Abstract
The base composition of peanut (var. NC-17) DNA determined from thermal denaturation profiles showed an average guanine plus cystosine content of 34% which was in close approximation to 36% guanine plus cytosine calculated from the buoyant density. Buoyant density also indicated the absence of satellite DNA. The genome size, 2.0 × 109 base pairs, as determined by reassociation kinetics of the single copy DNA was close to the genome size determined by cytophotometry, 2.1 × 109 base pairs. Peanut DNA averaging 450 to 600 base pairs long, reassociated in phosphate buffer and fractionated by hydroxylapatite, indicated a DNA genome composition of 36% nonrepetitive or single copy DNA; reassociation in formamide and followed by optical methods indicated the repetitive DNA possesses highly repeated, intermediately repeated and rarely repeated components of DNA with DNA sequences repeated on the average about 38,000, 6,700, and 200 times each. Different criteria of reassociation in formamide revealed further subdivisions of these four separate components of DNA. The DNA of above mentioned NC-17 variety compared to Florigiant variety showed no differences in thermal denaturation profiles, buoyant density, or in genome size.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendich A. J., Anderson R. S. Characterization of families of repeated DNA sequences from four vascular plants. Biochemistry. 1977 Oct 18;16(21):4655–4663. doi: 10.1021/bi00640a020. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Gene regulation for higher cells: a theory. Science. 1969 Jul 25;165(3891):349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: empirical equations describing DNA reassociation. Proc Natl Acad Sci U S A. 1976 Feb;73(2):415–419. doi: 10.1073/pnas.73.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Dutta S. K., Ojha M. Relatedness between major taxonomic groups of fungi based on the measurement of DNA nucleotide sequence homology. Mol Gen Genet. 1972;114(3):232–240. doi: 10.1007/BF01788892. [DOI] [PubMed] [Google Scholar]
- Flavell R. B., Bennett M. D., Smith J. B., Smith D. B. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem Genet. 1974 Oct;12(4):257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- Goldberg R. B. DNA sequence organization in the soybean plant. Biochem Genet. 1978 Feb;16(1-2):45–68. doi: 10.1007/BF00484384. [DOI] [PubMed] [Google Scholar]
- Green B. R., Gordon M. P. The satellite DNA's of some higher plants. Biochim Biophys Acta. 1967 Sep 26;145(2):378–390. doi: 10.1016/0005-2787(67)90056-1. [DOI] [PubMed] [Google Scholar]
- Gurley W. B., Hepburn A. G., Key J. L. Sequence organization of the soybean genome. Biochim Biophys Acta. 1979 Jan 26;561(1):167–183. doi: 10.1016/0005-2787(79)90500-8. [DOI] [PubMed] [Google Scholar]
- Ingle J., Timmis J. N., Sinclair J. The Relationship between Satellite Deoxyribonucleic Acid, Ribosomal Ribonucleic Acid Gene Redundancy, and Genome Size in Plants. Plant Physiol. 1975 Mar;55(3):496–501. doi: 10.1104/pp.55.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk J. T. Effect of methylation of cytosine residues on the buoyant density of DNA in caesium chloride solution. J Mol Biol. 1967 Aug 28;28(1):171–172. doi: 10.1016/s0022-2836(67)80087-1. [DOI] [PubMed] [Google Scholar]
- Knittel M. D., Black C. H., Sandine W. E., Fraser D. K. Use of normal probability paper in determining thermal melting values of deoxyribonucleic acid. Can J Microbiol. 1968 Mar;14(3):239–245. doi: 10.1139/m68-040. [DOI] [PubMed] [Google Scholar]
- LEUCHTENBERGER C. Quantitative determination of DNA in cells by Feulgen microspectrophotometry. Gen Cytochem Methods. 1958;1:219–278. [PubMed] [Google Scholar]
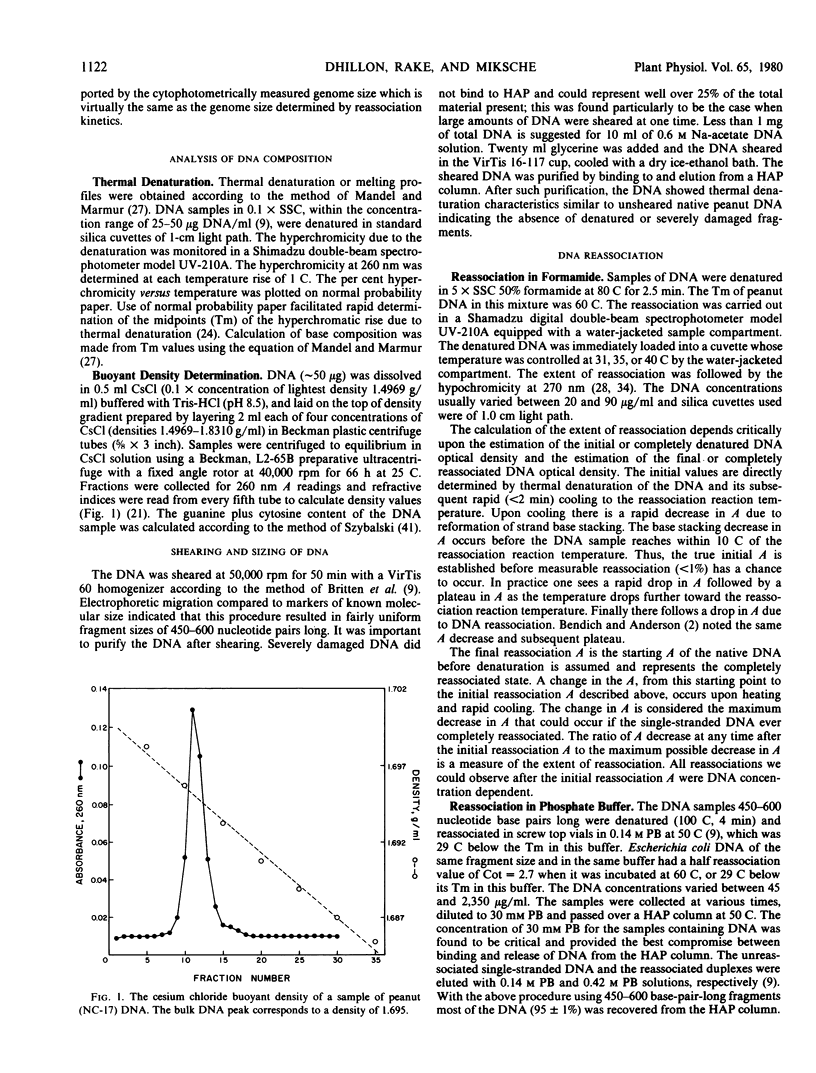
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Miksche J. P., Dhillon S. S., Berlyn G. P., Landauer K. J. Nonspecific light loss and intrinsic DNA variation problems associated with feulgen DNA cytophotometry. J Histochem Cytochem. 1979 Oct;27(10):1377–1379. doi: 10.1177/27.10.92496. [DOI] [PubMed] [Google Scholar]
- Murray M. G., Cuellar R. E., Thompson W. F. DNA sequence organization in the pea genome. Biochemistry. 1978 Dec 26;17(26):5781–5790. doi: 10.1021/bi00619a027. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Smith S. L., Wu J. R., Bonner J. Kinetic determination of the genome size of the pea. Plant Physiol. 1978 Jul;62(1):112–115. doi: 10.1104/pp.62.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rake A. V., Pollard E. C. The nature of residual Escherichia coli DNA after degradation induced by ionizing radiation. Radiat Res. 1972 May;50(2):334–338. [PubMed] [Google Scholar]
- Ris H., Kubai D. F. Chromosome structure. Annu Rev Genet. 1970;4:263–294. doi: 10.1146/annurev.ge.04.120170.001403. [DOI] [PubMed] [Google Scholar]
- Santiago L., Rake A. V. Rodent DNA reassociation kinetics. Biochem Genet. 1973 Jul;9(3):275–282. doi: 10.1007/BF00485740. [DOI] [PubMed] [Google Scholar]
- Smith M. J., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: reactivity of single-stranded tails in DNA-DNA renaturation. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4805–4809. doi: 10.1073/pnas.72.12.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson W. F. Aggregate formation from short fragments of plant DNA. Plant Physiol. 1976 Apr;57(4):617–622. doi: 10.1104/pp.57.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis J. N., Deumling B., Ingle J. Localisation of satellite DNA sequences in nuclei and chromosomes of two plants. Nature. 1975 Sep 11;257(5522):152–155. doi: 10.1038/257152a0. [DOI] [PubMed] [Google Scholar]
- VAN'T HOF J., SPARROW A. H. A relationship between DNA content, nuclear volume, and minimum mitotic cycle time. Proc Natl Acad Sci U S A. 1963 Jun;49:897–902. doi: 10.1073/pnas.49.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., Dure L. S., 3rd Developmental biochemistry of cotton seed embryogenesis and germination. VII. Characterization of the cotton genome. J Mol Biol. 1976 Mar 15;101(4):503–536. doi: 10.1016/0022-2836(76)90242-4. [DOI] [PubMed] [Google Scholar]



