Abstract
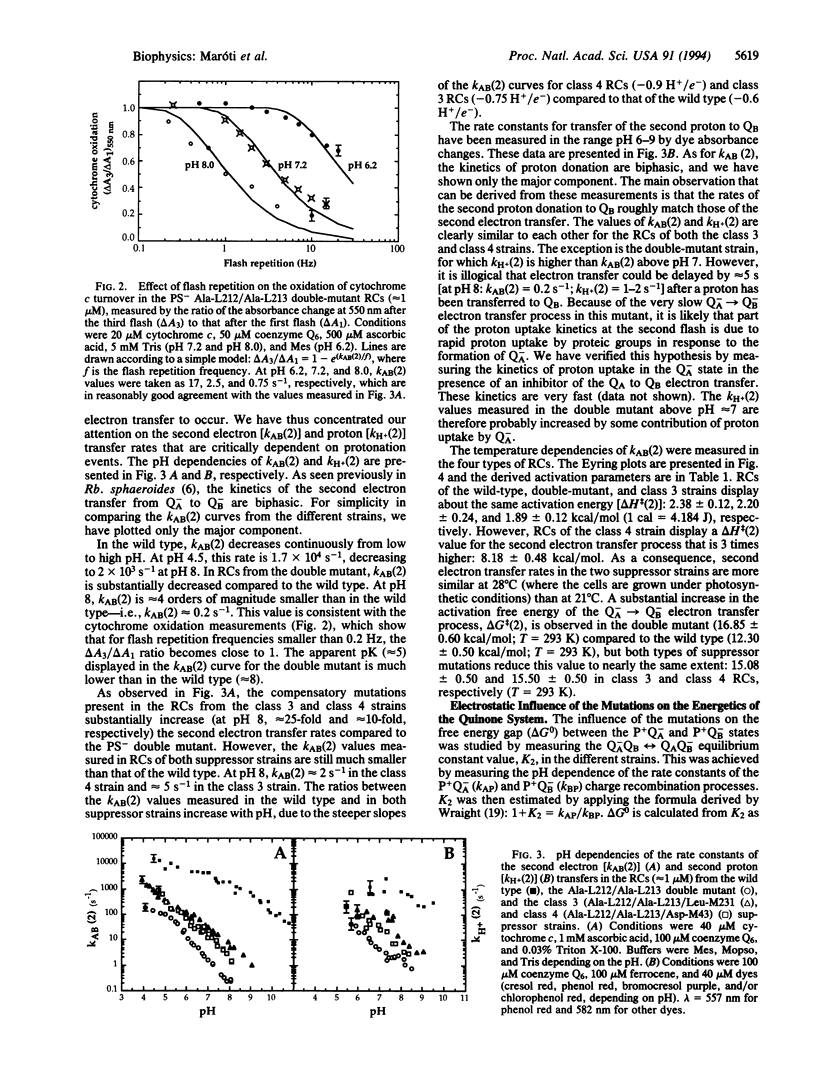
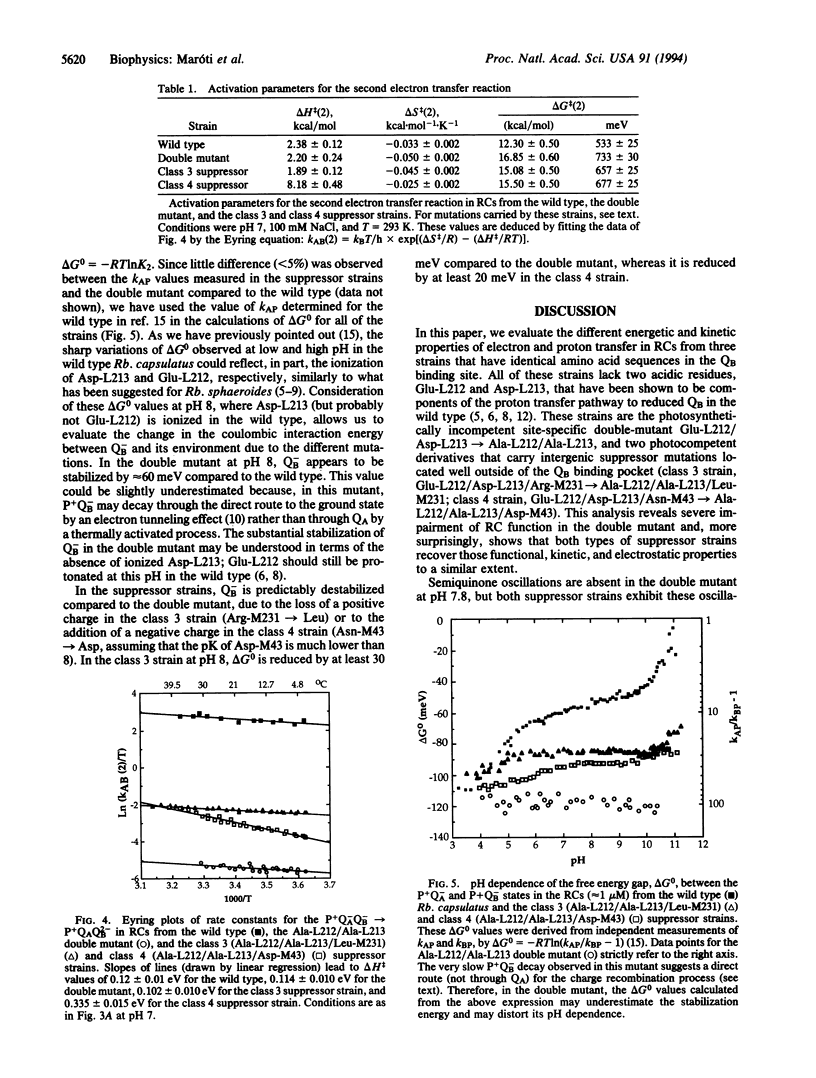
Light-induced charge separation in the photosynthetic reaction center results in delivery of two electrons and two protons to the terminal quinone acceptor QB. In this paper, we have used flash-induced absorbance spectroscopy to study three strains that share identical amino acid sequences in the QB binding site, all of which lack the protonatable amino acids Glu-L212 and Asp-L213. These strains are the photosynthetically incompetent site-specific mutant Glu-L212/Asp-L213-->Ala-L212/Ala-L213 and two different photocompetent derivatives that carry both alanine substitutions and an intergenic suppressor mutation located far from QB (class 3 strain, Ala-Ala + Arg-M231-->Leu; class 4 strain, Ala-Ala + Asn-M43-->Asp). At pH 8 in the double mutant, we observe a concomitant decrease of nearly 4 orders of magnitude in the rate constants of second electron and proton transfer to QB compared to the wild type. Surprisingly, these rates are increased to about the same extent in both types of suppressor strains but remain > 2 orders of magnitude smaller than those of the wild type. In the double mutant, at pH 8, the loss of Asp-L213 and Glu-L212 leads to a substantial stabilization (> or = 60 meV) of the semiquinone energy level. Both types of compensatory mutations partially restore, to nearly the same level, the original free energy difference for electron transfer from primary quinone QA to QB. The pH dependence of the electron and proton transfer processes in the double-mutant and the suppressor strains suggests that when reaction centers of the double mutant are shifted to lower pH (1.5-2 units), they function like those of the suppressor strains at physiological pH. Our data suggest that the main effect of the compensatory mutations is to partially restore the negative electrostatic environment of QB and to increase an apparent "functional" pK of the system for efficient proton transfer to the active site. This emphasizes the role of the protein in tuning the electrostatic environment of its cofactors and highlights the possible long-range electrostatic effects.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baciou L., Bylina E. J., Sebban P. Study of wild type and genetically modified reaction centers from Rhodobacter capsulatus: structural comparison with Rhodopseudomonas viridis and Rhodobacter sphaeroides. Biophys J. 1993 Aug;65(2):652–660. doi: 10.1016/S0006-3495(93)81114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. H., el-Kabbani O., Tiede D., Norris J., Schiffer M. Structure of the membrane-bound protein photosynthetic reaction center from Rhodobacter sphaeroides. Biochemistry. 1991 Jun 4;30(22):5352–5360. doi: 10.1021/bi00236a005. [DOI] [PubMed] [Google Scholar]
- Hanson D. K., Baciou L., Tiede D. M., Nance S. L., Schiffer M., Sebban P. In bacterial reaction centers protons can diffuse to the secondary quinone by alternative pathways. Biochim Biophys Acta. 1992 Sep 25;1102(2):260–265. doi: 10.1016/0005-2728(92)90108-e. [DOI] [PubMed] [Google Scholar]
- Hanson D. K., Tiede D. M., Nance S. L., Chang C. H., Schiffer M. Site-specific and compensatory mutations imply unexpected pathways for proton delivery to the QB binding site of the photosynthetic reaction center. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8929–8933. doi: 10.1073/pnas.90.19.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock M. L., McPherson P. H., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: replacement of serine-L223 by alanine inhibits electron and proton transfers associated with reduction of quinone to dihydroquinone. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6803–6807. doi: 10.1073/pnas.87.17.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock M. L., Rongey S. H., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: replacement of glutamic acid 212 in the L subunit by glutamine inhibits quinone (secondary acceptor) turnover. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6602–6606. doi: 10.1073/pnas.86.17.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongey S. H., Paddock M. L., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: second-site mutation Asn-M44-->Asp restores electron and proton transfer in reaction centers from the photosynthetically deficient Asp-L213-->Asn mutant of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1325–1329. doi: 10.1073/pnas.90.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E., Wraight C. A. Proton and electron transfer in the acceptor quinone complex of Rhodobacter sphaeroides reaction centers: characterization of site-directed mutants of the two ionizable residues, GluL212 and AspL213, in the QB binding site. Biochemistry. 1992 Jan 28;31(3):855–866. doi: 10.1021/bi00118a031. [DOI] [PubMed] [Google Scholar]
- Takahashi E., Wraight C. A. Small weak acids stimulate proton transfer events in site-directed mutants of the two ionizable residues, GluL212 and AspL213, in the QB-binding site of Rhodobacter sphaeroides reaction center. FEBS Lett. 1991 May 20;283(1):140–144. doi: 10.1016/0014-5793(91)80572-k. [DOI] [PubMed] [Google Scholar]
- Vermeglio A. Secondary electron transfer in reaction centers of Rhodopseudomonas sphaeroides. Out-of-phase periodicity of two for the formation of ubisemiquinone and fully reduced ubiquinone. Biochim Biophys Acta. 1977 Mar 11;459(3):516–524. doi: 10.1016/0005-2728(77)90050-0. [DOI] [PubMed] [Google Scholar]
- Wraight C. A. Electron acceptors of photosynthetic bacterial reaction centers. Direct observation of oscillatory behaviour suggesting two closely equivalent ubiquinones. Biochim Biophys Acta. 1977 Mar 11;459(3):525–531. doi: 10.1016/0005-2728(77)90051-2. [DOI] [PubMed] [Google Scholar]



