Abstract
Immunoproteasomes contain replacements for the three catalytic subunits of standard proteasomes. In most cells, oxidative stress and proinflammatory cytokines are stimuli that lead to elevated production of immunoproteasomes. Immune system cells, especially antigen-presenting cells, express a higher basal level of immunoproteasomes. A well-described function of immunoprotea-somes is to generate peptides with a hydrophobic C terminus that can be processed to fit in the groove of MHC class I molecules. This display of peptides on the cell surface allows surveillance by CD8 T cells of the adaptive immune system for pathogen-infected cells. Functions of immunoprotea-somes, other than generating peptides for antigen presentation, are emerging from studies in immunoproteasome-deficient mice, and are complemented by recently described diseases linked to mutations or single-nucleotide polymorphisms in immunoproteasome subunits. Thus, this growing body of literature suggests a more pleiotropic role in cell function for the immunoproteasome.
I. Introduction
While the first descriptions of the proteasome were published in the 1970s,1,2 it was not until the 1990s that the scientific community became aware of a specialized form of the proteasome3–6 that was later called the immunoproteasome (i-proteasome).7 The name i-proteasome was first given to 20S cores that had incorporated the low-molecular-weight proteins (LMPs) LMP2 and LMP7. These proteins were significantly upregulated in response to the major immunomodulatory cytokine interferon-gamma (IFN-γ). An additional factor influencing the choice of name was that the genes encoding LMP2 and LMP7 were located within the major histocompatibility complex (MHC) class II region. The third i-proteasome subunit, the multicatalytic endopeptidase complex subunit 1 (MECL-1), was discovered about a decade later.8–10 This subunit also responds to IFN-γ stimulation; however, the gene that encodes this protein lies outside the MHC class II region.
Proteasome subtypes are defined by their catalytic subunits. The standard proteasome catalytic subunits include β1, β2, and β5, which are constitutively expressed in all cells. The i-proteasome catalytic subunits, also known as the inducible subunits, are LMP2, MECL-1, and LMP7. An intermediate protea-some containing a mixture of both the standard and i-proteasome subunits has also been described. The most recent proteasome subtype to be discovered is the thymus-specific proteasome, which substitutes the β5 subunit with an alternate protein (β5t).11
The nomenclature for the various proteasome subunits is quite confusing because multiple names have been assigned to each subunit. This is due in part to contributions from multiple laboratories working on different organisms. Table I provides a list of names for the different catalytic subunits and should be a useful reference for future reading, as the nomenclature is not completely standardized. The UniProtKB nomenclature and gene names are also provided to aid in accessing online information. For the purpose of this chapter, we will use the common name for the standard and thymoproteasome subunits (β1, β2, β5, β5t) and the alternate names for the i-proteasome subunits (LMP2, LMP7, MECL-1).
TABLE I.
Nomenclature and Activity for Proteasome Catalytic Subunits
| Proteasome subunita |
Common nameb |
Alternative namesc | Gene | Activity |
|---|---|---|---|---|
| Beta type-6 | β1 | Y, Delta, LMP19, Pre3 | PSMB6 | Caspase-like |
| Beta type-9 | β1i | Lmp2, RING12, MC7 | PSMB9 | Chymotrypsin-like |
| Beta type-7 | β2 | Z, MC14, Lmp9, Pup1 | PSMB7 | Trypsin-like |
| Beta type-10 | β2i | MECL-1, Lmp10 | PSMB10 | Trypsin-like |
| Beta type-5 | β5 | X, epsilon, Lmp17, MB1, Pre2, Doa3, Prg1 |
PSMB5 | Chymotrypsin-like |
| Beta type-8 | β5i | Lmp7, RING10, Y2, C13 | PSMB8 | Chymotrypsin-like |
| Beta type-11 | β5t | Thymus-specific β5 | PSMB11 | Chymotrypsin-like |
UniProtKB nomenclature (www.uniprot.org).
Baumeister,12 names in italics are for yeast subunits.
Underlined alternative names will be used for the i-proteasome subunits.
II. Immunoproteasome Structure
The basic structure of all proteasome subtypes is essentially the same. Each 20S core particle is composed of four stacked rings of seven subunits each. The two outer rings contain the constitutively expressed α-subunits, which interact with regulatory complexes such as PA28 and PA700. The two inner rings contain the β-subunits. Three of the β-subunits in each ring perform distinct proteolytic activities. In the standard proteasome, activity of the catalytic sub-units β1, β2, and β5 have been classified as caspase-like, trypsin-like, and chymotrypsin-like for cleavage after acidic, basic, and hydrophobic amino acids, respectively (Table I). The standard catalytic subunits β1, β2, and β5 can be replaced in nascent proteasome cores by the inducible subunits LMP2, MECL-1, and LMP7, respectively. While the MECL-1 and LMP7 subunits perform the same type of activities as the β2 and β5, the LMP2 subunit performs chymotrypsin-like activity and cleaves after hydrophobic amino acids. The structural basis for this change is discussed in a later section. It has been suggested that the altered activity of the LMP2 subunit facilitates the generation of peptides for antigen presentation, which requires peptides with hydrophobic amino acids in the C-terminal position.
Each catalytic subunit is expressed with a propeptide that ranges in size from 20 to 72 amino acids (Table II). Cleavage of the propeptide is essential for maturation of the 20S core and activation of the catalytic threonine residue. Thus, the difference in the molecular mass of the unprocessed and the mature protein allows one to distinguish these two species on a high-percentage sodium dodecyl sulfate gel. A comparison of the amino acids and molecular mass of the unprocessed and mature proteins for humans and mice is provided in Table II . For most of the proteins, there is good agreement between the two species, except for the presence of the two isoforms of the β5 and LMP7 subunits that are unique to humans.
TABLE II.
Proteasome Catalytic Subunit Size
| Subunit name |
Primary Acc. (#)a |
Amino acids (#)b |
Molecular weight (Da)b |
Pro-pep (# AA) |
|||
|---|---|---|---|---|---|---|---|
| Human | Mouse | Human | Mouse | Human | Mouse | ||
| β1 | P28072 | Q60692 | 239/205 | 238/205 | 25358/21904 | 25379/21997 | 33/32 |
| LMP2 | P28065 | P28076 | 219/199 | 219/199 | 23264/21276 | 23397/21326 | 20 |
| β2 | Q99436 | P70195 | 277/234 | 277/234 | 29965/25218 | 29891/25252 | 43 |
| MECL-1 | P40306 | O35955 | 273/234 | 273/234 | 28936/24648 | 29063/24804 | 39 |
| β51c | P28074 | O55234 | 263/204 | 264/205 | 28480/22458 | 28532/22529 | 59 |
| β52c | P28074 | 203/144 | 21845/15822 | 9 | |||
| LMP71c | P28062 | P28063 | 276/204 | 276/204 | 30354/22660 | 30260/22650 | 72 |
| LMP72c | P28062 | 272/204 | 29770/22660 | 68 | |||
| β5t | A5LHX3 | Q8BG41 | 300/251 | 302/253 | 32530/27232 | 33220/27851 | 49 |
UniProtKB (www.uniprot.org).
Unprocessed protein/mature protein.
Two isoforms, indicated as 1 and 2, are known for β5 and β5i in humans.
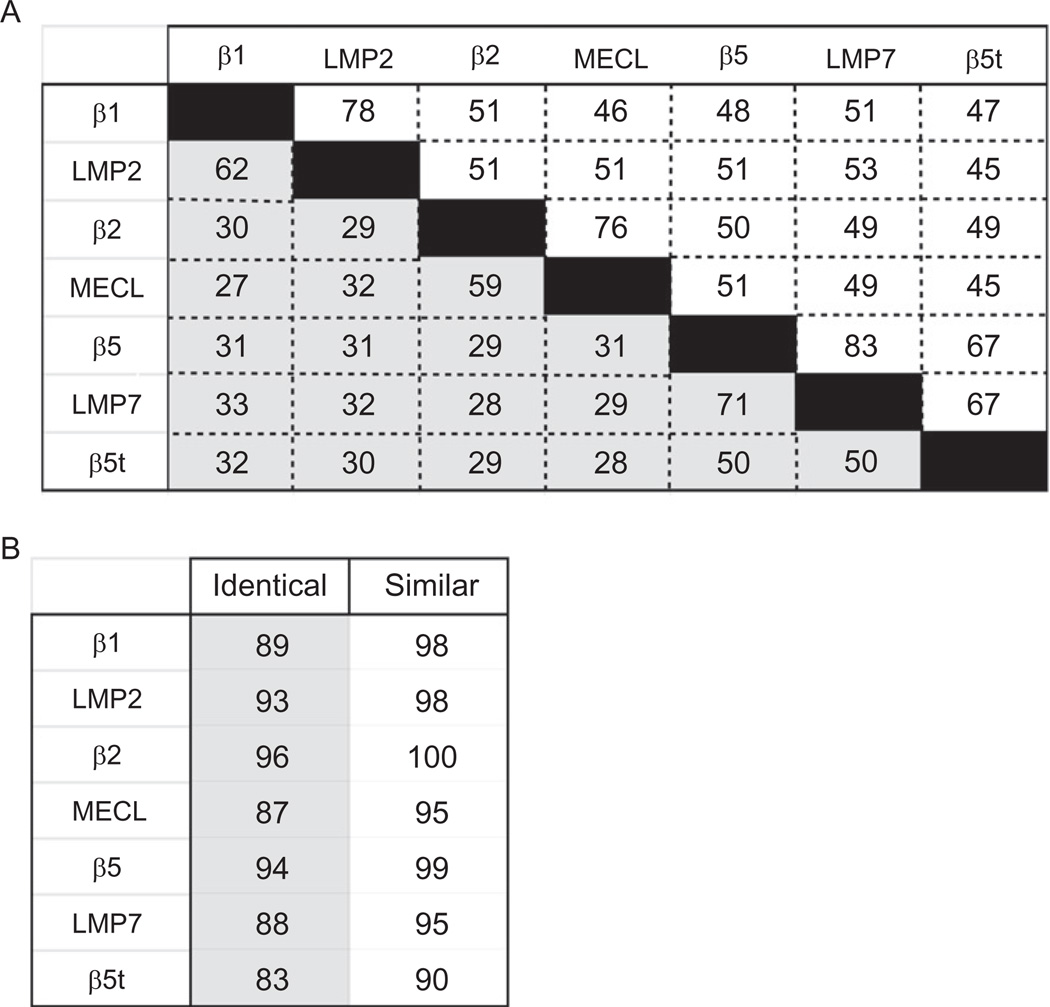
There is high sequence homology between the standard catalytic subunits and their i-proteasome correlates (Fig. 1A). The percentage of amino acid sequence identities is between 60% and 70%. When considering the conservative substitutions in each subunit, the amino acid sequence similarities range from 76% to 83%. The amino acid sequence for each subunit is also highly conserved between species. Sequence comparison for humans and mice shows that the primary sequence is 83–96% identical and 90–100% similar (Fig. 1B). Thus, it appears that there is strong evolutionary pressure to maintain the primary and secondary structure for all of the proteasome catalytic subunits.
Fig. 1.
Sequence homology for proteasome catalytic subunits. Percentage of the amino acid sequence that is identical (boxed in gray) or similar (boxed in white) for proteasome catalytic subunits. (A) Comparison of the primary sequence for catalytic subunits from mouse is shown. (B) Comparison of the primary sequence for each catalytic subunit comparing mouse versus human is shown. Percentages were obtained using the ClustalW algorithm from the UniProt Consortium.
The 20S is the predominant proteasome form in most cells and, in fact, may be the primary mechanism for the ATP-independent degradation of oxidized proteins following an oxidative insult.13,14 The core particle, containing all standard or i-proteasome catalytic subunits, or a mixture of both standard and i-proteasome subunits, has been described.15–17 Considering the various combinations of subunits, it is theoretically possible to generate 36 different 20S core complexes that differ in their proteolytic capacity and specificity. This is a dizzying number of proteasome subtypes when one also includes the different regulatory complexes that can associate with each end to form cores with PA700 or PA28 bound, or one molecule each of PA700 and PA28 to form the hybrid proteasome. Of particular importance to the i-proteasome is PA28. This regulatory complex contains seven proteins. In the cytosol, the α- and β-subunits form this complex, but the γ-subunit is present in the nucleus. Expression of PA28 is upregulated by IFN-γ, suggesting a role in regulating i-proteasome function. When this regulatory molecule binds to the outer ring of the α-subunits, it causes structural rearrangement of the N-terminal portion of the α-subunits that opens the central chamber and facilitates access of proteins to the catalytic core.18
III. Regulation of Gene Expression
The two genes that encode LMP2 and LMP7 were first discovered in the early 1980s by Monaco and McDevitt.19,20 These two genes were subsequently mapped to the MHC class II region, where they are clustered with the TAP-1 and TAP-2 genes. Like LMP2 and LMP7, the expression of TAP proteins is also upregulated by IFN-γ. The TAP proteins are transport molecules embedded in the endoplasmic reticulum (ER), where they shuttle peptides destined for MHC class I antigen presentation from the cytoplasm into the lumen of the ER. The MHC class II region is located on chromosome 6 in humans and chromosome 17 in mice.
The genomic organization of the gene for LMP2 is very similar for both humans and mice. The gene is approximately 2.3 Kb in size and contains six exons of identical length.21,22 The LMP2 gene is unusual in that it contains a bidirectional promoter that is shared with the TAP-1 gene.23 Both genes are divergently transcribed from a central promoter region of 539 base pairs that has no TATA box, but rather has several GC boxes that are the likely transcriptional start sites. Transcription is initiated at multiple sites for each gene, but remarkably, there is no overlap in transcripts between genes. The promoter region also contains binding sites for multiple transcription factors, including the interferon consensus sequence-2 and gamma interferon activated sequence elements that bind the dimer signal transducers and activators of transcription-1 (Stat-1) and interferon regulatory factor-1 (IRF-1).24 The Stat-1/IRF-1 dimers are the major transcription factors involved in IFN-γ signal transduction. Gene expression of LMP2 and LMP7 is strongly upregulated by IFN-γ.25 The promoter also contains additional binding sites for transcription factors, including 10 GC-rich regions, two cAMP responsive elements, two AP-2 elements, four Egr response elements, and three regions of consensus sequence for NF-κB binding. Transcription factors that have been associated with LMP2 regulation include NF-κB, Sp1, AP-1, cAMP responsive element binding protein (CREB), and, more recently, Zif268, also known as Egr1. 21, 23, 26 Both LMP2 and LMP7 were recently identified as target genes for Zif268, which suppresses transcription of these genes. It has been suggested that Zif268 may modulate the transcriptional response of Sp1 and NF-κB in the LMP2 and LMP7 genes since the binding sites of these three transcription factors lie in close proximity to each other.26
There are a few differences in the genomic organization of the LMP7 gene when comparing the human and mouse genomes.27 In the mouse, there are six exons, while the human gene contains seven exons. The difference occurs in the first human exon, which is modified by numerous insertions and deletions. This exon also contains two frame stop codons that results in the production of two different LMP7 isoforms in humans. In contrast, the mouse gene encodes for only one protein. The difference in amino acid sequence between the two LMP7 isoforms produced by the human gene is limited to the N-terminal portion of the protein, which is removed in the mature protein. Thus, while the catalytic activity is identical, there could be potential differences in assembly in the 20S core particle due to the altered amino acid composition of the N terminus. In both mouse and human, the 3′ end of the TAP-1 gene lies ~ 1 Kb upstream from the initial translational codon of LMP7. The promoter region contains a TATA box, two GC-rich regions, one cAMP regulatory element, four Sp1 sites, Egr response elements, and one region with the consensus sequence for NF-κB binding. Thus, similar transcription factors regulate both the LMP7 and LMP2 genes.26, 27
Discovery of the third i-proteasome gene for the MECL-1 subunit (PSMB10) occurred much later than the two i-proteasome genes encoded in the MHC class II region. The initial studies identified the MECL-1 subunit as a component of the i-proteasome based on its structural similarities to LMP2 and LMP7 and its responsiveness to IFN-γ.8, 10, 28 The gene for MECL-1 is found outside the MHC class II region and is expressed as a single copy. 9,29 In the mouse, it is located on chromosome 8 and in humans, on chromosome 16. Investigation of the genomic organization of the PSMB10 gene showed similarities between the mouse and human genome. Each gene contained eight exons of equal size with introns inserted at exactly the same place. The mouse gene was slightly larger than the gene in humans (2.5 vs. 2.3 Kb) due to a long interspersed repetitive DNA sequence known as L! that is located upstream of the promoter region. The promoter regions contain no CAAT or TATA boxes and, like other genes missing these elements, MECL-1 has multiple transcriptional start sites. The promoter also contains two elements that confer responsiveness to IFN and multiple additional regions that bind transcription factors, including Sp1, AP-1/2, and NF-κB.
The gene for the β5t subunit of the thymoproteasome was discovered by Murata and colleagues11 during a genome-wide database search for proteasome-related genes. The β5t gene is located adjacent to the β5 gene (PSMB5) and is encoded by a single exon. The β5 and β5t genes are transcribed in opposite directions and, in the mouse, are separated by 7.3 kb on chromosome 14. Based on Northern and Western blot analyses, expression of the β5t gene and gene product occurs exclusively in the thymus. The presence of the β5t gene was confirmed in both mice and humans.
Standard proteasomes are constitutively expressed in nearly all mammalian cells. In contrast, i-proteasome expression is generally lower under basal conditions, but can be significantly upregulated when cells are exposed to various factors, such as IFN-γ, or environmental stressors, such as oxidative stress. An exception to this generalization is the cells of the immune system, which can constitutively express i-proteasome at high levels. For example, cells in the spleen contain nearly all i-proteasome and low levels of the standard proteasome subunits.30
IFN-γ is the most well-studied factor used to investigate i-proteasome expression. In addition to affecting i-proteasome, IFN-γ also upregulated a host of other genes associated with generating peptides, including molecules from the MHC class I and II pathways, TAP and PA28. IFN-γ can also affect the population of proteasome subtypes present by causing dephosphorylation of the 20S, which favors dissociation with PA700 and association with PA28.31 Induction of i-proteasome subunit expression by IFN-γ has been well established for both cultured immune cells10 and cultured nonimmune cells, such as neurons32 and epithelial cells of the retina.33 This cytokine-induced expression results from binding of the Stat-1 and IRF-1 transcription factors to multiple IFN-γ consensus/activation sequences in the promoter region of the LMP2, LMP7, and MECL-1 genes.24,29 Other cytokines, such as IFNα/β, lipopolysac-charide, and TNFα also elicit an inflammatory response that involves increased i-proteasome expression.
The presence of binding sites for multiple transcription factors, such as CREB and NF-κB on the promoter region of i-proteasome genes suggests that additional cytokine-independent mechanisms of regulation are possible.26,27 For example, nitric oxide increases i-proteasome expression via the cGMP/ cAMP pathway.34 In the cascade of reactions that begins with elevated levels of nitric oxide, cGMP/cAMP activates the protein kinases A and G, which in turn phosphorylate CREB. Phosphorylation triggers the nuclear translocation of CREB and subsequent transcription of the i-proteasome subunits. This mechanism has been proposed to explain high basal i-proteasome expression in endothelial cells, which are exposed to constitutively high levels of nitric oxide.34
The ability of the cells to produce different subtypes of the proteasome, each possessing different catalytic properties and substrate preference, has several advantages. First, this plasticity allows the cell to respond to changing environmental conditions and adjust the proteolytic capacity to match each new challenge. Second, the presence of different subtypes increases the repertoire of substrates that can be degraded and peptides that are generated.
IV. Assembly of the 20S Core
Assembly of the 20S core is a chaperone-mediated process (reviewed in Ref. 35). The initial step involves the formation of the seven-member ring of α-subunits utilizing dimers of the proteasome-assembling chaperone (PAC1 and PAC2). The PAC1/2 chaperones function as scaffolding and prevent the α-subunits from associating with other nascent α-rings.36 PAC 3 and PAC 4 also assist with α-ring assembly and connect the end subunits together to form the heptameric ring. PAC3/4 also recruits and correctly positions β2, which is the first β-subunit of the standard proteasome to be incorporated into the nascent complex.36,37 PAC3/4 then dissociates from the complex and the remaining β-subunits are incorporated in a defined order (β3, β4, β5, β6, β1, and finally β7) with the assembled α-ring to form a half proteasome. Data from chimeric proteins containing different N-terminal propeptides have shown that the mutual interaction of the propeptides of the immature subunits drives the orderly assembly of the β-subunits.38 In the final step, two half proteasome core particles are combined with the aid of the chaperone proteasome maturation protein (POMP or proteassemblin, the mammalian orthologue to Ump1 in yeast). Formation of the complete 20S core initiates cleavage of the N-terminal propeptide from the three pairs of catalytic β-subunits, which exposes and activates the catalytic threonine residue (Thr1 in the mature 20S). The 20S proteasome then degrades the PAC and POMP chaperones, signaling successful assembly of the mature 20S core.
Although the general mechanism of 20S assembly is similar for all protea-some subtypes, the assembly of i-proteasome is favored over the standard proteasome for several reasons. In cells expressing both i-proteasome and standard proteasome subunits, the LMP2 subunit is the first subunit added, whereas the standard subunit homologue (β1) is incorporated much later in the assembly process. The presence of LMP2 facilitates incorporation of MECL-1. Data also support the idea that LMP2 is strictly required for MECL-1 incorporation, although this point remains controversial.38–41 The LMP7 subunit facilitates maturation of the nascent 20S core by assisting in the removal of the propeptides from LMP2 and MECL-1.39,40 The preferential incorporation of all three i-proteasome subunits, which is known as cooperative assembly, is also aided by the action of POMP. POMP expression is upregulated by IFN-γ, so POMP expression is concomitantly increased in tandem with the inducible subunits. Importantly, POMP binds the propeptide of LMP7 with greater affinity than that of β5, resulting in preferred biogenesis of the i-proteasome over formation of the standard proteasome.42
The rapid production of i-proteasome, estimated to be four times faster than the standard proteasome in response to IFN-γ, ensures that the cell can quickly expand that repertoire of peptides to aid in immune defense in a challenged organism.42 Conversely, the turnover of i-proteasome is also substantially faster compared with the standard proteasome. Pulse-chase experiments in T2 cells expressing either the standard or i-proteasome reported a half-life of 133 versus 27 h, respectively.42 Taken together, these data suggest that production and degradation of i-proteasome is a highly dynamic and transient process that permits the rapid response to environmental challenges and subsequent return to baseline levels that favor the standard proteasome subtype once the challenge has subsided. A physiologically relevant situation where the rapid adjustment in i-proteasome levels would be critical is in the initial stage of viral infection when cells are exposed to cytokines that are secreted by the innate immune system. As discussed later, i-proteasome plays a key role in regulating the cellular response to an inflammatory challenge.
While it was generally believed that the newly assembling proteasome cores preferentially incorporate all i-proteasome subunits, recent data have challenged this idea. Intermediate proteasomes containing a mixture of standard and inducible proteasome subunits have been reported in multiple cell types and tissues, including skeletal and cardiac muscle, liver, colon, small intestines, and IFN-γ-treated HeLa cells.15–17,41 In some tissues, intermediate proteasomes make up a significant portion of the proteasome population. Using newly developed antibodies that can distinguish specific subunits complexed in the 20S core, Guillaume and colleagues estimated the proportion of intermediate proteasome in normal human kidney and liver to be 30% and 50%, respectively, with the balance mainly standard proteasome.41 Intestines contained only intermediate and i-proteasome (containing only the inducible subunits), while heart contained mainly standard proteasome (83%) with the balance as intermediate and i-proteasome. This recent report is consistent with previous suggestions that the proteasome population is very diverse and that the distribution of subtypes is tissue specific.15–17,41
Knockout (KO) mice provide additional evidence that assembly of an intermediate proteasome is possible. For example, the content of LMP2 in lymphocytes, cultured retinal pigment epithelial cells, and retina of mice deficient in both LMP7 and MECL-1 is equivalent to that found in wild-type (WT) mice.30,38 Under conditions of stress, such as aging or exposure of cells to peroxide, WTcells respond by upregulating the inducible subunits.30 In cells and tissues of KO mice, this response was not replicated; that is, LMP2 expression was not increased in mice double-deficient in LMP7 and MECL-1.30 These data suggest that formation of the intermediate proteasome occurs under basal conditions. However, the full complement of i-proteasome subunits is required to obtain the maximum induction and incorporation of i-proteasome subunits into the core particle in response to stress.
V. Enzymatic Activity
As discussed earlier, the three standard catalytic subunits (β1, β2, β5) perform distinct proteolytic activities. Based on the proteolysis of model peptide substrates, the active sites have been classified as caspase-like, trypsin-like, and chymotrypsin-like for cleavage after acidic, basic, and hydrophobic amino acids, respectively. For the i-proteasome, some differences in catalytic activity and peptide generation have been noted (see discussion in section VII.A). Comparing activity of the β2 and β5 standard subunits with the corresponding i-proteasome subunits MECL-1 and LMP7, the specificity is generally the same. However, comparison of cleavage after branched chain and hydrophobic residues for standard and i-proteasome has not been consistent; both increased (reviewed in Ref. 35) and decreased30 activity have been reported for the i-proteasome. The discrepancy in data is due in part to heterogeneity of protea-some subtypes and cell-specific endogenous regulators in the tissue analyzed, and the inability of model peptide substrates to distinguish between proteasome subtypes. Cleavage after acidic residues, which is accomplished by the β1 standard subunit, is nearly abolished in the i-proteasome. Instead, there is a shift to chymotrypsin-like activity for LMP2, which promotes the generation of MHC class I-compatible peptides containing hydrophobic C-terminal anchors.
While the mechanism of hydrolysis (involving the active-site Thr1, Asp17, and Lys33) is the same for each subunit, the specificity of cleavage for each active site is determined by the amino acids that make up the S1 binding pocket (residues 20, 31, 35, 45, 49, 53, 115, 116),43,44 which is where protein substrates bind prior to cleavage (Fig. 2). Alignment of sequences comparing the standard with their i-proteasome subunit correlate generally shows high conservation of the amino acids that make up the binding pocket, except for LMP2 and β5t. LMP2 contains two prominent substitutions compared with residues in β1; the β1 Thr21 is replaced by Val and the Arg45 is replaced by Leu. These substitutions minimize the size of the S1 pocket of LMP2 and change the overall charge of the local environment from positive to neutral. These changes in primary sequence and charge state of the LMP2 binding pocket could explain the drastic reduction in the caspase-like activity of the i-proteasome. One other noteworthy change occurs in positions 115 and 116 in β5 and LMP7; substitution of Ser115 for a Glu and substitution of Glu116 for His in β5 and LMP7, respectively, would alter the size and polarity of the binding pocket. These structural changes could alter the substrate preference of each subunit and, consequently, result in the production of different peptides.
Fig. 2.
Sequence alignment of standard and immunoproteasome subunits. (A) Sequence from humans includes the last three residues of the propeptide, followed by residues 1–57 of the mature protein. The dash represents the site of autocatalytic cleavage, which is performed by the active-site Thr1. The conserved Gly adjacent to Thr1 is essential for efficient autocatalytic processing of the propeptide. Conserved residues essential for catalytic activity in the mature protein include Thr1, Asp17, and Lys33 (dark gray, white letters). The S1 binding pocket that imparts specificity of cleavage includes residues 20, 31, 35, 45, 49, and 53 (light gray). Note that in LMP 7, the murine sequence substitutes a Met in place of Val31 in humans. (B) Sequence of the β2 and MECL subunits highlighting four amino acids that contribute to the S1 binding pocket of the adjacent β1 or LMP 2 subunit. Residues 114, 116, 118, and 120 are highlighted in gray. Stars indicate nonconser-vative substitutions between the β2 and MECL subunits. The sequence of the β5 and LMP 7 subunits highlights two amino acid substitutions at positions 115 and 116, which are critical for substrate binding.
For the thymus-specific β5 subunit, β5t, sequence comparison with the two other β5 family members reveals five nonconservative substitutions for residues in the binding pocket (Fig. 2). While the majority of the amino acids in the S1 binding pocket of β5 and LMP7 are hydrophobic, hydrophilic residues populate the binding pocket of β5t.11 This difference in amino acid composition selectively reduces the chymotrypsin-like activity by 60–70%, but has no effect on the caspase-like or trypsin-like activities in the thymoproteasome.
Amino acids in subunits adjacent to the catalytic subunits also contribute to the S1-binding pocket and, thus, influence catalytic activity. Residues 114,116, 118, and 120 in β3 and β6, which are subunits constitutively present in all 20S core particles, contribute to the S1-pocket of subunits β2 and MECL and subunits β5 and LMP7, respectively (Fig. 2). These same residues in β2 and MECL contribute to the S1-pocket of β1 and LMP2. Comparison of residues in β2 and MECL reveals nonconservative substitutions at positions 114 (Tyr and His) and 120 (Asp and Ser). These differences in S1-pocket residues could have dramatic effects on the activity of the adjacent catalytic subunits (β1 or LMP2). This point is particularly relevant for the activity of the intermediate proteasomes, which contain a mixture of both standard and i-proteasome catalytic subunits.
Since i-proteasome, standard, and intermediate proteasomes produce some peptides that are unique to each subtype, 15,17 the repertoire of peptides generated within a cell is determined by the proteasome population that is present at any given time. Proteasome composition is quite dynamic, and changes dramatically due to challenges, such as cytokine or oxidative stress, in the cell environment. The diverse spectrum of peptides produced by different proteasome populations may be biologically important not only in producing different antigenic peptides, but also for generating biologically active peptides that regulate critical processes, such as cell signaling.45
VI. Immunoproteasome Knockout Mice
To clarify the physiological role of the i-proteasome subunits, mice deficient in one or more i-proteasome subunits have been generated through the targeted disruption of the gene. These KO mice were developed by immunologists to investigate the putative role of specific i-proteasome subunits in generating immunogenic peptides for antigen presentation. The LMP2−/− and LMP7−/− strains were developed in 1994 by two different laboratories.46,47 Development of the MECL-1−/− and the thymus-specific proteasome (β5t) strains occurred a decade later.11,48
For each single KO mouse, the gene disruption was accomplished using a deletion-type targeting vector containing a neomycin resistance cassette, which eliminates a portion of the gene upon homologous recombination in embryonic stem cells. Germline transmission of the targeted allele was observed in all KO mice. To generate the LMP2 KO mouse, an 800 base pair region spanning parts of exon 2 and intron 2 in the lmp2 gene was deleted.46 Even though the LMP2 and TAP-1 genes are in close proximity in the WT mouse, this genetic manipulation did not disrupt the TAP-1 gene in the mutant mouse. In the LMP7-deficient mouse, the targeting vector deleted exons 1–5, which corresponds to the first 247 of the 276 total amino acids in the protein. To produce the MECL-1 KO mouse, the targeting vector eliminated exons 5–7 and also disrupted the splicing of exon 8.48 KO of the thymus-specific protea-some (β5t) gene was accomplished by disrupting the β5t coding sequence with a vector containing the neomycin-resistant cassette and the cDNA encoding the Venus protein.11 The Venus protein in the mutant mouse was under the control of the β5t promoter, so expression of the Venus protein provided a means for identifying the β5t–expressing cells within the thymus. All strains of KO mice were viable, appeared healthy, and had no obvious gross physical abnormalities.
Double KO mice have also been developed by crossing MECL-1 with either LMP7−/− or LMP2−/− to produce mice with two i-proteasome subunits eliminated. Simultaneous elimination of both LMP7 and LMP2 genes by simply crossing single KO mice is highly unlikely due to the close proximity of these genes within the MHC class II locus. To overcome the limitations imposed by the gene structure, a new sequential deletion strategy was employed to generate mice double-deficient in both LMP7 and LMP2.49 This was accomplished by first introducing a construct to eliminate exon 1 in the PSMB8 gene; exons 2–5 were deleted by homologous recombination. After the neomycin resistance gene was removed by FLP recombinase activity, a second construct (phosphatase loxP–neo–loxP) was fused in frame to the start codon of the PSMB9 gene. This removed exon 1 by homologous recombination. Elimination of all i-proteasome subunits to generate a triple KO mouse was accomplished by crossing the double-deficient mice (LMP2−/− /LMP7−/−) with the MECL-1 KO mice.
Another approach to produce mice devoid of i-proteasome activity has been to combine the LMP2−/− /MECL-1−/− double KO with use of the LMP7-specific inhibitor, ONX 0914 (also known as PR-957). This approach produces a catalytically inactive i-proteasome.50 With the development of additional i-proteasome subunit-specific inhibitors, this strategy could be taken to investigate how enzymatic inactivity, rather than complete absence of specific subunits, affects cellular response to stimuli. The use of i-proteasome subunit-specific inhibitors helps eliminate some of the drawbacks of KO mice, which include the potential for compensatory mechanisms that would complicate interpretation of the response. Another limitation of KO mice is that genetic ablation of one or more i-proteasome subunits could alter the structure of the 20S core and affect activity or binding of regulatory molecules. The observation that KO mice do not replicate the phenotype of human diseases caused by mutations in specific i-proteasome subunits supports the idea that the presence of an inactive (mutant) protein may influence results in a way that is not consistent with the complete elimination of the protein. For example, LMP7 KO mice do not spontaneously develop the disease symptoms of the PSMB8-associated autoinflammatory syndromes,51–54 which will be discussed later.
In summary, i-proteasome KO mice have been extensively used by immunologists to investigate the putative role of specific i-proteasome subunits in immune function, emphasizing the generation of peptide substrates for loading into MHC class I. While the initial functional and phenotypic analysis of KO mice has reported surprisingly modest changes in immunologic-based measures, recent studies with KO mice have revealed novel roles for the i-proteasome that are unrelated to antigen presentation.
VII. Immunoproteasome Function
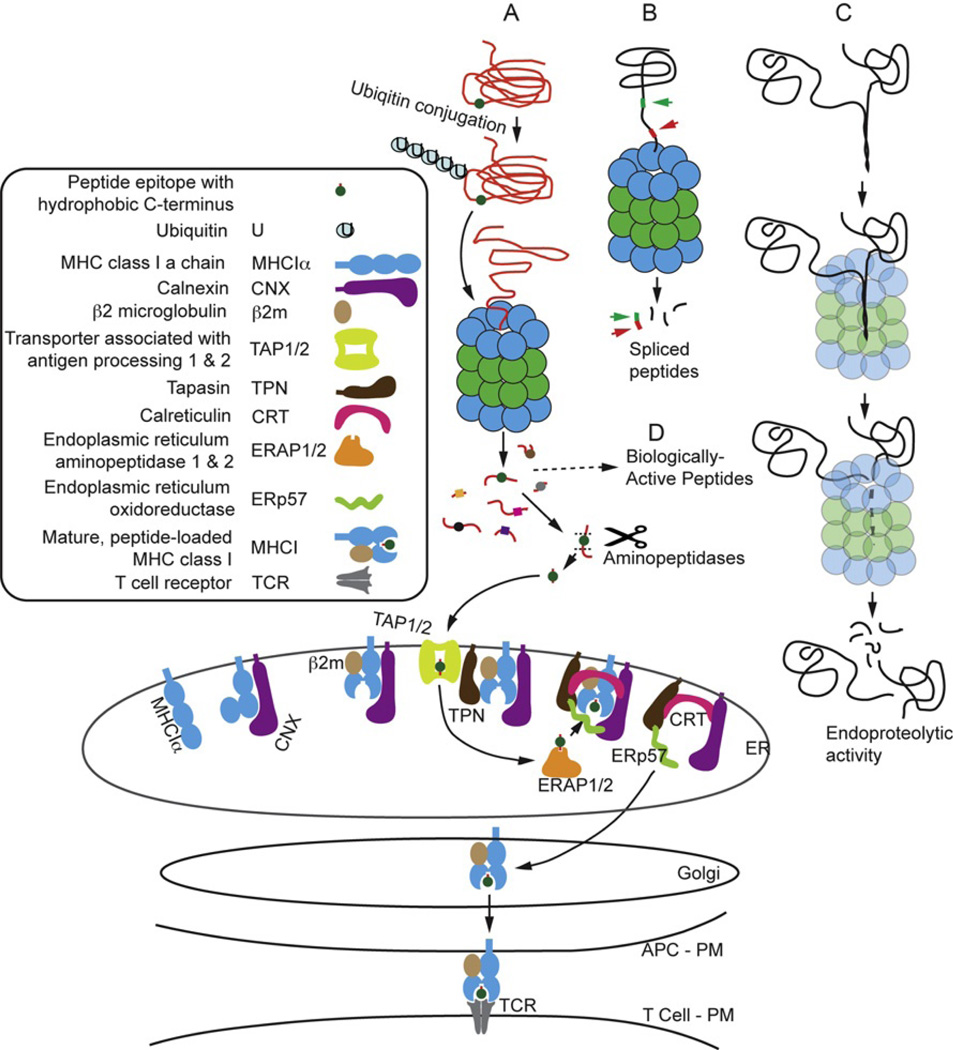
The structural and sequence similarities between the standard and i-proteasome cores strongly suggested that the generation of antigenic peptides for MHC class I presentation would be a shared function. A schematic representation of a minimal antigen-processing pathway for MHC class I-restricted epitopes derived from cytosolic proteins is shown in Fig. 3A.55 A representative protein with an epitope (round symbol) is ubiquitinated, targeting the protein to an i-proteasome 20S core. After proteolysis and ejection from the core, peptides are further cut by a battery of aminopeptidases, and arrive at TAP-1 and TAP-2, for transport into the ER. In the ER, the N terminus of peptides may be further clipped by ERAP1 or 2, in preparation for loading into the MHC class I complex. The MHC class I complex forms around nascent class I α chains chaperoned by calnexin (CNX), tapasin (TPN), and calreticulin (CRT), which also serve to promote proper folding of the MHC class I α chain and β2m chain, and enhance receptiveness of the groove for peptides. TPN promotes the association of the complex with TAP-1/2. Peptides displaying complementarity to the binding site in the class I heavy chain occupy the groove, which leads to dissociation of the heavy chain/peptide/β2m complex from the chaperones, and subsequent transport to the plasma membrane (PM). Peptides derived from membrane-associated proteins, and from proteins destined for secretion, are also generated in the ER, providing additional sequences for occupancy of the MHC class I groove. Novel peptides derived by splicing noncontiguous sequences have also been shown to be produced by the standard and i-proteasomes (Fig. 3B). These peptides can enter the MHC class I antigen presentation pathway, and have been shown to be rare targets of CD8 T cells.56 Since the only spliced peptides characterized to date were discovered as a result of MHC class I occupancy and antigen presentation leading to CD8 T cell responses, the frequency and significance of splicing reactions unrelated to antigen processing is unknown.
Fig. 3.
I-proteasome-dependent pathways of protein degradation. I-proteasome-dependent proteolysis in an antigen-presenting cell (APC) generates peptides and polypeptides for MHC class I-restricted antigen presentation, and for functions other than antigen presentation. (A) The antigen-processing pathway for degradation of a cytosolic protein leading to loading of peptides into MHC class I and antigen presentation to the T cell receptor (TCR). (B) Degradation of a protein by i-proteasome resulting in the generation of a spliced peptide candidate for antigen presentation. (C) I-proteasome-mediated endoproteolytic activity generating large, potentially biologically active polypeptides from a precursor protein. Small peptides may also be produced and would be available for antigen presentation. (D) Some of the small peptides produced by i-proteasome activity may have biological activity. PM, plasma membrane; ER, endoplasmic reticulum.
More recent evidence has expanded i-proteasome’s role to include regulation of key processes in maintaining cellular homeostasis and in responding to stress. The i-proteasome achieves these diverse functions through the selective degradation of protein substrates (Fig. 3C). For example, partial degradation of proteins, known as endoproteolysis, is one well-known mechanism for releasing active transcription factors of the NF-κB family.57 Degradation of a protein can stop signal transduction by uncoupling the pathway. Alternatively, degradation of an inhibitory protein may promote the activity of a signaling pathway. Peptides generated by either partial or complete degradation of proteins can be loaded into MHC class I molecules for recognition by CD8 T cells as part of immune surveillance. Degradation of proteins by i-proteasome could also produce biologically active peptides that regulate key cellular processes (Fig. 3D).45 The following section will discuss the diverse roles of i-proteasome in antigen processing and other nonimmune functions that have been recently discovered.
A. Antigen Processing and the Immune Response
T lymphocyte recognition of peptides generated from self and foreign proteins is essential for maintenance of the host organism in its niche. A major class of T lymphocytes, the CD8 T cells, recognizes peptides in the groove of MHC class I molecules. The peptides that are loaded into this groove are substantially provided by the proteolytic activity of proteasomes.58 The proteasomes access both endogenous and exogenous proteins through direct and cross-presentation pathways, respectively, in antigen-presenting cells of the immune system. Other MHC class I-positive cells process endogenous proteins through the direct pathway.59,60 The multiple proteasome subtypes efficiently perform the first step of antigen processing that provides the peptides of eight or nine residues ultimately required to fit into the groove of nascent MHC class I molecules in the ER.60 The cleft in MHC class I molecules includes accommodation for hydrophobic C-terminal anchor residues, which are generated mainly by the chymotrypsin-like activity of the β5/ LMP7 subunits. In the i-proteasome, the shift in proteolytic activity in LMP2 to chymotrypsin-like activity promotes the generation of peptides with a hydro-phobic C-terminal anchor, providing more, and different, peptides for occupancy of the peptide-binding cleft in MHC class I molecules.61–63 Extra residues on the N terminus are removed by proteases downstream of the proteasome (review Ref. 44). While there are a number of cytosolic proteases distinct from i-proteasome that also generate peptides, the i-proteasome is a major provider of peptides with a hydrophobic C terminus.
The location of the LMP2 and LMP7 i-proteasome subunits in the MHC region,4–6 and their production in response to proinflammatory cytokines, suggested their role in antigen processing, and led to the production of KO mice lacking one or both of these subunits to investigate subunit-specific differences. Although the KO mice lacked an obvious phenotype, more detailed analyses revealed differences. Mice deficient in LMP7 exhibited a modest reduction in surface expression of MHC class I, and resulted in mice with a reduced response to the male HY antigen.47 Deficiency in LMP2 was found to result in a reduced level of CD8 T cells and depressed the response to a nucleoprotein epitope of influenza A.46 Mice deficient in MECL-1 had a slightly reduced number of CD8 T cells, and a decreased response to two class I-restricted epitopes of lymphocytic choriomeningitis virus.48 The reduced response to the GP276–286 epitope was attributed to decreased precursor frequency of the specific T cells, rather than impaired production of the GP276–286 epitope. In summary, use of these KO mice, lacking specific subunits of the i-proteasomes, have demonstrated differences in the CD8 T cell responses, confirming their roles in antigen-specific interactions with microbial challenges. The combined results of a growing body of evidence show that the repertoires of peptides generated by i-proteasomes and standard proteasomes for both selection of the CD8 T cell repertoire and presentation by antigen-presenting cells in the periphery have substantial overlap, as well as distinct differences.46,48,64–67
van Helden and colleagues68 reported that MECL-1 and LMP7 deficiency resulted in reduced MHC class I expression, which made those cells susceptible to natural killer cell-mediated killing in mice whose immune system had been activated by a viral infection. The data are compelling, and several reports point to i-proteasome deficiency leading to the lack of suitable peptides for class I occupancy.47,69 The correlation between MECL-1 and LMP7 deficiency and reduced surface expression of MHC class I is based on two observations: that expression of MHC class I is dependent on occupancy of the cleft in the MHC α chain,70,71 and that chymotrypsin-like activity of the immune subunit provides a larger pool of precursors. While these hypotheses are sound, direct evidence for a shortage of i-proteasome-generated peptides being the rate-limiting step for maturation of MHC class I molecules and their expression on the cell surface is lacking, as total proteasome content is relatively constant, and standard proteasome would continue to provide peptide precursors, even if the repertoire was altered. Conversely, evidence continues to appear for other roles played by i-proteasome subunits. One of the most significant of these is the linkage of LMP2 to NF-κB.72 LPS stimulation of LMP2-deficient cells was less able to activate NF-κB due to reduced IκB degradation. As a transcription factor at the center of an important hub in innate and adaptive immune responses, NF-κB is positioned to have substantial influence on immunity, including the regulation of transcription of MHC class I.73
Although differences in antigen processing were found in the KO mice, there was no evidence for a global deficit in immune function; i-proteasome KO mice were almost without phenotype with respect to their overall health. The coordinated expression of the i-proteasome subunits following stimulation by TNFα and IFN-γ has led to speculation that loss or inhibition of all three subunits might yield a more definitive outcome. Mice deficient in two subunits, LMP2 and MECL-1, were examined in the absence and presence of ONX0914, an LMP7 inhibitor. Quantitative differences in the cytokine responses of splenocytes treated with polyclonal activators were found if the ONX0914 inhibitor was included.50 Comparison of responses to several epitopes of lymphocytic choriomeningitis virus by lymphocytes recovered from infected mice showed epitope-specific differences, but no widespread qualitative loss of responses.50
The absence of global effects on CD8 T cell responses in mice deficient in one or more i-proteasome subunits contrasts with several reports in which responses to specific epitopes are substantially altered, and can lead to differences in resistance to specific infectious diseases. Two examples clearly illustrate this point. Infection by the intracellular parasite Listeria monocytogenes strongly upregulates expression of i-proteasome subunits, including expression in liver cells.74 Despite the presence of cytotoxic T lymphocytes specific for the protective LLO296–304 epitope, clearance of the bacteria was substantially compromised in the liver of LMP7-deficient mice, but similar to WT in the spleen.75 Clearance of another parasite, Toxoplasma gondii, was also severely compromised in LMP7-deficient mice, and to a lesser degree in LMP2-deficient mice.76
Another type of proteasome was recently found to be specifically expressed in the cortical epithelial cells of the thymus (cortical thymic epithelial cell, cTEC).11 Like i-proteasomes, it contained the LMP2 and MECL-1 subunits, but contained a variant of β5, the β5t subunit, which substituted for the LMP7 subunit. The role of cTECs in positive selection of CD8 Tcells and the reduced production of CD8 T cells in β5t-deficient mice suggests a role for this subunit in generating the peptide repertoire available for MHC class I occupancy in CD8 T cell maturation (review Ref. 77). The function of immuno- and thymoproteasomes in generating the universe of CD8 T cell specificities for peptides, and their reciprocal role in generating the peptides that will occupy MHC class I on the cells of the body, that is, identifying them as potential targets for cytotoxic T lymphocytes, is increasingly established as a critical underpinning shaping self-recognition by CD8 T cells.
To date, the single and double i-proteasome-deficient mice have been examined for differences in antigen presentation, yielding modest differences overall. On the possibility that overlap in the cleavage specificities compensated for loss of subunits, triple i-proteasome-deficient (LMP2, LMP7, MECL-1) mice were made.49 The triple-deficient mice were viable, fertile, and immunocompetent. A decrease in CD8 Tcell numbers was found, as well as differences in antigen presentation. These were especially apparent when responses to immunodominant peptides by the corresponding T cell receptor (TCR) transgenic mice were examined. Mass spectrometry analysis of peptides collected from class I molecules expressed on the surface of the respective splenocytes showed that approximately one-third of the peptides were unique to the deficient mice, one-third each were associated with the WT MHC class I, and MHC class I occupied the overlap of peptides produced by standard and i-proteasomes. Identification of the peptides revealed that an average of approximately one peptide was found per protein of origin. Since pathogens usually contain a number of proteins, these results reveal the basis for the limited effect of i-proteasome deficiencies on CD8 T cell immune responses, despite the occasional substantial difference.
A number of studies have concentrated on the reactivity of TCR transgenic CD8 T cells to assess the role of i-proteasomes on the peptides that are presented in MHC class I. In instances in which the TCR in these transgenic mice is specific for a peptide that is differentially produced by i-proteasomes versus standard proteasomes, substantial differences in the T cells can be found by this strategy.78 Other studies have looked at the outcome of microbial infections, especially those in which a limited number of epitopes play a large role in the outcome of the immune response. Proteomic analysis of peptides eluted from MHC class I molecules (peptidomics) provides a powerful tool for assessing the diversity and identity of peptides in the class I cleft. Using WT and i-proteasome LMP2- and LMP7-deficient double KO mice, de Verteuil and colleagues isolated bone marrow dendritic cells that were induced to become mature and activated.79 Peptides were stripped from cell-surface MHC class I of dendritic cells and thymocytes, and assessed by mass spectrometry. Peptide diversity from WT dendritic cells was greater than that from WT thymocytes, and the proteins represented in MHC class I from each cell type differed, in part, with respect to cell functions. I-proteasome-deficient dendritic cells had a less diverse population of peptides in the cleft. In both cases, most proteins yielded only a single peptide sequence capable of loading into MHC class I, and there was substantial overlap between the dendritic cells and thymocytes. The presence or absence of i-proteasomes also altered mRNA levels of clusters of genes associated with dendritic cell function by unknown mechanisms that do not appear to be associated with peptide loading of MHC class I. Together, the data both reinforce the role of i-proteasome in generating peptides for class I, and also argue for functions distinct from peptide generation.
In addition to the induction of i-proteasome subunits by proinflammatory cytokines, adapter molecules are also induced, including PA28.8,80 PA28 alters the activity of both i-proteasome and standard proteasome, and leads to differences in the peptide output, without altering the active sites of the catalytic subunits. Effects on the production and availability of specific epitopes have been found. Among the examples are the TRP2288 and TRP2360 epitopes of a melanoma antigen.81,82 Flanking sequences adjacent to the epitope have been found to be required for the PA28 effect, but are not yet resolved at the molecular level.83 Although widely thought to support the i-proteasome function of generating epitopes for MHC class I, this function is not well supported by data. Other than rare examples, PA28 has not been shown to possess activities that significantly alter the production and repertoire of peptides for antigen presentation, and invites speculation that it alters immunoproteasome activity for another unknown purpose.
It is clear that standard, immuno- and thymoproteasomes, as well as hybrids containing a mixture of the catalytic subunits, provide peptides that go on to occupy the cleft in MHC class I molecules. The overlapping, but distinct, sets of peptides provide an enormous repertoire that enables the protective functions of adaptive immunity. However, analysis strategies affect the outcome of the investigations. For example, if the peptides produced by i-proteasome or standard proteasome or mixed proteasomes are assessed based on isolating them from cell-surface MHC class I molecules, the repertoire of peptides will necessarily reflect the selection pressure of the requirement for the peptides to form stable, mature MHC class I molecules. Due to this selection pressure, such an analysis provides limited information about the total repertoire of proteasome-produced peptides, and limits the extrapolation of the results to the total repertoire of proteins that may be substrates for degradation via proteasomes. The role of i-proteasome in providing a set of peptides for MHC class I occupancy that is distinct, though overlapping, from that generated by standard proteasome is clear. It is also inescapable that i-proteasome, like the standard proteasome, performs more functions than provision of peptides for CD8 T cell recognition of MHC class I complexes. Expression of i-proteasome in cells that have little, if any, expression of conventional MHC class I molecules, neurons, supports the search for functions that are distinct from outcomes due to differences in antigen processing. I-proteasome expansion of the peptide repertoire may be but one of its activities. The new challenge is to explain how the moderate differences in the activities of standard proteasome or i-proteasomes or mixed proteasomes can account for the diversity of differences that are increasingly being found, and which are not easily attributed to peptides in MHC class I. The following section reviews the accruing evidence that suggests new roles for i-proteasomes that appear unrelated to generating peptides for MHC class I occupancy.
B. Nonimmune Roles for I-proteasome
Existing evidence that i-proteasome performs functions that are unrelated to antigen processing includes its expression in uninjured, immune privileged tissues, such as the retina30,84–87 and brain.85,88,89 Localization of i-proteasome to synapses and Purkinje cells in the brain,88,89 and the photoreceptors and neurons in the outer plexiform layer in the retina,85 implies a role in normal, neuronal function. Evidence supporting this idea includes the significantly reduced retinal function, as assessed by electroretinography, measured in both single- (LMP2) and double-deficient (LMP7/MECL-1) i-proteasome KO mice.86 It is important to note that neurons express no, or at best minimal, levels of the MHC class I molecule that provides antigen presentation for CD8 T lymphocytes.
In addition to helping regulate cellular homeostasis, i-proteasome may also take part in the cellular response to stress and injury. This idea was initially based on the observation that i-proteasome expression was substantially upregulated following stress or injury in a number of cells that do not typically present antigen. For example, i-proteasome is significantly upregulated in the central nervous system in response to acute injury to the retina85 and brain,85,90 in diseased retina84 and brain,88,89 and aging.30,89,91 It is also found and in cultured cells exposed to oxidative stress.30,92 Low levels of i-proteasome subunits that are expressed in both cardiac and skeletal muscle are significantly upregulated with injury due to viral infection,93 ischemia reperfusion,94 and muscle atrophy due to aging95,96 or diabetes.97 These initial studies provided the rationale for more in-depth investigations using KO mice.
Mice deficient in i-proteasome have been invaluable in providing keen insight into functions that go beyond i-proteasome’s contribution to antigen processing and provide compelling evidence for its role in multiple cellular processes. One idea is that i-proteasome helps to protect against oxidative damage, in part, due to its enhanced ability to degraded oxidized protein substrates.97–99 Increased levels of oxidized proteins in the brain and liver of LMP2 KO mice100 and the greater sensitivity to an oxidative challenge exhibited by retinal pigment epithelial cells from mice double-deficient in LMP7 and MECL-1 subunits85 support a role for protecting against oxidative damage.
I-proteasome may also participate in regulating tumor development. Uterine leiomyosarcoma is a tumor not known to be associated with human papilloma virus. This cancer has a 5-year survival rate of 35%, so there is an urgent need to understand the factors that influence tumor development. LMP2-deficient mice develop this tumor starting at 6 months of age, reaching a 40% incidence at 14 months.101 Importantly, LMP2 expression is absent in leiomyo-sarcoma tumors in human patients, but present in the benign uterine tumor leiomyoma. To identify potential defects in LMP2-deficient cells, primary cultures of tumor cells and splenocytes from LMP2-deficient mice were investigated. The significant findings were that LMP2-deficient cells exhibited reduced expression of IRF-1, and its expression was not upregulated by IFN-γ. IRF-1 regulates the activity of proteins involved in cell-cycle progression and, hence, cell transformation and proliferation. Additional testing will be required to determine if this initial association between LMP2 and IRF-1 is the critical factor in the development of this devastating cancer.
The link between i-proteasome with lipid metabolism and diabetes is supported by the data from humans with mutations in the PSMB8 gene that encodes LMP7 and data from KO mice. Lipodystrophy is a prominent clinical symptom of several PSMB8-associated syndromes that have been recently described in humans.51–53 These patients have low levels of mature i-proteasome containing LMP7 due to a defect in i-proteasome assembly. To investigate the potential mechanism for lipodystrophy, Kitamura and colleagues used human and murine preadipocyte cell lines, which both contain high levels of LMP7, and tested the effect of knocking down LMP7 expression using siRNA against the PSMB8 gene.53 Downregulation of the PSMB8 mRNA levels inhibited adipocyte differentiation, suggesting that LMP7 is required for adipocyte maturation. Additionally, subcutaneous injection of PSMB8 siRNA into BALB/c mice resulted in atrophic adipose tissue (smaller total size and number of adipocytes) compared with control mice. Taken together, these data suggest that LMP7 is critically involved in adipocyte maturation and may help explain the progressive lipodystrophy exhibited by patients with PSMB8-associated syndromes.
Heart disease, a well-known complication of diabetes, leads to progressive loss of cardiac function, that is, decreased contractility and cardiac muscle mass.102–104 Using the streptozotocin (STZ) model of diabetes, Zu and colleagues showed that when STZ-treated mice developed diabetes, LMP7 levels increased in cardiac tissue, but LMP2 and MECL-1 decreased.105 Exposure of isolated hearts to high glucose replicated these changes in i-proteasome expression, suggesting that the high glucose associated with diabetes could alter proteasome composition in cardiac muscle. Of note, altered proteasome composition was also observed in diabetic skeletal muscle.97 To determine if the loss in LMP2 expression had physiological consequences for the heart, cardiac mass and function were measured in LMP2 KO mice. The cardiomyopathy observed in the LMP2 KO mouse was similar to the cardiac phenotype in WT diabetic heart. These results suggest that altered proteasome composition, which can occur due to metabolic changes associated with diabetes, can have detrimental effects on cardiac muscle mass and function.
Insulin-dependent diabetes mellitus and diabetes insipidus are both autoimmune diseases with well-known etiologies. Diabetes mellitus is caused by an autoimmune attack on the insulin-producing cells in the pancreas, and diabetes insipidus is caused by a reduction in the antidiuretic vasopressin due to the autoimmune killing of vasopressin-producing cells in the hypothalamus. CD8 T cells may be involved in the targeted cell killing of the insulin-producing beta cells in the pancreas that is the pathologic event in type 1 diabetes.106,107 To investigate the role of i-proteasome in progression of autoimmune disease, WT and i-proteasome KO mice lacking LMP7 and MECL-1 subunits were irradiated, and then given bone marrow transfer.108 Following bone marrow reconstitution from either WT or i-proteasome KO mice, i-proteasome-deficient mice exhibited symptoms of multiorgan autoimmune disease, including inflamed skin, diabetes insipidus, and a latent form of diabetes mellitus. The authors also supply supporting evidence in humans from their disease-association analysis using the Type 1 Diabetes Genetics Consortium Major Histocompatibility Complex fine-mapping data set. The analysis identified two single-nucleotide polymorphisms (SNPs) in the PSMB8 gene encoding LMP7 that were genetic risk factors for development of type 1 diabetes. These data support a role for i-proteasome in protecting from the onset and progression of autoimmune diseases, such as diabetes.
I-proteasome appears to also limit inflammatory damage, potentially by ridding the cell of damaged (i.e., misfolded, oxidized) proteins and/or by regulating the profiles of cytokines produced in response to an inflammatory challenge. Data from Seifert and colleagues suggest that the i-proteasome is more efficient at degrading the “defective ribosomal products” (DRIPS), which are misfolded and/or oxidized proteins that are produced during protein synthesis.99 Cytokine-induced inflammation significantly upregulates protein synthesis and production of DRIPS, which, if not cleared from the cell, can produce toxic protein aggregates. In i-proteasome-deficient cells, challenges with either lipopolysaccharide- or IFN-γ-induced aggresome-like structures and increased levels of oxidized and ubiquitinated proteins. In contrast, aggre-somes and oxidized proteins were much reduced in cells containing the full complement of i-proteasome subunits. In vivo experiments support the in vitro observations. In an experimental model of lipopolysaccharide-induced experimental autoimmune encephalitis, LMP7 KO mice exhibited a more severe clinical score, and higher levels of oxidized proteins and protein aggregates in the inflamed liver and brain.99
Other studies also support the idea that i-proteasome preserves protein homeostasis and viability in cells under an inflammatory challenge. In a murine model of acute enterovirus myocarditis, which is one of the most common viral diseases among young people, mice deficient in LMP7 exhibited severe myocardial destruction and large inflammatory foci in cardiac cells.93 Additionally, cardiomyocytes from the LMP7 KO mice accumulated substantially more oxidized and polyubiquitinated proteins compared with WT cells following exposure to IFN-γ.
In addition to enhanced proteolysis of damaged proteins, i-proteasome could also protect cells by regulating the profile of cytokines produced in response to an inflammatory challenge. Several cytokines that are secreted in innate and adaptive immune responses (i.e., IL-6, TNFα, IFN-γ, etc.) can amplify the inflammation in surrounding tissues and cause collateral damage. Endogenous regulatory mechanisms dampen the cytokine response to achieve a balance of destructive inflammation and effective control of infections. In cells from patients with PSMB8-associated mutations in LMP7, basal production of IL-6 was significantly higher compared to cells containing nonmutant LMP7.52,53 Stimulation with TNFα enhanced production of IL-6 in cultured fibroblasts from patients having Nakajo–Nashimura syndrome (NNS) who have lower levels of LMP7.52 In thioglycollate-elicited macrophages, exposure to lipopolysaccharide elicited lower IL-6 production in macrophages from LMP7/MECL-1 double-deficient i-proteasome mice.109 These examples show that the i-proteasome content can affect cytokine production in response to an inflammatory challenge.
Examples provided to this point have shown that i-proteasome deficiency is detrimental to the course of the disease, that is, experimental autoimmune encephalitis, acute enterovirus myocarditis, and PSMB8-associated syndromes. There are, however, several examples where i-proteasome deficiency or inactivation can be protective. Schmidt and colleagues used the dextran sodium sulfate-induced colitis model to examine the influence of LMP7 deficiency on intestinal inflammation.110 Several findings showed that LMP7 KO mice exhibited less inflammation in the tissue, produced lower levels of proinflammatory cytokines, and recruited fewer Gr-1-positive inflammatory cells and fewer cytokine-producing CD4 T cells. The positive effect of i-proteasome deficiency was replicated in a second study of experimentally induced colitis for mice with individual i-proteasome subunits removed (LMP2, LMP7, and MECL-1).111 A significant reduction in inflammatory markers was also found when the LMP7-specific inhibitor, PR-957, was used. This inhibitor was also successful in blocking cytokine production and reducing the inflammatory symptoms of experimental arthritis.112
Whether i-proteasome deficiency exerts a protective effect or exacerbates disease symptoms may depend on the underlying pathogenesis and/or tissue affected by the disease. What may be especially relevant to disease outcomes is how i-proteasome regulates specific cellular signaling pathways. There is a substantial body of literature that shows how i-proteasome can directly regulate several key signaling pathways, including NF-κB. The NF-κB pathway is one of the major mechanisms for responding to a broad range of stressors, such as cytokines, viral and bacterial products, toxic chemicals, UV light, and oxidative damage.113 NF-κB activation elicits rapid induction of early-response genes that help protect the cell from damage. Production of several proinflammatory cytokines is also regulated by NF-κB, so this pathway is particularly important for cells under an inflammatory challenge. Aberrant NF-κB regulation can lead to pathologies such as toxic shock, and neurodegenerative and inflammatory diseases.114
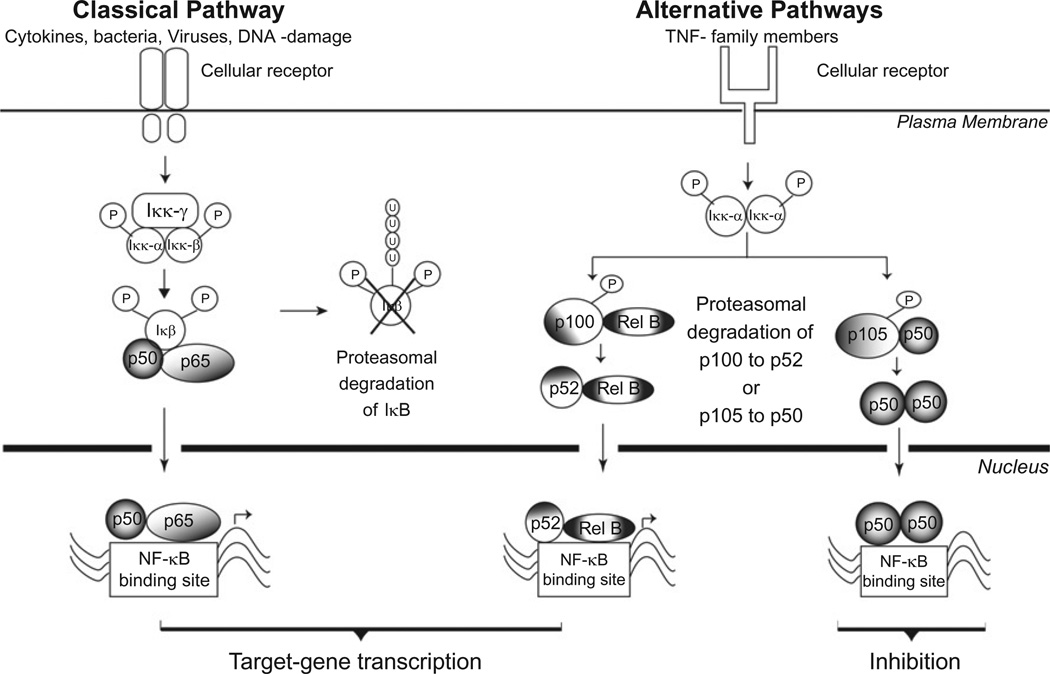
The NF-κB family consists of multiple proteins, including kinases, inhibitors (IκB), and transcription factors (p65, Rel-B, c-rel, p105/p50, p100/p52). Transcription factors associate to form homo- or heterodimers and are sequestered in the cytosol by inhibitory proteins (IκB) until upstream signaling events cause phosphorylation of select NF-κB family members (Fig. 4). Phosphorylation triggers ubiquitination, which signals selective proteolysis by the protea-some and allows for the subsequent translocation of the NF-κB dimer into the nucleus. Two separate pathways for proteasome-dependent activation include(1) the classical pathway, where the IκB inhibitory protein is degraded, thereby releasing the NF-κB dimer and allowing it to enter the nucleus, and (2) the alternative pathway, where proteolytic processing of the p100 or p105 precursor proteins removes the inhibitory portion of the protein via endoproteolysis, thereby generating the activated transcription factor p52 or p50.115 Defects in proteolysis of IκB and processing of p100/p105 have been reported in i-proteasome-deficient cells.
Fig. 4.
Proteasome-dependent activation of NF-κB. Receptor-mediated pathways of NF-κB activation include the classical pathway, which involves the binding of a ligand (i.e., cytokines, virus, or bacteria) to a receptor (i.e., IFN-γ or toll-like receptors), and subsequent activation of intracellular kinases that phosphorylate the inhibitory protein, IκB. Phosphorylation is the signal for targeted ubiquitination of IκB, followed by degradation by the proteasome. The release of the inhibitory protein from the transcription factor dimer exposes the nuclear localization signal, which facilitates the movement of the NF-κB dimer into the nucleus. The alternative pathway uses signals from ligand-bound TNF receptors, which trigger the phosphorylation of p100 or p105 by intracellular kinases and the subsequent endoproteolytic degradation of p100 or p105 to form the active transcription factor p52 or p50. Translocation and localization of heterodimers (i.e., p50/p65 or p52/ relB) to the NF-κB binding sites initiates transcription of the NF-κB-responsive gene. In contrast, binding of the p50 or p52 homodimers inhibits transcription of NF-κB responsive genes.
Inhibition and termination of the NF-κB signal is also regulated by protea-some. Dimers of p50 or p52, which are produced by proteasome-dependent processing of p105 and p100, are transcriptionally inactive and can prevent active transcription by blocking the NF-κB promoter region. Proteasome is also involved in terminating the signal, by degrading transcription factors (i.e., p65) that are docked on the promoter region.115
Evidence from i-proteasome-deficient mice has provided compelling support for the role of i-proteasome in NF-κB activation. In cell lines lacking LMP2, defects in the proteolytic processing of NF-κB precursors (p100/p105) to the active transcription factors (p52/p50), and decreased degradation of the NF-κB inhibitory protein IκBα, were observed.116–118 While findings reported from these initial studies were controversial,119,120 more recent studies have provided substantial evidence for i-proteasome regulation of NF-κB. For example, enhanced processing of NF-κB precursors p105 and degradation of IκBα were reported for i-proteasome isolated from the inflamed intestine of patients with Crohn’s disease.121 In TNFα-stimulated embryonic fibroblasts from LMP7 KO mice, reduced nuclear translocation of p65 suggested inhibited NF-κB activation.110 Findings consistent with these were found in studies of B cells from LMP2-deficient mice; delayed and less complete degradation of IκBα following lipopolysaccharide stimulation was observed.72
Recent evidence for regulation by i-proteasomes on events upstream of NF-κB was found using lipopolysaccharide to stimulate activated macrophages collected from i-proteasome-deficient mice.109 Lipopolysaccharide can induce production of the cytokine TNFα using the MyD88/TRAF6 pathway, while production of nitric oxide is accomplished via the TRIF/TRAF3 pathway. Both pathways ultimately intersect with and activate NF-κB. Stimulation by lipopolysaccharide alone resulted in a severalfold reduction in nitric oxide production for LMP2/LMP7 double KO macrophages but had no effect on TNFα production, suggesting defects in signaling through the TRIF/TRAF3 pathway. Western blots of key signaling molecules in this pathway also confirmed that LMP2/LMP7 double KO macrophages were unable to effectively signal via TRIF/TRAM.
I-proteasome may also regulate the signaling molecule phosphatase and tensin homologue deleted on chromosome 10 (PTEN). PTEN is a phosphatase that functions as a regulator of cell growth by inhibiting Akt signaling. Recent studies have shown that PTEN is an important determinant of cardiac muscle size.94,105 Notably, these studies also show that PTEN content in cardiac muscle is regulated by the presence of LMP2. The supporting data show that PTEN KO mice have larger hearts compared with WT mice, whereas hearts from LMP2 KO mice, which contain high PTEN, were significantly smaller and exhibited impaired contractility. Treatment of LMP2 KO hearts with a PTEN inhibitor improved cardiac function. PTEN is also involved in the cellular changes associated with cardioprotection following ischemic preconditioning. In WT mice, ischemic preconditioning led to lower PTEN content and activation of Akt. In LMP2 KO mice, PTEN content and Akt were unchanged, and functional measures of cardiac output showed no cardioprotection after ischemic preconditioning.
In summary, there is substantial data supporting a role for i-proteasome-mediated regulation of signaling pathways, including NF-κB and PTEN. Regulation may occur through degradation of proteins in the signaling cascade or through degradation of regulatory molecules that either inhibit or activate the signal. With most biological systems, there is considerable endogenous redundancy that serves to prevent catastrophic outcomes. This strategy is likely to apply to the functions provided by the i-proteasome, especially since i-proteasome deletion is not lethal. Its regulation may be subtle, perhaps by fine tuning the rate of signaling or downstream events, such as cytokine production. It is also highly likely that i-proteasome’s involvement also depends on the cell type and stimulation. We really are only beginning to understand some of the nuances in regulation by the i-proteasome.
VIII. Mutations and Linkage to Human Disease
The list of human diseases that have been linked to deregulation of the i-proteasome has grown exponentially over the past decade. However, the data supporting this link are often indirect. For example, increased i-proteasome expression, altered proteasome activity, and/or accumulation of ubiquitinated protein in the diseased tissue have been reported in multiple diseases, including several neurodegenerative diseases of the brain and retina.84,88,89 These diseases share oxidation and/or inflammation as part of the disease mechanism and, thus, changes in proteasome expression and activity are likely a consequence of the altered cellular condition rather than part of the primary disease mechanism. Experiments in cultured cells support this idea. Exposure of cells to sublethal levels of oxidative stress,30,92 inflammatory cytokines,32,33 or cellular expression of aggregate-prone proteins,32,122 which is an integral part of the mechanism of Huntington disease, all induce expression of i-proteasome. Therefore, one needs to be cautious in assuming that the correlation between altered i-proteasome expression or function and disease has mechanistic implications.
While the increased expression of i-proteasome in diseased tissue provides limited mechanistic information, it has opened the door to novel therapeutic options that use inhibitors (i.e., PR-957, PR-924, IPSI-001) that selectively target the i-proteasome catalytic subunits.123,124 This approach is designed to either specifically kill cells expressing high levels of i-proteasome, as is the goal with cancer, or inhibit signaling pathways, such as the proinflammatory NF-κB pathway in autoimmune disease. Since i-proteasome inhibitors should not affect the activity of the standard proteasome, which is constitutively present in all cells, i-proteasome inhibitors should be less toxic and have reduced off-target side effects compared with other broad-spectrum proteasome inhibitors, such as bortezomib and carfilozomib. Currently, preclinical studies with i-proteasome inhibitors are under way in the treatment of rheumatoid arthritis, inflammatory bowel disease, and cancer.111,112,125 Initial results have been promising. As a caveat, continued discovery of i-proteasome’s ever-expanding role in maintaining cellular homeostasis and in responding to stress suggests that evidence for unexpected side effects in off-target sites need to be vigilantly monitored.
Recent reports of the association between specific diseases and point mutations in i-proteasome subunits provide direct evidence that a loss in i-proteasome function can lead to disease. To date, discrete mutations in the LMP7 protein have defined a spectrum of PSMB8-associated autoinflammatory syndromes, which includes an inflammatory response in the absence of infection. These syndromes are distinguished from autoimmune disease because they primarily result from deregulation of the innate immune system rather than adaptive immunity. Notably, there is considerable overlap in symptoms between the different PSMB8-associated syndromes potentially because the primary cause is the altered function of the LMP7 protein. In some instances, the detailed structural analysis of the mutant protein has provided significant insight into the molecular details responsible for the development of the disease. Discovery of these PSMB8-associated syndromes highlights the profound effect i-proteasome dysfunction has on inflammation and function of specific organs.
One of the first described PSMB8-associated syndromes was an autosomal-recessive disease known as joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome (JMP).126 Patients with JMP exhibit mild metabolic disturbances (i.e., hypertriglycerides, low high-density lipoprotein cholesterol), elevated liver enzymes, muscle atrophy, hypergammaglobulinemia, and joint contractures mainly in the hands and feet. Genome-wide homozygosity mapping identified a missense mutation (c.224C>T) causing substitution at residue 75 (T75M) that resulted in loss of function for the LMP7 i-proteasome catalytic subunit due to a disruption in the tertiary structure.51
Another i-proteasome-associated inflammatory disorder is chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE) syndrome. The onset of CANDLE disease symptoms occurs within the first year of life and is characterized by recurrent fever, skin lesions, progressive lipodystrophy, and delayed development. The genetic cause was identified as two different mutations in the PSMB8 gene.54 In a subpopulation of patients, the missense mutation (c.224C>T) causing a methionine substitution at residue 75 (T75M) in the LMP7 protein was discovered. While there is some overlap in symptoms in patients with JMP and CANDLE, joint contracture and muscle atrophy is not present in CANDLE syndrome, which is distinguished by unique features, such as immature neutrophils in cellular infiltrates. The reason for the discrepancy in symptoms between patients harboring the same mutation is still unclear. A second mutation (c.405C>A) identified in CANDLE patients caused a substitution of a highly conserved cysteine at residue 135 to a stop codon (p.C135X), which resulted in truncation of 141 amino acids from the C-terminal end of the protein.
Patients with NNS exhibit periodic fever, skin rash, lipomuscular atrophy, and joint contractures due to a mutation (G201V) in LMP7.52 This mutation causes a conformational change that alters the catalytic binding pocket around residues Thr73 and Lys105, resulting in significantly decreased chymotrypsin-like activity. The loss in activity prevents the autolytic cleavage between Gly72 and Thr73, which is required for the LMP7 protein to be incorporated into the 20S core particle. Consequently, the assembly defect caused by the mutation-induced change in protein structure results in lower total content of the 20S and 26S proteasomes in cells normally expressing high levels of i-proteasome. This decrease in i-proteasome content was suggested to be responsible for the increased levels of oxidized and ubiquitinated proteins and activation of the p38 pathway in cells (monocytes, fibroblasts) from patients with NNS.
Autoinflammation and lipodystrophy are characteristics associated with Japanese autoinflamatory syndrome with lipodystrophy, which shares many of the characteristics exhibited by NNS patients but is due to a unique homozygous missense mutation (G197V) in the PSMB8 gene.53 The G197V mutation caused a reduction in cellular levels of the LMP7 protein, which consequently resulted in the accumulation of immature i-proteasome cores due to the loss of LMP7 processing. Disturbed adipocyte maturation is another key feature of this disease and highlights the widespread effect that i-proteasome defects can have on cells outside of the immune system.
One of the clinical features shared by several PSMB8-associated syndromes was deregulation of lipid processing, suggesting a potential role for i-proteasome in the metabolism and, more specifically, insulin regulation. In support of this idea, two SNPs in the PSMB8 gene (rs3763365, located 1048 base pairs downstream, and rs9276810 in intron 3) have been identified as an independent genetic risk factor for the development of type 1 diabetes.108
SNPs in the PSMB9 gene that encodes the LMP2 protein have also been associated with disease susceptibility. The SNP in exon 3 is a nonconservative base pair change that encodes two different amino acids (arginine (R) or histidine (H)) at position 60 in the LMP2 protein. Patients with the PSMB9 SNP rs17587 in exon 3, which results in an R60H substitution in LMP2, are at risk for developing ankylosing spondylitis, a chronic inflammatory joint disease that predominantly affects the spine.127 Genotype analysis showed an increased frequency and younger age at disease onset for patients with the R/R genotype.128 It was postulated that the excess bone growth associated with ankylosing spondylitis was likely due to altered NF-κB regulation by the mutant LMP2 protein.127 The NF-κB pathway affects osteoclastic activity by modulating the Wnt/β-catenin pathway, which is overactive in this disease due to low levels of the Wnt/β-catenin inhibitor, Dicckopf-1. This also raises the possibility that Dicckopf-1 could be a substrate for i-proteasome degradation. The R60H polymorphism in LMP2 has also demonstrated a strong association with psoriasis, another autoimmune disease.129
Specific LMP2 codon 60 polymorphisms can also confer resistance to disease. Genotype analysis of a large Italian population showed that individuals with the LMP2 H/H genotype had a reduced risk for developing multiple sclerosis, potentially due to the decreased production of immunodominant peptides from myelin basic protein.130 To experimentally test this idea, in vitro digestion of a 28-mer peptide from myelin basic protein (MBP102– 129) was performed using purified 20S i-proteasome containing LMP2 with either histidine or arginine in codon 60. Histidine-containing i-proteasomes demonstrated reduced degradation of the 28-mer peptide to the major immunodominant (MBP111–119) epitope. These results provide a potential mechanism that could explain the reduced risk for multiple sclerosis in individuals with the H/H genotype.
The strong association of the LMP2 codon 60 polymorphism with disease susceptibility has prompted a more in-depth examination of how this polymorphism can affect subunit structure and function.131 Structural considerations were studied using an in silico model that was based on the crystal structure of i-proteasome described by Unno and colleagues132 and taking into account the biochemical differences conferred by the two amino acids. In the mature protein, which has undergone proteolytic cleavage of the N terminus, the amino acid in codon 60 corresponds to residue 40. The crystal structure places this residue on the external surface of the core, close to the interface of the α- and β-rings but not in direct contact with either the α-subunits or the catalytic active site. The biochemical differences conferred by substitution of Arg for His could be quite substantial due to the increased size, which could introduce steric hindrance, and altered charge state of the amino acid from neutral (His) to positive (Arg). Based on these structural and biochemical considerations, three hypotheses were proposed to explain how the polymorphisms could alter i-proteasome function: (i) the residue at amino acid 40 could be the binding site for regulatory molecules (protein and nonprotein) whose affinity is dependent on the amino acid that is present; (ii) the difference in charge state could alter the electrical field at the catalytic site and affect activity; and (iii) the charge change could also affect the stability of the 20S core. The net charge of the α-subunits is − 12, and with arginine in residue 40, the net charge of the β-subunits is +12. The resultant electrical interaction between these opposing charge states likely contributes to the affinity between the two rings and aids in the assembly of the core particle. The presence of the neutral-charged His may have a negative effect on this process. The positive charge conferred by Arg at residue 40 also helps stabilize the structure via hydrogen bonds between water molecules and may also affect the positioning of the protein loops that form the channels through which substrates access the catalytic active sites in the core. On the other hand, the reduced hydrogen bonding and consequent greater structural flexibility conferred by the presence of the neutral His may help open the channel and permit greater access to the catalytic active site. These results suggest that the structural and biochemical changes caused by substitution of Arg or His at residue 40 in the LMP2 protein, which occurs as an SNP in the PSMB9 gene, can have a significant impact on i-proteasome function and may explain the disease susceptibility of specific genotypes.
Without a doubt, the number of human diseases directly linked to defects in i-proteasome will continue to grow. That information will greatly aid our understanding of i-proteasome’s ever-expanding role in cellular processes that are unrelated to the generation of peptides for antigen presentation.
Abbreviations
- IFN-γ
interferon-gamma
- LMP
low-molecular-weight protein
- MECL-1
Multi-catalytic endopeptidase complex subunit 1
- MHC class I
major histocompatibility complex class I
References
- 1.Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81:1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 2.Etlinger JD, Goldberg AL. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci USA. 1977;74:54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown MG, Driscoll J, Monaco JJ. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature. 1991;353:355–357. doi: 10.1038/353355a0. [DOI] [PubMed] [Google Scholar]
- 4.Glynne R, Powis SH, Beck S, Kelly A, Kerr LA, Trowsdale J. A proteasome-related gene between the two ABC transporter loci in the class II region of the human MHC. Nature. 1991;353:357–360. doi: 10.1038/353357a0. [DOI] [PubMed] [Google Scholar]
- 5.Kelly A, Powis SH, Glynne R, Radley E, Beck S, Trowsdale J. Second proteasome-related gene in the human MHC class II region. Nature. 1991;353:667–668. doi: 10.1038/353667a0. [DOI] [PubMed] [Google Scholar]
- 6.Ortiz-Navarrete V, Seelig A, Gernold M, Frentzel S, Kloetzel PM, Hammerling GJ. Subunit of the ‘20S’ proteasome (multicatalytic proteinase) encoded by the major histocompatibility complex. Nature. 1991;353:662–664. doi: 10.1038/353662a0. [DOI] [PubMed] [Google Scholar]
- 7.Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T, et al. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J Biochem. 1994;115:257–269. doi: 10.1093/oxfordjournals.jbchem.a124327. [DOI] [PubMed] [Google Scholar]
- 8.Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel PM. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur J Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi M, Ishibashi T, Tanaka K, Kasahara M. The mouse genes encoding the third pair of beta-type proteasome subunits regulated reciprocally by IFN-gamma: structural comparison, chromosomal localization, and analysis of the promoter. J Immunol. 1997;159:2760–2770. [PubMed] [Google Scholar]
- 10.Nandi D, Jiang H, Monaco JJ. Identification of MECL-1 (LMP-10) as the third IFN-gamma-inducible proteasome subunit. J Immunol. 1996;156:2361–2364. [PubMed] [Google Scholar]
- 11.Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 12.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 13.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 14.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J. 1998;335(Pt 3):637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahlmann B, Ruppert T, Kuehn L, Merforth S, Kloetzel PM. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J Mol Biol. 2000;303:643–653. doi: 10.1006/jmbi.2000.4185. [DOI] [PubMed] [Google Scholar]
- 16.Klare N, Seeger M, Janek K, Jungblut PR, Dahlmann B. Intermediate-type 20 S proteasomes in HeLa cells: “asymmetric” subunit composition, diversity and adaptation. J Mol Biol. 2007;373:1–10. doi: 10.1016/j.jmb.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 17.Drews O, Wildgruber R, Zong C, Sukop U, Nissum M, Weber G, et al. Mammalian protea-some subpopulations with distinct molecular compositions and proteolytic activities. Mol Cell Proteomics. 2007;6:2021–2031. doi: 10.1074/mcp.M700187-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Groettrup M, Kirk CJ, Basler M. Proteasomes in immune cells: more than peptide producers? Nat Rev Immunol. 2010;10:73–78. doi: 10.1038/nri2687. [DOI] [PubMed] [Google Scholar]
- 19.Monaco JJ, McDevitt HO. H-2-linked low-molecular weight polypeptide antigens assemble into an unusual macromolecular complex. Nature. 1984;309:797–799. doi: 10.1038/309797a0. [DOI] [PubMed] [Google Scholar]
- 20.Monaco JJ, McDevitt HO. The LMP antigens: a stable MHC-controlled multisubunit protein complex. Hum Immunol. 1986;15:416–426. doi: 10.1016/0198-8859(86)90019-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhou P, Zanelli E, Smart M, David C. Genomic organization and tissue expression of mouse proteasome gene Lmp-2. Genomics. 1993;16:664–668. doi: 10.1006/geno.1993.1245. [DOI] [PubMed] [Google Scholar]
- 22.Fruh K, Yang Y, Arnold D, Chambers J, Wu L, Waters JB, et al. Alternative exon usage and processing of the major histocompatibility complex-encoded proteasome subunits. J Biol Chem. 1992;267:22131–22140. [PubMed] [Google Scholar]
- 23.Wright KL, White LC, Kelly A, Beck S, Trowsdale J, Ting JP. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J Exp Med. 1995;181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Waters JB, Fruh K, Peterson PA. Proteasomes are regulated by interferon gamma: implications for antigen processing. Proc Natl Acad Sci USA. 1992;89:4928–4932. doi: 10.1073/pnas.89.11.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James AB, Conway AM, Morris BJ. Regulation of the neuronal proteasome by Zif268 (Egr1) J Neurosci. 2006;26:1624–1634. doi: 10.1523/JNEUROSCI.4199-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanelli E, Zhou P, Cao H, Smart MK, David CS. Genomic organization and tissue expression of the mouse proteasome gene Lmp-7. Immunogenetics. 1993;38:400–407. doi: 10.1007/BF00184520. [DOI] [PubMed] [Google Scholar]
- 28.Hisamatsu H, Shimbara N, Saito Y, Kristensen P, Hendil KB, Fujiwara T, et al. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J Exp Med. 1996;183:1807–1816. doi: 10.1084/jem.183.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruz M, Elenich LA, Smolarek TA, Menon AG, Monaco JJ. DNA sequence, chromosomal localization, and tissue expression of the mouse proteasome subunit lmp10 (Psmb10) gene. Genomics. 1997;45:618–622. doi: 10.1006/geno.1997.4977. [DOI] [PubMed] [Google Scholar]
- 30.Hussong SA, Kapphahn RJ, Phillips SL, Maldonado M, Ferrington DA. Immunoproteasome deficiency alters retinal proteasome’s response to stress. J Neurochem. 2010;113:1481–1490. doi: 10.1111/j.1471-4159.2010.06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bose S, Brooks P, Mason GG, Rivett AJ. gamma-Interferon decreases the level of 26 S proteasomes and changes the pattern of phosphorylation. Biochem J. 2001;353:291–297. doi: 10.1042/0264-6021:3530291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz-Hernandez M, Martin-Aparicio E, Avila J, Hernandez F, Lucas JJ. Enhanced induction of the immunoproteasome by interferon gamma in neurons expressing mutant Huntingtin. Neurotox Res. 2004;6:463–468. doi: 10.1007/BF03033282. [DOI] [PubMed] [Google Scholar]
- 33.Gregerson DS, Lew KL, McPherson SW, Heuss ND, Ferrington DA. RPE cells resist bystander killing by CTLs, but are highly susceptible to antigen-dependent CTL killing. Invest Ophthalmol Vis Sci. 2006;47:5385–5394. doi: 10.1167/iovs.06-0636. [DOI] [PubMed] [Google Scholar]
- 34.Kotamraju S, Matalon S, Matsunaga T, Shang T, Hickman-Davis JM, Kalyanaraman B. Upregulation of immunoproteasomes by nitric oxide: potential antioxidative mechanism in endothelial cells. Free Radic Biol Med. 2006;40:1034–1044. doi: 10.1016/j.freeradbiomed.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 35.Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 36.Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- 37.Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 38.De M, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. Beta 2 subunit propeptides influence cooperative proteasome assembly. J Biol Chem. 2003;278:6153–6159. doi: 10.1074/jbc.M209292200. [DOI] [PubMed] [Google Scholar]
- 39.Groettrup M, Standera S, Stohwasser R, Kloetzel PM. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc Natl Acad Sci USA. 1997;94:8970–8975. doi: 10.1073/pnas.94.17.8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, et al. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J Exp Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, et al. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci USA. 2010;107:18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heink S, Ludwig D, Kloetzel PM, Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc Natl Acad Sci USA. 2005;102:9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 44.Borissenko L, Groll M. Diversity of proteasomal missions: fine tuning of the immune response. Biol Chem. 2007;388:947–955. doi: 10.1515/BC.2007.109. [DOI] [PubMed] [Google Scholar]
- 45.Yewdell JW. Immunoproteasomes: regulating the regulator. Proc Natl Acad Sci USA. 2005;102:9089–9090. doi: 10.1073/pnas.0504018102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Kaer L, Ashton-Rickardt PG, Eichelberger M, Gaczynska M, Nagashima K, Rock KL, et al. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 47.Fehling HJ, Swat W, Laplace C, Kuhn R, Rajewsky K, Muller U, et al. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 48.Basler M, Moebius J, Elenich L, Groettrup M, Monaco JJ. An altered T cell repertoire in MECL-1-deficient mice. J Immunol. 2006;176:6665–6672. doi: 10.4049/jimmunol.176.11.6665. [DOI] [PubMed] [Google Scholar]
- 49.Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, Delgado JC, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. 2011;13:129–135. doi: 10.1038/ni.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basler M, Beck U, Kirk CJ, Groettrup M. The antiviral immune response in mice devoid of immunoproteasome activity. J Immunol. 2011;187:5548–5557. doi: 10.4049/jimmunol.1101064. [DOI] [PubMed] [Google Scholar]
- 51.Agarwal AK, Xing C, DeMartino GN, Mizrachi D, Hernandez MD, Sousa AB, et al. PSMB8 encoding the beta5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. Am J Hum Genet. 2010;87:866–872. doi: 10.1016/j.ajhg.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arima K, Kinoshita A, Mishima H, Kanazawa N, Kaneko T, Mizushima T, et al. Proteasome assembly defect due to a proteasome subunit beta type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc Natl Acad Sci USA. 2011;108:14914–14919. doi: 10.1073/pnas.1106015108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitamura A, Maekawa Y, Uehara H, Izumi K, Kawachi I, Nishizawa M, et al. A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J Clin Invest. 2011;121:4150–4160. doi: 10.1172/JCI58414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Ramot Y, Torrelo A, Paller AS, Si N, Babay S, et al. Mutations in PSMB8 cause CANDLE syndrome with evidence of genetic and phenotypic heterogeneity. Arthritis Rheum. 2011;64:895–907. doi: 10.1002/art.33368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 56.Dalet A, Stroobant V, Vigneron N, Van den Eynde BJ. Differences in the production of spliced antigenic peptides by the standard proteasome and the immunoproteasome. Eur J Immunol. 2011;41:39–46. doi: 10.1002/eji.201040750. [DOI] [PubMed] [Google Scholar]
- 57.Rape M, Jentsch S. Taking a bite: proteasomal protein processing. Nat Cell Biol. 2002;4:E113–E116. doi: 10.1038/ncb0502-e113. [DOI] [PubMed] [Google Scholar]
- 58.York IA, Goldberg AL, Mo XY, Rock KL. Proteolysis and class I major histocompatibility complex antigen presentation. Immunol Rev. 1999;172:49–66. doi: 10.1111/j.1600-065x.1999.tb01355.x. [DOI] [PubMed] [Google Scholar]
- 59.Brossart P, Bevan MJ. Presentation of exogenous protein antigens on major histocompatibility complex class I molecules by dendritic cells: pathway of presentation and regulation by cytokines. Blood. 1997;90:1594–1599. [PMC free article] [PubMed] [Google Scholar]
- 60.Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- 61.Cardozo C, Kohanski RA. Altered properties of the branched chain amino acid-preferring activity contribute to increased cleavages after branched chain residues by the “immunopro-teasome”. J Biol Chem. 1998;273:16764–16770. doi: 10.1074/jbc.273.27.16764. [DOI] [PubMed] [Google Scholar]
- 62.Chapiro J, Claverol S, Piette F, Ma W, Stroobant V, Guillaume B, et al. Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J Immunol. 2006;176:1053–1061. doi: 10.4049/jimmunol.176.2.1053. [DOI] [PubMed] [Google Scholar]
- 63.Lei B, Abdul Hameed MD, Hamza A, Wehenkel M, Muzyka JL, Yao XJ, et al. Molecular basis of the selectivity of the immunoproteasome catalytic subunit LMP2-specific inhibitor revealed by molecular modeling and dynamics simulations. J Phys Chem B. 2010;114:12333–12339. doi: 10.1021/jp1058098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sijts AJ, Ruppert T, Rehermann B, Schmidt M, Koszinowski U, Kloetzel PM. Efficient generation of a hepatitis B virus cytotoxic T lymphocyte epitope requires the structural features of immunoproteasomes. J Exp Med. 2000;191:503–514. doi: 10.1084/jem.191.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Hall T, Sijts A, Camps M, Offringa R, Melief C, Kloetzel PM, et al. Differential influence on cytotoxic T lymphocyte epitope presentation by controlled expression of either proteasome immunosubunits or PA28. J Exp Med. 2000;192:483–494. doi: 10.1084/jem.192.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morel S, Levy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin AL, et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 67.Chen W, Norbury CC, Cho Y, Yewdell JW, Bennink JR. Immunoproteasomes shape immu-nodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J Exp Med. 2001;193:1319–1326. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Helden MJ, de Graaf N, Bekker CP, Boog CJ, Zaiss DM, Sijts AJ. Immunoproteasome-deficiency has no effects on NK cell education, but confers lymphocytes into targets for NK cells in infected wild-type mice. PLoS One. 2011;6:e23769. doi: 10.1371/journal.pone.0023769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Graaf N, van Helden MJ, Textoris-Taube K, Chiba T, Topham DJ, Kloetzel PM, et al. PA28 and the proteasome immunosubunits play a central and independent role in the production of MHC class I-binding peptides in vivo. Eur J Immunol. 2011;41:926–935. doi: 10.1002/eji.201041040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cerundolo V, Elliott T, Elvin J, Bastin J, Rammensee HG, Townsend A. The binding affinity and dissociation rates of peptides for class I major histocompatibility complex molecules. Eur J Immunol. 1991;21:2069–2075. doi: 10.1002/eji.1830210915. [DOI] [PubMed] [Google Scholar]
- 71.Townsend A, Ohlen C, Foster L, Bastin J, Ljunggren HG, Karre K. A mutant cell in which association of class I heavy and light chains is induced by viral peptides. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):299–308. doi: 10.1101/sqb.1989.054.01.038. [DOI] [PubMed] [Google Scholar]
- 72.Hensley SE, Zanker D, Dolan BP, David A, Hickman HD, Embry AC, et al. Unexpected role for the immunoproteasome subunit LMP2 in antiviral humoral and innate immune responses. J Immunol. 2010;184:4115–4122. doi: 10.4049/jimmunol.0903003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Girdlestone J, Isamat M, Gewert D, Milstein C. Transcriptional regulation of HLA-A and -B: differential binding of members of the Rel and IRF families of transcription factors. Proc Natl Acad Sci USA. 1993;90:11568–11572. doi: 10.1073/pnas.90.24.11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khan S, van den Broek M, Schwarz K, de Giuli R, Diener PA, Groettrup M. Immunoprotea-somes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J Immunol. 2001;167:6859–6868. doi: 10.4049/jimmunol.167.12.6859. [DOI] [PubMed] [Google Scholar]
- 75.Strehl B, Joeris T, Rieger M, Visekruna A, Textoris-Taube K, Kaufmann SH, et al. Immu-noproteasomes are essential for clearance of Listeria monocytogenes in nonlymphoid tissues but not for induction of bacteria-specific CD8+ T cells. J Immunol. 2006;177:6238–6244. doi: 10.4049/jimmunol.177.9.6238. [DOI] [PubMed] [Google Scholar]
- 76.Tu L, Moriya C, Imai T, Ishida H, Tetsutani K, Duan X, et al. Critical role for the immuno-proteasome subunit LMP7 in the resistance of mice to Toxoplasma gondii infection. Eur J Immunol. 2009;39:3385–3394. doi: 10.1002/eji.200839117. [DOI] [PubMed] [Google Scholar]
- 77.Murata S, Takahama Y, Tanaka K. Thymoproteasome: probable role in generating positively selecting peptides. Curr Opin Immunol. 2008;20:192–196. doi: 10.1016/j.coi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 78.Osterloh P, Linkemann K, Tenzer S, Rammensee HG, Radsak MP, Busch DH, et al. Protea-somes shape the repertoire of Tcells participating in antigen-specific immune responses. Proc Natl Acad Sci USA. 2006;103:5042–5047. doi: 10.1073/pnas.0509256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Verteuil D, Muratore-Schroeder TL, Granados DP, Fortier MH, Hardy MP, Bramoulle A, et al. Deletion of immunoproteasome subunits imprints on the transcriptome and has a broad impact on peptides presented by major histocompatibility complex I molecules. Mol Cell Proteomics. 2010;9:2034–2047. doi: 10.1074/mcp.M900566-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dick TP, Ruppert T, Groettrup M, Kloetzel PM, Kuehn L, Koszinowski UH, et al. Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell. 1996;86:253–262. doi: 10.1016/s0092-8674(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 81.Sijts A, Sun Y, Janek K, Kral S, Paschen A, Schadendorf D, et al. The role of the proteasome activator PA28 in MHC class I antigen processing. Mol Immunol. 2002;39:165–169. doi: 10.1016/s0161-5890(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 82.Sun Y, Sijts AJ, Song M, Janek K, Nussbaum AK, Kral S, et al. Expression of the proteasome activator PA28 rescues the presentation of a cytotoxic T lymphocyte epitope on melanoma cells. Cancer Res. 2002;62:2875–2882. [PubMed] [Google Scholar]
- 83.Textoris-Taube K, Henklein P, Pollmann S, Bergann T, Weisshoff H, Seifert U, et al. The N-terminal flanking region of the TRP2360-368 melanoma antigen determines proteasome activator PA28 requirement for epitope liberation. J Biol Chem. 2007;282:12749–12754. doi: 10.1074/jbc.M611644200. [DOI] [PubMed] [Google Scholar]
- 84.Ethen CM, Hussong SA, Reilly C, Feng X, Olsen TW, Ferrington DA. Transformation of the proteasome with age-related macular degeneration. FEBS Lett. 2007;581:885–890. doi: 10.1016/j.febslet.2007.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferrington DA, Hussong SA, Roehrich H, Kapphahn RJ, Kavanaugh SM, Heuss ND, et al. Immunoproteasome responds to injury in the retina and brain. J Neurochem. 2008;106:158–169. doi: 10.1111/j.1471-4159.2008.05345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hussong SA, Roehrich H, Kapphahn RJ, Maldonado M, Pardue MT, Ferrington DA. A novel role for the immunoproteasome in retinal function. Invest Ophthalmol Vis Sci. 2011;52:714–723. doi: 10.1167/iovs.10-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kapphahn RJ, Bigelow EJ, Ferrington DA. Age-dependent inhibition of proteasome chymotrypsin-like activity in the retina. Exp Eye Res. 2007;84:646–654. doi: 10.1016/j.exer.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Diaz-Hernandez M, Hernandez F, Martin-Aparicio E, Gomez-Ramos P, Moran MA, Castano JG, et al. Neuronal induction of the immunoproteasome in Huntington’s disease. J Neurosci. 2003;23:11653–11661. doi: 10.1523/JNEUROSCI.23-37-11653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mishto M, Bellavista E, Santoro A, Stolzing A, Ligorio C, Nacmias B, et al. Immunoprotea-some and LMP2 polymorphism in aged and Alzheimer’s disease brains. Neurobiol Aging. 2006;27:54–66. doi: 10.1016/j.neurobiolaging.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 90.Yao X, Liu J, McCabe JT. Alterations of cerebral cortex and hippocampal proteasome subunit expression and function in a traumatic brain injury rat model. J Neurochem. 2008;104:353–363. doi: 10.1111/j.1471-4159.2007.04970.x. [DOI] [PubMed] [Google Scholar]
- 91.Gavilan MP, Castano A, Torres M, Portavella M, Caballero C, Jimenez S, et al. Age-related increase in the immunoproteasome content in rat hippocampus: molecular and functional aspects. J Neurochem. 2009;108:260–272. doi: 10.1111/j.1471-4159.2008.05762.x. [DOI] [PubMed] [Google Scholar]
- 92.Ding Q, Reinacker K, Dimayuga E, Nukala V, Drake J, Butterfield DA, et al. Role of the proteasome in protein oxidation and neural viability following low-level oxidative stress. FEBS Lett. 2003;546:228–232. doi: 10.1016/s0014-5793(03)00582-9. [DOI] [PubMed] [Google Scholar]
- 93.Opitz E, Koch A, Klingel K, Schmidt F, Prokop S, Rahnefeld A, et al. Impairment of immunoproteasome function by beta5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog. 2011;7:e1002233. doi: 10.1371/journal.ppat.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cai ZP, Shen Z, Van Kaer L, Becker LC. Ischemic preconditioning-induced cardioprotection is lost in mice with immunoproteasome subunit low molecular mass polypeptide-2 deficiency. FASEB J. 2008;22:4248–4257. doi: 10.1096/fj.08-105940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005;19:644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- 96.Husom AD, Peters EA, Kolling EA, Fugere NA, Thompson LV, Ferrington DA. Altered proteasome function and subunit composition in aged muscle. Arch Biochem Biophys. 2004;421:67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 97.Merforth S, Kuehn L, Osmers A, Dahlmann B. Alteration of 20S proteasome-subtypes and proteasome activator PA28 in skeletal muscle of rat after induction of diabetes mellitus. Int J Biochem Cell Biol. 2003;35:740–748. doi: 10.1016/s1357-2725(02)00381-3. [DOI] [PubMed] [Google Scholar]
- 98.Teoh CY, Davies KJ. Potential roles of protein oxidation and the immunoproteasome in MHC class I antigen presentation: the ‘PrOxI’ hypothesis. Arch Biochem Biophys. 2004;423:88–96. doi: 10.1016/j.abb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 99.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, et al. Immunoprotea-somes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142:613–624. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 100.Ding Q, Martin S, Dimayuga E, Bruce-Keller AJ, Keller JN. LMP2 knock-out mice have reduced proteasome activities and increased levels of oxidatively damaged proteins. Antioxid Redox Signal. 2006;8:130–135. doi: 10.1089/ars.2006.8.130. [DOI] [PubMed] [Google Scholar]
- 101.Hayashi T, Horiuchi A, Sano K, Hiraoka N, Kanai Y, Shiozawa T, et al. Molecular approach to uterine leiomyosarcoma: LMP2-deficient mice as an animal model of spontaneous uterine leiomyosarcoma. Sarcoma. 2011;2011:476498. doi: 10.1155/2011/476498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buchanan J, Mazumder PK, Hu P, Chakrabarti G, Roberts MW, Yun UJ, et al. Reduced cardiac efficiency and altered substrate metabolism precedes the onset of hyperglycemia and contractile dysfunction in two mouse models of insulin resistance and obesity. Endocrinology. 2005;146:5341–5349. doi: 10.1210/en.2005-0938. [DOI] [PubMed] [Google Scholar]
- 103.Aneja A, Tang WH, Bansilal S, Garcia MJ, Farkouh ME. Diabetic cardiomyopathy: insights into pathogenesis, diagnostic challenges, and therapeutic options. Am J Med. 2008;121:748–757. doi: 10.1016/j.amjmed.2008.03.046. [DOI] [PubMed] [Google Scholar]
- 104.Hu J, Klein JD, Du J, Wang XH. Cardiac muscle protein catabolism in diabetes mellitus: activation of the ubiquitin-proteasome system by insulin deficiency. Endocrinology. 2008;149:5384–5390. doi: 10.1210/en.2008-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zu L, Bedja D, Fox-Talbot K, Gabrielson KL, Van Kaer L, Becker LC, et al. Evidence for a role of immunoproteasomes in regulating cardiac muscle mass in diabetic mice. J Mol Cell Cardiol. 2010;49:5–15. doi: 10.1016/j.yjmcc.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mallone R, Martinuzzi E, Blancou P, Novelli G, Afonso G, Dolz M, et al. CD8+ T-cell responses identify beta-cell autoimmunity in human type 1 diabetes. Diabetes. 2007;56:613–621. doi: 10.2337/db06-1419. [DOI] [PubMed] [Google Scholar]
- 107.Skowera A, Ellis RJ, Varela-Calvino R, Arif S, Huang GC, Van-Krinks C, et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J Clin Invest. 2008;118:3390–3402. doi: 10.1172/JCI35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zaiss DM, Bekker CP, Grone A, Lie BA, Sijts AJ. Proteasome immunosubunits protect against the development of CD8 Tcell-mediated autoimmune diseases. J Immunol. 2011;187:2302–2309. doi: 10.4049/jimmunol.1101003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reis J, Hassan F, Guan XQ, Shen J, Monaco JJ, Papasian CJ, et al. The immunoproteasomes regulate LPS-induced TRIF/TRAM signaling pathway in murine macrophages. Cell Biochem Biophys. 2011;60:119–126. doi: 10.1007/s12013-011-9183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schmidt N, Gonzalez E, Visekruna A, Kuhl AA, Loddenkemper C, Mollenkopf H, et al. Targeting the proteasome: partial inhibition of the proteasome by bortezomib or deletion of the immunosubunit LMP7 attenuates experimental colitis. Gut. 2010;59:896–906. doi: 10.1136/gut.2009.203554. [DOI] [PubMed] [Google Scholar]
- 111.Basler M, Dajee M, Moll C, Groettrup M, Kirk CJ. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J Immunol. 2010;185:634–641. doi: 10.4049/jimmunol.0903182. [DOI] [PubMed] [Google Scholar]
- 112.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15:781–787. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 113.Chen F, Demers LM, Shi X. Upstream signal transduction of NF-kappaB activation. Curr Drug Targets Inflamm Allergy. 2002;1:137–149. doi: 10.2174/1568010023344706. [DOI] [PubMed] [Google Scholar]
- 114.Malek R, Borowicz KK, Jargiello M, Czuczwar SJ. Role of nuclear factor kappaB in the central nervous system. Pharmacol Rep. 2007;59:25–33. [PubMed] [Google Scholar]
- 115.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 116.Hayashi T, Faustman D. NOD mice are defective in proteasome production and activation of NF-kappaB. Mol Cell Biol. 1999;19:8646–8659. doi: 10.1128/mcb.19.12.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hayashi T, Faustman D. Essential role of human leukocyte antigen-encoded proteasome subunits in NF-kappaB activation and prevention of tumor necrosis factor-alpha-induced apoptosis. J Biol Chem. 2000;275:5238–5247. doi: 10.1074/jbc.275.7.5238. [DOI] [PubMed] [Google Scholar]
- 118.Hayashi T, Faustman DL. Selected contribution: association of gender-related LMP2 inacti-vation with autoimmune pathogenesis. J Appl Physiol. 2001;91:2804–2815. doi: 10.1152/jappl.2001.91.6.2804. [DOI] [PubMed] [Google Scholar]
- 119.Kessler BM, Lennon-Dumenil AM, Shinohara ML, Lipes MA, Ploegh HL. LMP2 expression and proteasome activity in NOD mice. Nat Med. 2000;6:1064. doi: 10.1038/80346. [Author reply 1065–1066] [DOI] [PubMed] [Google Scholar]
- 120.Runnels HA, Watkins WA, Monaco JJ. LMP2 expression and proteasome activity in NOD mice. Nat Med. 2000;6:1064–1065. doi: 10.1038/80349. [Author reply 1065–1066] [DOI] [PubMed] [Google Scholar]
- 121.Visekruna A, Joeris T, Seidel D, Kroesen A, Loddenkemper C, Zeitz M, et al. Proteasome-mediated degradation of IkappaBalpha and processing of p105 in Crohn disease and ulcer-ative colitis. J Clin Invest. 2006;116:3195–3203. doi: 10.1172/JCI28804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ding Q, Lewis JJ, Strum KM, Dimayuga E, Bruce-Keller AJ, Dunn JC, et al. Polyglutamine expansion, protein aggregation, proteasome activity, and neural survival. J Biol Chem. 2002;277:13935–13942. doi: 10.1074/jbc.M107706200. [DOI] [PubMed] [Google Scholar]
- 123.Lee W, Kim KB. The immunoproteasome: an emerging therapeutic target. Curr Top Med Chem. 2011;11:2923–2930. doi: 10.2174/156802611798281348. [DOI] [PubMed] [Google Scholar]
- 124.Kuhn DJ, Hunsucker SA, Chen Q, Voorhees PM, Orlowski M, Orlowski RZ. Targeted inhibition of the immunoproteasome is a potent strategy against models of multiple myeloma that overcomes resistance to conventional drugs and nonspecific proteasome inhibitors. Blood. 2009;113:4667–4676. doi: 10.1182/blood-2008-07-171637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Singh AV, Bandi M, Aujay MA, Kirk CJ, Hark DE, Raje N, et al. PR-924, a selective inhibitor of the immunoproteasome subunit LMP-7, blocks multiple myeloma cell growth both in vitro and in vivo. Br J Haematol. 2011;152:155–163. doi: 10.1111/j.1365-2141.2010.08491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garg A, Hernandez MD, Sousa AB, Subramanyam L, Martinez de Villarreal L, dos Santos HG, et al. An autosomal recessive syndrome of joint contractures, muscular atrophy, microcytic anemia, and panniculitis-associated lipodystrophy. J Clin Endocrinol Metab. 2010;95:E58–E63. doi: 10.1210/jc.2010-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haroon N, Maksymowych W, Rahman P, Tsui F, O’Shea F, Inman R. Radiographic severity in ankylosing spondylitis is associated with polymorphism in large multifunctional peptidase 2 (LMP2) in the SPARCC cohort. Arthritis Rheum. 2011 doi: 10.1002/art.33430. [DOI] [PubMed] [Google Scholar]
- 128.Vargas-Alarcon G, Gamboa R, Zuniga J, Fragoso JM, Hernandez-Pacheco G, Londono J, et al. Association study of LMP gene polymorphisms in Mexican patients with spondyloarthritis. Hum Immunol. 2004;65:1437–1442. doi: 10.1016/j.humimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 129.Kramer U, Illig T, Grune T, Krutmann J, Esser C. Strong associations of psoriasis with antigen processing LMP and transport genes TAP differ by gender and phenotype. Genes Immun. 2007;8:513–517. doi: 10.1038/sj.gene.6364404. [DOI] [PubMed] [Google Scholar]
- 130.Mishto M, Bellavista E, Ligorio C, Textoris-Taube K, Santoro A, Giordano M, et al. Immu-noproteasome LMP2 60HH variant alters MBP epitope generation and reduces the risk to develop multiple sclerosis in Italian female population. PLoS One. 2010;5:e9287. doi: 10.1371/journal.pone.0009287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mishto M, Santoro A, Bellavista E, Sessions R, Textoris-Taube K, Dal Piaz F, et al. A structural model of 20S immunoproteasomes: effect of LMP2 codon 60 polymorphism on expression, activity, intracellular localisation and insight into the regulatory mechanisms. Biol Chem. 2006;387:417–429. doi: 10.1515/BC.2006.056. [DOI] [PubMed] [Google Scholar]
- 132.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, et al. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]