Summary
Background
Sensitive and accurate methods to detect hematopoietic chimerism after hematopoietic stem cell transplantation (HSCT) are essential to evaluate engraftment and to monitor response to therapeutic procedures such as donor lymphocyte infusion. Continuous long-term follow up, however, requires large amounts of pre-HSCT samples limiting the application of many widely used techniques for sensitive chimerism monitoring.
Methods
DNAs from 42 normal healthy donors and 16 HSCT donor/recipient pairs were employed to validate the use of allele-specific insertion/deletion (indel) quantitative real-time polymerase chain reaction (qPCR) to quantify chimerism in samples with low amounts of DNA. Consequently, indel-qPCR analyses of samples from 16 HSCT patients were compared to short-tandem repeat (STR) specific PCR analyses.
Results
Typing with reduced amounts of input DNA (15 vs. 60 ng) allowed for the reliable distinction of positive (mean threshold cycle (ct) 28.05) and negative (ct >36) signals. The high informativity of primer/probe sets, with 12 out of 19 markers exceeding 20% informativity, was confirmed in our cohort (n = 74). Importantly, a fourfold reduction of input DNA compared to published protocols did not alter PCR efficiencies and allowed for a more sensitive detection of chimerism in 7 of 16 HSCT patients compared to results obtained by STR-PCR.
Conclusions
Our data suggest that indel-qPCR is a more sensitive technique for the detection of hematopoietic chimerism compared to STR-PCR and works efficiently for samples with low amounts of DNA.
Keywords: Stem cell transplantation, Chimerism, Molecular diagnostic techniques, Quantitative real-time polymerase chain reaction
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) following chemo- and/or radiotherapy is an established treatment option for many hematopoietic disorders. One goal of HSCT is to reconstitute normal hematopoiesis after myeloablative or non-myeloablative conditioning of the patient. Consequently, achievement of complete donor-derived hematopoiesis is one of the hallmarks of successful HSCT in hematopoietic malignancies. Moreover, several studies indicate that remaining or re-emerging recipient cells after ablation and transplantation are associated with a higher risk of disease relapse [1, 2, 3, 4, 5, 6, 7]. Therefore, the accurate and sensitive quantification of hematopoietic chimerism is a critical aspect of post-transplantation monitoring, since the results strongly influence the therapeutic approach such as donor lymphocyte infusions, additional chemotherapy, or re-transplantation [8].
To date several methods have been described to measure the relative contribution of donor and recipient cells to post-transplantation hematopoiesis. The most informative assays for chimerism quantification distinguish highly variable length polymorphisms of short-tandem repeats (STR or microsatellites) by polymerase chain reaction (PCR) [1, 9, 10]. Due to the high informativity and the feasibility of application, STR analysis is currently the standard molecular diagnostic tool for chimerism quantification after HSCT. One drawback of STR-PCR, however, is the relatively low sensitivity of 1–6% of cells of the minor genotype [11, 12, 13, 14, 15].
Several studies proposed real-time quantitative PCR (qPCR) based methods to detect short insertions/deletions (indel) or single nucleoide polymorphisms for chimerism quantification [5, 16, 17, 18, 19]. These methods allow for the detection and quantification of hematopoietic chimerism with sensitivities of or below 0.1% of the minor genotype. Alizadeh et al. [16] have proposed a set of 19 indel markers at 11 chromosomal loci for chimerism quantification by qPCR. The authors predicted that this selection of markers is sufficiently informative to analyze >90% of donor/recipient pairs. The sensitivity of indel-qPCR was at least one order of magnitude higher than previously published STR-PCR assays, allowing for the detection of recipient chimerism as low as 0.1%.
In contrast to STR-PCR-based methods, however, indel-qPCR is currently not widely utilized, probably due to the novelty of the method and the still restricted availability of commercial indelq-PCR assays. Therefore, some aspects of chimerism analysis by indel-qPCRs have yet to be validated in routine clinical application. One such aspect is a high amount of input DNA required for analysis compared to STR-PCR. This presents a challenge for long-term post-transplantation follow-up, because of the requirement of the same negative pre-transplant control sample for every analysis. Moreover, the applicability of these novel methods, especially in cases where DNA yield might be a limiting factor(e.g. in samples from cytopenic patients), has never been assessed. In order to address this concern, we validated the sensitivity and accuracy of indel-qPCR on spiked DNA preparations and performed a retrospective analysis of chimerism in samples with low DNA quantities from patients who underwent allogeneic HSCT for malignant diseases.
Patients and Methods
Patients and Normal Healthy Donors
Indel informativity and PCR efficiencies of primer/probe sets were established by analyzing 42 unrelated healthy blood donors and 16 HSCT donor/recipient pairs. The patient DNA samples for this study were selected from patients who had undergone allogeneic HSCT after myeloablative conditioning between 2005 and 2009 at the University Hospital Erlangen (Erlangen, Germany) by two criteria. First, a sufficient number of suitable pre- and post-HSCT samples had to be available. Second, at least one post-HSCT sample was required to exhibit mixed chimerism by STR-PCR analysis or another indication of a potential disease relapse. The final selection of 16 HSCT patients had been treated for acute lymphoblastic leukemia (ALL, n = 4), acute myeloid leukemia (AML, n = 5), multiple myeloma (MM, n = 4), and non-Hodgkin's lymphoma (NHL, n = 3). 14 transplants were from matched unrelated donors and two were from HLA-identical siblings.
Sample Preparation and DNA Extraction
The mononuclear cell (MNC) fraction of peripheral blood samples or bone marrow aspirates from patients was prepared by Ficoll density gradient centrifugation. Subsequently, genomic DNA was isolated from the MNCs with the EZ1 DNA blood 350μl Kit on a Geno-M6 automated DNA extractor (Qiagen, Hilden, Germany). DNA from normal healthy donors was prepared from peripheral blood or bone marrow aspirates by the QIAamp DNA Blood Midi Kit (Qiagen) according to the manufacturer's instructions. The concentration and purity of the DNA samples was determined by UV photometry.
STR-PCR
STR-PCR from patient DNA samples was carried out by utilizing the AmpFeSTR Profiler Plus ID PCR Amplification Kit (Applied Biosystems, Life Technologies, Foster City, CA, USA) according to the manufacturer's instructions. The multiplex PCR to measure multiple alleles in one reaction were carried out on a TProfessional Thermocycler (Biometra, Göttingen, Germany). Subsequently, STR fragment lengths were determined by running the PCR products on an ABI PRISM 3130 Genetic analyzer and analyzed by the GeneScan Analysis software (Applied Biosystems, Life Technologies).
Real-Time qPCR
Real-time qPCRs were set up with the TaqMan Gene Expression Mastermix Kit (Applied Biosystems, Life Technologies). For indel typing and chimerism analysis, reactions were prepared essentially as described in the literature [16]. TaqMan probes were ordered with a 5’ 6-carboxyfluorescein (FAM), and a 3’ 6-carboxy-tetramethyl-rhodamine (TAMRA) fluorophore label and standard PCR cycling conditions were used (2 min at 50 ° C, 10 min at 95 °C followed by 40 amplification cycles (95 °C for 45 s, 60 ° C for 60 s)). In contrast to the published protocol, the amount of DNA per reaction was reduced to 5 ng for indel typing and 20 ng for chimerism analysis. The reactions were scaled down to final volumes of 10 μl and 20 μl for indel typing and chimerism analysis, respectively. A no-template control reaction for each primer/probe set was performed in parallel to exclude possible contaminations. All real-time qPCR reactions were carried out in duplicate and run on a StepOne Real-Time PCR System (Applied Biosystems, Life Technologies). Subsequently, the reactions were analyzed with the StepOne Software (Applied Biosystems, Life Technologies). Duplicate reactions were repeated if the threshold cycle (ct) of duplicate reactions differed by more than 0.5 cycles.
Standard Curves
We determined the PCR efficiency of each indel-specific primer/probe set by performing standard curves. To this end, indel-marker-positive DNA from normal healthy donors was serially diluted twofold in marker-negative DNA 14 times to obtain a defined amount of total mixed DNA. The resulting DNA mixture with ratios of marker-positive to negative DNA from 1:0 to 1:8,000 was then subjected to real-time qPCR. We performed at least two standard curves for each primer/probe set with 15 ng and 60 ng of total DNA per PCR reaction. Finally, PCR efficiency was calculated from the slope of the standard curves by the StepOne Software. Standard curves for primer/probe set 5a were not performed because no DNA sample negative for marker 5a was present in our cohort.
Chimerism Analysis
Chimerism analysis by STR-PCR was performed by determining the peak area of donor- and recipient-specific STR alleles in post-HSCT samples with the GeneScan Analysis software. The peak areas were then used to calculate the level of donor and recipient chimerism according to published methods [20].
For chimerism analysis by indel-qPCR, indel typing reactions were performed with pre-HSCT samples of donors and recipients to determine informative markers. Subsequently, suitable markers were selected which were present in the recipient and absent in the donor. This setting allowed for the detection of low levels of chimerism. For chimerism analysis, at least two appropriate markers were chosen, if available. The ratio of recipient chimerism was then determined by the ΔΔct method according to the published protocol, using the GAPDH primer/probe set to normalize for the actual DNA amount [16]. In brief, indel-qPCR was carried out with pre-HSCT samples and subsequent patient samples. Subsequently, the ΔΔct ((ctmarker_post – ctGAPDH_post) –(ctmarker_pre – ctGAPDH_pre)) between pre- and post- HSCT samples was calculated. The percentages of recipient chimerism in the post-HSCT samples were calculated by the following formula, using the specific primer/probe efficiencies: % recipient chimerism = (1 + efficiency)–ΔΔct × 100%. Finally, the mean of all selected markers was calculated.
Results
Validation of Indel-Specific Real-Time qPCR
We established an indel-qPCR system for the monitoring of mixed chimerism in patients after HSCT as proposed by Alizadeh et al. [16]. First, we determined the informativity of the 19 indel markers used by analyzing the presence or absence of these markers in 42 normal healthy persons and 16 HSCT donor/recipient pairs. Because only a limited amount of DNA was available from some donor/recipient pairs, we performed the typing reaction with reduced amounts of DNA compared to the initially published by Alizadeh et al. [16] (5 ng instead of 100 ng). The mean ct value for positive signals in the typing reaction was 28.05 (table 1). Therefore, the reduced quantity of DNA still allowed for an unambiguous distinction between positive and negative (ct > 36) signals. Based on the results of all typing reactions, we calculated the theoretical informativity of each marker, defined as 50% of the probability of the presence a marker-positive and a marker-negative individual in any random pair chosen from our test population. The calculated informativity ranged from 0% for marker 5a to the theoretical maximum of 25% for marker 6, with a median informativity of 21.1%. Of note, 12 markers with informativities of at least 20% were present in our cohort from Southern Germany. The mean number of suitable markers which were present in the recipient and absent in the donor of the 16 HSCT pairs was 3.8 (range 1–6, not shown). These results are consistent with the results in the French patient cohort of Alizadeh et al. and strongly indicate the suitability of the marker selection for chimerism detection in different groups of patients and potentially even in related HSCT donor/recipient pairs. Indeed, we detected two or four informative indel markers in the two sibling pairs analyzed in our test population (table 2).
Table 1.
Markers forindel-qPCR
| indel marker | Ct typ. rxna, mean ± SD | Informativityb, % | PCR efficiencyc |
|
|---|---|---|---|---|
| 15 ng | 60 ng | |||
| S 01a | 28.22 ± 1.49 | 22.6 | 0.96 | 1.00 |
| S 01b | 28.45 ± 1.31 | 14.2 | 0.93 | 0.99 |
| S 02 | 28.76 ± 1.60 | 24.9 | 0.78 | 0.92 |
| S 03 | 28.22 ± 1.62 | 19.4 | 0.97 | 0.96 |
| S 04a | 28.78 ± 1.35 | 20.0 | 0.74 | 0.89 |
| S 04b | 28.65 ± 0.97 | 20.0 | 0.92 | 0.97 |
| S 05a | 28.00 ± 1.51 | 0.0 | n.d. | n.d. |
| S 05b | 28.23 ± 1.36 | 24.4 | 0.93 | 0.97 |
| S 06 | 27.63 ± 1.25 | 25.0 | 0.90 | 0.96 |
| S 07a | 27.98 ± 1.15 | 21.1 | 1.01 | 0.88 |
| S 07b | 29.24 ± 1.35 | 24.8 | 0.93 | 0.98 |
| S 08a | 27.90 ± 1.27 | 18.1 | 0.93 | 0.91 |
| S 08b | 28.22 ± 1.42 | 18.4 | 0.96 | 1.04 |
| S 09a | 27.06 ± 1.11 | 5.0 | 0.80 | 0.80 |
| S 09b | 29.08 ± 1.05 | 23.7 | 0.96 | 0.97 |
| S 10a | 27.89 ± 1.05 | 21.6 | 0.94 | 0.97 |
| S 10b | 26.97 ± 1.13 | 21.1 | 0.80 | 0.88 |
| S 11a | 27.73 ± 1.02 | 24.9 | 0.91 | 0.91 |
| S 11b | 27.53 ± 1.16 | 15.0 | 0.87 | 0.88 |
indel = Micro insertion/deletion polymorphism; typ. rxn = typing reaction; SD =standard deviation; n.d. = not determined.
Mean Ct value of all positive typing reactions performed with 5 ng template DNA (± SD).
Theoretical informativity of the marker among the 74 individuals analyzed.
PCR efficiencies deduced from standard curves performed with the indicated amounts of total DNA.
Table 2.
Patient and sample characteristics*
| Patient | Diagnosis | Number of samplesa | DNA, ng/µlb | First indication of recipient chimerism, days post HSCT |
Evidence for relapsec | Comment | |
|---|---|---|---|---|---|---|---|
| STR-PCR | indel-qPCR | ||||||
| 1 | AML | 6 | 3–475 | 1,273 | 1,273 | thrombocyte count ↓, blasts (BM/PB) | |
| 2 | AML | 11 | 9–176 | 208/569 | 208/569 | DEK-CAN fusion ↑, blasts(BM/PB) | 2× RL |
| 3 | AML | 8 | 10–172 | 853/1,105 | 231/1,008 | thrombocyte count ↓, blasts (BM) | 2× RL |
| 4 | ALL | 11 | 12–128 | 126/716 | 126/716 | blasts (BM/PB) | 2× RL, † |
| 5 | AML | 10 | 6–64 | 32/314 | 32/314 | blasts (BM/PB) | |
| 6 | AML | 2 | 5–61 | 97 | 97 | blasts (BM) | † |
| 7 | MM | 5 | 29–108 | – | 423 | clonal IgG ↑ | |
| 8 | MM | 3 | 19–46 | – | 111 | serum κ-chains ↑ | † |
| 9 | ALL | 3 | 3–250 | 105 | 105 | blasts (PB) | † |
| 10 | MM | 4 | 15–50 | 185 | 185 | clonal IgG ↑, plasma cell expansion (BM) | † |
| 11 | NHL | 5 | 10–189 | 29 | 29 | residual lymphoma | stable MC |
| 12 | ALL | 3 | 35–220 | 230 | 54 | BCR-ABL+ after HSCT | |
| 13 | MM | 5 | 18–238 | 243 | 148 | clonal IgG ↑, plasma cell expansion (BM) | HID |
| 14 | NHL | 6 | 20–105 | 41 | 41 | incompl. donor hematopoiesis | HID, stable MC |
| 15 | ALL | 4 | 35–259 | 86 | 36 | blasts (BM) | |
| 16 | NHL | 3 | 23–242 | 62 | 35 | blasts (PB) | |
AML = Acute myeloid leukemia; ALL = acute lymphoblastic leukemia; BM = bone marrow; HID = HLA-identical sibling donor; MC, mixed chimerism; MM = multiple myeloma; NHL= non-Hodgkin's lymphoma; PB = peripheral blood; STR = short-tandem-repeat; indel = micro insertion/deletion. polymorphism
= deceased.
Cases where indel-qPCR provided an earlier detection of recipient chimerism are highlighted in italics.
Number of post HSCT samples available for retrospective analysis by indel-qPCR.
DNA concentration range of the samples, 100μl of each sample were available in most cases.
Molecular and cytological evidence for relapse.
Next, we performed standard curves to calculate the PCR efficiency of each primer/probe set. The efficiencies were comparable between different markers and varied from 0.74 to 1.04 (table 1). In addition, we were able to confirm the linearity of the standard curves up to ct values of 36. Consequently, ct values greater than 36 were treated as ‘not determined’ for subsequent analyses.
Suitability of the Indel-Specific Real-Time qPCR for Samples with Low DNA Quantities
The available DNA quantities of the samples from our HSCT patient cohort showed considerable variation (1.8–285 pg total DNA per ml blood, not shown). The sample DNA concentrations ranged from 3 to 475 ng/µl (median concentration 58 ng/µl; table 2). Consequently, the amount of DNA available for analysis was limited in many samples. Published indel-qPCR protocols, however, require 100–500 ng of input DNA per reaction [16, 21, 22], precluding the analysis of several samples. Therefore, we wanted to assess the suitability of indel-qPCR for chimerism analysis with reduced amounts of input DNA. To this end, we performed additional standard curves with 15 ng of total DNA. Notably, we observed little variation of primer/probe efficiency compared to standard curve reactions performed with 60 ng of total DNA (table 1). Furthermore, 15 ng of marker-positive DNA resulted in a mean ct value of 25.79 (23.94–27.67; table 1). Consequently, 15 ng total DNA for each indel-qPCR reaction would be sufficient to detect hematopoietic chimerism of less than 1%, depending on the efficiencies of the primer/probe set used. These results suggested a higher sensitivity of the indel-qPCR compared to the widely used STR-PCR for chimerism analysis even in samples with low DNA content.
Comparison of Chimerism Quantification by STR-PCR and Indel-qPCR
Previously, hematopoietic chimerism of 16 HSCT patients had been quantified by STR-PCR of DNA isolated from the MNC fraction of bone marrow aspirates or peripheral blood. Sample collection started approximately 30 days after transplantation. Further bone marrow and/or blood samples for STR-PCR were collected subsequently at several time points. 14 of the patients in our cohort suffered from hematological relapse after HSCT. In these cases, relapse was diagnosed by cytological analysis and/or the reappearance of disease-specific molecular markers in addition to the STR-PCR chimerism analysis (table 2).
In this sample set, we assessed if the superior sensitivity of indel-qPCR would allow for earlier detection of hematopoietic chimerism after HSCT compared to STR-PCR by re-testing the samples with indel-qPCR.
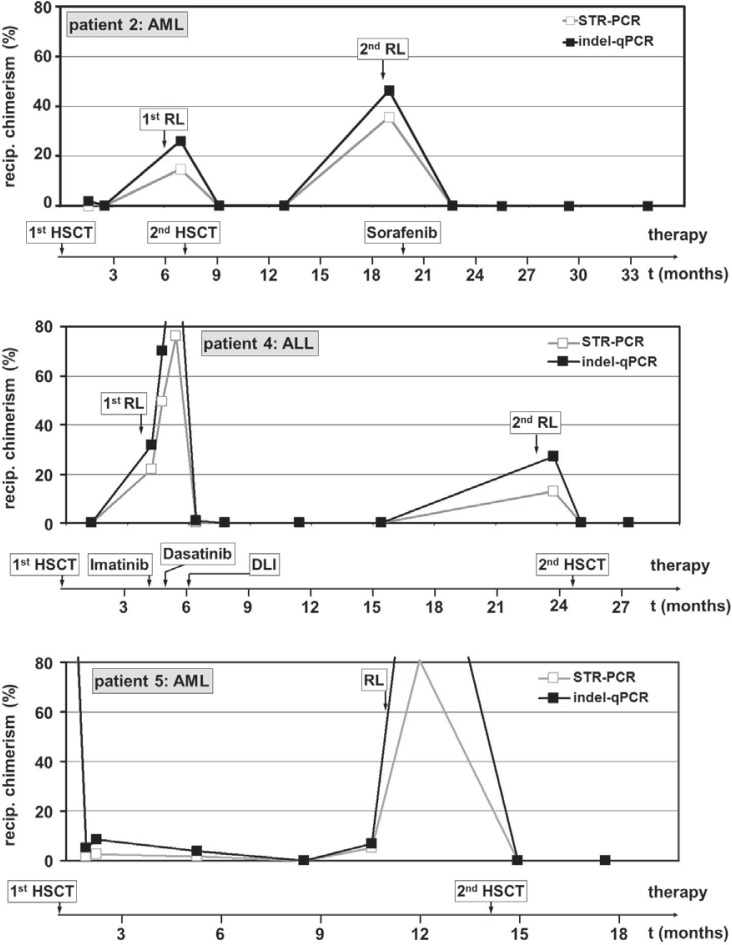
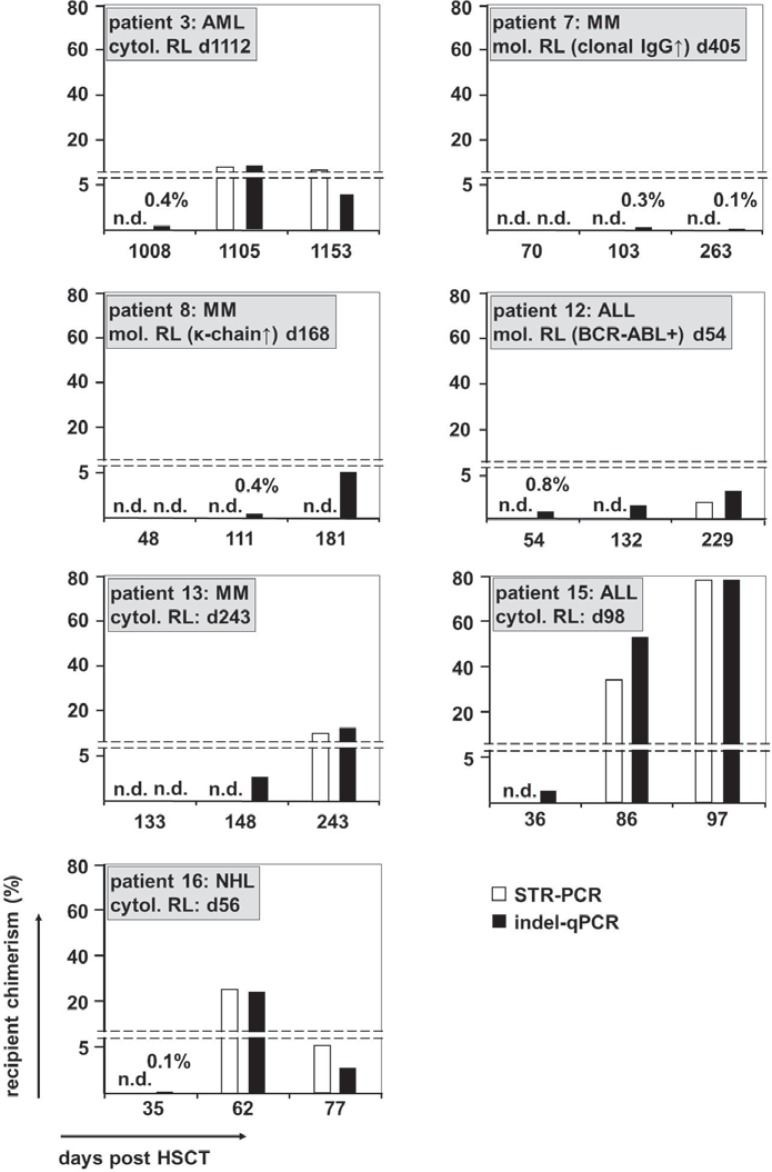
We compared the results of the indel-qPCR analysis to the results previously obtained by STR-PCR (see supplemental figures 1 and 2 (available at www.karger.com/?DOI=370255) for a graphical comparison of the 2 different methods). Chimerism analysis by indel-qPCR provided results, which were at least equivalently sensitive to previous data and fitted well to the clinical data of the patients (fig. 1). Of note, in most cases indel-qPCR analysis resulted in a slightly higher proportion of recipient hematopoiesis compared to STR-PCR (mean 4.0%, see also fig. 1 and 2). Most importantly, we unambiguously detected low-level hematopoietic chimerism in 6 of the 14 relapse patients at time points at which it was still undetected by STR-PCR (table 2, fig. 2). Another relapse patient (patient 16) also showed re-emergence of recipient hematopoiesis at an earlier time point in the indel-qPCR analysis. However, the ct values of the indel-qPCR were close to the detection limit and only 2 out of 4 markers analyzed provided positive results (not shown). Even so, subsequent samples unambiguously confirmed re-emergence of chimerism in this patient (fig. 2). The median period between the detection of chimerism by indel-qPCR and STR-PCR in the 7 cases of early chimerism detection was 95 days (range 27–176 days).
Fig. 1.
Follow-up of hematopoietic chimerism by STR-PCR and indel-qPCR in HSCT patients. The percentage of recipient chimerism determined by indel-qPCR and STR-PCR in samples taken after HSCT is shown for three representative patients with a long record of available samples (2, 4, and 5). The reappearance of blast cells in bone-marrow and/or peripheral blood samples (cytological relapse) and time-points of therapeutic interventions are indicated in the graph and the time scale, respectively. AML = Acute myeloid leukemia; ALL = acute lymphoblastic leukemia; MM = multiple myeloma; NHL = non-Hodgkin's lymphoma; STR = short-tandem-repeat; indel = micro insertion/deletion polymorphism; RL = relapse; HSCT = hematopoietic stem cell transplantation.
Fig. 2.
Comparison of the detection of emerging recipient chimerism by STR-PCR and indel-qPCR. The fraction of recipient chimerism in samples taken after HSCT is depicted for all 7 cases of earlier detection of re-emerging recipient hematopoiesis by indel-qPCR compared to STR-PCR. The underlying disease of each patient and the time-point of cytological or molecular relapse relapse (if available) is given for each patient. The percentage for samples with recipient chimerism <1% is specified above the respective bar graphs. AML = Acute myeloid leukemia; ALL = acute lymphoblastic leukemia; DLI = donor lymphocyte infusion; MM = multiple myeloma; n.d. = not detectable, NHL = non-Hodgkin's lymphoma; STR = short-tandem-repeat; indel = micro insertion/deletion; BM = bone marrow; PB = peripheral blood; cytol = cytological; mol = molecular; RL = relapse; HSCT = hematopoietic stem cell transplantation.
Of note, the diagnoses of the patients with earlier detection of chimerism by indel-qPCR were AML (n = 1), ALL (n = 2), MM (n = 3), and NHL (n = 1) (table 2, fig. 2). This indicates that indel-qPCR can be used for the quantification of chimerism after allogeneic HSCT in various hematopoietic malignancies. Taken together, these results confirmed the enhanced sensitivity of indel-qPCR compared to STR-PCR, even in samples with a limited amount of DNA.
Discussion
The aim of this study was to assess the sensitivity and accuracy of indel-qPCR chimerism monitoring following hematopoietic stem cell transplantation especially in cases where only a limited amount of DNA is available. Several lines of evidence highlight the importance of post-transplantation chimerism monitoring of patients who underwent HSCT. First, residual recipient hematopoiesis after myeloablative conditioning followed by HSCT correlates with graft failure and persisting disease [1, 23, 24]. Second, achievement of complete donor-derived hematopoiesis early after HSCT is associated with increased incidence of graft-versus-host disease [8, 23, 25, 26]. Last, some studies corroborated a higher risk for disease relapse in patients with mixed recipient chimerism after HSCT [4, 5, 27]. This finding, however, is controversial because some studies failed to demonstrate a correlation between mixed chimerism after HSCT and a higher risk for relapse [26, 28, 29]. Nevertheless, several groups established that increasing mixed chimerism is clearly associated with higher frequency of disease relapse in different hematopoietic malignancies [1, 2, 3, 5, 6, 7]. Taken together, these data illustrate that the routine assessment of chimerism is a useful approach to evaluate post-transplantation prognosis. Importantly, chimerism status may provide the rationale for post-transplantation therapy like modulation of immunosuppression or donor lymphocyte infusions [8]. Indeed, Bader et al. showed that the 3 year event-free survival was increased from 0 to 37% and from 0 to 36% after immunotherapy of patients with increasing recipient chimerism in pediatric ALL [30] and AML [31], respectively.
Recently, efforts to increase the sensitivity of chimerism detection have been undertaken. One strategy relies on the analysis of chimerism by STR-PCR in specific subsets of hematopoietic cells, resulting in high subset-specific sensitivity [32, 33, 34]. These analyses can be focused on cell subsets relevant for relapse or therapy efficacy. The cell-fractioning procedures, however, are time-consuming, require large amounts of cells, and may exhibit high variability in terms of yield, limiting the use of these methods for routine chimerism monitoring [34]. Another strategy concentrates on the improvement of the sensitivity of the detection technique. Consequently, indel-specific real-time qPCR methods have recently been developed which combine high informativity and high sensitivity [16, 21, 35]. Therefore, they seem to be suited for sensitive post-HSCT chimerism. Importantly, indel-qPCR does not depend on the presence of disease-specific markers. Therefore, this technique can provide a means to perform minimal residual disease monitoring in patients which cannot be analyzed by conventional minimal residual disease diagnostics because they lack specific markers.
In this study, we were able to retrospectively analyze 16 pre-selected donor/recipient pairs by an indel-qPCR as described by Alizadeh et al. [16]. With 12 of the 16 markers exceeding informativities of 20% in our German test cohort, our results confirm the conclusion by Alizadeh et al. that the proposed marker set is highly informative. These findings argue for the suitability of the selection of markers for chimerism analysis of a large proportion of HSCT patients. This is an important feature for routine diagnostics to ensure the broad applicability in clinical practice.
A potential shortcoming of indel-qPCR, however, is the requirement for high amounts of input DNA. Published protocols suggested 100–500 ng DNA per reaction to provide sensitivities of or below 0.1% [16, 21, 22]. Notably, chimerism monitoring by indel-qPCR requires pre-transplantation DNA samples of both donor and recipient for the calibration of each subsequent measurement. Consequently, the amount of DNA required for long-term follow-up after HSCT represents a limiting factor for the analysis according to published protocols. Although methods of DNA preparation and sample collection can be optimized for higher DNA quantities, samples may still provide only a low DNA yield due to several reasons. For example, disease-associated low blood cell counts may result in low DNA yield of peripheral blood and even bone marrow samples. This may occur in cytopenic patients as well as in HSCT patients suffering from certain non-malignant disorders like severe combined immunodeficiency or severe aplastic anemia. Moreover, cell fractioning procedures to determine lineage-specific chimerism may also provide only few cells for DNA extraction [32, 33, 34]. Our results indicate, however, that chimerism analysis by indel-qPCR is more sensitive compared to a commercial STR-PCR, even when using greatly reduced amounts of input DNA. This increase in sensitivity may reduce the need to fractionate hematopoietic cell subsets for standard high-sensitivity chimerism analysis. Additionally, indel-qPCR is not dependent on the expression of surface markers (e.g. CD34 in AMLs) routinely used for cell separation. Therefore, high-sensitivity chimerism quantification by indel-qPCR is more broadly applicable than by cell fractionation. Importantly, gaining or losing expression of surface markers (including CD34) on AML cells is not uncommon, especially in disease relapse [36]. Therefore, chimerism analysis, ideally, should not depend on the presence of certain markers. Nevertheless, indel-qPCR cannot fully replace lineage-specific chimerism quantification. Certain clinical questions, like the differentiation between lymphocytic and myeloid engraftment in AML or ALL patients after HSCT, can only be addressed by cell fractionation and analysis of subpopulations.
On average, we detected emerging recipient chimerism by our modified indel-qPCR protocol 95 days earlier in 7 out of 14 relapse patients than diagnosed by STR-PCR. Importantly, we observed no case in our study where detection of recipient by STR-PCR preceded the detection by indel-qPCR. These results suggest that enhanced sensitivity of chimerism analysis by indel-qPCR may result in earlier diagnosis of relapse. This increases the window for therapeutic interventions such as donor lymphocyte infusion. Nevertheless, additional prospective studies with defined and continuous sampling schemes and a larger patient cohort are needed to validate the data presented here.
Taken together, our results corroborate the high overall informativity of the indel markers employed in this study, arguing for the applicability of indel-qPCR to analyze the majority of donor/recipient pairs. Furthermore, our results provide evidence that indelq-PCR provides a higher sensitivity for the quantitative assessment of post-transplantation chimerism in samples with low DNA contents compared to STR-PCR. This gain in sensitivity can result in an earlier indication of disease relapse or graft failure and therefore provide better treatment options for the patient.
Disclosure Statement
The authors declare no competing financial interests.
Supplementary Material
Supplementary data
Acknowledgements
The authors are indebted to Birgit Lauer for expert technical support.
Imprint
ISSN Print Edition: 1660–3796
ISSN Online Edition: 1660–3818
Journal Homepage: http://www.karger.com/tmh
Publication Data: Volume 42, 2015 of ‘TRANSFUSION MEDICINE AND HEMOTHERAPY’ appears with 6 issues.
Copyright: © 2015 by S. Karger Verlag für Medizin und Naturwissenschaften GmbH, Freiburg (Germany). All rights reserved. No part of the journal may be reproduced in any form without the written permission of the publisher. This includes digitalisation and any further electronic computing, like saving, copying, printing or electronic transmission of digitalized material from this journal (online or offline). Authorization to photocopy items for internal or personal use of specific clients is granted by Karger.
Disclaimer: The statements and data contained in this publication are solely those of the individual authors and contributors and not of the publisher and the editor(s). The appearance of advertisements in the journal is not a warranty, endorsement, or approval of the products or services advertised or of their effectiveness, quality or safety. The publisher and the editor(s) disclaim responsibility for any injury to persons or property resulting form any ideas, methods, instructions or products referred to in the content or advertisements.
Distribution and Subscription: Karger offers three types of subscription: Print Only, Online Only and the combined Print + Online. The basic annual subscription rate is the same for all three delivery forms; however, a fee for the combined print and online subscription is levied, and there is a postage and handling charge for Print Only and Print + Online. Subscriptions run for a full calendar year. Prices are given per volume.
Print subscription: EUR 180.– + postage and handling.
Online subscription: EUR 180.–.
Combined (print + online) subscription: EUR 230.– + postage and handling.
For customers in Germany: Please turn to your bookshop or to S. Karger Verlag für Medizin und Naturwissenschaften GmbH
Wilhelmstr. 20A, 79098 Freiburg (Germany)
Tel. +49 761 45 20 70, Fax +49 761 45 20 714
E-mail information@karger.com
For customers in all other countries: Please contact your bookshop or
S. Karger AG
Allschwilerstr. 10, 4009 Basel (Switzerland)
Tel. +41 61 3 06 11 11, Fax +41 61 3 06 12 34
E-mail karger@karger.com
Advertising: Correspondence should be addressed to the publisher.
S. Karger Verlag für Medizin und Naturwissenschaften GmbH
Attn. Ellen Zimmermann (Head of Marketing)
E-mail e.zimmermann@karger.com
Price list No. 29 of January 1, 2015 is effective.
V.i.S.d.P. (Person responsible according to the German Press Law): Sibylle Gross
Type setting and printing: Kraft Druck GmbH, 76275 Ettlingen, Germany.
Bibliographic Services
Biological Abstracts
Current Contents/Clinical Medicine
Excerpta Medica/EMBASE
Medical Documentation Service
Reference Update
Research Alert
Science Citation Index
SCISEARCH Database
PubMed Central
References
- 1.Lawler M, Humphries P, McCann SR. Evaluation of mixed chimerism by in vitro amplification of dinucleotide repeat sequences using the polymerase chain reaction. Blood. 1991;77:2504–2514. [PubMed] [Google Scholar]
- 2.Bader P, Holle W, Klingebiel T, et al. Mixed hematopoietic chimerism after allogeneic bone marrow transplantation: the impact of quantitative PCR analysis for prediction of relapse and graft rejection in children. Bone Marrow Transplant. 1997;19:697–702. doi: 10.1038/sj.bmt.1700721. [DOI] [PubMed] [Google Scholar]
- 3.Barrios M, Jimenez-Velasco A, Roman-Gomez J, et al. Chimerism status is a useful predictor of relapse after allogeneic stem cell transplantation for acute leukemia. Haematologica. 2003;88:801–810. [PubMed] [Google Scholar]
- 4.Lamba R, Abella E, Kukuruga D, et al. Mixed hematopoietic chimerism at day 90 following allogenic myeloablative stem cell transplantation is a predictor of relapse and survival. Leukemia. 2004;18:1681–1686. doi: 10.1038/sj.leu.2403468. [DOI] [PubMed] [Google Scholar]
- 5.Koldehoff M, Steckel NK, Hlinka M, et al. Quantitative analysis of chimerism after allogeneic stem cell transplantation by real-time polymerase chain reaction with single nucleotide polymorphisms, standard tandem repeats, and Y-chromosome-specific sequences. Am J Hematol. 2006;81:735–746. doi: 10.1002/ajh.20693. [DOI] [PubMed] [Google Scholar]
- 6.Huisman C, de Weger RA, de Vries L, et al. Chimerism analysis within 6 months of allogeneic stem cell transplantation predicts relapse in acute myeloid leukemia. Bone Marrow Transplant. 2007;39:285–291. doi: 10.1038/sj.bmt.1705582. [DOI] [PubMed] [Google Scholar]
- 7.Horky O, Mayer J, Kablaskova L, et al. Increasing hematopoietic microchimerism is a reliable indicator of incipient AML relapse. Int J Lab Hematol. 2011;33:57–66. doi: 10.1111/j.1751-553X.2010.01249.x. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Wan LP, Qin YW, et al. Chimerism status is correlated to acute graft-versus-host disease after allogeneic stem cell transplantation. Int J Hematol. 2014 doi: 10.1007/s12185-014-1510-5. [DOI] [PubMed] [Google Scholar]
- 9.Scharf SJ, Smith AG, Hansen JA, et al. Quantitative determination of bone marrow transplant engraftment using fluorescent polymerase chain reaction primers for human identity markers. Blood. 1995;85:1954–1963. [PubMed] [Google Scholar]
- 10.Odriozola A, Riancho JA, Colorado M, Zarrabeitia MT. Evaluation of the sensitivity of two recently developed STR multiplexes for the analysis of chimerism after haematopoietic stem cell transplantation. Int J Immunogenet. 2013;40:88–92. doi: 10.1111/j.1744-313X.2012.01123.x. [DOI] [PubMed] [Google Scholar]
- 11.Acquaviva C, Duval M, Mirebeau D, et al. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Paris-Robert Debre experience. Leukemia. 2003;17:241–246. doi: 10.1038/sj.leu.2402762. [DOI] [PubMed] [Google Scholar]
- 12.Kreyenberg H, Holle W, Mohrle S, et al. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Tuebingen experience. Leukemia. 2003;17:237–240. doi: 10.1038/sj.leu.2402761. [DOI] [PubMed] [Google Scholar]
- 13.Koehl U, Beck O, Esser R, et al. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Frankfurt experience. Leukemia. 2003;17:232–236. doi: 10.1038/sj.leu.2402760. [DOI] [PubMed] [Google Scholar]
- 14.Chalandon Y, Vischer S, Helg C, et al. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Geneva experience. Leukemia. 2003;17:228–231. doi: 10.1038/sj.leu.2402758. [DOI] [PubMed] [Google Scholar]
- 15.Schraml E, Daxberger H, Watzinger F, Lion T. Quantitative analysis of chimerism after allogeneic stem cell transplantation by PCR amplification of microsatellite markers and capillary electrophoresis with fluorescence detection: the Vienna experience. Leukemia. 2003;17:224–227. doi: 10.1038/sj.leu.2402756. [DOI] [PubMed] [Google Scholar]
- 16.Alizadeh M, Bernard M, Danic B, et al. Quantitative assessment of hematopoietic chimerism after bone marrow transplantation by real-time quantitative polymerase chain reaction. Blood. 2002;99:4618–4625. doi: 10.1182/blood.v99.12.4618. [DOI] [PubMed] [Google Scholar]
- 17.Wilhelm J, Reuter H, Tews B, et al. Detection and quantification of insertion/deletion variations by allele-specific real-time PCR: application for genotyping and chimerism analysis. Biol Chem. 2002;383:1423–1433. doi: 10.1515/BC.2002.161. [DOI] [PubMed] [Google Scholar]
- 18.Hochberg EP, Miklos DB, Neuberg D, et al. A novel rapid single nucleotide polymorphism (SNP)-based method for assessment of hematopoietic chimerism after allogeneic stem cell transplantation. Blood. 2003;101:363–369. doi: 10.1182/blood-2002-05-1365. [DOI] [PubMed] [Google Scholar]
- 19.Eshel R, Vainas O, Shpringer M, Naparstek E. Highly sensitive patient-specific real-time PCR SNP assay for chimerism monitoring after allogeneic stem cell transplantation. Lab Hematol. 2006;12:39–46. [PubMed] [Google Scholar]
- 20.Pindolia K, Janakiraman N, Kasten-Sportes C, et al. Enhanced assessment of allogeneic bone marrow transplant engraftment using automated fluorescent-based typing. Bone Marrow Transplant. 1999;24:1235–1241. doi: 10.1038/sj.bmt.1702061. [DOI] [PubMed] [Google Scholar]
- 21.Maas F, Schaap N, Kolen S, et al. Quantification of donor and recipient hemopoietic cells by real-time PCR of single nucleotide polymorphisms. Leukemia. 2003;17:621–629. doi: 10.1038/sj.leu.2402856. [DOI] [PubMed] [Google Scholar]
- 22.Bai L, Deng YM, Dodds AJ, et al. A SYBR green-based real-time PCR method for detection of haemopoietic chimerism in allogeneic haemopoietic stem cell transplant recipients. Eur J Haematol. 2006;77:425–431. doi: 10.1111/j.1600-0609.2006.00729.x. [DOI] [PubMed] [Google Scholar]
- 23.Hill RS, Petersen FB, Storb R, et al. Mixed hematologic chimerism after allogeneic marrow transplantation for severe aplastic anemia is associated with a higher risk of graft rejection and a lessened incidence of acute graft-versus-host disease. Blood. 1986;67:811–816. [PubMed] [Google Scholar]
- 24.Mackinnon S, Barnett L, Heller G, O'Reilly RJ. Minimal residual disease is more common in patients who have mixed T-cell chimerism after bone marrow transplantation for chronic myelogenous leukemia. Blood. 1994;83:3409–3416. [PubMed] [Google Scholar]
- 25.Roy DC, Tantravahi R, Murray C, et al. Natural history of mixed chimerism after bone marrow transplantation with CD6-depleted allogeneic marrow: a stable equilibrium. Blood. 1990;75:296–304. [PubMed] [Google Scholar]
- 26.Bertheas MF, Lafage M, Levy P, et al. Influence of mixed chimerism on the results of allogeneic bone marrow transplantation for leukemia. Blood. 1991;78:3103–3106. [PubMed] [Google Scholar]
- 27.Miura Y, Tanaka J, Toubai T, et al. Analysis of donor-type chimerism in lineage-specific cell populations after allogeneic myeloablative and non-myeloablative stem cell transplantation. Bone Marrow Transplant. 2006;37:837–843. doi: 10.1038/sj.bmt.1705352. [DOI] [PubMed] [Google Scholar]
- 28.van Leeuwen JE, van Tol MJ, Joosten AM, et al. Mixed T-lymphoid chimerism after allogeneic bone marrow transplantation for hematologic malignancies of children is not correlated with relapse. Blood. 1993;82:1921–1928. [PubMed] [Google Scholar]
- 29.Ortega M, Escudero T, Caballin MR, et al. Follow-up of chimerism in children with hematological diseases after allogeneic hematopoietic progenitor cell transplants. Bone Marrow Transplant. 1999;24:81–87. doi: 10.1038/sj.bmt.1701816. [DOI] [PubMed] [Google Scholar]
- 30.Bader P, Kreyenberg H, Hoelle W, et al. Increasing mixed chimerism is an important prognostic factor for unfavorable outcome in children with acute lymphoblastic leukemia after allogeneic stem-cell transplantation: possible role for pre-emptive immunotherapy? J Clin Oncol. 2004;22:1696–1705. doi: 10.1200/JCO.2004.05.198. [DOI] [PubMed] [Google Scholar]
- 31.Bader P, Kreyenberg H, Hoelle W, et al. Increasing mixed chimerism defines a high-risk group of childhood acute myelogenous leukemia patients after allogeneic stem cell transplantation where pre-emptive immunotherapy may be effective. Bone Marrow Transplant. 2004;33:815–821. doi: 10.1038/sj.bmt.1704444. [DOI] [PubMed] [Google Scholar]
- 32.van Leeuwen JE, van Tol MJ, Bodzinga BG, et al. Detection of mixed chimaerism in flow-sorted cell subpopulations by PCR-amplified VNTR markers after allogeneic bone marrow transplantation. Br J Haematol. 1991;79:218–225. doi: 10.1111/j.1365-2141.1991.tb04525.x. [DOI] [PubMed] [Google Scholar]
- 33.Mattsson J, Uzunel M, Tammik L, et al. Leukemia lineage-specific chimerism analysis is a sensitive predictor of relapse in patients with acute myeloid leukemia and myelodysplastic syndrome after allogeneic stem cell transplantation. Leukemia. 2001;15:1976–1985. doi: 10.1038/sj.leu.2402311. [DOI] [PubMed] [Google Scholar]
- 34.Bornhauser M, Oelschlaegel U, Platzbecker U, et al. Monitoring of donor chimerism in sorted CD34+ peripheral blood cells allows the sensitive detection of imminent relapse after allogeneic stem cell transplantation. Haematologica. 2009;94:1613–1617. doi: 10.3324/haematol.2009.007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willasch A, Schneider G, Reincke BS, et al. Sequence polymorphism systems for quantitative real-time polymerase chain reaction to characterize hematopoietic chimerism-high informativity and sensitivity as well as excellent reproducibility and precision of measurement. Lab Hematol. 2007;13:73–84. [PubMed] [Google Scholar]
- 36.Baer MR, Stewart CC, Dodge RK, et al. High frequency of immunophenotype changes in acute myeloid leukemia at relapse: implications for residual disease detection (Cancer and Leukemia Group B Study 8361) Blood. 2001;97:3574–3580. doi: 10.1182/blood.v97.11.3574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data