Summary
Background
The D antigen is highly immunogenic, requiring only a small quantity of transfused red blood cells (RBCs) to cause alloimmunization in D– immunocompetent recipients. DEL was reported arousing alloimmunization to true Rh– patients. Molecular studies of the RHD gene have revealed that DEL individuals retain a grossly intact RHD gene or have a portion of RHD in their genomes. Avoiding immunization with clinically important antibodies is a primary objective in transfusion medicine.
Methods
In order to determine whether pregnant DEL women carrying an RhD+ fetus are at risk of anti-D alloimmunization, 808 Rh– pregnant women with a history of gestations or parturitions who regularly visited hospitals for their prenatal anti-D screening and postpartum care from January 2011 to December 2012 were investigated. Samples were analyzed for DEL by PCR with specific primers, PCR-sequence-specific primers (PCR-SSP), reverse transcription-PCR (RT-PCR), PCRrestriction fragment length polymorphism (PCR-RFLP), and by gene sequencing to characterize different alleles.
Results
Among the 808 Rh– pregnant women of our sample, 178 (22.0%) were typed as DEL; 168 DEL samples were confirmed to have the RHD (1,227 G>A) allele, 8 DEL samples were characterized by one base mutation of the RHD (3G >A) allele, and the remaining two DEL samples were determined to carry RHD-CE(4–9)-D or RHD-CE(2–5)-D. The observation of allo-anti-D in two prominent D epitope loss cases confirmed the partial nature of these DEL phenotypes.
Conclusions
In conclusion, evidence is provided that different DEL genotypes code either for partial or complete D antigen expression. It is suggested that the use of RhD+ RBCs in complete D antigen DEL patients does not induce adverse reaction.
Keywords: DEL variant, Pregnant women, Complete DEL, Partial DEL, Alloimmunization
Introduction
The risk of anti-D alloimmunization in D– healthy volunteers who received D+ red blood cells (RBCs) is higher than 80% [1, 2, 3, 4, 5, 6]. According to national transfusion guidelines from many countries, D– patients should receive RBCs from D– donors. Unfortunately, in some situations, such as massive transfusion or a shortage of D– RBCs, because Rh– persons represent about 0.3% in China, the transfusion of D+ RBCs to D– patients may be required. However, after transfusion of D+ RBCs to D– patients the anti-D alloimmunization is frequent [1]. Accordingly, a new strategy should be developed in order to optimize their use in China.
Routine serologic typing does not discriminate DEL from the D– phenotype, DEL phenotypes were determined on the basis of no agglutination in the indirect antiglobulin test (IAT) procedure and positive results with adsorption-elution techniques [7, 8]. The DEL phenotype arises from a mutation of the RHD gene, and more than 20 different DEL alleles have been described [4]. Molecular studies have indicated that DEL individuals retain a grossly intact D gene or a partial DEL with detectable D epitope (epD) loss [9, 10, 11, 12, 13]. D and CE polypeptides are coded for by the highly homologous RHD and RHCE genes. Partial D variants exhibit some degree of antigenic alteration derived from RHD/RHCE gene hybridization events, in most instances accompanied by a reduction in antigen density [14]. Amino acid substitutions in extracellular stretches of the D polypeptide lead to loss of one or more epDs, rendering also partial D individuals prone to anti-D alloimmunization upon contact with normal D+ RBCs expressing the complete set of epDs [14, 15]. These reports support the possibility that a partial DEL phenotype could theoretically induce anti-D alloimmunization [9, 11, 14]. The antibody is also capable of causing severe hemolytic disease of the fetus and newborn (HDFN) in sensitized pregnant DEL women carrying an RhD+ fetus [9]. A partial D-like epitope loss has been reported in DEL phenotypes to be associated with the RHD (IVS3 + 1G>A) allele, and individuals with this phenotype can make anti-D [11]. Although these anti-D antibodies may indicate a secondary immune response, a rapid primary anti-D immunization attributed to this DEL phenotype was still possible. Our study explored whether pregnant DEL women carrying an RhD+ fetus are at risk of anti-D alloimmunization.
Material and Methods
Study Population
The present study was conducted between January 2011 and December 2012 at the Department of Blood Transfusion of Affiliated Province Hospital of Anhui Medical University, the Department of Blood Transfusion of The First Affiliated Hospital of Anhui Medical University, and the Department of Blood Transfusion of The Third Affiliated Hospital of Anhui Medical University. The three hospitals are located in the capital cities of Anhui province and are the largest tertiary hospitals in Anhui, China. Medical services provided by the three hospitals laboratories include antenatal blood group and antibody screening on more than 150,000 pregnant women per year.
Inclusion Criteria
Only pregnant DEL women carrying Rh+ fetus who declared a history of gestations or parturitions were included. The disease can become progressively more severe in subsequent pregnancies where the fetus is D+. Pregnant women with autoantibodies, a history of blood transfusion, and receiving Rh immune globulin prophylaxis were excluded from the study groups, as were women who were alloimmunized to other blood group antigens. Our study was approved by the Scientific and Ethics Committee of Anhui Medical University. Informed consent was obtained from all participants.
Rh Phenotyping
During a 2-year study period, a total of 313,250 EDTA-anticoagulated blood samples from pregnant women were collected in the three hospitals. The D antigen was determined by direct agglutination with the monoclonal anti-D in saline according to the manufacturers’ instructions. Samples that were negative to anti-D in the direc t agglutination were retested by using the indirect antiglobulin test (IAT). Subsequently, all samples typed as D– with the IAT were retested for the DEL phenotype through an adsorption and elution test in tubes. Briefly, RBCs were incubated with an equal volume of polyclonal anti-D at 37 ° C for 1 h. The cells were then washed thoroughly. An eluate was prepared by using a heat elution technique. The eluates and final supernatants were used for the IATs. If the result was positive (≥1+), the sample was marked as DEL. RhC/c and E/e were determined by using monoclonal anti-C,c,E,e reagents (Gamma, Houston, TX, USA) according to the manufacturer's instructions.
Molecular Studies
Genomic DNA Extraction
Genomic DNA of DEL samples was extracted from 200 µl of EDTA-antico-agulated peripheral blood samples according to the manufacturer's recommendations (QIAamp DNA Blood Mini Kit, Qiagen GmbH, Hilden, Germany). To minimize the risk of contamination, DNA was isolated under laminar airflow, and aerosol-resistant tips were used. The optical density (OD) of the purified DNA was measured using an ultraviolet spectrophotometer. The DNA was extracted once from each sample and then stored at −20 ° C until further processing within 48 h.
RHD Genotyping
Amplification of RHD Exons
We devised RHD-specific primers based on intron sequences to amplify genomic DNA for 10 RHD exons 1–10. PCR was performed using a commercially available system (Expand High Fidelity Polymerase; Roche Applied Science, Basel, Switzerland). Cycling conditions for RHD exon 1 and 10 were denaturation at 95 °C for 40 s, annealing for 40 s at 64 ° C (exons 1–4, 6, 8–10), at 60 ° C (exons 5, 7) and at 57 °C (β-actin), and extension at 72 ° C for 1 min. The final extension was done at 72 °C for 10 min. Amplified DNA products were visualized by electrophoresis in a 1.5% agarose gel with ethidium bromide staining and photographed under UV light. β-actin gene was used as an internal control.
DNA Sequencing
The complete RHD exons 1–10 including adjacent intron regions were sequenced from PCR products using the respective PCR primers with an ABI PRISM 3730 automated sequencer (Applied Biosystems®; Life Technologies Carlsbad, CA, USA). The nucleotide and deduced amino acid sequences were analyzed and compared with the published sequences. D specificities of primers used for exon amplification and sequencing were identical and are listed in table 1.
Table 1.
Primers used for PCR and DNA sequencing
| Primer name* | Sequence 5′ to 3′ | Specificity | Genomic region | Position§ | Product size (bp) |
|---|---|---|---|---|---|
| E1-s (=E1-seq) | TCCATAGAGAGGCCAGCACAA | D | promoter | −152 to −132 | |
| E1-a | GCTATTTGCTCCTGTGACCACTT | D | intron 1 | 40–18 | 340 |
| E2-s | TGACGAGTGAAACTCTATCTCGAT | D | intron 1 | −1,064 to −1,041 | |
| E2-a (=E2-seq) | GGCATGTCTATTTCTCTCTGTCTAAC | D/CE | intron 2 | 355–330 | 1,606 |
| E3-s | GTCGTCCTGGCTCTCCCTCTCT | D | intron 2 | −29 to −8 | |
| E3-a (=E3-seq) | CTTTTCTCCCAGGTCCCTCCT | D/CE | intron 3 | 39–19 | 219 |
| E4-s | GCCGACACTCACTGCTCTTAC | D/CE | intron 3 | −36 to −16 | |
| E4-a (=E4-seq) | TGAACCTGCTCTGTGAAGTGC | D | intron 4 | 194–174 | 378 |
| E5-s | CTGCCAAAGCCTCTACCCG | D | intron 4 | −502 to −484 | |
| E5-a (=E5-seq) | GCTGACTCTCGCTCATGGT | D/CE | intron 5 | 315–297 | 984 |
| E6-s (=E6-seq) | CAGGGTTGCCTTGTTCCCA | D/CE | intron 5 | −95 to −77 | |
| E6-a | CTTCAGCCAAAGCAGAGGAGG | D | intron 6 | 41–21 | 274 |
| E7-s (=E7-seq) | CTACTCATAGTGTGGTCCGTAGACC | D | intron 6 | −280 to −256 | |
| E7-a | CAAATATTCACCGAAGCCTACTG | D/CE | intron 7 | 129–107 | 543 |
| E8-s | GGTCAGGAGTTCGAGATCAC | D | intron 7 | −594 to −575 | |
| E8-a (=E8-seq) | GATGGGGCACATAGACATCC | D/CE | intron 8 | 97–78 | 771 |
| E9-s (=E9-seq) | GGTCCAGGAATGACAGGGCT | D | intron 8 | −162 to −143 | |
| E9-a | CGCTGAGGACTGCAGATAGG | D | intron 9 | 294–275 | 530 |
| E10-s (=E10-seq) | CAAGAGATCAAGCCAAAATCAGT | D/CE | intron 9 | −67 to −45 | |
| E10-a | AGCTTACTGGATGACCACCA | D | 3UTR | 290–271 | 382 |
| β-actin-s | GGAAATCGTGCGTGACATT | – | – | – | |
| β-actin-a | CGTCATACTCCTGCTTGCTG | – | – | – | 473 |
s = Sense primer; a = antisense primer; seq = sequencing primer.
The positions of the synthetic oligonucleotides are indicated relative to their distances from the first nucleotide position of the start codon ATG for all primers in the promoter and in the exons or relative to their adjacent exon-intron boundaries for all other primers.
PCR-SSP and cDNA Sequencing to Determine RHD-CE-D Hybrid Alleles
For RHD and RHCE genotyping, testing for different partial and weak D variants and determination of the RHD zygosity of investigated and control blood samples, a PCR-SSP was performed with commercially available typing kits (CDE, weak D, RHd, Inno-Train). Kit ‘CDE’ is capable of properly identifying RHD-CE-D hybrid alleles by detecting RHD-specific DNA sequences in the 5′-untranslated region and exons 2, 3, 4, 5, 6, 7, 9, and 10 of RHD and RHC, RHc (intron1 and exon 2) and RHE, RHe (exon 5) of RHCE as described previously [16].
Total RNA was isolated with reagent (Trizol; InvitrogenTM, Life Technologies). RNA integrity and quantity was assessed by measuring the OD at 260 and 280 nm with the NANO DROP 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA). Reverse transcription with random hexamer primers was performed with the PrimeScriptTM RT Master Mix kit (Takala, Dalian, China) following manufacturer's instructions. Amplification of cDNA was performed on a thermocycler T Gradient (Biometra, Göttingen, Germany) with Primer a 5′-CACAGGATGAGCTCTAAGTAC-3’ located at the 5’ end of RHD/RHCE and Primer b 5′-TAAATGGTGAGATTCTCCTC-3’ located in RHD 3’ noncoding sequence or with Primer c 5′-CAAATCTGTCTCTGACCTTGTTTC-3’ located in RHCE 3’ noncoding sequence. Samples were submitted to an initial denaturation step (5 min at 94 °C), followed by 35 amplification cycles (45 s at 94 ° C; 45 s at 50 ° C; 1 min at 72 ° C) and a final elongation step of 10 min at 72 ° C. PCR products were sequenced by a genetic analyzer. Results were compared with the published sequences.
PCR-RFLP for RHD Zygosity Determination
According to the method published by Wagner and Flegel [17], a PCR amplification was performed by using the expand high-fidelity PCR system with primers rez7 (consensus, 5’ of the Rh box identity region) and rnb31 (specific for downstream of the Rh box, 3’ of the Rh box identity region). PCR products were digested with PstI for 3 h at 37 °C, and the fragments were resolved by electrophoresis on a 1% agarose gel.
Lookback
The antibody screening in the pregnant DEL women was performed using fully automated Johnson gel technology using erythrocyte screen cells (Shanghai Blood Center, Shanghai, China). Erythrocyte panels were performed to confirm allo-anti-D specificity (Immucor Inc, Norcross, GA, USA, or Sanquin Reagents, Amsterdam, the Netherlands). After confirmation of the maternal allo-anti-D, routine serological typing of her husband and the newborn was made. The newborn's total bilirubin concentration and reticulocyte count was measured, and an anti-D elution test and a direct antiglobulin test of cord blood were done.
Results
Serologic Studies
In this prospective study, the 808 blood samples of D– pregnant women reacted negatively with the IgM monoclonal anti-D in saline. Using the IAT and the adsorption and elution test, a total of 178 DEL samples was found. Their phenotypes of RhC/c and E/e were displayed in table 2.
Table 2.
The phenotypes of RhC/c and E/e in the 808 pregnant women samples
| Rh phenotypes | RhC/c and E/e phenotype |
|||||||
|---|---|---|---|---|---|---|---|---|
| Ccee | CCee | CCEe | CcEe | ccEe | ccEE | ccee | total | |
| Apparent D-* | 280 | 31 | 7 | 21 | 32 | 9 | 428 | 808 |
| DEL | 157 | 11 | 6 | 2 | 2 | 0 | 0 | 178 |
The apparent D negative phenotypes were determined by a microplate test.
Molecular Characterization of DELs
A total of 168 samples were determined by sequencing to carry the RHD 1227G>A allele, and the variant shows a silent nucleotide change at the exon boundary. Eight DEL samples were characterized by one base mutation of the RHD (3G>A) allele, codon 1–3 were changed from ATG to ATA. The remaining two DEL samples seemed to have RHD-CE-D hybrid alleles. According to RHD/CE PCR-SSP, one RHD-CE(4–9)-D and one RHD-CE(2–5)-D for RHD-CE-D hybrid alleles were identified as judged by testing for the RHD 5′-untranslated region and exons 2, 3, 4, 5, 6, 7, 9, and 10.
The 260/280 nm ratios of the samples were >1.8. Sample purity was confirmed by electrophoresis on an agarose gel. All samples contained 18S and 28S ribosomal RNA peaks with no visible degradation products (fig. 1A). The results of RT-PCR are shown in figure 1B. Nucleotide sequencing from their cDNA was performed to show that the two RHD-CE-D hybrid alleles did not harbor any additional variation. No novel allele was found in the retrospective study. Frequencies and respective 95% confidence intervals (95% CIs) for the alleles encountered in this study are given in table 3.
Fig. 1.
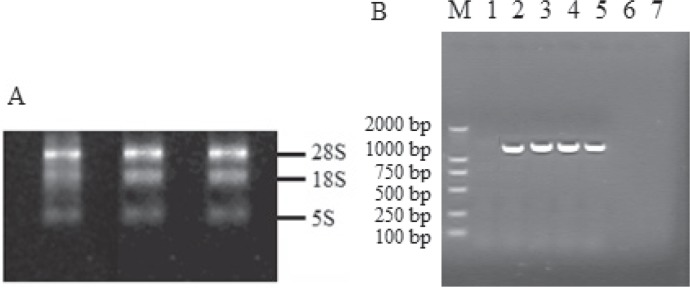
A The result of RNA electrophoresis. B The results of RT-PCR for the RHD-CE-D hybrid alleles. M: molecular marker; Lane 1: water control; Lane 2: amplicon of RHCE+ control with primer a and RHCE-specific reverse primer c (band of 1399 bp); Lane 3: amplicon of RHD+ control with primer a and RHD-specific reverse primer b (band of 1,446 bp); Lane 4–5: amplicons for RHD-CE(4–9)-D and RHD-CE(2–5)-D with primer a and RHD-specific reverse primer b (band of 1,446 bp); Lane 6–7: amplicons were lacking for RHD-CE(4–9)-D and RHD-CE(2–5)-D with primer a and RHCE-specific reverse primer c.
Table 3.
Estimated DEL allele frequencies and 95% CIs in 808 D– pregnant women
| Alleles observed | n | Estimate | 95% CI |
|
|---|---|---|---|---|
| lower | upper | |||
| RHD (1,227G>A) | 168 | 1:5 | 1:6 | 1:4 |
| RHD (3G>A) | 8 | 1:101 | 1:233 | 1:52 |
| RHD-CE(4–9)-D | 1 | 1:808 | 1:31,949 | 1:145 |
| RHD-CE(2–5)-D | 1 | 1:808 | 1:31,949 | 1:145 |
RHD Zygosity Determination
All 178 DEL samples presented with Rh box zygosity. 19 samples, including 16 samples with RHD (1,227G>A) allele and 3 samples carrying RHD (3G>A), were found to be RHD+/RHD+ homozygotes by PCR-RFLP analysis. The rest of the 159 samples including 2 samples with the RHD-CE-D hybrid allele, 5 samples with RHD (3G>A), 152 samples carrying RHD (1,227G>A) allele were RHD+/RHD– hemizygous. The associated serological data are described in table 4.
Table 4.
Results of PCR-RFLP analyses of the 178 DEL population
| DEL allele | RHD zygosity | RHCE haplotypes | Number of subjects |
|---|---|---|---|
| RHD (1227G>A) | 16 RHD+/RHD+ | Ccee | 12 |
| CCee | 2 | ||
| CCEe | 1 | ||
| CcEe | 1 | ||
| 152 RHD+/RHD- | Ccee | 138 | |
| CCee | 8 | ||
| CCEe | 3 | ||
| ccEe | 2 | ||
| CcEe | 1 | ||
| RHD (3G>A) | 3 RHD+/RHD+ | Ccee | 2 |
| CCEe | 1 | ||
| 5 RHD+/RHD- | Ccee | 3 | |
| CCee | 1 | ||
| CCEe | 1 | ||
| RHD-CE(4–9)-D | 1 RHD+/RHD- | Ccee | 1 |
| RHD-CE(2–5)-D | 1 RHD+/RHD- | Ccee | 1 |
| Total | 178 | ||
Complete DEL Is Not at Risk of Anti-D
There are 56 ethnic groups in China. More than 90% of the population is Han ethnicity; the rest of the population is composed of 55 minority groups. Of all 178 pregnant DEL women in this prospective study, 70 women were excluded from the study group (table 5) because they did not meet the above mentioned inclusion criteria. All 108 pregnant women were Han Chinese aged 21–41 years at the time of testing. In this cohort, 101 samples (93.5%) showed a RHD (1,227G>A) mutation, 5 samples (4.6%) were determined to carry RHD (3G>A) allele in RHD exon 1, and the remaining 2 samples (1.9%) showed the characteristics of the RHD-CE(4–9)-D or RHD-CE(2–5)-D hybrid gene. The 2 women carrying the RHD-CE-D hybrid developed allo-anti-D, and their children developed mild hemolytic disease of the newborn (table 6).
Table 5.
70 pregnant women were excluded from this study
| Exclusion criteria | Han | Tibetan | Uigur | Hui | Mongol | Sum |
|---|---|---|---|---|---|---|
| A history of blood transfusion | 6 | 1 | 1 | 8 | ||
| Mother receiving RhIG prophylaxis | 21 | 1 | 22 | |||
| If mother carries apparent D– fetus | 2 | 1 | 3 | |||
| Mother with autoantibodies | 2 | 1 | 3 | |||
| Mother with other blood group antibodies | 4 | 4 | ||||
| No a history of gestations or parturitions | 27 | 27 | ||||
| Non-Chinese Han population | 1 | 1 | 1 | 3 | ||
| Total | 62 | 2 | 1 | 2 | 3 | 70 |
Table 6.
Anti-D alloimmunization among two RHD-CE-D hybrid allele pregnant women
| Pregnant women | Newborn | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| DEL allele | n | ABO | zygosity* | anti-D titer | TBIL#, μmol/l | Ret countΔ, % | ABO | DAT | anti-A/B |
| RHD-CE(4-9)-D | 1a | A | RHD+/RHD- | 256 | 185 | 8.88 | AB | + | – |
| RHD-CE(2-5)-D | 1 | O | RHD+/RHD- | 128 | 190 | 8.04 | O | + | – |
Presence (+) or absence (–) of the RHD gene.
TBIL concentration 24 h after delivery.
Ret count 7 h after delivery.
A 26-year-old Chinese Han woman carrying RHD-CE(4–9)-D allele. Routine serotyping demonstrated an A phenotype and genotyping showed hemizygous. Her husband was group B. She was found to be sensitized to the D antigen, apparently after a previous first-trimester miscarriage, with an anti-D titer 256 using an IAT. More detailed maternal anti-D did not react with self RBCs, excluding an autoantibody. A live female infant was delivered at 39 weeks of gestation with a birth weight of 3500 g, the newborn was not anemic. However, the newborn TBIL concentration 24 h after delivery was 185 μmol/l. The reticulocyte count was 8.88% at 7 h after delivery and decreased to 2.44% by 120 h. The infant's phenotype was AB, D+C+c+E–e+. The D antigen was detected with immunoglobulin M anti-D in an immediate-spin tube test indicating normal levels of expression. Cord blood serology showed a positive direct antiglobulin test, and anti-D was eluted from the cord RBCs. It was noted that the infant was AB while the mother was group A; however, anti-B was excluded as the cause of HDFN because anti-B was not detected in the eluate from the cord RBCs. The newborn's hyperbilirubinemia was attributed to maternal anti-D reacting against the infant's D+ RBCs.
Prospective Transfusion Policy of DEL Patients in Chinese Han
Indications for mandatory use of D– RBCs: DEL patients with anti-D antibody or carrying partial DEL allele. Indications for recommended use of D– RBCs: women of childbearing age carrying a complete DEL allele, under-age children (<18 years). Indications for acceptable use of D+ RBCs: massive transfusion patients, adult men (>18 years) and women of non-childbearing age with no detectable anti-D antibody in case of insufficient D– RBCs in stock.
Discussion
If RBC units of D+ were transfused to DEL recipients, the clinical consequences are of current interest in China. Molecular studies have shown that a heterogeneous array of variant RHD alleles can result in the DEL phenotype [10, 18, 19, 20, 21, 22, 23]. DEL derives from several mechanisms, including splice-site mutation, missense mutation, RHD-CE-D hybrid, frame shift mutation, and a long deletion of the RHD gene [14, 24, 25, 26, 27, 28]. RHD (IVS3 + 1G>A) is a DEL allele that has only been observed in Caucasians. RHD (1,227G>A) and RHD (IVS3 + 1G>A) belong to splice-site mutation type [11, 12]. RHD (M295I, 885G>T) is a missense mutation, RHD (X418 L) is frame shift mutation [12, 29]. RHD (del Ex8) and RHD (del Ex9) alleles are classified as long deletions of the RHD gene [26, 28], and RHD (del Ex9) is now determined to be nonexistent but resulted from a misinterpretation of data on RHD (1,227G>A) [12, 27]. RHD-CE(4–9)-D belongs to RHD-CE-D hybrid gene [14, 30, 31, 32]. Körmöczi and colleagues [9] proposed that DEL phenotypes should be divided into two subtypes: complete types where the majority of epDs are conserved such as RHD (1,227G>A) and partial D-like variants with characteristic epD loss caused by either RHD-CE-D hybrid genes or RHD point mutations RHD (IVS3 + 1G>A) affecting extracellular RhD loops. The distinction between complete and partial D is of clinical importance, because only partial D individuals exhibit a tendency to anti-D alloimmunization [9, 11, 14].
So far, no report was available that pregnant women with RHD-CE-D hybrid DEL genes carrying D+ fetus developed allo-anti-D. In this prospective study, the two pregnant women who were typed as DEL carrying RHD-CE(4–9)-D and RHD-CE(2–5)-D had anti-D alloimmunization. One might speculate that a limited variety of antibody epitope specificity in the two variants is associated with a lower propensity for hemolytic activity because the HDFN was rather mild in relation to the anti-D titer in the two cases reported here. Anti-D alloimmunization in DEL individuals may in fact not be a very rare case; a predisposition to allo-anti-D formation requires epD loss and may therefore likely be partial DEL type-specific [9]. However, this speculation requires confirmation by more case reports because the correlation between anti-D titer and severity of HDFN is weak. Anti-D often persists for a long time and may even be found several decades after initial formation. Whether the two pregnant DEL women elicited primary immunization or secondary immune response is not yet proven as an unnoticed and uncompleted D+ pregnancy could not be excluded as the primary immunization event. In particular, the RHD+/RHD– hemizygote has the potential of stimulating anti-D at least in secondary immunizations. A few instances of primary immunization have also been reported. In the two anti-D cases, a positive direct antiglobulin test was detected in the newborns. Of these one received small-volume RBC transfusions and phototherapy, the other one was treated only with phototherapy. The RHD (1,227G>A) DEL phenotypes type may not be prone to anti-D alloimmunization after exposure to ‘regular’ D+ RBCs [10, 33]. Shao et al. [10] reported no anti-D isoimmunization among 44 women with RHD (1,227G>A) who had not received prophylactic anti-D, providing supportive evidence that the epDs are conserved. So we propose to identify patients carrying RHD (1,227G>A) and RHD (3G>A) DEL alleles as these patients can receive Rh+ units if necessary. Likewise, pregnant women carrying complete DEL variants RHD (3G>A) and RHD (1,227G>A) might not be stimulated to make allo-anti-D in response to an Rh+ fetus. Wagner et al. [15] found a new RHD allele in a lookback initiated after an unexplained anti-D immunization event. The DEL phenotype expressed by this RHD (IVS5–38del4) allele elicited a rapid anti-D immune response. However, Von Zabern et al. [34] reported four samples carrying RHD (IVS5–38del4) that does not cause a DEL phenotype; the deletion has no influence on D antigen expression at all, and an association of this 4 bp deletion with the DEL phenotype was excluded.
Our prior policy concerned the use of D– RBCs in DEL recipients and insufficient D– RBCs in stock. The implementation of the new policy will result in a significant change in the use of D– RBCs. The new policy will reduce the consumption of D– RBCs by about 1,560 units/year in the three hospitals. The number of units saved is significant given the demand to obtain Rh– blood for transfusion recipients who are truly at risk of making allo-anti-D. The DEL variant is linked to the C antigen, which permits the rapid exclusion of the DEL variant during recipient pre-transfusion testing. In those apparent Rh–/C+ individuals, the RHD (1,227 G>A) and RHD (3G>A) alleles, which were the two most frequent DEL alleles in Chinese Han ethnicity, could be detected by PCR-SSP simply and precisely.
Anti-D in pregnant women with the partial DEL phenotype has caused HDFN, but there are no universally accepted guidelines for Rh immunoprophylaxis in DEL variant women. The clinical management of patients carrying DEL phenotypes is based on the predicted clinical relevance: it is recommended that patients carrying complete DEL types could be transfused with D+ blood. Likewise, pregnant women should not receive anti-D prophylaxis (RhIg) if they carry one of these complete DEL types. In contrast, patients and pregnant women who carry one of the partial DEL types should receive transfusion of D– blood or receive RhIg or both.
To the best of our knowledge this is the first paper in which a transfusion policy concerning the use of D+ RBCs in DEL patients is prospectively evaluated. In conclusion, our study shows that the use of D+ RBCs in selected DEL patients carrying complete epDs is justified and may be highly beneficial for institutions.
Disclosure Statement
The authors declare that they have no conflict of interest in the subject matter of the manuscript. The authors did not receive any financial support in addition to the project grant mentioned.
Acknowledgment
We thank Dr. B.-L. Wang for his excellent technical and financial assistance and valuable advice during the preparation of this manuscript. This work was supported by the National Natural Science Foundation of the People's Republic of China (no. 30972815).
References
- 1.Gonzalez-Porras JR, Graciani IF, Perez-Simon JA, Martin-Sanchez J, Encinas C, Conde MP, Nieto MJ, Corral M. Prospective evaluation of a transfusion policy of D+ red blood cells into D– patients. Transfusion. 2008;48:1318–1324. doi: 10.1111/j.1537-2995.2008.01700.x. [DOI] [PubMed] [Google Scholar]
- 2.Cook K, Rush B. Rh(D) immunization after massive transfusion of Rh(D)-positive blood. Med J Aust. 1974;1:166–168. doi: 10.5694/j.1326-5377.1974.tb50783.x. [DOI] [PubMed] [Google Scholar]
- 3.Gunson HH, Stratton F, Phillips PK. The use of modified cells to induce an anti-Rh response. Br J Haematol. 1971;21:683–694. doi: 10.1111/j.1365-2141.1971.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 4.Urbaniak SJ, Robertson AE. A successful program of immunizing Rh-negative male volunteers for anti-D production using frozen/thawed blood. Transfusion. 1981;21:64–69. doi: 10.1046/j.1537-2995.1981.21181127486.x. [DOI] [PubMed] [Google Scholar]
- 5.Keith L, Berger GS, Pollack W. The transfusion of Rh-positive blood into Rh-negative women. Am J Obstet Gynecol. 1976;125:502–506. doi: 10.1016/0002-9378(76)90365-3. [DOI] [PubMed] [Google Scholar]
- 6.Pollack W, Ascari WQ, Crispen JF, O'Connor RR, Ho TY. Studies on Rh prophylaxis. II. Rh immune prophylaxis after transfusion with Rh-positive blood. Transfusion. 1971;11:340–344. doi: 10.1111/j.1537-2995.1971.tb04425.x. [DOI] [PubMed] [Google Scholar]
- 7.Krog GR, Clausen FB, Berkowicz A, Jørgensen L, Rieneck K, Nielsen LK, Dziegiel MH. Is current serologic RhD typing of blood donors sufficient for avoiding immunization of recipients? Transfusion. 2011;51:2278–2285. doi: 10.1111/j.1537-2995.2011.03156.x. [DOI] [PubMed] [Google Scholar]
- 8.Flegel WA, von Zabern I, Wagner FF. Six years’ experience performing RHD genotyping to confirm D– red blood cell units in Germany for preventing anti-D immunizations. Transfusion. 2009;49:465–471. doi: 10.1111/j.1537-2995.2008.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Körmöczi GF, Gassner C, Shao CP, Uchikawa M, Legler TJ. A comprehensive analysis of DEL types: partial DEL individuals are prone to anti-D alloimmunization. Transfusion. 2005;45:1561–1567. doi: 10.1111/j.1537-2995.2005.00584.x. [DOI] [PubMed] [Google Scholar]
- 10.Shao CP, Xu H, Xu Q, Sun GD, Li JP, Zhang BW, Liang XH, Liu Z, Zhou Y, Li D, Zhuang NB. Antenatal Rh prophylaxis is unnecessary for ‘Asia type’ DEL women. Transfus Clin Biol. 2010;17:260–264. doi: 10.1016/j.tracli.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Gardener GJ, Legler TJ, Hyett JA, Liew YW, Flower RL, Hyland CA. Anti-D in pregnant women with the RHD (IVS3 + 1G>A) associated DEL phenotype. Transfusion. 2012;52:2016–2019. doi: 10.1111/j.1537-2995.2011.03538.x. [DOI] [PubMed] [Google Scholar]
- 12.Li Q, Hou L, Guo ZH, Ye LY, Yue DQ, Zhu ZY. Molecular basis of the RHD gene in blood donors with DEL phenotypes in Shanghai. Vox Sang. 2009;97:139–146. doi: 10.1111/j.1423-0410.2009.01181.x. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Ye LY, Guo ZH, Zhang YX, Wang LL, Zhu ZY. Molecular basis of D variants between Uigur and Han blood donors in Xinjiang. Transfus Med. 2008;18:199–203. doi: 10.1111/j.1365-3148.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- 14.Ouchari M, Jemni Yacoub S, Houissa B, Abdelkefi S, Chakroun T, Bouslama M, Jerray I, Belhedi S, Hmida S. System RH screening of partials D with RHD/RHCE hybrid gene. Transfus Clin Biol. 2013;20:35–39. doi: 10.1016/j.tracli.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Wagner T, Körmöczi GF, Buchta C, Vadon M, Lanzer G, Mayr WR, Legler TJ. Anti-D immunization by DEL red blood cells. Transfusion. 2005;45:520–526. doi: 10.1111/j.0041-1132.2005.04256.x. [DOI] [PubMed] [Google Scholar]
- 16.Körmöczi GF, Förstemann E, Gabriel C, Mayr WR, Schönitzer D, Gassner C. Novel weak D types 31 and 32: adsorption-elution-supported D antigen analysis and comparison to prevalent weak D types. Transfusion. 2005;45:1574–1580. doi: 10.1111/j.1537-2995.2005.00580.x. [DOI] [PubMed] [Google Scholar]
- 17.Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood. 2000;95:3662–3668. [PubMed] [Google Scholar]
- 18.Shao CP, Xiong W, Zhou YY. Multiple isoforms excluding normal RhD mRNA detected in Rh blood group Del phenotype with RHD 1227A allele. Transfus Apher Sci. 2006;34:145–152. doi: 10.1016/j.transci.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Ye L, Yue D, Wo D, Ding X, Guo S, Li Q, Guo Z, Zhu Z. Molecular bases of unexpressed RHD alleles in Chinese D– persons. Transfusion. 2009;49:1655–1660. doi: 10.1111/j.1537-2995.2009.02181.x. [DOI] [PubMed] [Google Scholar]
- 20.Ye LY, Guo ZH, Li Q, Zhu ZY. Molecular and family analyses revealed two novel RHD alleles in a survey of a Chinese RhD-negative population. Vox Sang. 2007;92:242–246. doi: 10.1111/j.1423-0410.2006.00886.x. [DOI] [PubMed] [Google Scholar]
- 21.Ye SH, Wu DZ, Wang MN, Wu XY, Xu HG, Xu H, Shao CP. A comprehensive investigation of RHD polymorphisms in the Chinese Han population in Xi'an. Blood Transfus. 2013;11:1–9. doi: 10.2450/2013.0121-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silvy M, Simon S, Gouvitsos J, Di Cristofaro J, Ferrera V, Chiaroni J, Bailly P. Weak D and DEL alleles detected by routine SNaPshot genotyping: identification of four novel RHDalleles. Transfusion. 2011;51:401–411. doi: 10.1111/j.1537-2995.2010.02830.x. [DOI] [PubMed] [Google Scholar]
- 23.Sun CF, Chou CS, Lai NC, Wang WT. RHD gene polymorphisms among RhD-negative Chinese in Taiwan. Vox Sang. 1998;75:52–57. [PubMed] [Google Scholar]
- 24.Shao CP, Maas JH, Su YQ, Köhler M, Legler TJ. Molecular background of Rh D-positive, D-negative, D(el) and weak D phenotypes in Chinese. Vox Sang. 2002;83:156–161. doi: 10.1046/j.1423-0410.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 25.Yan L, Wu J, Zhu F, Hong X, Xu X. Molecular basis of D variants in Chinese persons. Transfusion. 2007;47:471–477. doi: 10.1111/j.1537-2995.2006.01138.x. [DOI] [PubMed] [Google Scholar]
- 26.Richard M, Perreault J, Constanzo-Yanez J, Khalifé S, St-Louis M. A new DEL variant caused by exon 8 deletion. Transfusion. 2007;47:852–857. doi: 10.1111/j.1537-2995.2007.01199.x. [DOI] [PubMed] [Google Scholar]
- 27.Luettringhaus TA, Cho D, Ryang DW, Flegel WA. An easy RHD genotyping strategy for D-East Asian persons applied to Korean blood donors. Transfusion. 2006;46:2128–2137. doi: 10.1111/j.1537-2995.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- 28.Chang JG, Wang JC, Yang TY, Tsan KW, Shih MC, Peng CT, Tsai CH. Human Rh Del is caused by a deletion of 1013 bp between introns 8 and 9 including exon 9 of RHD gene. Blood. 1998;92:2602–2604. [PubMed] [Google Scholar]
- 29.Gassner C, Doescher A, Drnovsek TD, Rozman P, Eicher NI, Legler TJ, Lukin S, Garritsen H, Kleinrath T, Egger B, Ehling R, Körmöczi GF, Kilga-Nogler S, Schoenitzer D, Petershofen EK. Presence of RHD in serologically D–, C/E+ individuals: a European multicenter study. Transfusion. 2005;45:527–538. doi: 10.1111/j.0041-1132.2004.04211.x. [DOI] [PubMed] [Google Scholar]
- 30.Szulman A, Nardozza LM, Barreto JA, Araujo Júnior E, Moron AF. Investigation of pseudogenes RHDψ and RHD-CE-D hybrid gene in D-negative blood donors by the real time PCR method. Transfus Apher Sci. 2012;47:289–293. doi: 10.1016/j.transci.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Gunel T, Kalelioglu I, Gedikbasi A, Ermis H, Aydinli K. Detection of fetal RHD pseudogene (RHDψ) and hybrid RHD-CE-Ds from RHD-negative pregnant women with a free DNA fetal kit. Genet Mol Res. 2011;10:2653–2657. doi: 10.4238/2011.October.26.1. [DOI] [PubMed] [Google Scholar]
- 32.Martin FO, de Menezes SS, Chiba AK, Langhi DM, Jr, Nardozza LM, Chiattone CS, Bordin JO. RHD gene polymorphisms in alloimmunized RhD-negative individuals with high rate of racial admixture. Transfus Apher Sci. 2013;48:113–116. doi: 10.1016/j.transci.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Shao CP. Transfusion of RhD-positive blood in ‘Asia type’ DEL recipients. N Engl J Med. 2010;362:472–473. doi: 10.1056/NEJMc0909552. [DOI] [PubMed] [Google Scholar]
- 34.von Zabern I, Flegel WA. IVS5–38del4 deletion in the RHD gene does not cause a DEL phenotype: relevance for RHD alleles including DFR-3. Transfusion. 2007;47:1552–1555. doi: 10.1111/j.1537-2995.2007.01353.x. [DOI] [PubMed] [Google Scholar]


