Abstract
Although trastuzumab has succeeded in breast cancer treatment, acquired resistance is one of the prime obstacles for breast cancer therapies. There is an urgent need to develop novel HER2 antibodies against trastuzumab resistance. Here, we first rational designed avidity-imporved trastuzumab and pertuzumab variants, and explored the correlation between the binding avidity improvement and their antitumor activities. After characterization of a pertuzumab variant L56TY with potent antitumor activities, a bispecific immunoglobulin G-like CrossMab (Tras-Permut CrossMab) was generated from trastuzumab and binding avidity-improved pertuzumab variant L56TY. Although, the antitumor efficacy of trastuzumab was not enhanced by improving its binding avidity, binding avidity improvement could significantly increase the anti-proliferative and antibody-dependent cellular cytotoxicity (ADCC) activities of pertuzumab. Further studies showed that Tras-Permut CrossMab exhibited exceptional high efficiency to inhibit the progression of trastuzumab-resistant breast cancer. Notably, we found that calreticulin (CRT) exposure induced by Tras-Permut CrossMab was essential for induction of tumor-specific T cell immunity against tumor recurrence. These data indicated that simultaneous blockade of HER2 protein by Tras-Permut CrossMab could trigger CRT exposure and subsequently induce potent tumor-specific T cell immunity, suggesting it could be a promising therapeutic strategy against trastuzumab resistance.
Keywords: calreticulin exposure, CrossMab, HER2-overexpressing breast cancer, pertuzumab, trastuzumab, T cell immunity
Abbreviations: ADCC; antibody-dependent cellular cytotoxicity; CDR, complementarity determining region; CH1, constant heavy chain 1; CL, constant light chain; CRT, calreticulin; FCM, flow cytometry; HER, human epidermal growth factor receptor; HER2-ECD, extracellular domain of HER2; LDH, lactate dehydrogenase; mAb, monoclonal antibody; PBMCs, peripheral blood mononuclear cells; PI3K, phosphatidylinositol 3-kinase; SEC, size-exclusion chromatography
Introduction
Approximately 20%–25% of invasive breast cancers have overexpression of the human epidermal growth factor receptor (HER) 2 tyrosine kinase receptor.1,2 Vast number of studies have demonstrated that elevation of HER2 expression levels are associated with reduced disease-free and overall survival in metastatic breast cancer,2,3 and therapeutic strategies are being developed to target this oncoprotein.4,5 Trastuzumab, a recombinant humanized monoclonal antibody (mAb) directed against an extracellular region of HER2,6 was the first HER2-targeted therapy approved for the treatment of HER2-overexpressing breast cancer. It has revolutionized the approach to treat patients with HER2-positive breast cancer and the prognosis of the disease.7,8 Nevertheless, not all patients benefit from trastuzumab. Around 15% of women relapse after trastuzumab-based therapy, indicating the presence of trastuzumab resistance.9.
Moreover, despite widespread use of trastuzumab in breast cancer treatment, the efficacy remains variable and often modest, which does not benefit from the latest technologies available for engineering more potent reagents.4,5 The pursuit of improved reagents to replace trastuzumab is intense, with several candidates currently under clinical evaluation.5 Pertuzumab is another ErbB2-specific humanized antibody that binds to a distinct epitope from trastuzumab.10,11 Its mechanism of action is complementary to trastuzumab, inhibiting ligand-dependent HER2-HER3 dimerization and reducing signaling via intracellular pathways such as phosphatidylinositol 3-kinase (PI3K/Akt). It has shown antitumor activities in both the metastatic and the neoadjuvant settings and is now being tested as adjuvant therapy.12,13 The combination therapy of trastuzumab and pertuzumab that has complementary mechanisms of action synergistically inhibits the in vitro and in vivo growth of ErbB2-overexpressing breast cancer cell lines.14–16 Notably, the addition of pertuzumab after progression to ongoing trastuzumab in xenografts synergistically increases tumor inhibition compared with trastuzumab alone.17 In line with these observations, a phase II trial of pertuzumab and trastuzumab combination therapy in patients with HER2-overexpressing breast cancer indicated that the combination of pertuzumab and trastuzumab was active and well tolerated in patients with HER2-positive breast cancer who had experienced progression during prior trastuzumab therapy.18 Very recently, the combination of pertuzumab, trastuzumab, and docetaxel, as compared with placebo, trastuzumab and docetaxel, when used as first-line treatment for HER2-positive breast cancer, significantly prolonged progression-free survival in clinical trials.13 All of these experimental and clinical data revealed that HER2 still can be considered as a valid therapeutic target even after breast cancer have progressed on multiple HER2-directed therapies and that simultaneous blockade of HER2 protein by trastuzumab and pertuzumab may overcome trastuzumab resistance. Additionally, complex diseases are often multifactorial in nature, and involve redundant or synergistic action of disease mediators, including crosstalk between their signaling networks.19,20 Thus, design of novel antibodies with the ability to bind more than one effector molecule or binding site may have the potential to provide better clinical efficacy and/ or reach a broader patient population than inhibition of a single target site by mAbs.
To our knowledge, the classical IgG architecture as it was selected during evolution has many advantages for the therapeutic application of bispecific antibodies.21,22 The Fc part is identical to that of a conventional IgG antibody, resulting in IgG-like pharmacokinetic properties and retained effector functions such as the mediation of ADCC through FcγRIIIa binding. IgG-like size and molecular weight are expected to result in IgG-like diffusion, tumor penetration, and accumulation in comparison with bispecific tetravalent antibodies of higher molecular weight. Considering these benefits, we first converted the HER2 antibody trastuzumab and pertuzumab into an IgG-like bispecific antibody (Tras-Per CrossMab) by using CrossMab technology23 and evaluated its antitumor activities. Then, we rational designed trastuzumab and pertuzumab variants with different binding avidities by using the computational method we have previously developed24 and investigated the correlation between the binding avidity of anti-HER2 antibodies and their antitumor activities. Based on these avidity-improved HER2 antibodies, Tras-Permut CrossMab was characterized with potent antitumor protections against breast cancer. Further studies showed that CRT exposure triggered by Tras-Permut CrossMab was important for induction of tumor-specific T cell immunity against tumor recurrence, suggesting that it would be a promising therapeutic agent for combating trastuzumab resistance in breast cancer.
Results
Design and characterization of Tras-Per CrossMab
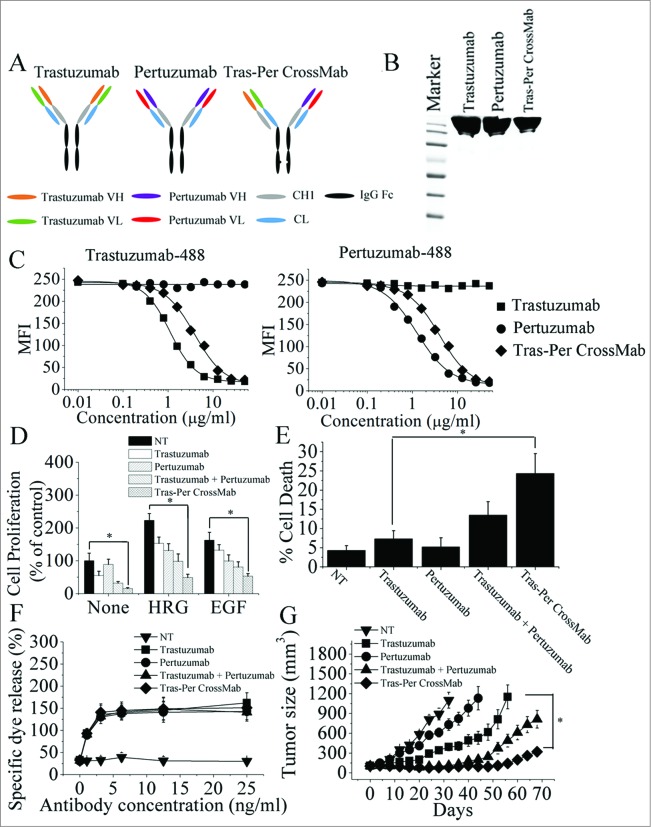
Based on CrossMab technology recently reported,23,25 we designed an IgG-like bispecific CrossMab (Tras-Per CrossMab) that deviates only minimally from the naturally occurring HER2 antibody trastuzumab and pertuzumab. As shown in Fig. 1A, the constant heavy chain 1 (CH1) of pertuzumab was replaced with the constant light chain (CL) of antibody, generating a polypeptide chain made of pertuzumab HV-CL-Hinge-CH2 and CH3. To generate Tras-Per CrossMab, exchange of CH1 and CL domains of pertuzumab could be essential for correct association of the light chain and the cognate heavy chain of the half IgG of pertuzumab in Tras-Per CrossMab. Hetero-dimerization of the heavy chain of trastuzumab and pertuzumab was achieved by using the KiH method.26,27 The resulting highly purified Tras-Per CrossMab was assessed on SDS/PAGE (Fig. 1B). Then, competitive binding assays were conducted to examine the relative binding affinity of Tras-Per CrossMab for the trastuzumab and pertuzumab epitope on HER2 protein. The results showed that Tras-Per CrossMab retained the full binding activities of both parental antibodies (Fig. 1C). The relative binding affinity of CrossMab for the trastuzumab or pertuzumab binding epitope was similar to that of trastuzumab or pertuzumab, respectively. Furthermore, the avidity constant (Kd) of CrossMab for the extracellular domain of HER2 (HER2-ECD) was determined by an ELISA (Table 1). The data showed that the binding avidity of CrossMab was much higher than that of pertuzumab.
Figure 1.
Characterization of Tras-Per CrossMab. (A), schematic diagram of the Fab domain exchange resulting in the generation of Tras-Per bispecific antibody when combined with the KiH technology. (B), sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS/PAGE) analysis of CD20-Flex BiFP. Samples of trastuzumab, pertuzumab, and Tras-Per CrossMab were assessed by SDS/PAGE analysis under non-reducing conditions. (C), competitive binding assay. Trastuzumab, pertuzumab, and Tras-Per CrossMab were evaluated for their ability to compete with Alexa Fluor 488-labeled trastuzumab or Alexa Fluor 488-labeled pertuzumab for binding to BT-474 cells. (D), MTS assay examining the proliferation effects of 100 nmol/L of control IgG, trastuzumab, pertuzumab, trastuzumab combined with pertuzumab, or Tras-Per CrossMab on breast cancer cell BT-474 or in the absence or presence of ErbB ligand (EGF or HRG). Results are shown as percentage of control cell proliferation. Error bars, SD. *p < 0.05. (E), in the absence of ligands, cell death induced by HER2 antibodies (10 μg/mL) were assessed by staining with SYTOX® Red and FCM. *p < 0.05. (F), in vitro ADCC analysis of trastuzumab, pertuzumab, trastuzumab in combination with pertuzumab, or Tras-Per CrossMab. PBMCs (effector cells) were added with BT-474 cells (target cells) into 96-well plates containing anti-HER2 antibodies at different concentrations. All experiments were performed in triplicate. (G), mice with BT-474 xenograft tumors were treated for the duration of the study with control IgG, pertuzumab, trastuzumab, trastuzumab + pertuzumab or Tras-Per CrossMab (2 mg/kg). Points, mean tumor volume (mm3) (n = 10 mice/group); bars, SD. *p < 0.05, Mann–Whitney test.
Table 1.
Binding avidities of CrossMab for HER2-ECD
| Antibodies | Kd (nM) | SD |
|---|---|---|
| Tras-Per CrossMab | 2.57 | 0.68 |
| Tras-Permut CrossMab | 0.53 | 0.08 |
Experimental error is the SD from three independent trials.
Potent antitumor activities of Tras-Per CrossMab against breast cancer
We first evaluated the ability of trastuzumab, pertuzumab, trastuzumab plus pertuzumab, and Tras-Per CrossMab to inhibit the in vitro proliferation of breast cancer cells. As shown in Fig. 1D, the trastuzumab was much more effective than pertuzumab in suppressing breast cancer cell proliferation in the absence of ErbB ligand (Fig. 1D). In contrast, after HRG- and EGF-stimulation, pertuzumab exhibited a greater anti-proliferative activity than trastuzumab in breast cancer (Fig. 1D). Interestingly, Tras-Per CrossMab exhibited greater anti-proliferative activity than trastuzumab or pertuzumab in the absence or presence of ErbB ligand (Fig. 1D). Then, the cell death induced by Tras-Per CrossMab was studied. In the absence of ligands, cells were treated with anti-HER2 antibodies or a control IgG for 5 d and were stained with SYTOX® Red to further assess cell death. After treatment with Tras-Per CrossMab, the percentages of cells staining positive for SYTOX® Red increased more than 3-fold compared with that of trastuzumab (Fig. 1E). Moreover, in the presence or absence of HER2 ligands, the similar results can be observed, suggesting that HER2 ligands could not play an important role in induction of cell death by these HER2 antibodies (Data not shown). To evaluate the ADCC activities of Tras-Per CrossMab, a standard lactate dehydrogenase (LDH) assay was performed. Purified human peripheral blood mononuclear cells (PBMCs) from healthy donors were used as effector cells and breast cancer cells were used as target. Assays were conducted at effector/target (E/T) ratios of 25:1 using antibody concentrations ranging from 1 to 25 ng/mL (Fig. 1F). To facilitate comparative analysis of the data, we plotted the concentrations of antibody vs. the present of specific dye release at E/T ratios of 25:1. Our data showed a clear antibody concentration-dependent susceptibility to ADCC in trastuzumab, pertuzumab or Tras-Per CrossMab treatment group. Furthermore, Tras-Per CrossMab exhibited similar high level of ADCC activities compared with trastuzumab, pertuzumab, and trastuzumab plus pertuzumab. These data suggested that Tras-Per CrossMab with the advantages of both trastuzumab and pertuzumab has the ability to initiate potent cell death, ADCC and anti-proliferation activities against breast cancer, showing superior antitumor efficacy than the parental intact IgG architecture therapeutic antibody trastuzumab or pertuzumab.
The therapeutic efficacy of Tras-Per CrossMab was determined in nude mice bearing established BT-474 xenograft tumors. In agreement with previous report, we found that trastuzumab suppressed tumor growth better than pertuzumab in the BT-474 xenograft model (Fig. 1G). Although treatment with trastuzumab or pertuzumab could not inhibit the BT-474 tumor growth, both of these two anti-HER2 mAbs significantly delayed the BT-474 tumor progression (Fig. 1G). Further study showed that trastuzumab plus pertuzumab was more efficient in inhibition of the BT-474 tumors progression than either of these mAbs alone. In accordance with these observations, Tras-Per CrossMab had the capacity to inhibit tumor growth in vivo much more effectively than each of these mAbs alone. Moreover, Tras-Per CrossMab exhibited more prominent effects in inhibition of in vivo tumor growth than combination treatment of trastuzumab and pertuzumab, suggesting that the IgG-like bispecific antibody Tras-Per CrossMab converted from trastuzumab and pertuzumab may have the potential to be developed as a therapeutic antibody with potent antitumor activities against breast cancer.
The correlation between binding avidity improvement of HER2 antibodies and their antitumor activities
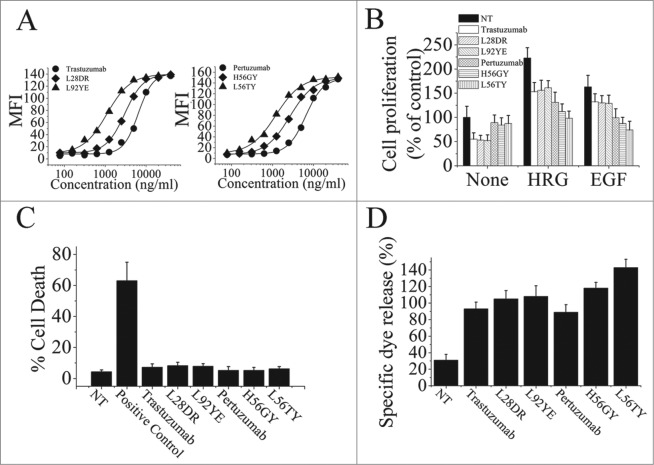
To further investigate the relationship between the binding avidity of HER2 antibodies and their antitumor activities, we first used the computational method previously described24,28 to design the trastuzumab and pertuzumab variants with different binding avidities. The binding of these trastuzumab or pertuzumab variants to the HER2-overexpressing human breast cancer cell line SK-BR-3 were determined by flow cytometry (Fig. 2A). Next, the results of binding avidity analysis by ELISA were shown in Table 2, which was consistent with the data obtained by flow cytometry assays. The anti-proliferation effects of these avidity-improved variants were subsequently analyzed (Fig. 2B). Our data clearly showed that, in the absence of ErbB ligand, binding avidity improvement of both trastuzumab and pertuzumab could not enhance their anti-proliferative activities. However, the increase in binding avidity of pertuzumab significantly improved its anti-proliferative activities in the presence of ErbB ligand. Furthermore, the important role of pertuzumab binding avidity in enhancement of its anti-proliferation effects was subsequently confirmed by using these pertuzumab variants with different avidity-improved levels. We found that, although the anti-proliferative activities of trastuzumab exhibited a avidity-independent pattern in the absence or presence of ErbB ligand, the increase of pertuzumab anti-proliferative activities was in an avidity-dependent manner (Fig. 2B). Then, the cell death induced by these avidity-improved variants in the absence of ligands was evaluated. As shown in Fig. 2C, the increase of cell death induced by trastuzumab variants or pertuzumab variants was not observed (Fig. 2C). Moreover, our data showed that improvement of pertuzumab binding avidity could significantly increase its ADCC activities, although the enhancement of ADCC was not observed in these avidity-improved trastuzumab variants (Fig. 2D).
Figure 2.
The correlation of trastuzumab and pertuzumab binding avidity-improvement and their antitumor activities. (A), antigen binding activities of trastuzumab and pertuzumab variants on SK-BR-3 cells were determined by FCM. (B), MTS assay examining the proliferation effects of 100 nmol/L of trastuzumab variants or pertuzumab variants in breast cancer cell BT-474 with the absence or presence of ErbB ligand (EGF or HRG). Results are shown as percentage of control cell proliferation. Error bars, SD. (C), in the absence of ligands, cell death induced by trastuzumab variants or pertuzumab variants were assessed by staining with SYTOX® Red and FCM. (D), In vitro ADCC analysis of trastuzumab and pertuzumab variants with binding avidity improvement. In the absence of ligands, PBMCs were added with BT-474 cells into 96-well plates containing trastuzumab or pertuzumab variants. Data are mean ± SD of at least three experiments.
Table 2.
Binding avidities of Her2 antibody variants for HER2-ECD
| Antibody | KdWT/Kdmutant | Antibody | KdWT/Kdmutant |
|---|---|---|---|
| Trastuzumab KdWT = 0.16 ± 0.02 nM | Pertuzumab KdWT = 8.62 ± 0.43 nM | ||
| L28DR | 1.86 ± 0.09 | H56GY | 3.21 ± 0.12 |
| L92YE | 9.75 ± 1.64 | L56TY | 8.43 ± 1.12 |
Experimental error is the SD from three independent trials.
Optimization of Tras-Per CrossMab by introducing a single point mutation L56TY
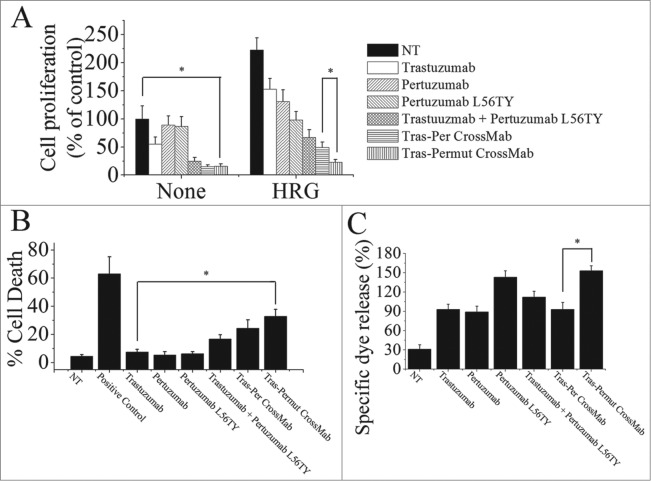
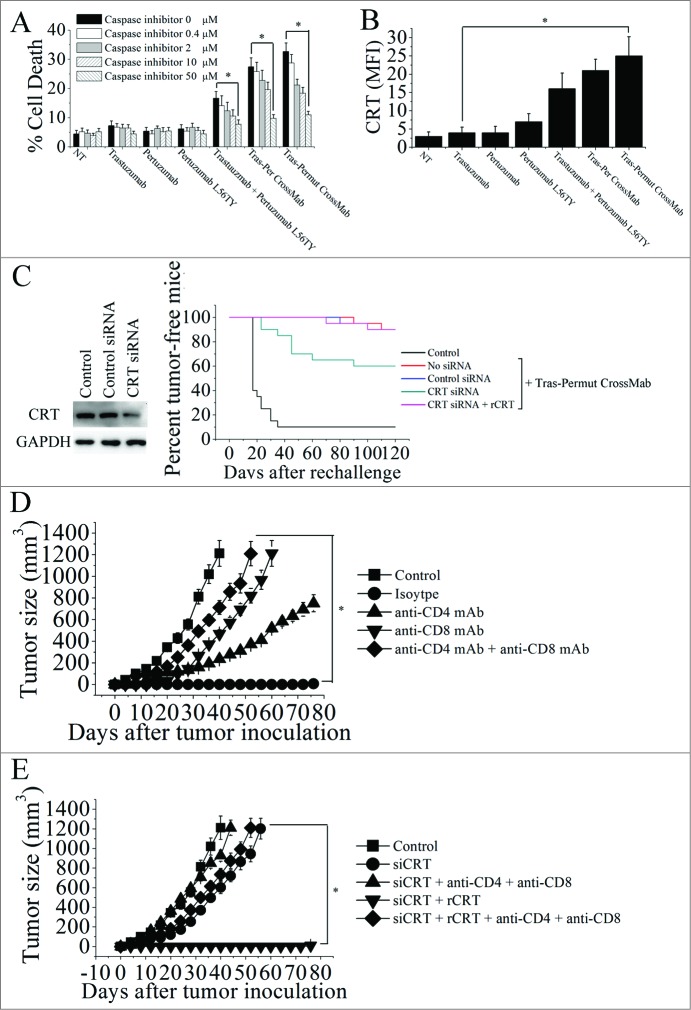
To further investigate whether introduction of a single point mutation L56TY in the complementarity determining region (CDR) of pertuzumab light chain in Tras-Per CrossMab could markedly improve the antitumor activities as the parental antibody pertuzumab, we first evaluated the anti-proliferation effects of these antibodies. As shown in Fig. 3A, introducing the single point mutation L56TY (Tras-Permut CrossMab) could not significantly increase the anti-proliferative activities as compared with Tras-Per CrossMab in the absence of ErbB ligand. In line with our expectation, after stimulation of ErbB ligand, Tras-Permut CrossMab showed more potent proliferation inhibition against breast cancer than Tras-Per CrossMab (Fig. 3A). Compared with Tras-Per CrossMab, the cell death induced by Tras-Permut CrossMab was slightly increased (Fig. 3B). Further studies showed that cell death induced by Tras-Permut CrossMab was caspase-dependent cell death, which could be inhibited by the caspase inhibitor in a dose-dependent manner (Fig. S2). Then, the ADCC activity of Tras-Permut CrossMab was evaluated. In line with the experiments we have mentioned above, introduction of the single point mutation L56TY in Tras-Permut CrossMab could significantly improve the ADCC activities as the same increased level of pertuzumab L56TY, which showed more significant ADCC activities than that of Tras-Per CrossMab (Fig. 3C). To further evaluate the inhibition of HER2 signaling by Tras-Permut CrossMab, we determined the disruption of HER2/HER3 heterodimerization by Tras-Permut CrossMab and subsequently examined the Src phosphorylation on Y416 and Akt phosphorylation on S473. Our data indicated that Tras-Permut CrossMab was more effective than trastuzumab or pertuzumab in disrupting ligand-dependent HER2/HER3 association, which consequently inhibited downstream signaling cascades including Src and PI3K/Akt with more efficiency (Fig. S1).
Figure 3.
The in vitro antitumor effects of Tras-Permut CrossMab. (A), the anti-proliferation effect of Tras-Permut CrossMab (100 nmol/L) was evaluated by MTS assay. Mean ± SD (n = 3). *p < 0.05. (B), induction of cell death by Tras-Permut CrossMab (10 μg/mL) in the absence of ligands was assessed by staining with SYTOX® Red and FCM. The graphs are representative of at least three experiments, each of which showed similar results. *p < 0.05. (C), ADCC activity against BT-474 cells using human PBMCs as effector cells at E/T ratio of 25:1. The ADCC activity of Tras-Permut CrossMab was measured using a standard LDH assay as described in “ADCC assays.” Data are mean ± SD (n = 3). *p < 0.05.
The in vivo antitumor activities of Tras-Permut CrossMab
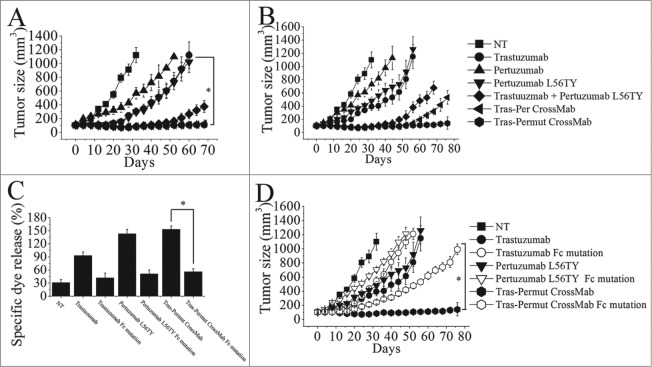
The therapeutic efficacy of Tras-Permut CrossMab was further evaluated in nude mice bearing established BT-474 xenograft tumors. In line with our in vitro experiments, pertuzumab variant with binding avidity improvement (pertuzumab L56TY) suppressed tumor growth better than the parental antibody pertuzumab in the BT-474 xenograft model (Fig. 4A). Although, combination of trastuzumab with pertuzumab L56TY exhibited more prominent effects in inhibition of the BT-474 tumor growth than either of these mAbs alone, both of these two CrossMab showed more significant inhibition in the in vivo tumor progression than the combination therapy (Fig. 4A). To further evaluate the in vivo antitumor activities of Tras-Permut CrossMab, we decreased the dose of HER2 antibodies in nude mice bearing established BT-474 xenograft tumors. Our data clearly showed that Tras-Permut CrossMab treatment resulted in complete regression of BT-474 tumors, whereas no tumor eradication was observed in tumor-bearing mice treated with trastuzumab, pertuzumab or trastuzumab plus pertuzumab L56TY. Remarkably, although both of these two CrossMab exhibited similar in vivo antitumor activities in the high dosage, Tras-Permut CrossMab could still provide the protection to eradicate breast cancer in the low dose (Fig. 4B).
Figure 4.
The therapeutic effect of of Tras-Permut CrossMab in breast cancer (BT-474) xenograft tumor models. Mice with BT-474 xenograft tumors were treated for the duration of the study with high dosage (10 mg/kg), (A) or low dosage (2 mg/kg), (B) of control IgG, trastuzumab, pertuzumab, pertuzumab L56TY, trastuzumab + pertuzumab L56TY, Tras-Per CrossMab or Tras-Permut CrossMab. Points, mean tumor volume (mm3) (n = 10 mice/group); bars, SD. *p < 0.05, Mann–Whitney test. (C), the ADCC activity of Tras-Permut CrossMab was measured using a standard LDH assay as described in “ADCC assays.” Data are mean ± SD (n = 3). *p < 0.05. (D), the role of Fc part of HER2 antibodies in their therapeutic effects. Tumor-bearing mice were treated for the duration of the study with low dosage (2 mg/kg) of trastuzumab, trastuzumab Fc mutation, pertuzumab L56TY, pertuzumab L56TY Fc mutation, Tras-Permut CrossMab and Tras-Permut CrossMab Fc mutation. Points, mean tumor volume (mm3) (n = 10 mice/group). Data are shown as means ± SEM. *p < 0.05, Mann–Whitney test.
Recent studies indicated that induction of FcR-mediated cytotoxicity played an important role in mediating tumor regression in trastuzumab therapy.29,30 To further determine the role of FcR-mediated cytotoxicity in triggering the in vivo antitumor protections of the CrossMab, the Fc part of antibody was introduced the following mutations (E236P/L237V/L238A/ΔG239 + A329G/A332S/P333S) as previously described to decrease the ADCC activity of antibody.31,32 In line with our expectation, introducing mutations in the Fc part of trastuzumab could decrease its ADCC activities and the in vivo antitumor activities when compared with the parental antibody trastuzumab (Fig. 4C and D). Decrease of ADCC activities in pertuzumab L56TY by introducing Fc part mutations could also reduce its antitumor protections. Moreover, introduction of these mutations in the Fc part of Tras-Permut CrossMab could markedly decrease the inhibition of the BT-474 tumor growth in vivo, suggesting that FcR-mediated cytotoxicity plays an essential role in anti-HER2 antibody therapy.
The important role of calreticulin exposure induced by Tras-Permut CrossMab in induction of antitumor T cell immunity against breast cancer
Induction of immunogenic cancer cell death should be one of the aims of antitumor therapy, because it would allow the immune system to contribute through a “bystander effect” to the eradication of therapy-resistant cancer cells and cancer stem cells.33–36 Previous studies have revealed that CRT exposure during cell death induced by anthracyclines could trigger the immune response, and the surface exposure of CRT can be considered as an event that occurs only in immunogenic cancer cell death.33 Thus, we first evaluated the cell death induced by these HER2 antibodies (Fig. 5A). Although trastuzumab, pertuzumab, and pertuzumab L56TY could not induce potent cell death as described above, the increase of cell death was observed after treatment with trastuzmab plus pertuzumab L56TY. Notably, both Tras-Per CrossMab and Tras-Permut CrossMab could induce potent cell death (Fig. 5A). To further investigate whether the caspase was involved in the CrossMab-induced cell death, the cell-permeable caspase inhibitor ZVAD-FMK was used, and our experimental results revealed that ZVAD-FMK over a range of concentrations from 50 to 0.4 μM was able to prevent the CrossMab-induced cell death (Fig. 5A). These results indicated that the Tras-Per CrossMab could efficiently activate caspase-dependent pathways in breast cancer. Then, the CRT exposure after treatment with these HER2 antibodies was detected by flow cytometry. As shown in Fig. 5B, CRT exposure can be detected after treatment with both Tras-Per CrossMab and Tras-Permut CrossMab. To further explore the role of CRT exposure in the therapeutic efficacy of Tras-Permut CrossMab, we used a specific siRNA to downregulate CRT expression of 4T1-Her2 cells. As shown in Fig. 5C, CRT siRNA could significantly reduce the expression of CRT and markedly decrease the capacity of Tras-Permut CrossMab to protect these mice from tumor recurrence. More importantly, addition of recombinant CRT protein (rCRT), which has been reported to bind to the surface of the tumor cells,33 could restore the tumor suppression ability of Tras-Permut CrossMab.
Figure 5.
The calreticulin exposure induced by Tras-Permut CrossMab is essential for induction of antitumor T cell immunity against breast cancer. (A), the inhibition of cell death by caspase inhibitor Z-VAD-fmk in the range from 50 μM to 0.4 μM were evaluated after 5 d (in the absence of ligands). Mean ± SD (n = 3). *p < 0.05. (B), the surface exposure of CRT was determined by immunofluorescence cytometry 48 h after treatment with HER2 antibodies. The antibody-untreated group stained with an anti-CRT antibody was used as the negative controls. *p < 0.05. (C), in vivo antitumor protection depends on CRT. 4T1-HER2 cells were transfected with the indicated siRNAs, then treated with rCRT and/or Tras-Permut CrossMab. The antitumor response was measured by challenging BALB/c mice simultaneously with Tras-Permut CrossMab-treated tumor cells in one flank and untreated, live tumor cells in the opposite flank (n = 20). The siRNA-transfected cells were measured by immunoblotting. (D), BALB/c mice were inoculated with Matrigel-mixed 4T1-HER2 cells and then treated with Tras-Permut CrossMab or control Ig (2 mg/kg). After the first inoculation, the CrossMab-treated mice were secondarily inoculated with Matrigel-mixed 4T1-HER2 cells in the opposite flank (n = 25). Some mice were treated with anti-CD4 mAb, anti-CD8 mAb, anti-CD4 and anti-CD8 mAbs or isotype Ig. Naive mice were inoculated with Matrigel-loaded 4T1-HER2 cells as the control. *p < 0.05, Mann–Whitney test. (E), after injection of siCRT-treated 4T1-HER2 cells (containing 0.1mL Matrigel) into the inguinal mammary fat pads of female BALB/c mice, the mice were treated with Tras-Permut CrossMab (2 mg/kg). After the first challenge, the CrossMab-treated mice were subsequently inoculated with Matrigel-mixed 4T1-HER2 cells in the opposite flank (n = 25). Some mice were treated with rCRT and/or anti-CD4 and anti-CD8 mAbs (n = 25). Naive mice were inoculated with Matrigel-loaded 4T1-HER2 cells as the control. The primary tumor material is examined through measurement of tumor size. *p < 0.05, Mann–Whitney test.
Numbers of studies have identified that CRT can be regarded as a key feature determining anticancer immune responses and consequently inducing tumor-specific T cell immunity against tumor recurrence.33,37 To determine whether Tras-Permut CrossMab treatment enabled the induction of tumor-specific T cell immunity against tumor recurrence, CD8+ and CD4+ specific antibodies were employed. After complete eradication of 4T1-Her2 cells in BALB/c mice by Tras-Permut CrossMab, the tumor-free mice was re-challenged with 4T1-Her2 cells on the opposite flank. In the CrossMab treamtent group, the mice rejected the re-challenged 4T1-Her2 cells (Fig. 5D). Depletion of either CD8+ T cells or CD4+ T cells abrogated the secondary rejection, although some suppression of tumor growth was still observed in the CD4+ T cell-depleted mice (Fig. 5D). To further explore the correlation between CRT exposure and tumor-specific T cell immunity, CRT-decreased 4T1-Her2 cells were used (Fig. 5E). In agreement with the results mentioned above, decrease of CRT in 4T1-Her2 cells could significantly abrogate the inhibition of tumor progression in the opposite flank, although addition of rCRT could recover the protection against tumor recurrence (Fig. 5E). Furthermore, depletion of both CD8+ T cells and CD4+ T cells could potently compromise the restore of antitumor protection induced by Tras-Permut CrossMab. These results indicated that CRT exposure during cell death induced by Tras-Permut CrossMab could play an essential role in the induction of tumor-specific T cell immunity against tumor recurrence.
The potent antitumor activities of Tras-Permut CrossMab against trastuzumab-resistance
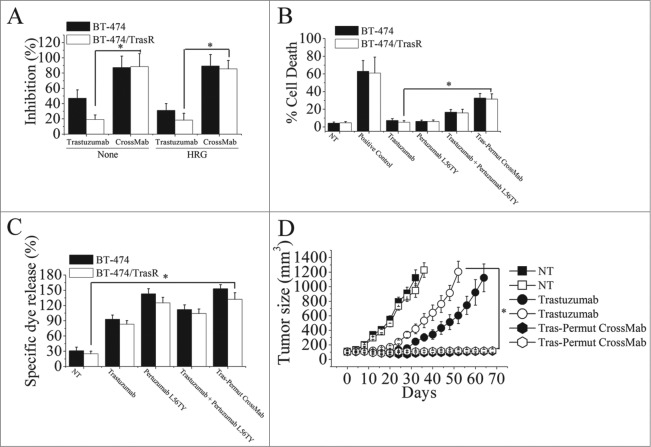
Trastuzumab-resistant breast cancer cell line BT-474/TraR was developed as previously described.14 We first evaluated the anti-proliferation effects of Tras-Permut CrossMab against trastuzumab-resistant breast cancer. Our results showed that, although trastuzumab-resistant breast cancer could significantly resist the anti-proliferative activities of trastuzumab, pertuzumab still exhibited potent anti-proliferative activities against trastuzumab-resistant breast cancer in the presence of ErbB ligand (Data not shown). As shown in Fig. 6A, we used percentage of inhibition to describe the anti-proliferation effects of Tras-Permut CrossMab in both trastuzumab-sensitive and -resistant breast cancer. These are no differences between the percentage of inhibition and control cell proliferation in the presence or absence of ligands. Notably, Tras-Permut CrossMab could more effectively inhibit the proliferation of trastuzumab-resistant breast cancer compared with trastuzumab in the absence or presence of ErbB ligand (Fig. 6A). Further investigation indicated that, although slight cell death can be observed after treatment with trastuzumab in both BT-474 and BT-474/TraR cells, Tras-Permut CrossMab could induce high level of cell death in trastuzumab-resistant tumor cells, which was comparable to that in trastuzumab-sensitive breast cancer (Fig. 6B). We also used the Annexin V/PI staining assay to evaluate the induction of cell death against Trastuzumab-resistant cells. The similar results can be observed between these two different methods in the presence or absence of ligands (Data not shown). The ADCC activities of HER2 antibodies in trastuzumab-resistant breast cancer were subsequently evaluated. As shown in Fig. 6C, although anti-proliferation effects of trastuzumab were not observed in trastuzumab-resistant breast cancer, trastuzumab could still induce potent ADCC activities in both trastuzumab-sensitive and -resistant breast cancer. In accordance with our expectation, the marked ADCC activities of Tras-Permut CrossMab can be found in both BT-474 and BT-474/TrasR cells (Fig. 6C).
Figure 6.
Combating trastuzumab-resistance by Tras-Permut CrossMab. (A), MTS assay examining the proliferation effects of Tras-Permut CrossMab (100 nmol/L) in both trastuzumab-sensitive and -resistant breast cancer cells (BT-474 and BT-474/TrasR) in the absence or presence of ErbB ligand (HRG). Results are shown as percentage of cell proliferation inhibition. Error bars, SD. Tras-Permut CrossMab was abbreviated as CrossMab. *p < 0.05. (B), Induction of cell death against trastuzumab-resistant breast cancer was assessed by staining with SYTOX® Red and FCM (In the absence of HER2 ligands, the concentration of antibodies is 10 μg/mL). Data are mean ± SD (n = 3). *p < 0.05. (C), ADCC activities of trastuzumab, pertuzumab L56TY, trastuzumab in combination with pertuzumab L56TY, or Tras-Permut CrossMab against both trastuzumab-sensitive and -resistant breast cancer. Mean ± SD values from four separate experiments. *p < 0.05. (D), tumor volume of trastuzumab-sensitive and -resistant BT-474 breast tumor xenografts after treatment with control IgG (10 mg/kg), trastuzumab (10 mg/kg), or Tras-Per CrossMab (10 mg/kg). Data are shown as means ± SEM. *p < 0.05, Mann–Whitney test.
Then, nude mice bearing established BT-474/TraR xenograft tumors were used to investigate the in vivo antitumor activities of Tras-Permut CrossMab against trastuzumab-resistant breast cancer. As shown in Fig. 6D, trastuzumab treatment could delay the trastuzumab-sensitive tumor progression as compared with the control, but tumor growth was more rapid in the group of mice inoculated with BT-474/TraR cells than that of inoculated with BT-474 cells. In both trastuzumab-sensitive and -resistant xenograft tumor models, Tras-Permut CrossMab exhibited remarkable antitumor activities against breast cancer (p < 0.05 for compared with the control IgG treatment group), suggesting that it could be a promising agent for next generation of target therapy against breast cancer.
Discussions
Although anti-HER2 mAb trastuzumab has revolutionized the treatment of breast cancer,6 the acquired resistance is one of the prime obstacles for breast cancer treatment. As we known, cancer is usually multifactorial in nature, involving a redundancy of disease-mediating ligands and receptors, as well as crosstalk between signal cascades.19,20 A targeted therapeutic agent inhibiting one key pathway in a tumor may not completely shut off a hallmark capability, allowing some cancer cells to survive with residual function until they or their progeny eventually adapt to the selective pressure imposed by the therapy being applied. Therefore, blockade of multiple target function domains or proteins may result in improved therapeutic efficacy, which can be achieved by use of the dual targeting strategies applying bispecific antibodies that have emerged as an alternative to combination therapy. Although, previous preclinical studies showed that inhibition of HER1 enhanced the response to trastuzumab in HER1-HER2 co-expressing cells,38,39 clinical activity of selective HER1 inhibitors in patients with breast cancer has been disappointing, either as single agents,40,41 or in combination with chemotherapy (in patients unselected for HER2 status),42,43 or in combination with trastuzumab in patients with HER2-positive breast cancer. Thus, attention has shifted to other members of the HER family, especially HER3. Although HER3 has only weak intrinsic tyrosine kinase activity,44 HER2-HER3 heterodimerization is the most potent mitogenic signaling pair in the HER family,45 and HER3 is now recognized as having a critical role as a co-receptor for amplified HER2.46 Although HER2 is the target protein of trastuzumab and pertuzumab, it has been demonstrated that pertuzumab with different binding site on HER2 protein could efficiently inhibit ligand-induced HER2/HER3 dimerization, whereas trastuzumab has only a minor effect in the presence of a ligand.47 Furthermore, growing clinical evidence revealed that combination of trastuzumab and pertuzumab could significantly increase progression-free survival.15,16 These findings indicated that block the domain IV and domain II of HER2, which is respectively the binding epitope of trastuzumab and pertuzumab, could be a promising strategy for next generation of target therapy against breast cancer. In the present study, we first converted the anti-HER2 mAb trastuzumab and pertuzumab into an IgG-like bispecific antibody (Tras-Per CrossMab) by using CrossMab technology.23 It has been demonstrated that Tras-Per CrossMab with the IgG architecture could bind the target protein with intact binding affinity compared with the parental antibodies (Fig. 1). Further studies showed that Tras-Permut CrossMab with the advantages of both trastuzumab and pertuzumab could not only inhibit the proliferation of breast cancer in the absence of ligands as trastuzumab, but also disrupt HER2-HER3 heterodimerization and consequently suppress the proliferation of tumor cells in the presence of ligands (Figs. 1 and S1). To further evaluate the in vivo antitumor effects of Tras-Permut CrossMab, the tumor-bearing mice were treated with high dose (10 mg/kg) or low dose (2 mg/kg) of Tras-Permut CrossMab as described above. Our results showed that Tras-Permut CrossMab exhibited potent antitumor activities against breast cancer in both low and high dosages. Even in the high-dose group, we have not observed any adverse side effects in the in vivo experiments.
As we known, binding avidity of antibody is a very important characteristic for antibody functions, which has close correlation with detection limits, drug dosages and drug efficacy.48 Based on the computational method we have previously developed,24 we have improved the binding avidity of trastuzumab and pertuzumab and subsequently investigated the relationship between the binding avidity of these HER2 antibodies and their antitumor activities. In agreement with the previous findings,28,49 binding avidity improvement of HER2 antibodies could enhance their ADCC activities. However, our results indicated that improvement the binding avidity of trastuzumab could not significantly increase its anti-proliferation effects and induction of cell death (Fig. 2). Intriguingly, we found that enhancement of pertuzumab binding avidity could markedly increase its anti-proliferative activities in the presence of ErbB ligand, although the improvement of its anti-proliferation effects was not observed in the absence of ErbB ligands. In our opinions, increase of pertuzumab binding avidity could bind the targeted site more tightly, which endows the pertuzumab with capacity to more efficiently disrupt ligand-induced HER2/HER3 dimerization and to more significantly enhance its anti-proliferative activities. These expectations have been further validated by our co-immunoprecipitation assays. Vast number of previous studies have demonstrated that, although trastuzumab and pertuzumab bind the same target protein, the mechanisms of action of these 2 HER2 antibodies are different.29,50 In line with their findings, our results showed that binding avidity improvement of these 2 antibodies exhibited different changes in their antitumor activities, suggesting that the killing mechanisms of trastuzumab and pertuzumab are distinct, and improvement of pertuzumab binding avidity can potently enhance its antitumor activities both in vitro and in vivo.
To further evaluate the important role of immune responses induced by Tras-Permut CrossMab, CD8+ T cells and CD4+ T cells were depleted by specific antibodies. Our data clearly showed that depletion of both CD8+ T cells and CD4+ T cells markedly reduced the antitumor protection of Tras-Permut CrossMab, indicating that antitumor specific T cell immunity induced by Tras-Permut CrossMab is important for the promising antitumor efficacy of Tras-Permut CrossMab. Further investigation revealed that CRT is exposed on the surface of dying cell induced by Tras-Permut CrossMab. As we known, apoptosis is associated with a series of subtle alterations in the plasma membrane that render the dying cells palatable to phagocytic cells.51 Such “eat me” signals, including the translocation of CRT from inside the cell to the surface, could elicit the recognition and removal of apoptotic cells by professional and nonprofessional phagocytes, which might consequently trigger antitumor immune responses.33 In the present study, potent caspase-dependent cell death and CRT exposure can be observed after treatment with Tras-Permut CrossMab, indicating that Tras-Permut CrossMab treatment could trigger CRT translocation to the surface during the cell death. The most striking findings in our present study were that CRT downregulation by siRNA could markedly reduce the antitumor protection of Tras-Permut CrossMab, which can be recovered by addition of rCRT. Depletion of both CD8+ T cells and CD4+ T cells could significantly decrease the antitumor protection of Tras-Permut CrossMab restored by rCRT treatment. In accordance with our findings, Obeid and his colleagues have revealed that CRT could be a key feature determining anticancer immune responses and delineate a possible strategy for immunogenic therapy in future.33 To further validate whether Tras-Permut CrossMab could induce potent antitumor activities against trastuzumab resistance, trastuzumab resistant breast cancer cells were used. Both our in vitro and in vivo experimental data showed that Tras-Permut CrossMab with marked ADCC, cell death and anti-proliferation effects had the potential to be regard as a promising therapeutic agent to combat trastuzumab resistance.
Materials and Methods
Cell lines, antibodies, and animals
The human breast cancer cell lines BT-474 and SK-BR-3 cells and murine mammary carcinoma 4T1 cells were obtained from the American Type Culture Collection. 4T1-HER2 cells were produced by transfected 4T1 cells with human HER2 proteins. Trastuzumab-resistant cell line (BT-474/TrasR) was generated from BT-474 cells as described previously.14 All the cell lines were authenticated twice by morphologic and isoenzyme analyses during the study period. Cell lines were routinely checked for contamination by Mycoplasma using Hoechst staining and consistently found to be negative. Lenti-Concentin Virus Precipitation Solution (EMB810A-1) was purchased from ExCell Biology. Six-week-old female BALB/c mice and five-week-old female BALB/c nude mice were obtained from the Beijing Experimental Animal Center of Chinese Academy of Sciences. All animals were treated in accordance with guidelines of the Committee on Animals of the PLA general hospital. The study using human PBMCs from the donors was approved by the Institutional Review Board of the PLA general hospital.
Construction, expression, and purification of Tras-Per CrossMab
Tras-Per CrossMab was constructed by using the method as previously described.23,25 Tras-Permut CrossMab was generated by introducing the point mutation Thr56Tyr in the pertuzumab light-chain CDR of CrossMab. The CrossMabs were produced by transient expression in HEK293F suspension cells from three expression plasmids at equimolar ratios. The CrossMab was obtained in high purity via standard protein A affinity chromatography followed by size-exclusion chromatography (SEC).
Competitive binding assay
Cells at 1 × 106 cells/mL were incubated with a subsaturating concentration of the indicated Alexa Fluor 488-conjugated anti-HER2 mAbs and increasing concentrations of purified competing antibodies for 1 h at 4°C. Then, the cells were washed and analyzed by flow cytometry using a FACScan flow cytometer (Becton Dickinson). The IC50 values of competitors were calculated using a 4-variable algorithm.
Binding activity assays
The affinities of anti-HER2 antibodies for HER2-ECD were determined as described previously.24 Briefly, each mAb was incubated with increasing concentrations of HER2-ECD for an hour. The concentration of free antibody was then measured by ELISA using immobilized HER2-ECD and was used to calculate affinity (Kd).
Computational redesign of the binding avidity of trastuzumab and pertuzumab
The crystal structure of the trastuzumab Fab-HER2 complex (PDB code 1N8Z) and pertuzumab Fab-HER2 complex (PDB code 1S78) were used in our present studies. Hydrogen-atom positions were assigned using the Biopolymer module of Insight II (Accelrys). The computational mutation was carried out on trastuzumab or pertuzumab. Docking was performed using Monte Carlo Simulated Annealing52,53 for random generation of a maximum of 60 structures through the Affinity module of Insight II (CVFF force field54). The lowest energy complexes presenting lower root mean square deviation were selected for the binding-free energy calculations. Briefly, the protein–protein complexes generated were minimized using the CHARMM force field (CHARMM version 34b1 program55) and the Generalized Born with a simple Switching implicit solvent model.56 Finally, the binding-free energy was calculated using the Molecular Mechanics Poisson-Boltzmann surface area (MM/PBSA) method.57 The simulation procedure was described in detail in our previous manuscript.24,28
Cell death assay
Cells were seeded at 5 × 104 cells/well in 12-well dishes. After 5 d, cells were treated with HER2 antibodies, trastuzumab combined with pertuzumab at a fixed 1:1 ratio, Tras-Per CrossMab , or Tras-Permut CrossMab for 5 d After washing, cells were treated with SYTOX® Red (Life technologies) for 15 min and analyzed by flow cytometry (FCM). In some cases, the caspase inhibitor Z-VAD-FMK was added with a range from 50 to 0.4 μM concentration. After 5 d, cells were treated with SYTOX® Red (Life technologies) for 15 min and analyzed by FCM.
Cell proliferation assay
Cells were incubated with different concentrations of recombinant HER2 mAbs for 2 h, followed by the addition of ErbB ligands or not. Recombinant human EGF and HRG were added at a final concentration of 5 and 1 nmol/L, respectively. After an additional 4-d incubation, cell proliferation was determined by CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS assay) Kit (Promega).
ADCC assays
ADCC assays were performed as described previously.28 Briefly, the cells were incubated with antibodies for 1 h in phenol red-free Dulbecco modified Eagle medium culture medium in a 5% CO2 incubator at 37°C, followed by the addition of human PBMCs as effector cells. After an additional incubation for 4 h at 37°C, the cell lysis was determined by measuring the amount of LDH released into the culture supernatant. Maximum LDH release was determined by lysis in 0.2% Triton X-100.
siRNAs and manipulation of surface CRT
siRNA heteroduplexes specific for CRT(sense strand: 5′-rCrCrGrCUrGrGrGUrCrGrArAUrCrRrAr ATT-3′), and an unrelated control (5′-rGrCrCrGrGUrAUrGrCrCrGrGUUrArArGUTT-3′) were designed as previous described.33 4T1-HER2 cells were transfected by siRNAs at a final concentration of 100 nM using HiPerFect (Qiagen). Thirty-six hours after transfection, cells were assessed for total CRT content by immunoblotting.
Fluorescence detection of cell surface CRT
Cells were first washed with FACS buffer (1× PBS, 5% fetus bovine serum, and 0.1% sodium azide) and then incubated with rabbit anti-mouse CRT antibody (1:100) in FACS buffer at 4°C for 30 min. Cells reacted with anti-rabbit IgG (H+L) Alexa fluor 488-conjugates (1:500) in FACS buffer at 4°C for 30 min. After washing three times with FACS buffer, surface CRT was detected by FCM.
In vivo therapy study
For BT-474 or BT-474-TraR xenograft studies, female BALB/c nude mice were implanted with 0.72 mg 60-d release 17b-estradiol pellets (Innovative Research of America). After 6 d, 1 × 107 BT-474 or BT-474-TraR cells were injected into the mammary fat pad in a 1:1 PBS:Matrigel suspension (BD Matrigel; BD Biosciences). When tumor volumes reached an average of approximately 100 mm3, the mice were randomly divided into groups of 10 mice each. Treatments consisted of twice weekly intravenous injection of different anti-HER2 mAbs for 4 consecutive weeks. Control mice were given vehicle (IgG) alone. Tumors were measured with digital calipers and tumor volumes were calculated by the formula: volume 1⁄4 length (width)2/2.
Assessment of tumor-specific T-cell immunity
After being administered with Tras-Permut CrossMab, the antibody-treated tumor-free mice were rechallenged with 4T1-HER2 cells subcutaneously in the opposite flank. Some groups of these mice were intraperitoneally administered with anti-CD4 and/or anti-CD8 mAb after the rechallenge. Effective depletion of CD4+ and/or CD8+ T cells was verified by FCM.
Statistical analysis
Statistical analysis was conducted by Student unpaired t test to identify significant differences unless otherwise indicated. Differences were considered significant at p < 0.05.
Supplementary Material
Authors’ Contributions
Conception and design: F. Zhang, L. Zhao, S.C. Jiao, Y. Hu, J.L. Yang.
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): F. Zhang, J. Zhang, L.C. Zhao, M.Y. Liu, R.X. LingHu, F. Feng, X.D. Gao.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J. Zhang, L.C. Zhao, M.Y. Liu, R.X. LingHu, F. Feng, X.D. Gao.
Writing, review, and/or revision of the manuscript: F. Zhang, L. Zhao, S.C. Jiao, Y. Hu, J.L. Yang.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): F. Zhang, L. Zhao, S.C. Jiao, Y. Hu, J.L. Yang.
Study supervision: L. Zhao, Y. Hu, J.L. Yang.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81402552, 81301956), Innovation Program of Shanghai Municipal Education Commission (2015z90030001), the Natural Science Foundation of Beijing, China (7154238), Roche's Scientific Corporation Program, Project of PLA Eleventh-Five Year Research Program of China, Research Fund of Ministry of Public Health, and the Young Scholar Program of Second Military Medical University (2014QN01).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989; 244:707-12; PMID:; http://dx.doi.org/ 10.1126/science.2470152 [DOI] [PubMed] [Google Scholar]
- 2. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235:177-82; PMID:; http://dx.doi.org/ 10.1126/science.3798106 [DOI] [PubMed] [Google Scholar]
- 3. Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, Ma Y, Hung G, Robinson RA, Harris C, El-Naggar A, et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol 1997; 15:2894-904; PMID: [DOI] [PubMed] [Google Scholar]
- 4. Emde A, Köstler WJ, Yarden Y, on behalf of AROME. Therapeutic strategies and mechanisms of tumorigenesis of HER2-overexpressing breast cancer. Crit Rev Oncol Hematol 2012; 84:e49-57; PMID:; http://dx.doi.org/ 10.1016/j.critrevonc.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol 2012; 9:16-32; http://dx.doi.org/ 10.1038/nrclinonc.2011.177 [DOI] [PubMed] [Google Scholar]
- 6. Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89:4285-9; PMID:; http://dx.doi.org/ 10.1073/pnas.89.10.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marty M. Randomized phase II trial of the efficacy and safety of trastuzumab combined with docetaxel in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer administered as first-line treatment: the M77001 study group. J Clin Oncol 2005; 23:4265-74; PMID:; http://dx.doi.org/ 10.1200/JCO.2005.04.173 [DOI] [PubMed] [Google Scholar]
- 8. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001; 344:783-92; PMID:; http://dx.doi.org/ 10.1056/NEJM200103153441101 [DOI] [PubMed] [Google Scholar]
- 9. Nahta R, Esteva FJ. HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast Cancer Res 2006; 8:215; PMID:; http://dx.doi.org/ 10.1186/bcr1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 2003; 5:317-28; http://dx.doi.org/ 10.1016/S1535-6108(04)00083-2 [DOI] [PubMed] [Google Scholar]
- 11. Cho H-S, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 2003; 421:756-60; PMID:; http://dx.doi.org/ 10.1038/nature01392 [DOI] [PubMed] [Google Scholar]
- 12. Cortes J, Fumoleau P, Bianchi GV, Petrella TM, Gelmon K, Pivot X, Verma S, Albanell J, Conte P, Lluch A, et al. Pertuzumab monotherapy after trastuzumab-based treatment and subsequent reintroduction of trastuzumab: activity and tolerability in patients with advanced human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol 2012; 30:1594-600; PMID:; http://dx.doi.org/ 10.1200/JCO.2011.37.4207 [DOI] [PubMed] [Google Scholar]
- 13. Baselga J, Cortes J, Kim S-B, Im S-A, Hegg R, Im Y-H, Roman L, Pedrini JL, Pienkowski T, Knott A, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med 2012; 366:109-19; PMID:; http://dx.doi.org/ 10.1056/NEJMoa1113216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li B, Meng Y, Zheng L, Zhang X, Tong Q, Tan W, Hu S, Li H, Chen Y, Song J, et al. Bispecific antibody to ErbB2 overcomes trastuzumab resistance through comprehensive blockade of ErbB2 heterodimerization. Cancer Res 2013; 73:6471-83; PMID:; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0657 [DOI] [PubMed] [Google Scholar]
- 15. Scheuer W, Friess T, Burtscher H, Bossenmaier B, Endl J, Hasmann M. Strongly enhanced antitumor activity of trastuzumab and pertuzumab combination treatment on HER2-positive human xenograft tumor models. Cancer Res 2009; 69:9330-6; PMID:; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-4597 [DOI] [PubMed] [Google Scholar]
- 16. Nahta R, Hung M-C, Esteva FJ. The HER-2-targeting antibodies trastuzumab and pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res 2004; 64:2343-6; PMID:; http://dx.doi.org/ 10.1158/0008-5472.CAN-03-3856 [DOI] [PubMed] [Google Scholar]
- 17. Friess T. Combination treatment with erlotinib and pertuzumab against human tumor xenografts is superior to monotherapy. Clin Cancer Res 2005; 11:5300-9; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-04-2642 [DOI] [PubMed] [Google Scholar]
- 18. Baselga J, Gelmon KA, Verma S, Wardley A, Conte P, Miles D, Bianchi G, Cortes J, McNally VA, Ross GA, et al. Phase II trial of pertuzumab and trastuzumab in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer that progressed during prior trastuzumab therapy. J Clin Oncol 2010; 28:1138-44; PMID:; http://dx.doi.org/ 10.1200/JCO.2009.24.2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 20. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100:57-70; PMID:; http://dx.doi.org/ 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 21. Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol 2006; 6:343-57; PMID:; http://dx.doi.org/ 10.1038/nri1837 [DOI] [PubMed] [Google Scholar]
- 22. Sliwkowski MX, Mellman I. Antibody therapeutics in cancer. Science 2013; 341:1192-8; PMID:; http://dx.doi.org/ 10.1126/science.1241145 [DOI] [PubMed] [Google Scholar]
- 23. Schaefer WW, Regula JTJ, Bähner MM, Schanzer JJ, Croasdale RR, Dürr HH, Gassner CC, Georges GG, Kettenberger HH, Imhof-Jung SS, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A 2011; 108:11187-92; PMID:; http://dx.doi.org/ 10.1073/pnas.1019002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li B, Zhao L, Wang C, Guo H, Wu L, Zhang X, Qian W, Wang H, Guo Y. The protein-protein interface evolution acts in a similar way to antibody affinity maturation. J Biol Chem 2010; 285:3865-71; PMID:; http://dx.doi.org/ 10.1074/jbc.M109.076547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao L, Tong Q, Qian W, Li B, Zhang D, Fu T, Duan S, Zhang X, Zhao J, Dai J, et al. Eradication of non-Hodgkin lymphoma through the induction of tumor-specific T cell immunity by CD20-Flex BiFP. Blood 2013; 122:4230-6; PMID:; http://dx.doi.org/ 10.1182/blood-2013-04-496554 [DOI] [PubMed] [Google Scholar]
- 26. Atwell SS, Ridgway JBJ, Wells JAJ, Carter PP. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J Mol Biol 1997; 270:26-35; http://dx.doi.org/ 10.1006/jmbi.1997.1116. [DOI] [PubMed] [Google Scholar]
- 27. Ridgway JB, Presta LG, Carter P. “Knobs-into-holes” engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng 1996; 9:617-21; PMID:; http://dx.doi.org/ 10.1093/protein/9.7.617 [DOI] [PubMed] [Google Scholar]
- 28. Li B, Zhao L, Guo H, Wang C, Zhang X, Wu L, Chen L, Tong Q, Qian W, Wang H, et al. Characterization of a rituximab variant with potent antitumor activity against rituximab-resistant B-cell lymphoma. Blood 2009; 114:5007-15; PMID:; http://dx.doi.org/ 10.1182/blood-2009-06-225474 [DOI] [PubMed] [Google Scholar]
- 29. Stern HM. Improving treatment of HER2-positive cancers: opportunities and challenges. Sci Transl Med 2012; 4:127rv2-127rv2; PMID: [DOI] [PubMed] [Google Scholar]
- 30. Park S, Jiang Z, Mortenson ED, Deng L, Radkevich-Brown O, Yang X, Sattar H, Wang Y, Brown NK, Greene M, et al. The therapeutic effect of anti-HER2/neu antibody depends on both innate and adaptive immunity. Cancer Cell 2010; 18:160-70; PMID:; http://dx.doi.org/ 10.1016/j.ccr.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Armour KL, Clark MR, Hadley AG, Williamson LM. Recombinant human IgG molecules lacking Fcgamma receptor I binding and monocyte triggering activities. Eur J Immunol 1999; 29:2613-24; PMID:; http://dx.doi.org/ 10.1002/(SICI)1521-4141(199908)29:08%3c2613::AID-IMMU2613%3e3.0.CO;2-J [DOI] [PubMed] [Google Scholar]
- 32. Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem 2001; 276:6591-604; PMID:; http://dx.doi.org/ 10.1074/jbc.M009483200 [DOI] [PubMed] [Google Scholar]
- 33. Obeid M, Tesniere A, Ghiringhelli F, Fimia GM, Apetoh L, Perfettini J-L, Castedo M, Mignot G, Panaretakis T, Casares N, et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med 2007; 13:54-61; PMID:; http://dx.doi.org/ 10.1038/nm1523 [DOI] [PubMed] [Google Scholar]
- 34. Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol 2006; 6:715-27; PMID:; http://dx.doi.org/ 10.1038/nri1936 [DOI] [PubMed] [Google Scholar]
- 35. Lake RA, van der Most RG. A better way for a cancer cell to die. N Engl J Med 2006; 354:2503-4; PMID:; http://dx.doi.org/ 10.1056/NEJMcibr061443 [DOI] [PubMed] [Google Scholar]
- 36. Steinman RM, Mellman I. Immunotherapy: bewitched, bothered, and bewildered no more. Science 2004; 305:197-200; PMID:; http://dx.doi.org/ 10.1126/science.1099688 [DOI] [PubMed] [Google Scholar]
- 37. Chao MP, Jaiswal S. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci Transl Med 2010; 2:63ra94–4; PMID:21178137; http://dx.doi.org/10.1126%2Fscitranslmed.3001375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. D'Alessio A, De Luca A, Maiello MR, Lamura L, Rachiglio AM, Napolitano M, Gallo M, Normanno N. Effects of the combined blockade of EGFR and ErbB-2 on signal transduction and regulation of cell cycle regulatory proteins in breast cancer cells. Breast Cancer Res Treat 2010; 123:387-96; PMID:; http://dx.doi.org/ 10.1007/s10549-009-0649-x [DOI] [PubMed] [Google Scholar]
- 39. O'Donovan N, Byrne AT, O'Connor AE, McGee S, Gallagher WM, Crown J. Synergistic interaction between trastuzumab and EGFR/HER-2 tyrosine kinase inhibitors in HER-2 positive breast cancer cells. Invest New Drugs 2011; 29:752-9; PMID:; http://dx.doi.org/ 10.1007/s10637-010-9415-5 [DOI] [PubMed] [Google Scholar]
- 40. Green MD, Francis PA, Gebski V, Harvey V, Karapetis C, Chan A, Snyder R, Fong A, Basser R, Forbes JF, et al. Gefitinib treatment in hormone-resistant and hormone receptor-negative advanced breast cancer. Ann Oncol 2009; 20:1813-7; PMID:; http://dx.doi.org/ 10.1093/annonc/mdp202 [DOI] [PubMed] [Google Scholar]
- 41. Dickler MN, Cobleigh MA, Miller K D, Klein PM, Winer EP. Efficacy and safety of erlotinib in patients with locally advanced or metastatic breast cancer. Breast Cancer Res Treat 2009; 115:115-21; PMID:; http://dx.doi.org/ 10.1007/s10549-008-0055-9 [DOI] [PubMed] [Google Scholar]
- 42. Gioulbasanis I, Saridaki Z, Kalykaki A, Vamvakas L, Kalbakis K, Ignatiadis M, Amarantidis K, Kakolyris S, Georgoulias V, Mavroudis D. Gefitinib in combination with gemcitabine and vinorelbine in patients with metastatic breast cancer pre-treated with taxane and anthracycline chemotherapy: a phase I/II trial. Anticancer Res 2008; 28:3019-25; PMID: [PubMed] [Google Scholar]
- 43. Mayer EL, Partridge AH, Harris LN, Gelman RS, Schumer ST, Burstein HJ, Winer EP. Tolerability of and adherence to combination oral therapy with gefitinib and capecitabine in metastatic breast cancer. Breast Cancer Res Treat 2009; 117:615-23; PMID:; http://dx.doi.org/ 10.1007/s10549-009-0366-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arteaga CL, O'Neill A, Moulder SL, Pins M, Sparano JA, Sledge GW, Davidson NE. A phase I-II study of combined blockade of the ErbB receptor network with trastuzumab and gefitinib in patients with HER2 (ErbB2)-overexpressing metastatic breast cancer. Clin Cancer Res 2008; 14:6277-83; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-08-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J 1996; 15:2452-67; PMID: [PMC free article] [PubMed] [Google Scholar]
- 46. Campbell MR, Amin D, Moasser MM. HER3 comes of age: new insights into its functions and role in signaling, tumor biology, and cancer therapy. Clin Cancer Res 2010; 16:1373-83; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Molina MA, Codony-Servat J, Albanell J, Rojo F, Arribas J, Baselga J. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res 2001; 61:4744-9; PMID: [PubMed] [Google Scholar]
- 48. Marshall SA, Lazar GA, Chirino AJ, Desjarlais JR. Rational design and engineering of therapeutic proteins. Drug Discov Today 2003; 8:212-21; PMID:; http://dx.doi.org/ 10.1016/S1359-6446(03)02610-2 [DOI] [PubMed] [Google Scholar]
- 49. Tang Y, Lou J, Alpaugh RK, Robinson MK, Marks JD, Weiner LM. Regulation of antibody-dependent cellular cytotoxicity by IgG intrinsic and apparent affinity for target antigen. J Immunol 2007; 179:2815-23; PMID:; http://dx.doi.org/ 10.4049/jimmunol.179.5.2815 [DOI] [PubMed] [Google Scholar]
- 50. Nahta R, Yu D, Hung M-C, Hortobagyi GN, Esteva FJ. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol 2006; 3:269-80; PMID:; http://dx.doi.org/ 10.1038/ncponc0509 [DOI] [PubMed] [Google Scholar]
- 51. Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ 2009; 16:3-11; PMID:; http://dx.doi.org/ 10.1038/cdd.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Savini L, Gaeta A, Fattorusso C, Catalanotti B, Campiani G, Chiasserini L, Pellerano C, Novellino E, McKissic D, Saxena A. Specific targeting of acetylcholinesterase and butyrylcholinesterase recognition sites. Rational design of novel, selective, and highly potent cholinesterase inhibitors. J Med Chem 2003; 46:1-4; PMID:; http://dx.doi.org/ 10.1021/jm0255668 [DOI] [PubMed] [Google Scholar]
- 53. Gemma S, Gabellieri E, Huleatt P, Fattorusso C, Borriello M, Catalanotti B, Butini S, De Angelis M, Novellino E, Nacci V, et al. Discovery of huperzine A-tacrine hybrids as potent inhibitors of human cholinesterases targeting their midgorge recognition sites. J Med Chem 2006; 49:3421-5; PMID:; http://dx.doi.org/ 10.1021/jm060257t [DOI] [PubMed] [Google Scholar]
- 54. Dauber Osguthorpe P, Roberts VA, Osguthorpe DJ, Wolff J, Genest M, Hagler AT. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins 1988; 4:31-47; PMID:; http://dx.doi.org/ 10.1002/prot.340040106 [DOI] [PubMed] [Google Scholar]
- 55. Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 1983; 4:187-217; http://dx.doi.org/ 10.1002/jcc.540040211 [DOI] [Google Scholar]
- 56. Im W, Lee MS, Brooks CL. Generalized born model with a simple smoothing function. J Comput Chem 2003; 24:1691-702; PMID:; http://dx.doi.org/ 10.1002/jcc.10321 [DOI] [PubMed] [Google Scholar]
- 57. Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 2000; 33:889-97; PMID:; http://dx.doi.org/ 10.1021/ar000033j [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.