Abstract
Medical message boards are online resources where users with a particular condition exchange information, some of which they might not otherwise share with medical providers. Many of these boards contain a large number of posts and contain patient opinions and experiences that would be potentially useful to clinicians and researchers. We present an approach that is able to collect a corpus of medical message board posts, de-identify the corpus, and extract information on potential adverse drug effects discussed by users. Using a corpus of posts to breast cancer message boards, we identified drug event pairs using co-occurrence statistics. We then compared the identified drug event pairs with adverse effects listed on the package labels of tamoxifen, anastrozole, exemestane, and letrozole. Of the pairs identified by our system, 75–80% were documented on the drug labels. Some of the undocumented pairs may represent previously unidentified adverse drug effects.
Keywords: data mining, information extraction, medical message board, drug adverse effect
1. Introduction
Internet message boards provide a rich data resource for a variety of purposes. Many boards contain a large number of candid messages which can be used to learn more about a given population without relying on costly methods of data collection, such as focus groups. For example, this medium has been intensively explored by marketing researchers. Glance, et al. [1] have developed a comprehensive system that crawls message boards and derives several metrics about consumer products that characterize how users rate those products and how frequently people discuss them. Feldman, et al. [2] have also published a system that extracts comparisons between different products and the attributes that they are compared on. Other systems have focused on different forms of web content such as product reviews in order to extract people’s sentiment about certain products [3;4] or have relied on the pages produced by Google searches to extract the reputation of a product [5].
Even though it is well known that the lay public frequently uses online resources such as message boards to seek and exchange medical information [9–13], medical message boards have been examined to a much lesser degree. Malouf, Davidson, and Sherman [14] applied sentiment analysis to epilepsy blogs in order to extract patient preferences for different seizure disorder drugs. Pharmacovigilance researchers have also applied data mining methods in order to extract signals of possible adverse drug events from databases such as the Adverse Event Reporting System (AERS) [15–17]. Electronic health records have also been examined to identify similar signals [18] as well as to generate biomedical hypotheses [19]. Though there is a history of data mining in the medical domain, it has yet to exploit the knowledge that can be derived from online user-generated content.
The burgeoning of medical message boards provides evidence of the increasing frequency of discussions about health-related matters in society at large, and the boards in turn provide a fertile source of data for identifying and evaluating the advice, concerns and opinions that are sought and provided by message board users. Users of medical message boards often ask questions and seek advice about topics that they may be hesitant to discuss with their health care providers. One such topic is concern about adverse effects experienced with some medications, especially those prescribed for serious conditions such as cancer, where patient anxiety may be heightened by the characteristics of the disease and the long-term exposure to potentially toxic drugs.
Although adverse events should be reported through available channels, such as the AERS of the U.S. Food and Drug Administration, many patients do not do so, perhaps because of ignorance of these channels, embarrassment, perceptions of negative provider attitude, their extreme illness, etc. Instead, they often use informal networks such as internet message boards to report and discuss adverse events. However, these data remain largely untapped by researchers in medical and healthcare domains.
To date, there are no reports in the medical or social sciences literature that apply methods to extract information from the text of medical discussion board content to identify and analyze reports of adverse drug events. While there are several studies evaluating discussions in message boards and chat rooms, these have used qualitative research methods [20–24]. Reasons for this include the unstructured nature of message board text, the use of nonstandard abbreviations, a wide variation in the use of syntax and spelling, the temporal relationship between posts in a single message thread, and references within messages to other threads.
Many systems have focused on extracting information from online resources other than message boards, particularly blogs. Although blogs are another venue where users may share personal experiences and opinions, they consist of a single main author privileged to post new topics. Readers of the blog may or may not be permitted to comment on the topics that the main author posts. On the other hand, all message board users are equally privileged to introduce new topics and reply to posts made by other users. These blog-focused systems are able to retrieve posts containing opinions about current events [6], identify album and song titles and users’ sentiment regarding them [7], and cluster blog posts by content [8]. However, the tasks that these systems were designed to perform are not focused on extracting medical information from these posts, and blogs are structurally different than message boards, necessitating the use of techniques specific to medical message boards.
Effective identification of adverse events related to medications or therapeutics has tremendous public health implications. Using the growing online media to generate appropriate medical hypotheses presents great potential to help address this issue. We present here methods that we used to identify such information, specifically self-reported adverse events that may be associated with four hormonal medications that are commonly used in the treatment of breast cancer.
2. Methods
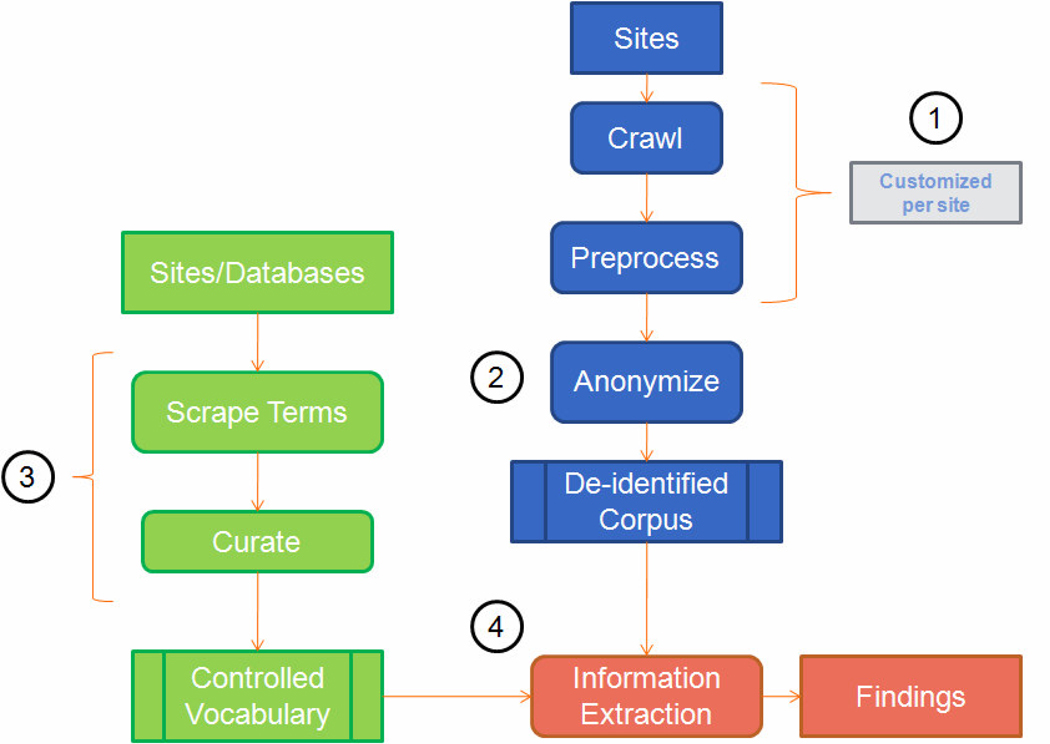
In order to address the unique difficulties posed by medical message boards, our system was structured as illustrated in Figure 1.
Figure 1.
Overview of the system architecture. 1) Corpus generation; 2) Removal of personal identifiers; 3) Construction of controlled vocabulary; 4) Information extraction
In step 1, the system downloaded message post pages from a set of message board sites and removed content unrelated to the posts from these pages. In step 2, the de-identification module removed personal identifiers (e.g., e-mail addresses, phone numbers, and usernames) from these posts. In Step 3 we developed the controlled vocabulary by scraping drug and side effect terms from various databases and websites using automated scripts in the Scrape Terms process. Afterwards, terms that were not indicative of a drug or event/symptom were removed by hand in the curation process. In step 4, all terms in the controlled vocabulary were identified in our de-identified corpus and any pair of terms that co-occurred at a statistically significant rate was treated as a “finding”.
2.1. Step 1: Corpus development
The breast cancer message board corpus was developed in three steps: message board identification, download, and anonymization. A collection of breast cancer message boards was identified by manually searching for large message boards specifically devoted to breast cancer. A custom-built web crawler, a program to browse the Internet and download pages, was used to download messages from each message board. Since each board was structured differently, the crawler had to be customized separately for each site in order to make sure that only message post and index page links within that board were followed. These saved pages were “cleaned” by extracting the following fields from each message:
Author (replaced with an anonymous identifier in the anonymization step)
Thread ID (the unique identifier for this message’s thread)
Time Posted
Message Body
Message Subject
This cleaning step was necessary since much of the text on a webpage is unrelated to the users’ posts. Such extraneous text may include the header, footer, navigation bar, and Javascript for the page. After preprocessing, only about 48% of the tokens, defined as strings of characters delimited by whitespace, in the original HTML pages were kept to generate the corpus. The final corpus contained over 1.1 million messages, comprising over 100 million words; the average message entry is 99 tokens long, with standard deviation of 135 tokens. This is because a large proportion of the messages tend to be very short (1–4 tokens), with a very long tail of increasingly long messages.
The following sites were used to generate the corpus: breastcancer.org, komen.org, csn.cancer.org, bcsupport.org, healthboards.com, cancercompass.com, webmd.com, dailystrength.org, revolutionhealth.com, ehealthforum.com, oprah.com. Most messages from the breast cancer corpus are from breastcancer.org (70%), komen.org (16.5%), and csn.cancer.org (9.2%). Each of the other sites was responsible for 1% or less of the total entries. This Zipfian distribution of messages over sites, where a select few sites contain many messages and a long list of sites contain much fewer, is to be expected.
2.2. Step 2: Anonymization
In order to de-identify the messages, we created an anonymizer to remove the following information:
Email Addresses
Phone Numbers
Uniform Resource Locators (URLs)
Social Security Numbers (SSNs)
Usernames
Proper Names
Although the breast cancer message board corpus consisted of posts that were made publicly available online, in order to protect the identities of the authors, we removed these possible identifiers. Although our approach relies chiefly on co-occurrence statistics, we also referred back to the original de-identified messages in order to verify that pairs found to be statistically significant were actually related within the messages. The University of Pennsylvania institutional review board requires that message board posts be de-identified before being used for research purposes (either analyzed qualitatively or extracting co-occurrence statistics.)
Email addresses, phone numbers, URLs, and SSNs were easily removed using regular expressions, a tool used to recognize strings of characters, since these types of identifying information all have predictable structures. There seemed to be very few instances of these. However, usernames and proper names were more difficult to remove since message posts often contain spelling errors, non-standard constructions, inconsistent capitalization, and a wide variety of nicknames and usernames that would not be found in list of proper names. We first used the 2008 Stanford Named Entity Recognizer (NER) [25] trained on a combination of the Conference on Natural Language Learning (CoNLL) 03, Message Understanding Conference (MUC) 6, MUC-7, and Automatic Content Extraction (ACE) 08 corpora to find proper names.
The performance of the Stanford NER was evaluated, using the precision, recall, and F-score metrics:
| (1) |
| (2) |
| (3) |
Over a random sample of 500 messages containing 523 names identified by a human coder, the Stanford NER exhibited mediocre performance, achieving precision of 69.6%, recall of 77.6%, and an F-score of 73.4%. This is likely due to the fact that it was not trained on message board text and relied on features that may be indicative of proper names in less noisy text, but not in message board text (e.g., capitalization). In addition, the Stanford NER was not designed to identify usernames, which could be very different from proper names. Thus, we had to design and implement our own system to remove both proper names and usernames [26].
To do so, we trained a conditional random field (CRF) [27] over a 1000 message sample from the breast cancer corpus consisting of 91,344 tokens, of which 822 were proper names and 682 were usernames. Proper names and usernames were manually identified and tagged by a human coder in order to form this training set. Each token was described by a feature vector. Some examples of features in this vector were whether the token belonged to a particular dictionary (e.g., proper names, common English words, and very common English words), whether the token matched a username in this thread, the position of the token in the message, and the case of the token. The features for the previous and following two tokens were also included in each token’s feature vector.
To remove proper names and usernames from a particular message, we tokenized the message and ran the CRF over the feature vectors for these tokens. These vectors included features that are useful to named entity recognition across domains (e.g., is the token title case, does it belong to a list of names, is it a possible misspelling of a name) as well as features that take advantage of the structure of message boards and message board posts (e.g., does the token often occur near the beginning or end of posts, does the token have a high tf-idf value out of all tokens in a particular message board when treating entire message boards as documents). Any token with a predicted probability of being a name greater than 0.05 was replaced with an anonymous tag. This 0.05 threshold was chosen in order to maximize the recall of the anonymizer without removing too many non-name tokens. Testing this system over 500 messages, containing 483 total names, sampled from the breast cancer corpus, yielded a precision of 67.4%, recall of 98.1%, and F-score of 79.9% for removing names. Our anonymizer demonstrates precision comparable to the Stanford NER’s, a much higher recall (98.1% compared to 77.6%), and is designed to remove usernames as well. A sample message from the de-identified corpus is provided below:

Block 1. Sample message after anonymization. The message was altered to prevent searching on the Internet for its original posting and subsequent identification of the poster.
2.3. Step 3: Controlled vocabulary
As mentioned above, patients posting on medical message boards often use lay vocabulary to describe the symptoms they are experiencing or the medicines they are taking. In order to extract useful information from these posts, a controlled vocabulary of lay medical terms was constructed. Websites and databases containing lists of dietary supplements, pharmaceuticals, and adverse events were scraped for terms to populate the vocabulary. Since most of these lists of terms had a regular structure (e.g., each term was marked with an explicit HTML tag on a website or located in a specific column of a database), we were able to collect the terms using simple scripts, which were programmed, for example, to save all terms that occur in a particular field of a database or in a particular list on a webpage. Terms that were likely to result in false positives (e.g., event: boil, shake) were manually removed from the vocabulary. The medical vocabulary consisted of the following:
Dietary supplements: hand-compiled by one of the authors (JM), who has expertise in complementary and alternative medicine: 507 terms
Pharmaceuticals: Cerner Multum’s Drug Lexicon, (Denver, CO): 16,383 terms
Events (terms that could either refer to an indication for a drug or an adverse event caused by a drug): http://www.medicinenet.com/ and adverse events listed in the AERS database over the period 2004 through the second quarter of 2009; not specific to breast cancer: 26,817 terms
We then augmented all of these lists using the Consumer Health Vocabulary (CHV) provided by the Consumer Health Vocabulary Initiative1, in order to produce a vocabulary closer to the lay vocabulary that would be used by patients on a discussion board. This also provided us with a way of classifying several different terms as being instances of a more general term (e.g., turmeric, tumeric [sic], and curcumin are three different ways that a user may refer to curcumin).
2.4. Step 4: Information Extraction
After the anonymized corpus and controlled vocabularies were generated, we generated frequency counts of each vocabulary term in the corpus, in order to establish which terms are talked about the most. This is similar to the Buzz count used in Glance, et al. [1]. We retrieved the single term counts by counting the number of messages in which each term occurred. Each token was first stemmed using a Porter stemmer, an algorithm meant to remove inflection from a word, from the Natural Language Toolkit (NLTK) [28] before matching it to a term in the controlled vocabulary.
We also extracted association rules between pairs of terms. By association rules we mean pairs of terms that co-occur within 20 tokens more frequently than would be expected if the terms were distributed independently. These rules have no causal direction, but simply suggest that there is a correlation between the presence of one term and the presence of the other. In order to generate all possible association rules between terms, all terms in the controlled vocabulary occurring in the corpus were identified. Any two terms co-occurring within a window of 20 tokens apart were treated as a possible association rule; this window seemed to produce the best precision and recall of valid association rules over a random sample of 500 messages.
Our approach of relying on co-occurrences of terms to generate association rules has been used in several other systems [2;5;14;18;29;30]. For each association rule (X, Y) output by the query script, we constructed a 2 by 2 table of the occurrence of X and Y and calculated a one-tailed Fisher’s exact P-value for that rule, expressing the likelihood that these two terms co-occurred independently by chance. All association rules with p-values greater than the Simes-corrected [31] 0.05 threshold were then determined to be non-significant. We corrected the 0.05 threshold in order to account for multiple testing bias, and in particular chose Simes correction since it is not as strict as Bonferroni correction. Even though Simes correction is not as conservative as Bonferroni correction, the number of statistically significant association rules generated per drug was relatively low and could be analyzed by a single person for possible signals (median of two statistically significant rules per drug, with a maximum of 95). However, rules with very low counts were not ignored only because they were infrequent; these low count pairs could potentially signal a rare, but very real relationship between the two terms.
3. Results
3.1 Validating the system
In order to validate our system, we selected four of the most commonly used drugs to treat breast cancer: tamoxifen (Nolvadex), anastrozole (Arimidex), letrozole (Femara), and exemestane (Aromasin). Tamoxifen has long been the standard treatment for hormonally-responsive breast cancer. Anastrozole, letrozole, and exemestane are aromatase inhibitors, more recently developed, that are also used to treat hormonally-responsive breast cancer. We chose these four drugs because they have similar indications and were frequently mentioned in the breast cancer corpus. Table 1 shows the top 10 association rules by count identified by our system for tamoxifen.
Table 1.
Top ten rules returned for tamoxifen ranked by count. Drug+/Event+ is the number of messages that contain the drug and event co-occurring within a 20 token window, Drug+/Event− is the number of messages containing only the drug and not the event, Drug−/Event+ is the number of messages containing only the event and not the drug, and Drug−/Event− is the number of messages mentioning neither the drug nor the event. P-values in this list are very close to 0 or 1.
| Event | Drug− /Event− |
Drug− /Event+ |
Drug+ /Event− |
Drug+ /Event+ |
P-Value |
|---|---|---|---|---|---|
| hot flashes | 1,186,668 | 10,809 | 30,306 | 1,949 | 0 |
| breast cancer | 1,135,252 | 62,225 | 30,914 | 1,341 | 1 |
| menopause | 1,189,938 | 7,539 | 31,236 | 1,019 | 0 |
| pain | 1,114,656 | 82,821 | 31,547 | 708 | 1 |
| weight gain | 1,193,000 | 4,477 | 31,751 | 504 | 4.47E-143 |
| joint pain | 1,192,247 | 5,230 | 31,780 | 475 | 3.12E-104 |
| uterine cancer | 1,196,619 | 858 | 31,881 | 374 | 6.03E-276 |
| fatigue | 1,182,660 | 14,817 | 31,973 | 282 | 1 |
| night sweat | 1,195,587 | 1,890 | 31,981 | 274 | 1.40E-100 |
| depression | 1,191,644 | 5,833 | 32,001 | 254 | 1.53E-12 |
| weight loss | 1,193,207 | 4,270 | 32,055 | 200 | 9.69E-13 |
For each of these four drugs, we compiled a documented list of all adverse events (AEs) believed to occur from the drug. This list was compiled from all of the AEs mentioned in tables and notes contained in the drug label.
For each of the four drugs, we compiled a list of significantly associated events returned by our system. We then evaluated our system’s performance by comparing our system list against the documented list for that particular drug using the precision and recall metrics. In this context, precision was defined as the proportion of the events we found that occur in the documented list as AEs:
| (4) |
Recall was defined as the proportion of documented AEs occurring in the system list:
| (5) |
This method of evaluation is similar to that used to evaluate a pharmacovigilance system over electronic health records by Wang, et al. [18].
3.2. Identifying rare and novel events
In addition to evaluating our system for its ability to identify many of the AEs listed on the drug label, we also investigated the events returned by our system that were either documented as rare AEs or were not listed on the drug label at all. For example, our system identified many AEs that were documented as rare events for tamoxifen; these included “fatty liver”, “uterine cancer”, “stroke”, and “pulmonary embolism”. We referred back to the messages where the terms occurred in order to determine the context in which authors mentioned these events. A few anecdotes from these messages are listed below. Note that the majority of message board users referred to the aromatase inhibitors by their brand names and these anecdotes reflect that.

Block 2. Anecdotes of rare (as defined by label) AEs occurring with tamoxifen. Very few authors mentioned that they developed uterine cancer, and that they were simply scared of developing it. For the other rare events, authors had mentioned that they actually experienced these AEs.
Although it is mentioned as a very rare AE on the label, “uterine cancer” co-occurred 374 times with tamoxifen in our breast cancer corpus. This does not necessarily suggest that it is a more common AE than the label states, but simply that people frequently talk about it. Most of these messages demonstrated anxiety about taking tamoxifen because of this side effect, rather than having actually developed uterine cancer. However, for other AEs, such as “pulmonary embolism” and “fatty liver”, authors mentioned that they had actually developed the conditions.
There were a few AEs that were reported with one of these four drugs that were not listed on the drug label. Many of these were false positives (i.e., AE and drug mentions were unrelated in the posts, determined by human). However, there were a few significantly associated events (with p-value less than Simes-corrected .05) that message board authors claimed to have actually occurred. The following undocumented AEs were found to be based on instances where message board authors claimed to have actually experienced the AE.
tamoxifen: weight gain
anastrozole: chapped lips, dry eye, lupus, conjunctivitis, fibromyalgia
exemestane: dry eye, high cholesterol, vaginal dryness
letrozole: mood swings, vaginal discharge
Not all of these AEs may be truly caused by the drug, but, at the very least, they may give practitioners a better idea of common perceptions that patients hold about these drugs. A few examples of posts where authors mentioned experiencing these AEs are listed below in Block 3.

Block 3. Sample anecdotes of undocumented AEs that message board authors claimed to have experienced from tamoxifen, anastrozole, exemestane, and letrozole.
4. Discussion
4.1 Contributions to low recall
Our system results in relatively low recall of drug AEs documented on the label, which seems particularly striking for more common side effects, given that these AEs are very common and one would expect authors to mention these symptoms frequently in their posts. However, many of these common AEs are mentioned very frequently throughout the entire corpus. One example is fatigue, a known AE of anastrozole, but also a potential effect of cancer itself. The terms “fatigue” and “arimidex” co-occurred 210 times within the breast cancer corpus. However, each of these terms occurred in 15,099 and 17,124 messages, respectively, yielding a non-significant p-value. In order for the association rule “arimidex-fatigue” to be significant, these two terms would have had to co-occur at a much higher frequency than what was observed. Our system seems better suited to finding rare AEs that are related to a specific drug, such as “uterine cancer” for “tamoxifen”. However, identifying new adverse events is probably a more appropriate goal than measuring the frequency of known side effects.
It is also important to note that multiple event terms returned by our system may match just a single AE in the documented AE list. For example, if the documented AE list contained “pain in extremity”, then the terms “finger pain” and “toe pain” in the system’s returned list would be considered members of the documented AE list. This also contributes to our system’s low recall.
The low recall against the documented list is due mostly to the fact that the documented list is long. Some of the terms in this list are pervasive within posts (e.g., fatigue, nausea, pain), and do not co-occur frequently enough with the specific drug to suggest a significant association rule. However, other AEs in the documented list are never mentioned by authors at all. Some examples of terms that are absent from our breast cancer corpus but are documented AEs for one or more of the four drugs we investigated are “increased bilirubin”, “increased creatinine”, “thrombocytopenia”, “increased alkaline phosphate”, and “stevens-johnson syndrome”. Many of these are laboratory abnormalities that might not be noticed or known by patients.
4.2 Undocumented events returned by system
Our system exhibited relatively high precision in the AEs that it returned. However, on average, 23% of the drug-event rules identified by our system were undocumented on the label. Some of these undocumented AEs appeared to be unrelated to the drug. These events tended to co-occur with the drug very infrequently (generally less than 10 times), and occur very infrequently throughout the corpus. Because the number of co-occurrences between the drug and event was so small, it was simple for us to refer directly to the messages where these terms co-occurred and determine whether the author reported the event as being an AE of the drug.
It is possible that some of the AEs returned could have been influenced by “spam” posts in our corpus. However, we consider this unlikely. Out of a random sample of 200 messages from the breast cancer corpus, only one message (0.5%) could have been construed as “spam”. This message was a request for donations to a group dedicated to providing financial assistance to economically disadvantaged women with breast cancer. We believe that the low level of spam in this corpus is due to message board users monitoring the content of their respective boards and removing messages that are posted by spammers. Thus, it is unlikely that these undocumented drug AEs were due to spam messages.
Some pairs that our system identified were considered to be false positives since they were not specifically documented as, but were similar to, AEs on the drug label. “arimidex-gout” was one of these pairs. Although gout is not a documented AE of anastrozole, it is a case of acute arthritis. The messages that mentioned this event noted remedies for gout that may be effective in treating aromatase inhibitor-induced arthritis. “Menopause” is an event that was returned by all four drugs. Authors used the term “menopause” to describe the AEs that they were experiencing as menopausal, particularly hot flashes; they did not specifically claim that a particular drug had induced menopause.
Our system does not determine the kind of relationship (indication or side effect) between the drug and event for each association rule. Rather, it suggests only that the drug and event terms are correlated. For example, the events “bone density decreased”, “bone loss”, and “osteoporosis” were all significantly associated with tamoxifen. However, tamoxifen is known to increase bone density [32]. Referring back to the original messages where these event terms and “tamoxifen” co-occurred revealed that post authors were glad that tamoxifen prevented or remedied these events. Some terms were actually contraindications for the drug, such as “Factor V Leiden mutation” (associated with an increased risk of blood clots) for tamoxifen. Even though these terms were related to the drug, they are not caused by the drugs and were thus treated as false positives.
The remainder of the undocumented events consisted of messages where the author claimed to have experienced the AE in response to the drug. Block 3 contains several examples of these undocumented AEs. Some of these AEs may be common knowledge in the medical community even though they are not listed on the label. For example, it is known that tamoxifen tends to make it more difficult for patients to lose weight [33] even though neither “weight gain” nor “difficulty to lose weight” is mentioned as an AE on the tamoxifen label. Other AEs mentioned in the corpus may not be commonly accepted as an AE of that particular drug. Controlled studies would be needed to verify that these undocumented AEs actually occur from a particular drug. However, these anecdotes may serve as signals that could be rigorously verified in controlled studies.
4.3 Further work
We understand that medical message board corpora are very different from clinical trial data, health care data, and databases of reported adverse events. Medical message boards provide a community and support for their members. Post authors not only communicate how they are feeling and any AEs they have experienced, but also AEs that they are worried about or that a friend may have claimed to experience. However, given that we have shown that our system is able to reliably extract known AEs for several drugs, it may have potential utility for identifying undocumented AEs for dietary supplements that breast cancer patients use as well.
Extending the system to determine the relation between the drug and event, for example, indication or AE, would be useful for other drugs as well as nutritional supplements and herbal preparations. We will also improve our system to extract other types of information from medical message boards. Currently our system is able to identify documented AEs of drugs with high precision by the frequency that they co-occur. However, the current system cannot determine what type of speech act the author used for each specific case where they co-occurred. For example, the author could be claiming that they actually experienced the AE, claiming that their friend experienced the AE, wondering if a drug causes that particular AE, or are just worried about a specific AE. Refining this system to identify these speech acts would enable our system to be used as an alternative to focus groups. Another possible route of exploration would be to extend our system to identify instances of drug non-adherence, and identify the AEs that led to this decision.
It would be interesting to know how people generally feel about a particular drug, based on their posts on message boards. Do they like a drug? Do they hate it? Why? Extending our system to perform a sentiment analysis on the breast cancer corpus would allow researchers and medical practitioners to get a sense of what people think about certain drugs.
We also intend to apply our system to medical message board corpora for conditions other than breast cancer, in order to evaluate its ability to generalize to other conditions. Currently we have compiled corpora of obesity, diabetes, and arthritis message board posts. These are ideal candidates for our method since they contain a large number of messages because they are all chronic conditions with a high prevalence. We expect similar results to those generated from the breast cancer corpus. In addition, we will continue to update the system’s controlled vocabulary based on how well it identifies relevant instances of drug and event mentions.
6. Conclusion
We have designed a system to identify signals of potential adverse events that overcomes many of the difficulties associated with medical message boards and is able to extract useful information from them. Our system is able to generate a corpus of medical message board text by downloading the message pages, extracting the relevant fields from the pages, and programmatically removing identifying information from the documents. These de-identified documents are then searched for terms occurring in the controlled vocabulary in order to extract association rules that may signify meaningful relationships between these terms.
We demonstrated the efficacy of this system by extracting the significant drug-event association rules for four hormonal breast cancer treatment drugs from a corpus of breast cancer message board posts and compared this list to a list of all AEs that were documented on each drug label. Although our system’s recall over these documented AE lists was relatively low (average of 35.1%), the precision was relatively high (average of 77.0%), suggesting that the terms in the significant association rules returned are not only correlated, but represent a real semantic relationship as well.
In addition, we verified that some of the undocumented AEs considered significant by our system signaled an AE that message board authors claimed to have experienced from the drug. Whether or not the AE was actually caused by the drug is unknown. However, it may signal a true AE from that drug, and may be worth further investigation. Even if the AE is not caused by the drug, it is useful to know what symptoms patients believe (correctly or not) are being caused by the drug.
In future work, we plan to extend the system to identify the speech act that was used for each drug-side effect mention and to use this system to identify undocumented adverse effects from dietary supplements.
-
>
We develop a system to extract drug adverse events from medical message boards.
-
>
We validate system over a corpus of breast cancer message posts.
-
>
75–80% of returned adverse events are documented on the drug label.
-
>
Some of the undocumented adverse events referred to actual users experiencing them.
-
>
This could be used as a tool to identify signals of drug adverse events.
Table 2.
Recall refers to the proportion of terms in the documented list that matched terms in the system list, over the total number of terms in the documented list. Precision refers to the proportion of terms in the system list that matched terms in the documented list over the total number of terms in the system list. ‘N’ refers to the size of the denominator for each value calculated.
| Recall | Precision | |||
|---|---|---|---|---|
| Value | N | Value | N | |
| tamoxifen (Nolvadex) | 42.4% | 66 | 79.1% | 62 |
| anastrozole (Arimidex) | 36.8% | 76 | 75.0% | 55 |
| exemestane (Aromasin) | 25.0% | 64 | 75.8% | 33 |
| letrozole (Femara) | 35.6% | 59 | 78.0% | 41 |
| All four drugs | 35.1% | 265 | 77.0% | 191 |
Acknowledgment
This project is supported by the National Library of Medicine (RC1LM010342). Access to the Lexicon database was supported by the National Center for Research Resources (5Kl2RR024132). We thank Cristin Freeman, MPH for providing access to the Lexicon database and for sharing her expertise throughout the project. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health. This study was approved by the Institutional Review Board at the University of Pennsylvania.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contributions
JHH, JM, LU, SHennessy, and SHill conceptualized the research. JHH, LU, and SHill designed the system methodology. JM reviewed the drug and supplement vocabularies. SHennessy, JM, and CL interpreted the drug-symptom findings. AB constructed the message board corpus and implemented the system. AC contributed to the validation of the system. AB and JHH wrote the manuscript. All authors contributed to revision and approved this manuscript.
Reference List
- 1.Glance N, Hurst M, Nigam K, Siegler M, Stickton R, Tomokiyo T. Deriving marketing intelligence from online discussion; Proceedings of the eleventh ACM SIGKDD international Conference on Knowledge Discovery in Data Mining; 2005. pp. 419–428. [Google Scholar]
- 2.Feldman R, Fresco M, Goldenberg J, Netzer O, Ungar L. Extracting Product Comparisons from Discussion Boards; Proceedings of the 2007 Seventh IEEE International Conference on Data Mining; 2007. pp. 469–474. [Google Scholar]
- 3.Chklovski T. Deriving quantitative overviews of free text assessments on the web; IUI '06: Proceedings of the 11th international conference on Intelligent user interfaces; 2006. pp. 155–162. [Google Scholar]
- 4.Dave K, Lawrence S, Pennock DM. Mining the peanut gallery: opinion extraction and semantic classification of product reviews; Proceedings of the 12th international conference on World Wide Web; 2003. pp. 519–528. [Google Scholar]
- 5.Morinaga S, Yamanishi K, Fukushima T. Mining product reputations on the web. KDD '02; Proceedings of the eighth ACM SIGKDD international conference on Knowledge; 2002. pp. 341–349. [Google Scholar]
- 6.Mishne G. Using blog properties to improve retrieval; Proceedings of the International Conference on Weblogs and Social Media (ICWSM); 2007. [Google Scholar]
- 7.Gruhl D, Nagarajan M, Pieper J, Robson C. Context and domain knowledge enhanced entity spotting in informal text. 8th International Semantic Web Conference (ISWC'09); Proceedings of the 8th International Semantic Web Conference (ISWC'09); 2009. pp. 260–276. [Google Scholar]
- 8.Hayes C, Avesani P, Bojars U. From web to social web: discovering and deploying user and content profiles. Springer-Verlag; 2007. An analysis of bloggers, topics, and tags for a blog recommender system; pp. 1–20. [Google Scholar]
- 9.Cousineau TM, Rancourt D, Green TC. Web chatter before and after the Women's Health Initiative results: a content analysis of on-line menopause message boards. Journal of Health Communication. 2006;11(2):133–147. doi: 10.1080/10810730500526661. [DOI] [PubMed] [Google Scholar]
- 10.Donelle L, Hoffman-Goetz L. Health literacy and online health discussions of North American Black women. Women & Health. 2008;47(4):71–90. doi: 10.1080/03630240802100689. [DOI] [PubMed] [Google Scholar]
- 11.Gooden RJ, Winefield HR. Breast and prostate cancer online discussion boards: a thematic analysis of gender differences and similarities. Journal of Health Psychology. 2007;12(1):103–114. doi: 10.1177/1359105307071744. [DOI] [PubMed] [Google Scholar]
- 12.Meric F, Bernstam EV, Mirza NQ, Hunt KK, Ames FC, Ross MI, et al. Breast cancer on the world wide web: cross sectional survey of quality of information and popularity of websites. BMJ. 2002;324(7337):577–581. doi: 10.1136/bmj.324.7337.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz PN, Stava C, Beck ML, Vassilopoulou-Sellin R. Internet message board use by patients with cancer and their families. Clinical Journal of Oncology Nursing. 2003;7(6):663–667. doi: 10.1188/03.CJON.663-667. [DOI] [PubMed] [Google Scholar]
- 14.Malouf R, Davidson B, Sherman A. Mining web texts for brand associations; Proceedings of the AAAI- 2006 spring symposium on computational approaches to analyzing weblogs; 2006. pp. 125–126. [Google Scholar]
- 15.Almenoff JS, LaCroix KK, Yuen NA, Fram D, DuMouchel W. Comparative Performance of Two Quantitative Safety Signalling Methods: Implications for Use in a Pharmacovigilance Department. Drug Safety. 2006;29(10):875–887. doi: 10.2165/00002018-200629100-00005. [DOI] [PubMed] [Google Scholar]
- 16.DuMouchel W, Pregibon D. Empirical bayes screening for multi-item associations; Proceedings of the Seventh ACM SIGKDD international Conference on Knowledge Discovery and Data Mining; 2001. pp. 67–76. [Google Scholar]
- 17.Wilson A, Thabane L, Holbrook A. Application of data mining techniques in pharamcovigilance. British Journal of Clinical Pharmacology. 2004;57(2):127–134. doi: 10.1046/j.1365-2125.2003.01968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Hripcsak G, Markatou M, Friedman C. Active Computerized Pharmacovigilance Using Natural Language Processing, Statistics, and Electronic Health Records: A Feasibility Study. Journal of the American Medical Informatics Association. 2005;16(3):328–337. doi: 10.1197/jamia.M3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman JA, Chu WW, Parker DS, Goldman RM. Term Domain Distribution Analysis: a Data Mining Tool for Text Databases. Methods of Information in Medicine. 2010;2010(2):96–101. [PubMed] [Google Scholar]
- 20.Armstrong N, Powell J. Patient perspectives on health advice posted on Internet discussion boards: a qualitative study. Health Expectations. 2009;12(3):313–320. doi: 10.1111/j.1369-7625.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nahm ES, Resnick B, DeGrezia M, Brotemarkle R. Use of discussion boards in a theory-based health web site for older adults. Nursing Research. 2009;58(6):419–426. doi: 10.1097/NNR.0b013e3181bee6c4. [DOI] [PubMed] [Google Scholar]
- 22.Copelton DA, Valle G. "You don't need a prescription to go gluten-free": the scientific self-diagnosis of celiac disease. Social Science & Medicine. 2009;69(4):623–631. doi: 10.1016/j.socscimed.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Habermeyer E, Habermeyer V, Jahn K, Domes G, Nagel E, Herpertz SC. [An internet based discussion board for persons with borderline personality disorders moderated health care professionals]. [German] Psychiatrische Praxis. 2009;36(1):23–29. doi: 10.1055/s-2008-1067508. [DOI] [PubMed] [Google Scholar]
- 24.Shigaki CL, Smarr KL, Yang G, Donovan-Hanson K, Siva C, Johnson RA, et al. Social interactions in an online self-management program for rheumatoid arthritis. Chronic Illness. 2008;4(4):239–246. doi: 10.1177/1742395308097862. [DOI] [PubMed] [Google Scholar]
- 25.Finkel JR, Grenager T, Manning C. Incorporating non-local information into information extraction systems by Gibbs sampling. ACL 2005. In the Proceedings of the 43rd Annual Meeting of the Association for Computational Linguistics; 2005. pp. 363–370. [Google Scholar]
- 26.Benton A, Hill S, Ungar L, Chung A, Leonard C, Freeman C, et al. A system for de-identifying medical message board text; International Conference on Machine Learning and Applications 2010; 2010. [Google Scholar]
- 27.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. Eighteenth International Conference on Machine Learning. In the Proceedings of the Eighteenth International Conference on Machine Learning. 2001:282–289. [Google Scholar]
- 28.Bird S. NLTK: the natural language toolkit. Annual meeting of the ACL. In the Proceedings of the COLING/ACL on Interactive presentation sessions; 2004. pp. 69–72. [Google Scholar]
- 29.Friedman C. Discovering Novel Adverse Drug Events Using Natural Language Processing and Mining of the Electronic Health Record. In the Proceedings of the 12th Conference on Artificial Intelligence in Medicine: Artificial Intelligence in Medicine; 2009. pp. 1–5. [Google Scholar]
- 30.Eysenbach G. Infodemiology and infoveillance: framework for an emerging set of public health methods to analyze search, communication and publication behavior on the internet. Journal of Medical Internet Research. 2009;11(1) doi: 10.2196/jmir.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika. 1986;73(3):751–754. [Google Scholar]
- 32.Resch A, Biber E, Seifert M, Resch H. Evidence that tamoxifen preserves bone density in late postmenopausal women with breast cancer. Acta Oncologica. 1998;37(7–8):661–664. doi: 10.1080/028418698430007. [DOI] [PubMed] [Google Scholar]
- 33.Hoskin PJ, Ashley S, Yarnold JR. Weight gain after primary surgery for breast cancer - effect of tamoxifen. Breast Cancer Research and Treatment. 22(2):129–132. doi: 10.1007/BF01833342. 992. [DOI] [PubMed] [Google Scholar]