Abstract
Background
Eph receptors, comprising the A- and B- subfamilies, are the largest family of receptor tyrosine kinases in the mammalian genome, and their function is critical for morphogenesis in a variety of contexts. Whereas signaling through B-type Ephs has been demonstrated to play a role in cleft lip and palate (CL/P), the involvement of A-type Ephs has not been examined in this context notwithstanding a recent genome-wide association study that identified the EPHA3 locus as a candidate for non-syndromic CL/P.
Results
Here we present a systematic analysis of the gene expression patterns for the nine EphA receptors at progressive stages of mouse development and find that EphA3, EphA4 and EphA7 exhibit restricted overlapping patterns of expression during palate development. We find that homozygous mutation of EphA3 or compound homozygous mutation of EphA3 and EphA4 in mice does not result in defective midfacial development, supporting the possibility of redundant function with EphA7. We also document previously undescribed expression patterns in other tissues of the craniofacial complex including the lacrimal duct and salivary glands.
Conclusions
Together, these results are consistent with the hypothesis that mutations in EPHA family genes may cause CL/P and also suggest that functional redundancy between family members may be at play.
Keywords: Eph, ephrin, palate, lip, craniofrontonasal syndrome
Introduction
Eph receptors are the largest known group of receptor tyrosine kinases and, along with their membrane bound ephrin ligands, contribute to a number of developmental processes, including boundary formation, cell migration, and axon guidance (Bush and Soriano, 2010; Klein, 2012). Both receptor and ligand are membrane-bound and cell-to-cell contact is required for activation. The Eph receptors are divided into two classes, the A-type and the B-type. In mice, there are nine A-type Ephs (EphA1-8 and EphA10) and five B-type Ephs (EphB1-4 and EphB6). Both classes of receptors have similar structures but differ in their binding affinities for ephrin ligands. In general, the A-type receptors bind GPI anchored A-type ephrin ligands promiscuously, and the B-type receptors bind transmembrane B-type ephrin ligands; known exceptions to this are EphA4, which can bind to both classes of ephrin, and EphB2, which can bind to ephrin-A5 (Blits-Huizinga et al., 2004; Gale et al., 1996; Himanen et al., 2004). Full activation of Eph/ephrin signaling requires the formation of higher order oligomers, and it has been shown in cell culture that EphA and EphB receptors can hetero-oligomerize and cross-phosphorylate, suggesting this mode of signaling cross-talk must also be considered in vivo (Janes et al., 2012, 2011).
The mammalian palate consists of a primary and secondary part and its development occurs between E10.5 and E15.5 in mice. Development of the midface begins with formation of the maxillary process (MXP) and frontonasal process (FNP) which are composed of mesenchyme derived from the neural crest, and bound externally by a thin layer of epithelium derived from the ectoderm. The FNP is separated into medial (MNP) and lateral nasal processes (LNP) by the formation of the nasal pits and the subsequent fusion of the MNP with the LNP and MXP is required for development of the intact upper lip and primary palate. The secondary palatal shelves form around E11.5 as outgrowths from the MXP. These palatal shelves grow vertically alongside the tongue before elevating to a horizontal position at E14 and fusing to form the intact secondary palate by E15.5. Improper growth or fusion of the palatal shelves can result in a cleft secondary palate phenotype (Bush and Jiang, 2012; Ferguson, 1988; Gritli-Linde, 2007).
The B-type Eph/ephrins have been implicated in cleft lip and palate; specifically, mutations in EFNB1 cause craniofrontonasal syndrome (CFNS), a human congenital disorder that includes cleft palate, frontonasal dysplasia, and craniosynostosis of the coronal sutures (Twigg et al., 2004; Wieland et al., 2004). Efnb1-deficient mice phenocopy most aspects of CFNS, including cleft palate, which is caused by defective outgrowth of the secondary palatal shelves and reduced anterior palatal mesenchyme cell proliferation (Bush and Soriano, 2010; Compagni et al., 2003; Davy et al., 2004). B-type ephrin signaling has also been implicated in palate fusion. Ephrin-B2 is highly expressed in the pre-fusion epithelium of the palatal shelves, and its pharmacologic disruption in palatal explant culture results in failure of secondary palate fusion (Benson and Serrano, 2012; San Miguel et al., 2011). Cleft palate has also been reported in efnB2LacZ knock-in mice, though the developmental basis of this phenotype has not been reported (Dravis and Henkemeyer, 2011). EphB receptors functioning in secondary palate development have also been identified. Mice harboring compound homozygous mutation of EphB2 and EphB3 exhibit a cleft palate phenotype described to be very similar to the efnb1 null phenotype (Orioli et al., 1996; Risley et al., 2009).
The involvement of A-type Ephs and ephrins in lip and palate development is unknown. Recently, a genome-wide meta-analysis of non-syndromic CL/P indicated association with a single nucleotide polymorphism (SNP) 3kb upstream of the EPHA3 gene, suggesting that regulation of EPHA3 could be significant for CLP (Ludwig et al., 2012). It has been demonstrated that EphA4 and ephrin-A4 are required for proper boundary formation in the coronal sutures to prevent craniosynostosis (Merrill et al., 2006; Ting et al., 2009), and expression of multiple family members has been reported in the developing tooth (Luukko et al., 2005) but their expression and function have not been assessed in lip or palate development.
Here, we characterize the expression patterns of the A-type Eph receptors during craniofacial development and find that several receptors exhibit highly restricted expression patterns in this context. Overlapping patterns of expression of EphA3, EphA4, and EphA7 were detected in the developing palate and nasal structures, however, no overt midfacial phenotype was observed in either EphA3−/− or EphA3−/−; EphA4−/− mice, suggesting redundant function of multiple Eph receptors during palate development.
Results and Discussion
EphA3, EphA4, and EphA7 are highly expressed during secondary palate development
To investigate the expression patterns of the EphA receptors, in-situ hybridization was performed at key midface developmental stages. To examine early primary palate development, stages E9.5, E10.5 and E11.5 were studied via whole mount in-situ hybridization. Of the nine receptors examined, three (EphA3, EphA4, and EphA7) exhibited prominent expression patterns in the craniofacial region at these stages. Whereas at all three stages, expression of EphA3 was detected in the second branchial arch (Fig. 1A, D, G), at E10.5 and E11.5 EphA3 was also strongly expressed in the proximal part of the mandibular component of the first branchial arch (Fig. 1D, G, J). EphA4 was expressed in the MXP at E10.5 and E11.5, as well as in the mandibular prominence (MDP) at E11.5 (Fig. 1E, H, K). EphA7 was present in the MXP at E10.5 and E11.5, and its expression in the LNP and MNP was apparent at E11.5 (Fig. 1F, I, L). In-situ hybridization of frontal sections at E12.5, revealed that expression of all three of these receptors was maintained in the LNP; expression of EphA4 and EphA7 was also apparent in the MNP at this stage (Fig. 2A, B, C).
Figure 1.
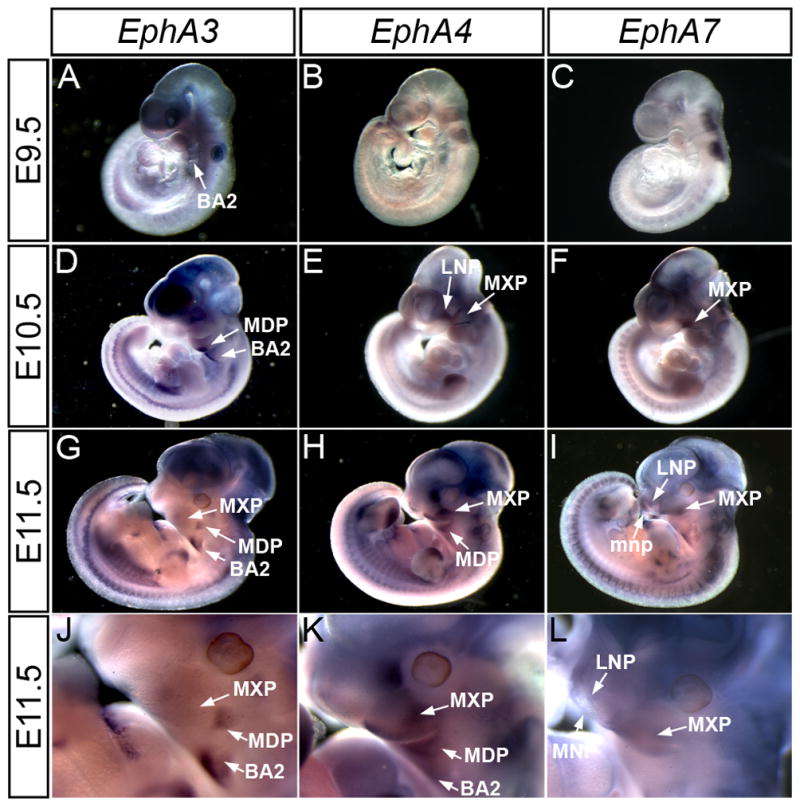
EphA3, EphA4, and EphA7 exhibit elevated expression in the craniofacial region of E9.5, E10.5 and E11.5 mouse embryos. Lateral views of whole mount in-situ hybridization show expression of EphA3 (A, D, G) in the second branchial arch and MDP, EphA4 (B, E, H) in the LNP, MXP, and MDP and EphA7 (C, F, I) in the LNP, MDP and MXP. (J, K, L) Higher magnification of craniofacial region in E11.5 embryos shows expression patterns in MXP, LNP, and BA2. BA2, secondary branchial arch; LNP, lateral nasal prominence; MNP, medial nasal prominence; MDP, mandibular prominence; MXP, maxillary prominence.
Figure 2.
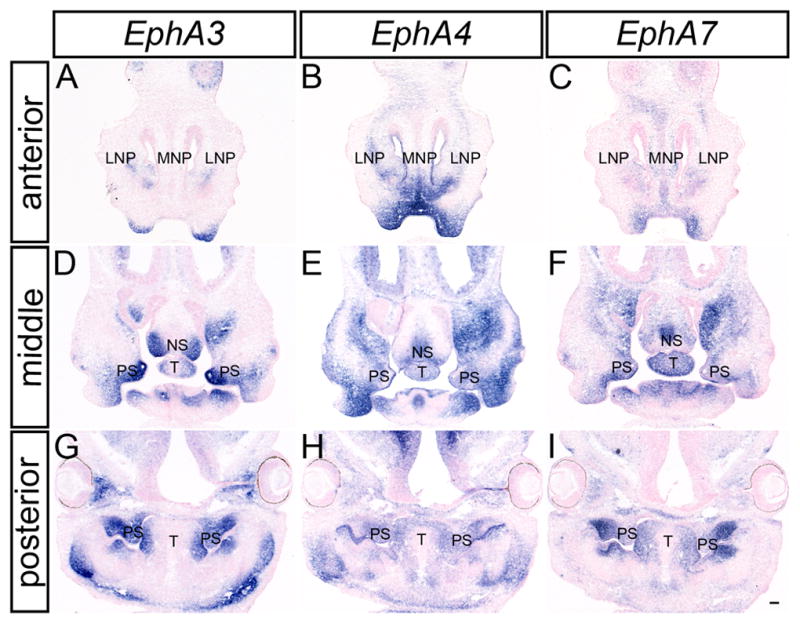
EphA3, EphA4 and EphA7 expression in the craniofacial region of E12.5 mouse embryos. In-situ hybridization of frontal sections of E12.5 embryos for EphA3 (A, D, G), EphA4 (B, E, H), and EphA7 (C, F, I) probes show overlapping expression of all three receptors in the LNP, MNP, MXP, nasal septum, palatal shelves and tongue. MXP, maxillary prominence; LNP, lateral nasal prominence; MNP, medial nasal prominence; MXP, maxillary prominence; NS, nasal septum; PS, palatal shelves; T, tongue. Scale bar = 100μm.
These three receptors were also expressed at robust levels during secondary palate development. EphA3 was observed in the palatal shelf mesenchyme along the entire antero-posterior axis at each stage of palatal development (Fig. 2A, D, G; 3 A, D; 4 A, D). Whereas EphA3 exhibited highly elevated expression throughout the secondary palate mesenchyme, it was most abundantly expressed in the lateral aspect of the palatal shelves at pre-elevation stages (Fig. 2G; Fig. 3D) and along the oral side after elevation occurred (Fig. 4A, D). In anterior sections, EphA3 was expressed in a restricted pattern in the lateral nasal septum and lateral parts of the maxillary prominence (Fig. 2D; Fig. 3A; Fig. 4A). EphA3 expression was detected in the lateral tongue mesenchyme at all stages examined (Fig. 2D, G; Fig 3A, D; Fig. 4A, D). Although EphA4 exhibited broader patterns of expression than EphA3 at all stages examined (Fig. 2B, E, H; Fig. 3B, E; Fig. 4B, E), its expression overlapped that of EphA3, with slightly elevated lateral expression in the palatal shelves at E13.5 (Fig. 3E) and expression restricted to the oral-side mesenchyme of the secondary palate at E14.5 (Fig 4E). EphA7 was also prominently expressed in the palatal shelf mesenchyme at all stages examined, in a pattern very similar to that of EphA3 (Fig. 2I; Fig. 3F; Fig. 4F). The expression of EphA3 and EphA7 was restricted to the palatal mesenchyme lingual to the molar tooth buds; similarly delimited expression was also detected in the mandibular mesenchyme immediately lingual to the molar tooth buds. Expression of EphA7 in the nasal septum was widespread, with a region of more robust expression in the medial nasal septum (Fig. 2F; Fig. 3C; Fig. 4C). At all three stages, EphA7 is also expressed in the lateral maxillae (Fig. 2F; Fig. 3C; Fig. 4C) and the lateral tongue mesenchyme (Fig. 2F, I; Fig. 3C, F; Fig. 4C, F).
Figure 3.
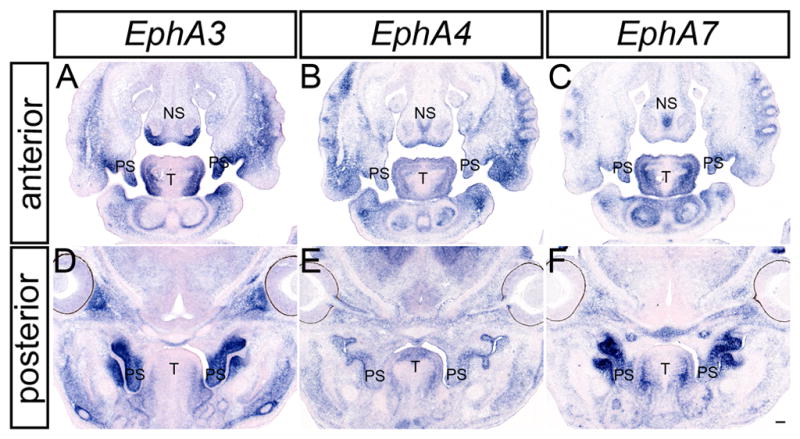
EphA3, EphA4 and EphA7 expression in the craniofacial region of E13.5 mouse embryos. In-situ hybridization analysis of frontal sections of E13.5 mouse embryos for EphA3(A, D), EphA4 (B, E), and EphA7 (C, F) show overlapping expression of all three receptors in the nasal septum, secondary palatal shelves and tongue. NS, nasal septum; PS, palatal shelves; T, tongue. Scale bar = 100μm.
Figure 4.
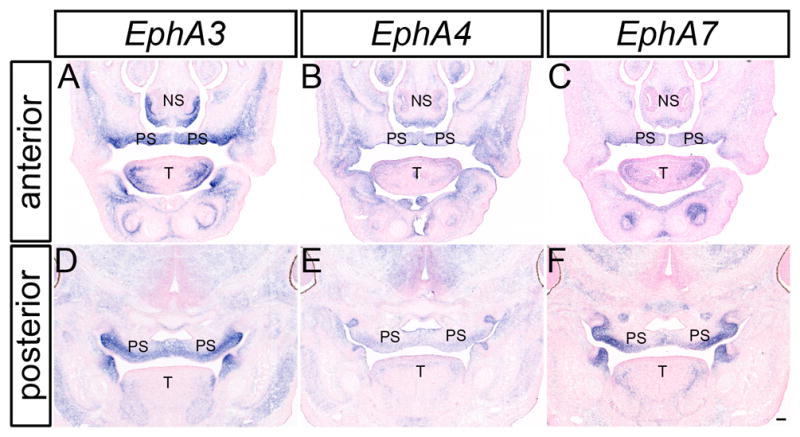
EphA3, EphA4 and EphA7 expression in the craniofacial region of E14.5 mouse embryos. In-situ hybridization analysis of frontal sections of E13.5 mouse embryos for EphA3(A, D), EphA4 (B, E), and EphA7 (C, F) show overlapping expression of all three receptors in the nasal septum, secondary palatal shelves and tongue. NS, nasal septum; PS, palatal shelves; T, tongue. Scale bar = 100μm.
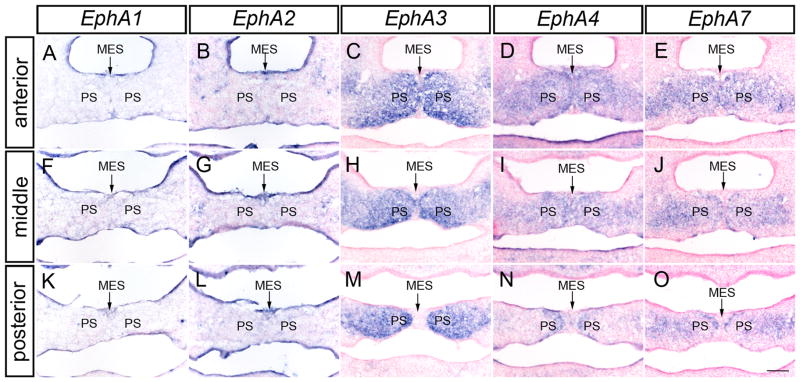
We next wanted to examine whether EphA receptors exhibited patterns of expression that would suggest a role in palatal fusion. We found that though EphA1 and EphA2 were expressed throughout the palatal epithelium at E13.5 (Fig. 6A, B), their expression was not elevated in the medial edge epithelium (MEE) or midline epithelial seam (MES) at pre-fusion or fusion stages (Fig. 5A, B, F, G, K, L and data not shown). Similarly, close examination of section in-situ hybridization of E14.5 embryos undergoing palatal fusion revealed EphA3, EphA4 and EphA7 expression expressed at elevated levels in the mesenchyme of the fusing palatal shelves and reduced or absent in the epithelial cells (Fig. 5C–E, H–J, and M–O, arrows). Notably, this mesenchymal expression appeared strongest directly adjacent to the MES, whereas its expression was weaker in more lateral regions. In total, these expression patterns are highly supportive of a potential role for these Eph receptors in the development of the secondary palate.
Figure 6.
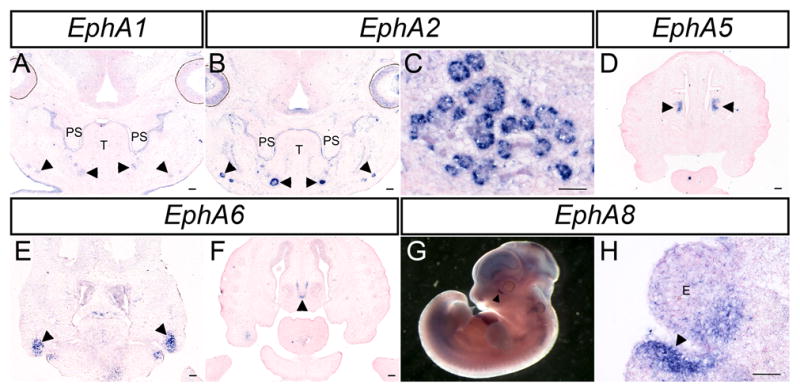
EphA gene expression in other tissues of the craniofacial complex. (A–F,H) In-situ hybridization performed on frontal sections at indicated stages. (A) Expression of EphA1 and (B) EphA2 in the palatal shelf epithelium and submandibular glands (arrowheads in A, B) at E13.5. (C) EphA2 is also expressed in the submandibular gland at E14.5. (D) At E14.5 EphA5 is expressed in the mesenchyme of the nasal conchae. (E,F) EphA6 is expressed at E12.5 in the MXP (arrowheads in E) and in the nasal septum cartilage at E13.5 (arrowhead in F). (G,H) EphA8 expression is seen surrounding the nasolacrimal groove at E11.5 (arrowhead in G) and E12.5 (arrowhead in H). PS, palatal shelves; T, tongue; E, eye. Scale bars = 100μm.
Figure 5.
EphA receptor genes are expressed in the fusing secondary palate. Expression of (A, F, K) EphA1, (B, G, L) EphA2 is not present in the MES and MEE. (C, H, M) EphA3, (D, I, N) EphA4, and (E, J, O) EphA7 are expressed in the mesenchyme of the fusing palate at E14.5 but distinctly missing from the MES and MEE. MES, midline epithelial seam; PS, palatal shelves. Scale bar = 100μm.
Other EphA expression patterns in the craniofacial region
Other Eph receptors also display distinct patterns of expression during craniofacial development. EphA2 was highly expressed in the epithelium of the submandibular gland at E13.5 and E14.5 (Fig. 6B, C), where EphA1 and EphA8 were expressed at lower levels (Data not shown). EphA5 expression was detected in the mesenchyme of the concha of the nasal cavity (Fig. 6H arrows). EphA6 was detected at E12.5 in the maxillary prominence as well as in a highly restricted manner in the nasal septum cartilage at E13.5 and E14.5 (Fig. 6I arrows; Fig. 6J, K). EphA6 is also expressed in a highly restricted manner in the nasal septum cartilage at E13.5 and E14.5 (Fig. 6J, arrow). Beginning at E11.5, EphA8 was highly elevated in the mesenchyme surrounding the forming nasolacrimal groove (Fig. 6A, B arrows). We were unable to detect EphA10 in craniofacial regions at any stage examined (data not shown).
Analysis of midfacial development in EphA3−/− and EphA3−/−; EphA4−/− mutant embryos
Based on these expression patterns, we sought to determine whether loss of EphA receptor function resulted in a cleft palate phenotype. Although EphA3−/− homozygous mutant mice have been reported to exhibit defects in the atrial septa and atrioventricular endocardial cushions, the secondary palate had not been reported (Stephen et al., 2007). While our analysis of EphA3−/− mutant embryos (n = 6) yielded the previously reported heart phenotype (Stephen et al., 2007), no midfacial phenotype was observed (Fig. 7C, G). The highly overlapping expression of EphA4, EphA7, and EphA3 suggested the possibility of functional redundancy. Targeted homozygous disruption of EphA4 results in viable and fertile mice that exhibit defects in the corticospinal tract and hindlimb innervation as well as defects of the coronal suture (Dottori et al., 1998; Helmbacher et al., 2000; Ting et al., 2009). Interestingly though, a recent report of a fetus with cleft palate and spina bifida that harbored a deletion of the genomic region including the EPHA4 and PAX3 genes suggested that EphA4 might be involved in palate development (Goumy et al., 2014). We therefore asked whether EphA3 might be redundant with EphA4 in the development of the secondary palate. EphA3−/−; EphA4−/− mice displayed normal development of the midface (n = 5), however, with no evidence of cleft lip or cleft secondary palate (Fig. 7D, H).
Figure 7.
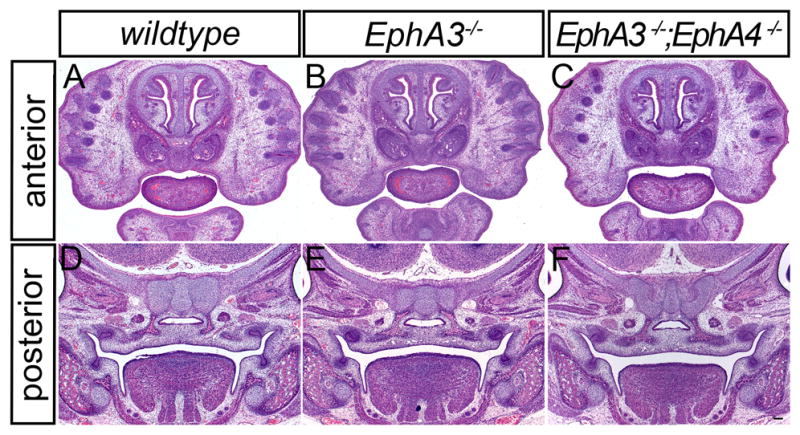
Histological analysis of EphA3 and EphA4 mutant embryos. Comparison of frontal sections of a wild-type E14.5 embryo (A, D), with EphA3−/− homozygous mutant (B, E) and EphA3−/−; EphA4−/− compound mutant embryos (C, F) shows no overt midfacial phenotype. Scale bar = 100μm.
Conclusion
Here we describe previously unreported patterns of expression for A-type Eph receptors during craniofacial development. Amongst these, the strong, restricted expression of EphA2 in the branching salivary gland indicates a possible developmental role for EphA2 during salivary gland morphogenesis, perhaps similar to its role in branching morphogenesis of the mammary gland (Vaught et al., 2009). The expression patterns of EphA3 and EphA7 are highly suggestive of a role for these receptors during secondary palate formation. Our analysis of EphA3−/− mutant embryos did not reveal a cleft palate phenotype and it has been reported that most EphA7−/− mutants reach adulthood without morphological defects, while approximately one quarter exhibit anencephaly (Holmberg et al., 2000; Rashid et al., 2005). The highly overlapping expression of EphA3 and EphA7 in the secondary palate implies redundant function, however; our ongoing studies will test this possibility.
Experimental Procedures
Mice
All experiments involving mice were performed in accordance with protocols approved by the University of California at San Francisco Institutional Animal Care and Use Committee. Embryos for gene expression analysis were derived from an F2 intercross of C57BL/6J and 129S4 inbred mice. EphA4 mutant mice (EphA4tm1.1Bzh, MGI ID: 4430285) in the 129S7 genetic background were obtained from Jackson labs and backcrossed ten generations to C57BL/6J (Herrmann et al., 2010). EphA4lox/lox mice were crossed with Beta-Actin-CreTg/+ (Tg(ACTB-cre)2Mrt, MGI ID: 2176050) (Lewandoski et al., 1997) coisogenic in the C57Bl/6J background to generate EphA4Δ/+ mice. EphA3+/− (EphA3tm1Abn, MGI ID: 2681606) mice were obtained in the 129S1 background and backcrossed two generations to C57BL/6J before crossing with EphA4Δ/+ mice to generate compound mutants for analysis (Vaidya et al., 2003).
Molecular cloning of probes
Murine EphA receptor gene sequences were found using NCBI. Gene specific primers (sequences available upon request) were designed to amplify exonic probe sequences that would avoid sequences similarity between family members. Resultant RT-PCR products were cloned into pBluescript for EphA1, EphA2, EphA3, EphA5, EphA7, and EphA8. The EphA1 probe consists of a 398bp KpnI/EcoRI fragment spanning exons 16 and 17. The EphA2 probe consists of a 627bp SacII/KpnI fragment spanning the second and third exon. The EphA3 probe contains a 627bp XhoI/EcoRI fragment covering exons 16 and 17. The EphA5 probe comprises of a 493bp KpnI/BamHI fragment spanning exons three through six. The EphA7 probe is made up of a 653bp SacI/HindIII fragment spanning exons three through six of both variants. The EphA8 probe consists of a 686bp KpnI/XhoI fragment spanning exons three and four. The EphA6 and EphA10 RT-PCR products were cloned into pGEM-TEasyVector (Promega). The EphA6 probe contains a 557 RT-PCR fragment from the first to the third exon and the EphA10 probe consists of a 389bp RT-PCR fragment spanning exons 3 of both variants. The EphA4 probe consists of a 402bp BamHI/AvaI fragment covering exons 10 through 12 subcloned into the pGem4 plasmid.
In-situ hybridization
Embryos for whole mount in-situ hybridization were fixed in 4% PFA in PBS overnight and dehydrated into PBS/methanol. For in-situ hybridization on sections, embryos were fixed in 4% PFA in PBS overnight and graded through sucrose/OCT before embedding in OCT for cryosectioning. All sections were cut at a thickness of 12μm. In-situ hybridization was carried out according to standard protocols. Sections were counterstained with Nuclear Fast Red.
Histology
Embryos were fixed overnight in Bouin’s fixative and graded through ethanol and histoclear before embedding in paraffin. Sections were cut at a thickness of 7μm and stained with hematoxylin and eosin.
Acknowledgments
The EphA4 in-situ probe was kindly provided by Rulang Jiang. EphA3 mutant mice were generously provided by Sam Pfaff with permission from Arthur Brown. We are grateful to Craig Miller, Nancy Ann Oberheim, and Audrey O’Neill for helpful advice, discussions and comments on the manuscript. This work was supported by grants R00DE020855 and R01DE023337 from the National Institute of Dental and Craniofacial Research to J.O.B.
References
- Benson MD, Serrano MJ. Ephrin regulation of palate development. Craniofacial Biol. 2012;3:376. doi: 10.3389/fphys.2012.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blits-Huizinga CT, Nelersa CM, Malhotra A, Liebl DJ. Ephrins and their receptors: binding versus biology. IUBMB Life. 2004;56:257–65. doi: 10.1080/15216540412331270076. [DOI] [PubMed] [Google Scholar]
- Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Dev Camb Engl. 2012;139:231–243. doi: 10.1242/dev.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Soriano P. Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries. Genes Dev. 2010;24:2068–2080. doi: 10.1101/gad.1963210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagni A, Logan M, Klein R, Adams RH. Control of skeletal patterning by ephrinB1-EphB interactions. Dev Cell. 2003;5:217–30. doi: 10.1016/s1534-5807(03)00198-9. [DOI] [PubMed] [Google Scholar]
- Davy A, Aubin J, Soriano P. Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev. 2004;18:572–83. doi: 10.1101/gad.1171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M, Hartley L, Galea M, Paxinos G, Polizzotto M, Kilpatrick T, Bartlett PF, Murphy M, Köntgen F, Boyd AW. EphA4 (Sek1) receptor tyrosine kinase is required for the development of the corticospinal tract. Proc Natl Acad Sci U S A. 1998;95:13248–13253. doi: 10.1073/pnas.95.22.13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravis C, Henkemeyer M. Ephrin-B reverse signaling controls septation events at the embryonic midline through separate tyrosine phosphorylation-independent signaling avenues. Dev Biol. 2011;355:138–151. doi: 10.1016/j.ydbio.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MW. Palate development. Dev Camb Engl. 1988;103(Suppl):41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Gale NW, Holland SJ, Valenzuela DM, Flenniken A, Pan L, Ryan TE, Henkemeyer M, Strebhardt K, Hirai H, Wilkinson DG, Pawson T, Davis S, Yancopoulos GD. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron. 1996;17:9–19. doi: 10.1016/s0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- Goumy C, Gay-Bellile M, Eymard-Pierre E, Kemeny S, Gouas L, Déchelotte P, Gallot D, Véronèse L, Tchirkov A, Pebrel-Richard C, Vago P. De novo 2q36.1q36.3 interstitial deletion involving the PAX3 and EPHA4 genes in a fetus with spina bifida and cleft palate. Birt Defects Res A Clin Mol Teratol. 2014:1–5. doi: 10.1002/bdra.23246. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Helmbacher F, Schneider-Maunoury S, Topilko P, Tiret L, Charnay P. Targeting of the EphA4 tyrosine kinase receptor affects dorsal/ventral pathfinding of limb motor axons. Dev Camb Engl. 2000;127:3313–3324. doi: 10.1242/dev.127.15.3313. [DOI] [PubMed] [Google Scholar]
- Herrmann JE, Pence MA, Shapera EA, Shah RR, Geoffroy CG, Zheng B. Generation of an EphA4 conditional allele in mice. Genes N Y N 2000. 2010;48:101–105. doi: 10.1002/dvg.20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen JP, Chumley MJ, Lackmann M, Li C, Barton Wa, Jeffrey PD, Vearing C, Geleick D, Feldheim Da, Boyd AW, Henkemeyer M, Nikolov DB. Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci. 2004;7:501–509. doi: 10.1038/nn1237. [DOI] [PubMed] [Google Scholar]
- Holmberg J, Clarke DL, Frisén J. Regulation of repulsion versus adhesion by different splice forms of an Eph receptor. Nature. 2000;408:203–206. doi: 10.1038/35041577. [DOI] [PubMed] [Google Scholar]
- Janes PW, Griesshaber B, Atapattu L, Nievergall E, Hii LL, Mensinga A, Chheang C, Day BW, Boyd AW, Bastiaens PI, Jorgensen C, Pawson T, Lackmann M. Eph receptor function is modulated by heterooligomerization of A and B type Eph receptors. J Cell Biol. 2011;195:1033–45. doi: 10.1083/jcb.201104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes PW, Nievergall E, Lackmann M. Concepts and consequences of Eph receptor clustering. Semin Cell Dev Biol. 2012;23:43–50. doi: 10.1016/j.semcdb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Klein R. Eph/ephrin signalling during development. Dev Camb Engl. 2012;139:4105–9. doi: 10.1242/dev.074997. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, Becker J, Herberz R, Alchawa T, Nasser E, Böhmer AC, Mattheisen M, Alblas MA, Barth S, Kluck N, Lauster C, Braumann B, Reich RH, Hemprich A, Pötzsch S. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Publ Group. 2012;44:968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukko K, Løes S, Kvinnsland IH, Kettunen P. Expression of ephrin-A ligands and EphA receptors in the developing mouse tooth and its supporting tissues. Cell Tissue Res. 2005;319:143–152. doi: 10.1007/s00441-004-0951-1. [DOI] [PubMed] [Google Scholar]
- Merrill AE, Bochukova EG, Brugger SM, Ishii M, Pilz DT, Wall Sa, Lyons KM, Wilkie AOM, Maxson RE. Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum Mol Genet. 2006;15:1319–1328. doi: 10.1093/hmg/ddl052. [DOI] [PubMed] [Google Scholar]
- Orioli D, Henkemeyer M, Lemke G, Klein R, Pawson T. Sek4 and Nuk receptors cooperate in guidance of commissural axons and in palate formation. EMBO J. 1996;15:6035–6049. [PMC free article] [PubMed] [Google Scholar]
- Rashid T, Upton AL, Blentic A, Ciossek T, Knöll B, Thompson ID, Drescher U. Opposing gradients of ephrin-As and EphA7 in the superior colliculus are essential for topographic mapping in the mammalian visual system. Neuron. 2005;47:57–69. doi: 10.1016/j.neuron.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Risley M, Garrod D, Henkemeyer M, McLean W. EphB2 and EphB3 forward signalling are required for palate development. Mech Dev. 2009;126:230–239. doi: 10.1016/j.mod.2008.10.009. [DOI] [PubMed] [Google Scholar]
- San Miguel S, Serrano MJ, Sachar A, Henkemeyer M, Svoboda KKH, Benson MD. Ephrin reverse signaling controls palate fusion via a PI3 kinase-dependent mechanism. Dev Dyn Off Publ Am Assoc Anat. 2011;240:357–364. doi: 10.1002/dvdy.22546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen LJ, Fawkes AL, Verhoeve A, Lemke G, Brown A. A critical role for the EphA3 receptor tyrosine kinase in heart development. Dev Biol. 2007;302:66–79. doi: 10.1016/j.ydbio.2006.08.058. [DOI] [PubMed] [Google Scholar]
- Ting MC, Wu NL, Roybal PG, Sun J, Liu L, Yen Y, Maxson RE., Jr EphA4 as an effector of Twist1 in the guidance of osteogenic precursor cells during calvarial bone growth and in craniosynostosis. Development. 2009;136:855–64. doi: 10.1242/dev.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SRF, Kan R, Babbs C, Bochukova EG, Robertson SP, Wall Sa, Morriss-Kay GM, Wilkie AOM. Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc Natl Acad Sci U S A. 2004;101:8652–8657. doi: 10.1073/pnas.0402819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya A, Pniak A, Lemke G, Brown A. EphA3 null mutants do not demonstrate motor axon guidance defects. Mol Cell Biol. 2003;23:8092–8098. doi: 10.1128/MCB.23.22.8092-8098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaught D, Chen J, Brantley-Sieders DM. Regulation of mammary gland branching morphogenesis by EphA2 receptor tyrosine kinase. Mol Biol Cell. 2009;20:2572–2581. doi: 10.1091/mbc.E08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland I, Jakubiczka S, Muschke P, Cohen M, Thiele H, Gerlach KL, Adams RH, Wieacker P. Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am J Hum Genet. 2004;74:1209–15. doi: 10.1086/421532. [DOI] [PMC free article] [PubMed] [Google Scholar]