 9.Tsukasaki M. Expression of POEM, a positive regulator of osteoblast differentiation, is suppressed by TNF-alpha. Biochem. Biophys. Res. Commun. 2011;410:766–770. doi: 10.1016/j.bbrc.2011.06.048. [DOI] [PubMed] [Google Scholar]
- 10.Jazayeri J.A., Carroll G.J., Vernallis A.B. Interleukin-6 subfamily cytokines and rheumatoid arthritis: role of antagonists. Int. Immunopharmacol. 2010;10:1–8. doi: 10.1016/j.intimp.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Senaldi G. Novel neurotrophin-1/B cell-stimulating factor-3: a cytokine of the IL-6 family. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11458–11463. doi: 10.1073/pnas.96.20.11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards C.D. The enigmatic cytokine oncostatin M and roles in disease. ISRN Inflamm. 2013;2013:1–23. doi: 10.1155/2013/512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka M., Miyajima A. Oncostatin M, a multifunctional cytokine. Rev. Physiol. Biochem. Pharmacol. 2003;149:39–52. doi: 10.1007/s10254-003-0013-1. [DOI] [PubMed] [Google Scholar]
- 14.Li W.Q., Dehnade F., Zafarullah M. Oncostatin M-induced matrix metalloproteinase and tissue inhibitor of metalloproteinase-3 genes expression in chondrocytes requires Janus kinase/STAT signaling pathway. J. Immunol. 2001;166:3491–3498. doi: 10.4049/jimmunol.166.5.3491. [DOI] [PubMed] [Google Scholar]
- 15.Heymann D., Rousselle A.V. Gp130 cytokine family and bone cells. Cytokine. 2000;12:1455–1468. doi: 10.1006/cyto.2000.0747. [DOI] [PubMed] [Google Scholar]
- 16.Shih C., Yen C.C. Effect of oncostatin-M on proliferation and activity in osteoblastic MC3T3-E1 cells. Zhonghua Yi Xue Za Zhi (Taipei) 1999;62:710–716. [PubMed] [Google Scholar]
- 17.Bellido T., Borba V.Z., Roberson P., Manolagas S.C. Activation of the Janus kinase/STAT (signal transducer and activator of transcription) signal transduction pathway by interleukin-6-type cytokines promotes osteoblast differentiation. Endocrinology. 1997;138:3666–3676. doi: 10.1210/endo.138.9.5364. [DOI] [PubMed] [Google Scholar]
- 18.Bellido T., O’Brien C.A., Roberson P.K., Manolagas S.C. Transcriptional activation of the p21(WAF1, CIP1, SDI1) gene by interleukin-6 type cytokines. A prerequisite for their pro-differentiating and anti-apoptotic effects on human osteoblastic cells. J. Biol. Chem. 1998;273:21137–21144. doi: 10.1074/jbc.273.33.21137. [DOI] [PubMed] [Google Scholar]
- 19.Chipoy C. Downregulation of osteoblast markers and induction of the glial fibrillary acidic protein by oncostatin M in osteosarcoma cells require PKCdelta and STAT3. J. Bone Miner Res. 2004;19:1850–1861. doi: 10.1359/JBMR.040817. [DOI] [PubMed] [Google Scholar]
- 20.Xiao F., Wang H., Fu X., Li Y., Ma K., Sun L., Gao X., Wu Z. Oncostatin M inhibits myoblast differentiation and regulates muscle regeneration. Cell Res. 2011;21:350–364. doi: 10.1038/cr.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A., Lin Z., SenBanerjee S., Jain M.K. Tumor necrosis factor alpha-mediated reduction of KLF2 is due to inhibition of MEF2 by NF-kappaB and histone deacetylases. Mol. Cell. Biol. 2005;25:5893–5903. doi: 10.1128/MCB.25.14.5893-5903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sunadome K., Yamamoto T., Ebisuya M., Kondoh K., Sehara-Fujisawa A., Nishida E. ERK5 regulates muscle cell fusion through Klf transcription factors. Dev. Cell. 2011;20:192–205. doi: 10.1016/j.devcel.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S.Q. Interleukin 29 enhances expression of Toll receptor 3 and mediates antiviral signals in human keratinocytes. Inflamm. Res. 2011;60:1031–1037. doi: 10.1007/s00011-011-0364-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dose-dependent effects of OSM on Npnt mRNA expressions in C2C12 cells. C2C12 cells were treated with 0, 1, 10, or 100 ng/ml of OSM for 24 h. Results are shown as the mean ± SD from 3 samples as compared to the level with 0 ng/ml of OSM. ∗∗P < 0.01, ANOVA relative to the level with 0 ng/ml of OSM.


