Abstract
Hydrolysis of the N-glycosyl bond between a nucleobase and deoxyribose leaves an abasic site within duplex DNA. The abasic site can react with exocyclic amines of nucleobases on the complementary strand to form interstrand DNA-DNA cross-links (ICLs). We find that several enzymes from the base excision repair (BER) pathway protect an abasic site on one strand of a DNA duplex from cross-linking with an amine on the opposing strand. Human alkyladenine DNA glycosylase (AAG) and E. coli 3-methyladenine DNA glycosylase II (AlkA) accomplish this by binding tightly to the abasic site and sequestering it. AAG protects an abasic site opposite T, the product of its canonical glycosylase reaction, by a factor of ~10-fold, as estimated from its inhibition of the reaction of an exogenous amine with the damaged DNA. Human apurinic/apyrimidinic site endonuclease 1 (APE1) and E. coli endonuclease III (Nth) both decrease the amount of ICL at equilibrium by generating a single strand DNA nick at the abasic position as it is liberated from the cross-link. The reversibility of the reaction between amines and abasic sites allows BER enzymes to counter the potentially disruptive effects of this type of cross-link on DNA transactions.
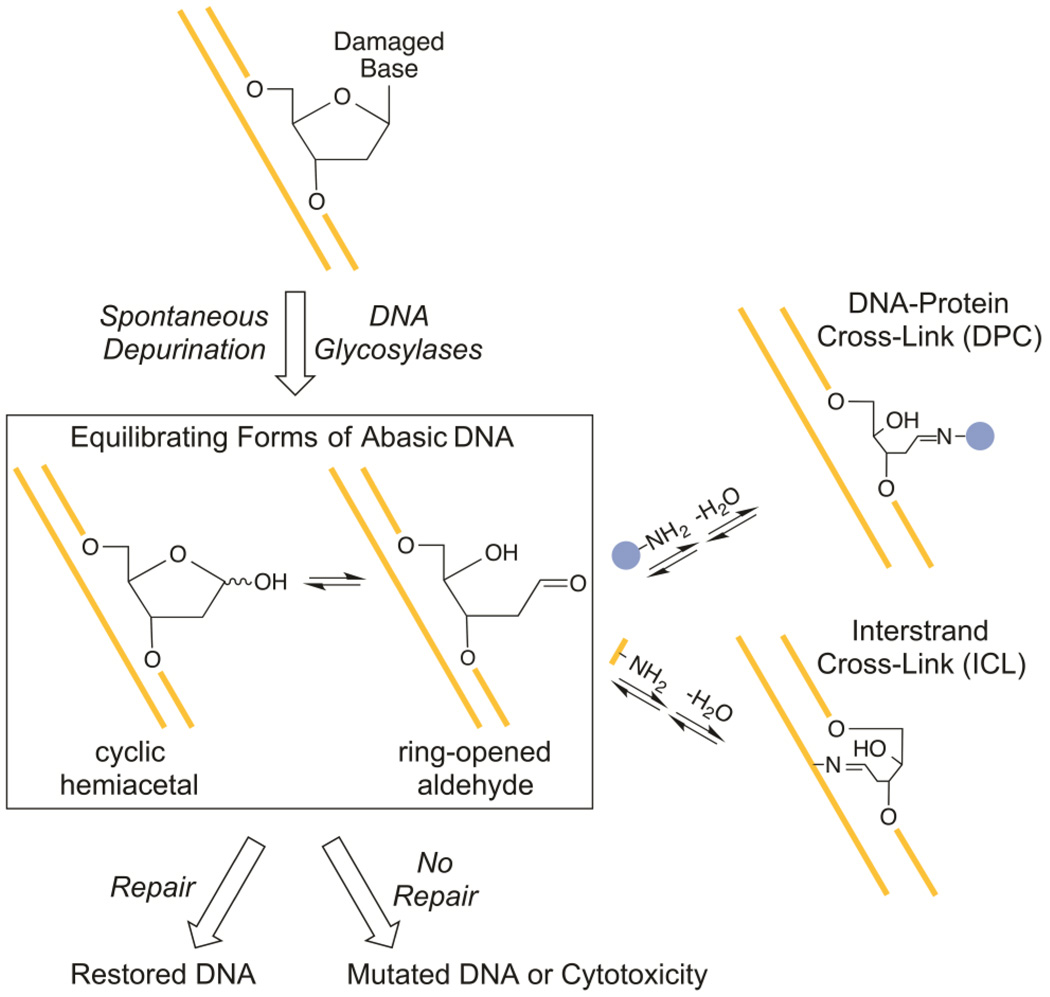
Abasic sites form within duplex DNA both spontaneously and due to the activity of DNA glycosylases during the course of cellular BER (Figure 1). DNA repair enzymes normally act on abasic DNA to restore the original sequence, but abasic lesions that persist can interfere with DNA replication, transcription, and topoisomerase activity, resulting in mutagenesis or toxicity.1 The open chain aldehyde form of the abasic site is susceptible to attack by amine nucleophiles.2 There have been many reports of reactions between abasic sites and amine groups of various proteins to form DNA-protein cross-links (DPCs; Figure 1).3–22 Although DPCs between abasic sites and active site amines are well-characterized intermediates in the mechanisms of bifunctional DNA glycosylases/lyases that participate in DNA repair,23 the biological relevance of DPCs that involve other proteins is unknown. Some of these DPCs may result from side reactions between the reactive abasic moiety and opportunistic amine groups of proteins that bind to DNA or are abundant. Abasic sites have also been shown to react with exocyclic amines of nucleobases on the complementary DNA strand to form ICLs (Figure 1).24–26 Cross-links that involve the exocyclic amines of G, 2-aminopurine (P), and A have been characterized, and unidentified amines have been implicated in ICLs detected in various DNA duplexes known to contain abasic sites.27–31 The presence of such ICLs and DPCs in DNA may obstruct its repair, replication, recombination, and transcription.
Figure 1.
Possible fates of abasic sites within duplex DNA. Abasic sites can react with an amine group of a protein (blue) to form a DPC or with an exocyclic amine of a nucleobase (yellow) to form an ICL.
As intermediates along the BER pathway, abasic sites within duplex DNA are the products or substrates of many repair enzymes.1 We therefore tested the effects of several BER enzymes on the stability of a cross-link between an abasic site and the exocyclic amine of a nucleobase on the opposing strand. These studies provide a starting point for understanding how chemically analogous ICLs and DPCs may be avoided or reversed in vivo.
The DNA glycosylases AAG and AlkA bind to their abasic DNA products with high affinity.32, 33 We investigated whether or not the propensity of abasic DNA to form an ICL was altered by these proteins and discovered that cross-linking was prevented when either glycosylase was present in excess. Pentafluorobenzylhydroxylamine (PFBHA) was used as a proxy for the amine nucleophile of an opposing nucleobase to quantitatively probe protection of several different abasic DNA intermediates by AAG and AlkA.
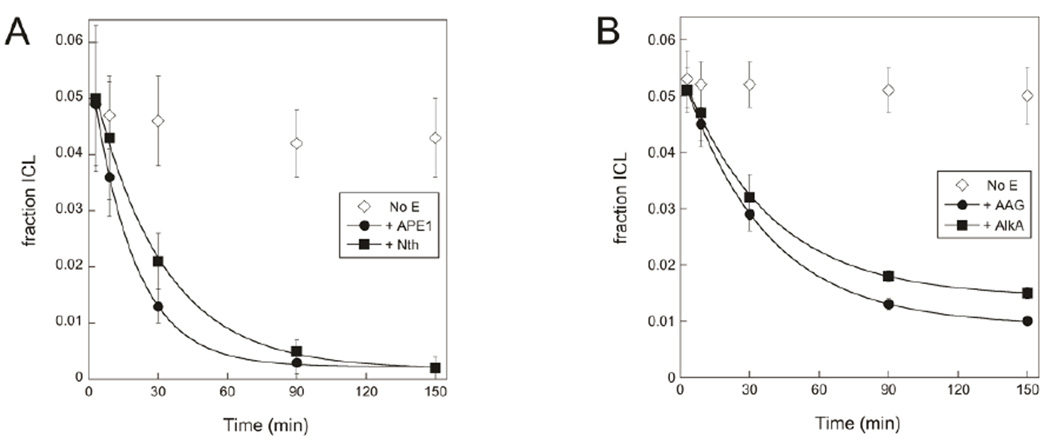
We also considered the effect of repair enzymes that use abasic DNA as a substrate on the lifetime of an ICL. APE1 and Nth both reacted irreversibly with the free abasic DNA as it was released from the cross-link. By consuming the free abasic DNA, these enzymes shifted the equilibrium for cross-linking toward the unlinked species and eliminated the ICL.
Experimental Procedures
Reagents
Uracil DNA glycosylase (UDG; 5000 units/mL) was from New England Biolabs. O-(2,3,4,5,6-Pentafluorobenzyl)hydroxylamine hydrochloride (PFBHA) was from Sigma-Aldrich.
Purification of Recombinant Proteins
Truncated human AAG lacking the first 79 amino acids and full-length human APE1 were expressed in E. coli and purified as previously described.34, 35 Full-length E. coli Nth was purified as previously described36 and stored in 25 mM NaHEPES (pH 7.5), 100 mM NaCl, 10% glycerol (v/v), 0.1 mM EDTA, 1 mM DTT at −80 °C. Nth was buffer exchanged into 10 mM NaMES (pH 6.5), 100 mM NaCl, and 1 mM TCEP using a NAP-5 gel filtration column (GE Healthcare) prior to use in experiments. Full-length E. coli AlkA was expressed in BL21 (DE3) E. coli cells as a C-terminal 6-His-tagged protein from a modified pET24 vector that encoded a TEV protease cleavage site. Cultures were grown at 37 °C in LB+0.1% glucose to mid-log phase, transferred to an ice bath to cool quickly to below room temperature, and then transferred to a 16 °C incubator for induction with 0.1 mM IPTG for ~16 hrs. Cells were lysed in buffer A [100 mM Tris-Cl, pH 7.4, 5% (v/v) glycerol] containing 300 mM NaCl, 5 mM β-mercaptoethanol, and 1 mM PMSF, and then centrifuged. 10% polyethyleneimine was slowly added to the supernatant at 4 °C to a final concentration of 0.2%, and the solution was stirred for 10 min to precipitate contaminating DNA prior to centrifugation. The supernatant was passed over a NTA-Ni2+ agarose column that had been equilibrated with buffer A. After the column had been washed with several column volumes of buffer A, buffer A containing 500 mM NaCl, and buffer A containing 15 mM imidazole, AlkA was eluted from the column with an imidazole gradient (from 15 to 500 mM). Peak fractions were pooled and supplemented to contain 5 mM β;-mercaptoethanol and 1 mM EDTA. AlkA was treated with TEV protease (mass ratio of 100:1, fusion protein to protease) for ~16 hrs at 16 °C to remove the His tag. The reaction was loaded onto a Source S cation exchange column that had been equilibrated with 50 mM Tris-Cl, pH 7.4, 5% (v/v) glycerol, and 25 mM NaCl, and AlkA was eluted from the column with a NaCl gradient (from 25 to 500 mM). Peak fractions were dialyzed into storage buffer containing 50 mM NaHEPES (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, and 1 mM DTT and stored at −80 °C.
Synthesis and Purification of Oligonucleotides
The 35mer DNA oligonucleotides shown in Figure 2 were synthesized by Integrated DNA Technologies or the Keck Center at Yale University. Oligonucleotides 1, 3, and 4 were synthesized using standard protecting groups and deprotected according to the supplier's recommendations. The oligonucleotides were desalted using Sephadex G-25 and purified using denaturing polyacrylamide gel electrophoresis as previously described.37 Oligonucleotide concentrations were determined from their absorbances at 260 nm using calculated extinction coefficients prior to annealing. Oligonucleotide 2, which contains a centrally located abasic site, was prepared by incubating 25 nmol of 1 with 20 units of UDG for 4 hrs at 37 °C and then removing the UDG by phenol-chloroform extraction. A NAP-5 gel filtration column (GE Healthcare) was then used to exchange oligonucleotide 2 into a storage buffer containing 10 mM NaMES (pH 6.5) and 40 mM NaCl. Denaturing polyacrylamide gel electrophoresis of UDG-treated samples that had been subjected to alkaline hydrolysis at 70 °C showed that >97% of the 35mer was cleaved into 17mer, indicating that the conversion of 1 to 2 was essentially complete (data not shown). The duplex DNA oligonucleotides 1/3, 2/3, and 2/4 were prepared by combining 1 or 2 with a 1.25-fold excess of 3 or 4. The 3'-fluorescein reporter group of 1 and 2 was coupled to the DNA via a phosphorothioate linkage to minimize degradation due to the known 3'-exonuclease activity of APE1.38
Figure 2.
Sequences of oligonucleotides used in this study. The abbreviations ab and P denote abasic and 2-aminopurine residues, respectively, and -S-(6-fam) indicates the location of a phosphorothioate linked to a fluorescein label.
DNA Interstrand Cross-Linking Reactions
Reactions were carried out at 37 °C in 50 mM NaMES (pH 6.5), 100 mM NaCl, 0.1 mg/mL BSA, 1 mM EDTA, and 1 mM TCEP. Typical reactions contained 0.25 µM DNA and 0–1.5 µM AAG, AlkA, APE1, or Nth. APE1 and Nth reactions included 0.25 mM MgCl2. Reactions were initiated by adding DNA to the remaining reaction components in a final volume of 20–40 µL. Two different procedures were used to quench the cross-linking reactions, and the ICL was stable to both treatments. For preliminary reactions such as those in Figure 3, aliquots were withdrawn at various times and quenched with sodium hydroxide to give a final concentration of 0.2 M. The quenched samples were heated at 50 °C for 10 min to quantitatively cleave abasic sites. Samples were mixed with an equal volume of formamide prior to gel analysis. Alternatively, aliquots were withdrawn at various times and quenched with sodium borohydride (NaBH4) to give a final concentration of 0.1 M. The 0.5 M NaBH4 quench solution was stored in 10 mM sodium hydroxide at −20 °C to retain its potency. The quenched samples were incubated on ice for 30 min, to quantitatively reduce and thereby stabilize abasic sites, and then mixed with 2.5 volumes of formamide prior to gel analysis. Samples were run on 20% (w/v) polyacrylamide sequencing gels containing 6.6 M urea. Gels were scanned using a Typhoon Trio imager (GE Healthcare), and emission was measured with a 520BP40 filter after excitation of the fluorescein label at 488 nm. The data were converted to fraction ICL (fraction ICL = [ICL]/([ICL] + [ab-DNA])) and then fit by a single exponential. For appearance of ICL, fraction ICL = end point(1-exp(-kobst)), where kobs is the observed rate constant and t is time. For disappearance of ICL, fraction ICL = (fraction ICL)init(exp(-kobst)). In all cases, the nonlinear least-squares fit was good (R > 0.98).
Figure 3.
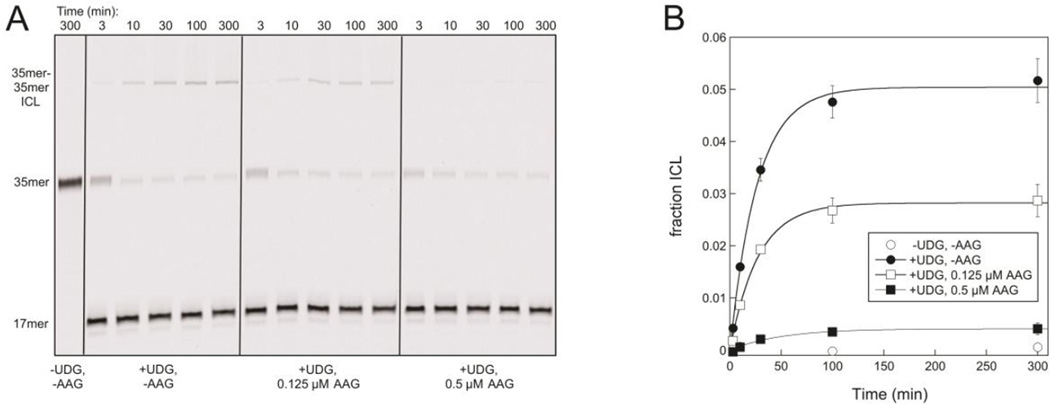
AAG prevents an ICL from forming between an abasic site and P located on opposing strands of a 35mer DNA duplex. A. A 35mer-35mer ICL builds up over time in reactions that contain 0.25 µM duplex 1/3 treated with UDG, but AAG reduces the overall amount of ICL that accumulates. Reactions were carried out at 37 °C and pH 6.5, and samples were quenched and subjected to mild alkaline hydrolysis prior to separation on a 20% denaturing polyacrylamide gel. The slightly higher levels of 35mer present at the earliest time points in reactions containing UDG reveal that the conversion of intact 1/3 to abasic 2/3 is not quite complete at these times. B. The fraction of ICL at time points in A and duplicate reactions is shown. AAG reduces the amount of ICL that forms, with only trace amounts detected when the AAG concentration exceeds the DNA concentration. Lines are exponential fits to the data and give end points of 5% ICL in the absence of AAG, 2.8% ICL in the presence of 0.125 µM AAG, and 0.4% ICL in the presence of 0.5 µM AAG.
Analysis of Equilibrium Cross-Linking
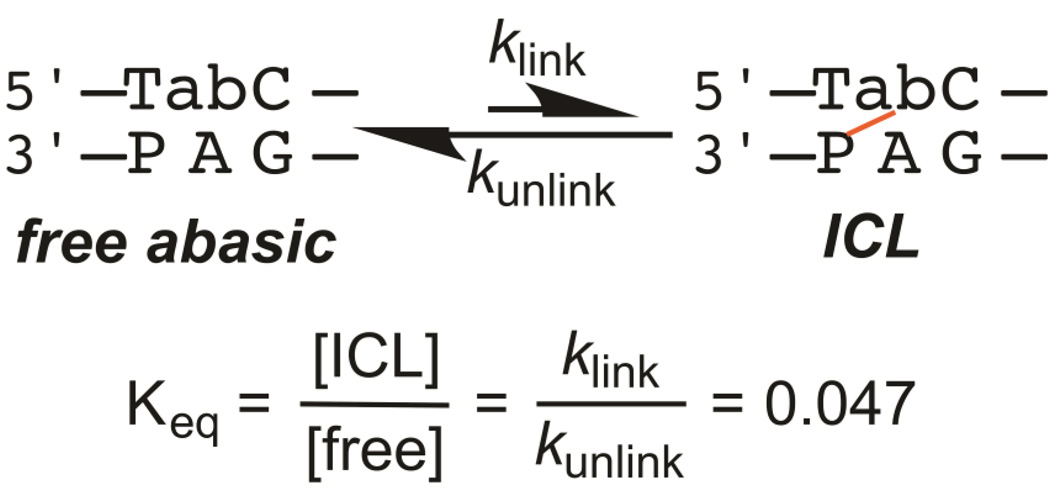
The observed rate constant, kobs, for the approach to equilibrium of a reversible reaction is equal to the sum of its forward and reverse rate constants, kf and kr (kobs = kf + kr), and Keq = kf/kr.39, 40 The rate constants for cross-linking, kf = klink = 0.001 min−1, and for unlinking, kr = kunlink = 0.033 min−1, were calculated from these relationships and the measured values of kobs= 0.034 min−1 and Keq= 0.047 (see Results).
Reactions of PFBHA With Abasic DNA
Reactions were carried out at 37 °C in 50 mM NaMES (pH 6.5), 100 mM NaCl, 0.1 mg/mL BSA, 1 mM EDTA, and 1 mM TCEP. Typical reactions contained 0.25 µM abasic 35mer DNA, 0.5–5 mM PFBHA, and 0–1.5 µM AAG or AlkA. Reactions were initiated by adding PFBHA to the remaining reaction components in a final volume of 20–40 µL. Aliquots were withdrawn at various times and quenched with NaBH4 as described above. Quenched samples were mixed with 2.5 volumes of formamide that contained 1.38 µM of an unlabeled 49mer oligonucleotide that hybridizes with 3 or 4 (5'-GACATGATTGCCCGATAGCATCCTICCTTCTCTCCATGCGTCAATTGTC-3', I = inosine) and then heated at 70 °C for 2 min prior to separation on 20% (w/v) polyacrylamide sequencing gels containing 6.6 M urea. The presence of 1 µM unlabeled 49mer (~20 equivalents) reduced smearing of the fluorescently labeled DNA bands on gels, presumably through competitive hybridization with the unlabeled 35mer strand. Consistent with this model, the presence of the 49mer did not affect the appearance of DNA bands from reactions that contained only single stranded DNA (data not shown). The 49mer competitor used in our experiments was chosen because it was already available in the lab, and no other potential competitors were tested. To separate the abasic 35mer DNA from its PFBHA adduct as cleanly as possible, gels were run for extended periods of time before being scanned and analyzed as described above. Nevertheless, control reactions lacking PFBHA had an average background corresponding to 15% PFBHA-DNA, so this value was subtracted from each data point before fitting the data to a single exponential: fraction PFBHA-DNA = 1-exp(-kobst), where kobs is the observed rate constant and t is time.
Results
DNA Glycosylases Prevent Interstrand Cross-Linking of DNA
Gates and coworkers have shown that abasic sites in duplex DNA can form ICLs with exocyclic amines of G, P, or A at certain positions in the opposing strand.24–26 Based on these studies, we paired the uracil-containing oligonucleotide 1 with a P-containing complementary strand 3 (Figure 2) and confirmed that an ICL formed within duplex 1/3 only after the uracil had been excised by catalytic amounts of UDG, leaving an abasic site (Figure 3A, left). We then investigated if ICL formation was affected by the presence of the BER glycosylase AAG, which binds to its abasic DNA product with high affinity.32 The ICL could only be detected when the DNA concentration exceeded the AAG concentration (Figure 3); all of the abasic sites are expected to be bound to AAG when AAG is in excess, preventing the exocyclic amine of P from initiating a cross-link. Similar results were obtained in the presence of AlkA, a DNA glycosylase from E. coli that also binds tightly to its abasic DNA product (Figure S1).33
Characterization of the Duplex 2/3 Cross-Linking Reaction
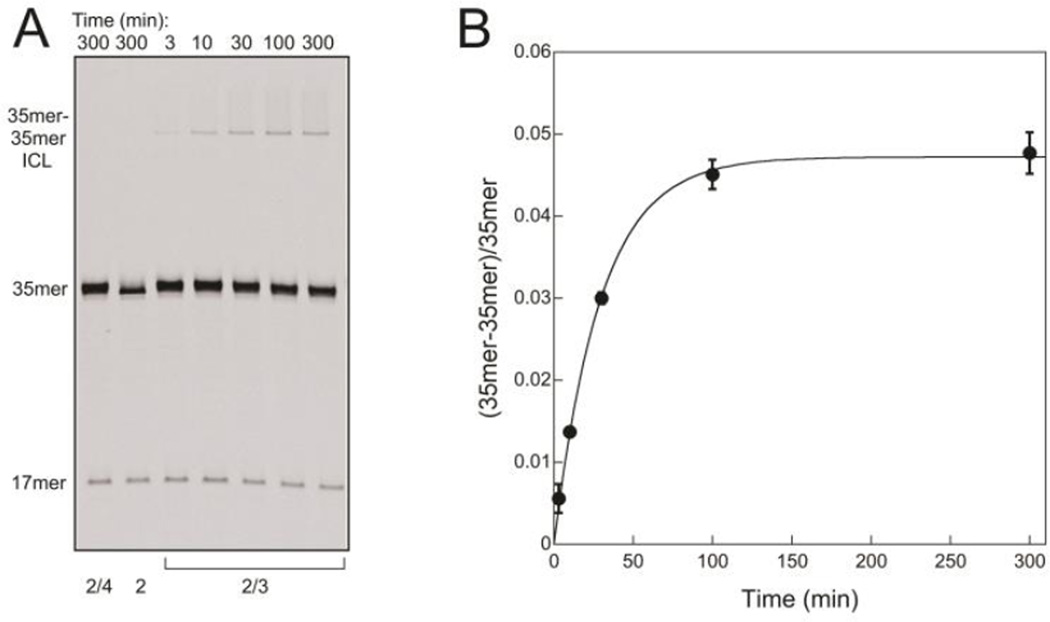
To investigate the cross-linking reaction in greater detail, the abasic site was first generated within single stranded DNA by treating 1 with UDG to remove uracil. The resulting oligonucleotide 2 was purified and used alone or with a complementary strand in a duplex. The ICL forms over time in reactions containing newly annealed duplex 2/3, which has P base-paired to the T that is located on the 5'-side of the abasic site (Figure 4A, right). The cross-linking reaction requires the P-containing opposing strand, for ICL is not detected in reactions of duplex 2/4 (no P in the opposing strand) nor in reactions of single-stranded 2 (no opposing strand) (Figure 4A, left). The ratio of cross-linked to unlinked DNA at each time point in the reaction of duplex 2/3 is plotted in Figure 4B, revealing that the reaction proceeds to an end point ratio of 0.047 with t1/2~20 min at 37 °C.
Figure 4.
An ICL forms between an abasic site and P located on opposing strands of the 35mer DNA duplex 2/3. A. The amount of 35mer-35mer ICL increases with time in a reaction that contains duplex 2/3 (lanes 3–7). No ICL forms in control reactions containing duplex 2/4 (lane 1) or single stranded 2 (lane 2). Reactions contained 0.25 µM DNA and were carried out at 37 °C and pH 6.5. Samples were reduced with NaBH4 prior to separation on a 20% denaturing polyacrylamide gel. The 17mer that is visible in all lanes arises from nonenzymatic β-elimination of 2 that occurs during preparation and storage of this oligonucleotide, and its amount (6%) remains constant during the reaction courses. B. The ratio of ICL to unlinked 2/3 at time points in A and duplicate reactions is shown. An end point ratio of 0.047 and kobs of 0.034 min−1 for the approach to the end point were obtained from an exponential fit to the data.
Results described below show that the cross-linking reaction is reversible. Therefore, we interpret the end point to represent the equilibrium between formation (klink) and breakdown (kunlink) of the cross-link, corresponding to Keq = 0.047 (Figure 5).39, 40 Measurement of the equilibrium end point and kobs for the approach to equilibrium (Figure 4B) allows the rate constants for cross-linking, klink = 0.001 min−1, and for unlinking, kunlink = 0.033 min−1, to be calculated (see Experimental Section). These results indicate that the cross-linked species is less stable than the free abasic species for duplex 2/3, and the ~5% cross-link at equilibrium falls within the 2–80% yield range that has been measured for other ICLs between abasic sites and exocyclic amines of nucleobases on the opposing strand.24–26 The kinetics and thermodynamics of cross-linking between abasic sites and exocyclic amines appear to vary widely, and the sequence determinants that account for this variation are not yet known. Despite its somewhat low yield, the duplex 2/3 cross-link is convenient for studying the effects of BER enzymes on cross-linking reactions because of its relatively fast approach to equilibrium (t1/2~20 min at 37 °C).
Figure 5.
The equilibrium between the species with and without an ICL for duplex 2/3. The Keq value shown is for the duplex at 37 °C and pH 6.5, from Figure 4B.
BER Enzymes Deplete ICLs in DNA
APE1 and Nth were added to reactions containing the preformed duplex 2/3 ICL to investigate the effect of BER enzymes that react with abasic DNA. The amount of ICL in the reaction decreased upon addition of either enzyme (Figure 6A), and in each case the rate of disappearance of the ICL closely matched kunlink, the rate constant for unlinking, reported above. This result suggests that Nth and APE1 bind and then consume free abasic DNA as it is liberated from the exocyclic amine, shifting the equilibrium away from the ICL. Although we considered the possibility that APE1 would nick the cross-linked DNA 5' to the abasic nucleotide, no evidence for such an activity was observed.
Figure 6.
ICL breakdown due to BER enzymes. A. Duplex 2/3 ICL disappears over time in reactions containing 0.5 µM APE1 or 0.5 µM Nth; exponential fits to the data give values of kobs of 0.05 min−1 and 0.03 min−1 for the reactions containing APE1 and Nth, respectively. B. Duplex 2/3 ICL disappears with kobs= 0.03 min−1 in reactions containing either 0.5 µM AAG or 0.5 µM AlkA. For both A and B, BER enzymes were added to 0.25 µM duplex 2/3 at equilibrium at 37 °C and pH 6.5, and samples were reduced with NaBH4 prior to separation on 20% denaturing polyacrylamide gels (Figure S2). APE1 and Nth reactions included 0.25 mM MgCl2.
Similarly, AAG and AlkA were added to reactions containing the preformed duplex 2/3 ICL to investigate the effect of BER enzymes that bind tightly to abasic DNA but do not consume it. The ICL disappeared in the presence of either AAG or AlkA with an observed rate constant of 0.03 min−1, the same as kunlink, the rate constant for unlinking of the duplex 2/3 ICL (Figure 6B). AAG and AlkA appear to bind to the abasic site as it becomes available, and the glycosylase-bound abasic DNA is subsequently protected from cross-linking.
Oxyamine Reactivity as a Model for Interstrand Cross-Linking of DNA
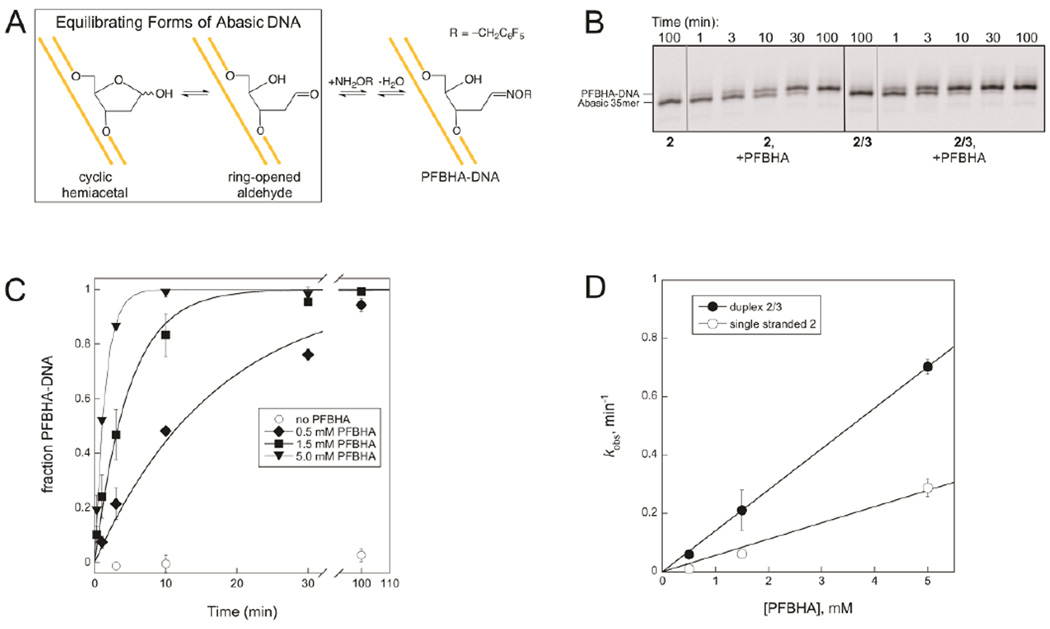
As described above, the duplex 2/3 ICL does not form in the presence of excess AAG or AlkA (Figure 3 and Figure S1), and the disappearance of the ICL upon addition of AAG and AlkA is limited by the unlinking reaction, which is glycosylase-independent (Figure 6B). Therefore, although our results indicate that these DNA glycosylases protect abasic DNA from cross-linking by binding to it, it is difficult to get a quantitative estimate for their protective effects. To overcome this limitation, we turned to the reaction of abasic DNA with the oxyamine PFBHA (Figure 7A). The reactions of cross-linking nucleobases or PFBHA with abasic DNA both feature amine nucleophiles. However, unlike the amine group of the cross-linking nucleobase, the amine group of PFBHA is located on a separate molecule from the abasic site, so its concentration can be varied to alter the rate of its second order reaction with the abasic aldehyde and to drive the reaction to completion.
Figure 7.
Reaction of PFBHA and abasic DNA. A. The oxyamine adduct PFBHA-DNA is the product of the reaction between PFBHA and abasic DNA. This reaction is analogous to the well-characterized reaction of methoxyamine with abasic DNA48–50 but has the advantage that the larger PFBHA adduct is more easily separated from the abasic DNA substrate on a gel. B. PFBHA reacts with 2 or 2/3 to form PFBHA-DNA. No adduct forms in the absence of PFBHA. Reactions contained 0.25 µM DNA and were carried out at 37 °C and pH 6.5 with or without 1.5 mM PFBHA. Samples were reduced with NaBH4 before separation on a 20% denaturing polyacrylamide gel. This image displays only the region of the gel where the 35mer and the PFBHA-35mer appear; A full image of the same gel (Figure S3) shows that ICL formation is minimal in reactions of duplex 2/3 when PFBHA is present, because PFBHA outcompetes the exocyclic amine for reaction with the abasic site at the concentrations of PFBHA used. C. The kobs for formation of PFBHA-DNA increases with PFBHA concentration in duplex 2/3 reactions. Values of kobs at each PFBHA concentration were determined from exponential fits to the data and are plotted in D. D. Linear fits for the dependence of kobs on PFBHA concentration give second order rate constants for the reaction of PFBHA with 2 of 0.06 mM−1min−1 and with 2/3 of 0.14 mM−1min−1.
The progress of reactions of PFBHA with abasic DNA can be followed by separating the DNA substrate from its PFBHA-DNA product using denaturing PAGE (Figure 7B). The adduct is only detected in reactions that contain both PFBHA and an abasic site within either single stranded DNA (2) or duplex DNA (2/3). The fraction of DNA that has reacted with PFBHA at each time point in reactions containing duplex 2/3 and different concentrations of PFBHA is plotted in Figure 7C, and kobs values obtained from these curves and analogous curves give second order rate constants of 0.06 mM−1min−1 and 0.14 mM−1min−1 for the reactions of PFBHA with single stranded 2 and duplex 2/3, respectively (Figure 7D). The ~2-fold difference between these rate constants suggests that the abasic site is more accessible to PFBHA in duplex 2/3 than in single stranded 2, perhaps because duplex DNA is more rigid than single stranded DNA.
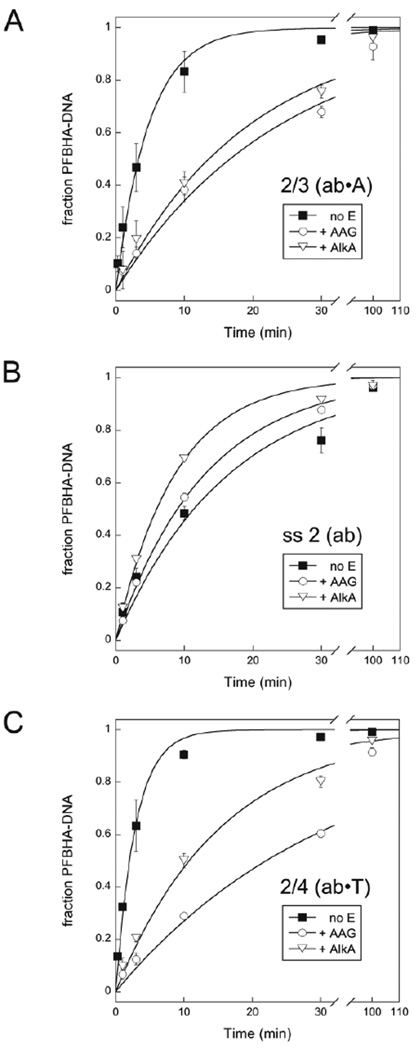
If abasic DNA is bound to a DNA glycosylase, its availability for reaction with an amine is expected to change. We probed protection of the abasic site in different DNA contexts by comparing the reactivity of several oligonucleotides with PFBHA in the presence and absence of AAG and AlkA (Figure 8). Both AAG and AlkA slow the reaction of PFBHA with duplex 2/3 when the glycosylases are present in 6-fold excess (Figure 8A). AAG protects duplex 2/3 by a factor of 5, and AlkA protects it by a factor of 4 (Table 1). Similar protection factors are obtained at lower glycosylase concentrations (Figure S4 and Table S1). In contrast, AAG and AlkA do not slow the reaction of PFBHA with single stranded 2 (Figure 8B), presumably because AAG and AlkA have low affinity for single stranded DNA. Since damaged purines within duplex DNA are the preferred substrates for AAG,41 its typical products contain an abasic site opposite a pyrimidine. We therefore investigated protection of duplex 2/4, which has T opposite the abasic site (Figure 8C). AAG protects duplex 2/4 by a factor of 11, and AlkA protects it by a factor of 5 (Table 1).
Figure 8.
AAG and AlkA protect abasic DNA in a duplex from PFBHA. Reactions of duplex 2/3 (A), single stranded 2 (B), and duplex 2/4 (C) contained 0.25 µM DNA, 1.5 mM PFBHA, and 1.5 µM AAG, 1.5 µM AlkA, or no added enzyme. Samples from reactions performed at 37 °C and pH 6.5 were reduced with NaBH4 prior to separation on 20% denaturing polyacrylamide gels. Exponential fits to the data are shown, and kobs values from the fits are recorded in Table 1.
Table 1.
AAG and AlkA Protect Abasic DNA in a Duplex from PFBHA.
| DNA | DNA glycosylase | kobs (min−1)a | Protection factorb |
|---|---|---|---|
| 2/3 | none | 0.21 | (1) |
| AAG | 0.042 | 5 | |
| AlkA | 0.051 | 4 | |
| 2 | none | 0.061 | (1) |
| AAG | 0.077 | 0.8 | |
| AlkA | 0.12 | 0.5 | |
| 2/4 | none | 0.35 | (1) |
| AAG | 0.032 | 11 | |
| AlkA | 0.065 | 5 |
Observed rate constants for reactions of 1.5 mM PFBHA with 0.25 µM DNA and 1.5 µM AAG, 1.5 µM AlkA, or no added enzyme were measured at 37 °C and pH 6.5. Measurements are the average of at least two independent reactions, and the standard deviation is ≤10%.
Protection factor is defined as kobs for the reaction in the absence of glycosylase divided by kobs for the reaction in the presence of glycosylase.
Discussion
An estimated 104 abasic sites arise from spontaneous depurination per human cell per day,1 and the repair activity of DNA glycosylases increases this number (Figure 1). Both the abasic sites themselves and the products of their potential side reactions can interfere with the normal processing of DNA. Abasic sites within duplex DNA are the substrates or products of many BER enzymes, so these enzymes are expected to be at the frontline of the cellular response to such damage. We assessed the effects of several BER enzymes on the cross-linking reaction between an abasic site and the exocyclic amine of a nucleobase on the opposing DNA strand. The recent characterizations of ICLs between abasic sites and the exocyclic amines of G, P and A show that this type of cross-link occurs in multiple sequence contexts and may be relatively common.24–26 Consistent with this, the ICL we report for duplex 2/3 is the second cross-link to be found within a 5'-Tab/3'-PA sequence,25 despite the otherwise unrelated neighboring DNA in the two examples.
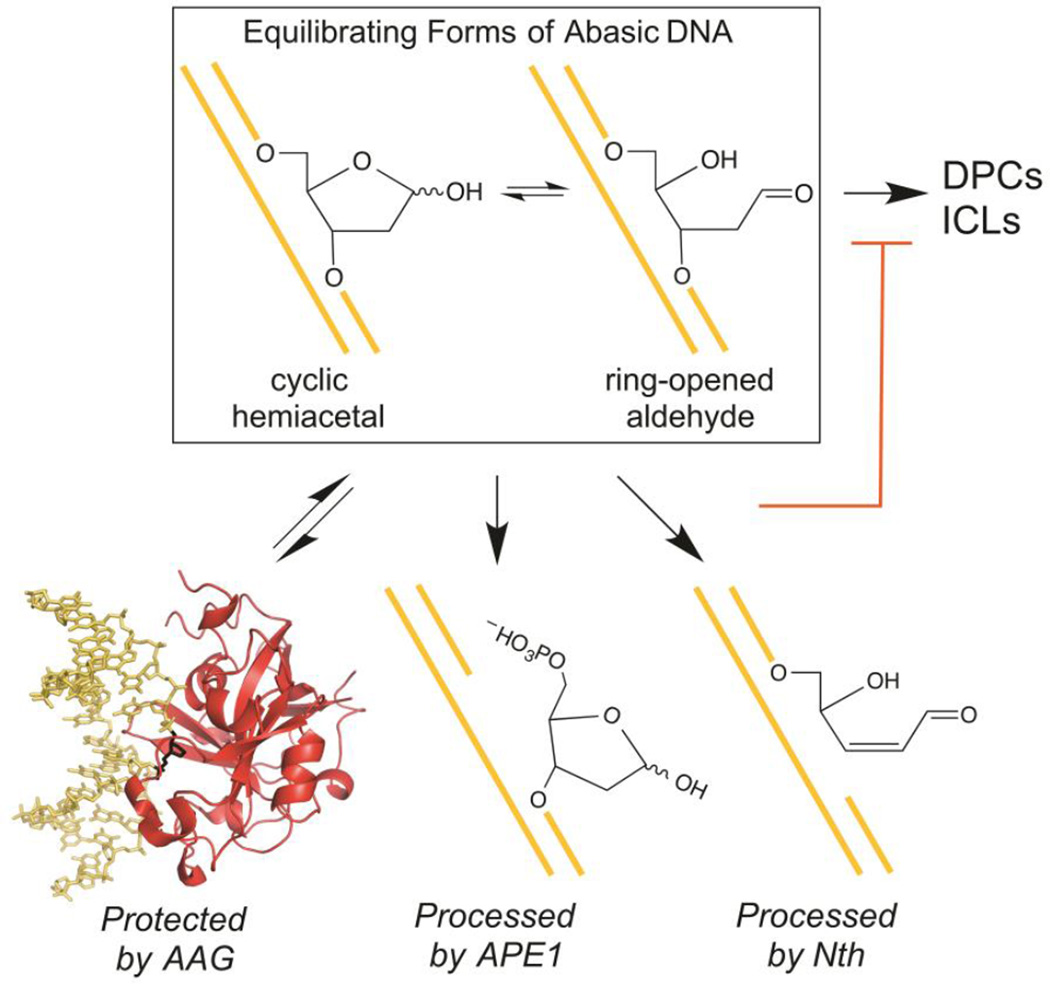
Our model for how the BER enzymes AAG, AlkA, APE1, and Nth prevent cross-links between abasic sites and amines is summarized in Figure 9. Although the model is based on our results with a 2-aminopurine cross-link, it should be generally applicable to other amine ICLs and DPCs: the exocyclic amines of 2-aminopurine and the natural nucleobases are chemically analogous and are similar to the amines of Lys side chains and protein NH2-termini.
Figure 9.
BER enzymes prevent abasic sites in duplex DNA from cross-linking by binding tightly to the free abasic sites (AAG, AlkA) or by reacting irreversibly with them (APE1, Nth). An abasic pyrrolidine nucleotide is flipped into the active site of AAG in the structure that is shown.51
Many DNA glycosylases, including AAG and AlkA, bind to their abasic DNA products with high affinity.32, 33, 42–46 Tight binding has been proposed to shield this reactive DNA intermediate from side reactions until it is further processed by downstream enzymes of the BER pathway.1 The observed protection of abasic sites from interstrand cross-linking (Figure 3 and Figure S1) and from reaction with PFBHA (Figure 8 and Table 1) by AAG and AlkA provides experimental evidence for this idea. In the same way, these and other DNA glycosylases are expected to prevent bound abasic sites from forming inappropriate DPCs with reactive protein amine groups and from reacting with other biological amines (Figure 9). Interestingly, AAG and AlkA do not always protect abasic DNA. The reactivity of abasic DNA with small alcohols is greatly increased when the abasic site is bound to AAG or AlkA, in which case the glycosylases catalyze the formation of alcohol-DNA adducts.47
The duplex 2/3 cross-link breaks down in the presence of AAG, AlkA, APE1, or Nth with an observed rate constant that closely matches the rate constant for its unlinking reaction (Figure 6). This strongly suggests that each of these enzymes interacts with the abasic site as it is freed from the cross-link and does not act directly on the cross-linked DNA. AAG and AlkA appear to bind to the abasic site and protect it from subsequent cross-linking. APE1 and Nth instead nick the DNA at the free abasic site, shifting the cross-linking equilibrium away from the ICL (Figure 9).
Although the reversibility of the ICL reaction allows unlinked abasic sites in DNA to be protected or further processed by the BER enzymes we surveyed, it is important to note that the known ICLs have a significant lifetime once formed. For example, the half-time of the duplex 2/3 unlinking reaction is ~20 min, and the half-times estimated from published data for three ICLs between an abasic site and G or A range from 260–7500 min (Table S2). In the absence of an active mechanism for their breakdown, such ICLs could persist long enough to obstruct DNA processes including replication and transcription. It remains possible that other enzymes repair such damage by reacting directly with cross-links between abasic sites and amines.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the O'Brien lab for helpful discussions and critical reading of the manuscript.
Funding
This work was supported by NIH grant RO1 GM108022 to P.J.O.
Abbreviations
- ICL
interstrand DNA-DNA cross-link
- BER
base excision repair
- AAG
human alkyladenine DNA glycosylase (also known as MPG, methylpurine DNA glycosylase)
- AlkA
E. coli 3-methyladenine DNA glycosylase II
- APE1
human apurinic/apyrimidinic site endonuclease 1
- Nth
E. coli endonuclease III
- DPC
DNA-protein cross-links
- P
2-aminopurine
- PFBHA
pentafluorobenzylhydroxylamine
- UDG
uracil DNA glycosylase.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Figures S1–S4 and Tables S1–S2, as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
References
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- 2.Lhomme J, Constant JF, Demeunynck M. Abasic DNA structure, reactivity, and recognition. Biopolymers. 1999;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 3.Permana PA, Ho DK, Cassady JM, Snapka RM. Mechanism of action of the antileukemic xanthone psorospermin: DNA strand breaks, abasic sites, and protein-DNA cross-links. Cancer Res. 1994;54:3191–3195. [PubMed] [Google Scholar]
- 4.Yacoub A, Augeri L, Kelley MR, Doetsch PW, Deutsch WA. A Drosophila ribosomal protein contains 8-oxoguanine and abasic site DNA repair activities. Embo J. 1996;15:2306–2312. [PMC free article] [PubMed] [Google Scholar]
- 5.Bogenhagen DF, Pinz KG. The action of DNA ligase at abasic sites in DNA. J Biol Chem. 1998;273:7888–7893. doi: 10.1074/jbc.273.14.7888. [DOI] [PubMed] [Google Scholar]
- 6.Nicolas E, Beggs JM, Haltiwanger BM, Taraschi TF. A new class of DNA glycosylase/apurinic/apyrimidinic lyases that act on specific adenines in single-stranded DNA. J Biol Chem. 1998;273:17216–17220. doi: 10.1074/jbc.273.27.17216. [DOI] [PubMed] [Google Scholar]
- 7.Zharkov DO, Grollman AP. MutY DNA glycosylase: base release and intermediate complex formation. Biochemistry. 1998;37:12384–12394. doi: 10.1021/bi981066y. [DOI] [PubMed] [Google Scholar]
- 8.Wang YX, Neamati N, Jacob J, Palmer I, Stahl SJ, Kaufman JD, Huang PL, Winslow HE, Pommier Y, Wingfield PT, Lee-Huang S, Bax A, Torchia DA. Solution structure of anti-HIV-1 and anti-tumor protein MAP30: structural insights into its multiple functions. Cell. 1999;99:433–442. doi: 10.1016/s0092-8674(00)81529-9. [DOI] [PubMed] [Google Scholar]
- 9.Nicolas E, Beggs JM, Taraschi TF. Gelonin is an unusual DNA glycosylase that removes adenine from single-stranded DNA, normal base pairs and mismatches. J Biol Chem. 2000;275:31399–31406. doi: 10.1074/jbc.M004505200. [DOI] [PubMed] [Google Scholar]
- 10.Postel EH, Abramczyk BM. Escherichia coli nucleoside diphosphate kinase is a uracil-processing DNA repair nuclease. Proc Natl Acad Sci U S A. 2003;100:13247–13252. doi: 10.1073/pnas.2333230100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rieger RA, Zaika EI, Xie W, Johnson F, Grollman AP, Iden CR, Zharkov DO. Proteomic approach to identification of proteins reactive for abasic sites in DNA. Mol Cell Proteomics. 2006;5:858–867. doi: 10.1074/mcp.M500224-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Nazarkina ZK, Khodyreva SN, Marsin S, Lavrik OI, Radicella JP. XRCC1 interactions with base excision repair DNA intermediates. DNA Repair (Amst) 2007;6:254–264. doi: 10.1016/j.dnarep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Ilina ES, Lavrik OI, Khodyreva SN. Ku antigen interacts with abasic sites. Biochim Biophys Acta. 2008;1784:1777–1785. doi: 10.1016/j.bbapap.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Khodyreva SN, Prasad R, Ilina ES, Sukhanova MV, Kutuzov MM, Liu Y, Hou EW, Wilson SH, Lavrik OI. Apurinic/apyrimidinic (AP) site recognition by the 5'-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1) Proc Natl Acad Sci U S A. 2010;107:22090–22095. doi: 10.1073/pnas.1009182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller TA, Meek K, Hausinger RP. Human AlkB homologue 1 (ABH1) exhibits DNA lyase activity at abasic sites. DNA Repair (Amst) 2010;9:58–65. doi: 10.1016/j.dnarep.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5'-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci U S A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestryakov P, Zharkov DO, Grin I, Fomina EE, Kim ER, Hamon L, Eliseeva IA, Petruseva IO, Curmi PA, Ovchinnikov LP, Lavrik OI. Effect of the multifunctional proteins RPA, YB-1, and XPC repair factor on AP site cleavage by DNA glycosylase NEIL1. J Mol Recognit. 2012;25:224–233. doi: 10.1002/jmr.2182. [DOI] [PubMed] [Google Scholar]
- 19.Zhou C, Sczepanski JT, Greenberg MM. Mechanistic studies on histone catalyzed cleavage of apyrimidinic/apurinic sites in nucleosome core particles. J Am Chem Soc. 2012;134:16734–16741. doi: 10.1021/ja306858m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sczepanski JT, Zhou C, Greenberg MM. Nucleosome core particle-catalyzed strand scission at abasic sites. Biochemistry. 2013;52:2157–2164. doi: 10.1021/bi3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Ory A, Zafra O, de Vega M. Efficient processing of abasic sites by bacterial nonhomologous end-joining Ku proteins. Nucleic Acids Res. 2014;42:13082–13095. doi: 10.1093/nar/gku1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prasad R, Horton JK, Chastain PD, 2nd, Gassman NR, Freudenthal BD, Hou EW, Wilson SH. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic Acids Res. 2014;42:6337–6351. doi: 10.1093/nar/gku288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun B, Latham KA, Dodson ML, Lloyd RS. Studies on the catalytic mechanism of five DNA glycosylases. Probing for enzyme-DNA imino intermediates. J Biol Chem. 1995;270:19501–19508. doi: 10.1074/jbc.270.33.19501. [DOI] [PubMed] [Google Scholar]
- 24.Dutta S, Chowdhury G, Gates KS. Interstrand cross-links generated by abasic sites in duplex DNA. J Am Chem Soc. 2007;129:1852–1853. doi: 10.1021/ja067294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson KM, Price NE, Wang J, Fekry MI, Dutta S, Seiner DR, Wang Y, Gates KS. On the formation and properties of interstrand DNA-DNA cross-links forged by reaction of an abasic site with the opposing guanine residue of 5'-CAp sequences in duplex DNA. J Am Chem Soc. 2013;135:1015–1025. doi: 10.1021/ja308119q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price NE, Johnson KM, Wang J, Fekry MI, Wang Y, Gates KS. Interstrand DNA-DNA cross-link formation between adenine residues and abasic sites in duplex DNA. J Am Chem Soc. 2014;136:3483–3490. doi: 10.1021/ja410969x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freese E, Cashel M. Crosslinking of deoxyribonucleic acid by exposure to low pH. Biochim Biophys Acta. 1964;91:67–77. doi: 10.1016/0926-6550(64)90171-9. [DOI] [PubMed] [Google Scholar]
- 28.Burnotte J, Verly WG. Crosslinking of methylated DNA by moderate heating at neutral pH. Biochim Biophys Acta. 1972;262:449–452. doi: 10.1016/0005-2787(72)90488-1. [DOI] [PubMed] [Google Scholar]
- 29.Goffin C, Verly WG. Interstrand DNA crosslinks due to AP (apurinic/apyrimidinic) sites. FEBS Lett. 1983;161:140–144. doi: 10.1016/0014-5793(83)80747-9. [DOI] [PubMed] [Google Scholar]
- 30.Goffin C, Bricteux-Gregoire S, Verly WG. Some properties of the interstrand crosslinks in depurinated DNA. Biochim Biophys Acta. 1984;783:1–5. doi: 10.1016/0167-4781(84)90071-x. [DOI] [PubMed] [Google Scholar]
- 31.Prakash AS, Gibson NW. Sequence-selective depurination, DNA interstrand cross-linking and DNA strand break formation associated with alkylated DNA. Carcinogenesis. 1992;13:425–431. doi: 10.1093/carcin/13.3.425. [DOI] [PubMed] [Google Scholar]
- 32.Abner CW, Lau AY, Ellenberger T, Bloom LB. Base excision and DNA binding activities of human alkyladenine DNA glycosylase are sensitive to the base paired with a lesion. J Biol Chem. 2001;276:13379–13387. doi: 10.1074/jbc.M010641200. [DOI] [PubMed] [Google Scholar]
- 33.Zhao B, O'Brien PJ. Kinetic mechanism for the excision of hypoxanthine by Escherichia coli AlkA and evidence for binding to DNA ends. Biochemistry. 2011;50:4350–4359. doi: 10.1021/bi200232c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 35.Baldwin MR, O'Brien PJ. Human AP endonuclease 1 stimulates multiple-turnover base excision by alkyladenine DNA glycosylase. Biochemistry. 2009;48:6022–6033. doi: 10.1021/bi900517y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldwin MR, O'Brien PJ. Nonspecific DNA binding and coordination of the first two steps of base excision repair. Biochemistry. 2010;49:7879–7891. doi: 10.1021/bi100889r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hedglin M, O'Brien PJ. Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry. 2008;47:11434–11445. doi: 10.1021/bi801046y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson DM, 3rd, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 39.Jencks WP. Catalysis in Chemistry and Enzymology. New York: Dover; 1987. [Google Scholar]
- 40.Fersht A. Structure and Mechanism in Protein Science. New York: W. H. Freeman and Company; 1999. [Google Scholar]
- 41.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J Biol Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 42.Waters TR, Swann PF. Kinetics of the action of thymine DNA glycosylase. J Biol Chem. 1998;273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 43.Porello SL, Leyes AE, David SS. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 44.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J Biol Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- 45.O'Neill RJ, Vorob'eva OV, Shahbakhti H, Zmuda E, Bhagwat AS, Baldwin GS. Mismatch uracil glycosylase from Escherichia coli: a general mismatch or a specific DNA glycosylase? J Biol Chem. 2003;278:20526–20532. doi: 10.1074/jbc.M210860200. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen HS, Sundheim O, Gilljam KM, Slupphaug G, Krokan HE, Kavli B. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Admiraal SJ, O'Brien PJ. DNA-N-glycosylases process novel O-glycosidic sites in DNA. Biochemistry. 2013;52:4066–4074. doi: 10.1021/bi400218j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talpaert-Borle M, Liuzzi M. Reaction of apurinic/apyrimidinic sites with [14C]methoxyamine. A method for the quantitative assay of AP sites in DNA. Biochim Biophys Acta. 1983;740:410–416. doi: 10.1016/0167-4781(83)90089-1. [DOI] [PubMed] [Google Scholar]
- 49.Liuzzi M, Talpaert-Borle M. A new approach to the study of the base-excision repair pathway using methoxyamine. J Biol Chem. 1985;260:5252–5258. [PubMed] [Google Scholar]
- 50.Vasseur JJ, Rayner B, Imbach JL. Preparation of a short synthetic apurinic oligonucleotide. Biochem Biophys Res Commun. 1986;134:1204–1208. doi: 10.1016/0006-291x(86)90378-5. [DOI] [PubMed] [Google Scholar]
- 51.Lau AY, Schärer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.