Abstract
Moonlighting proteins serve one or more additional functions in addition to their canonical roles. Moonlighting functions arise when an adventitious interaction between a protein and a new partner improves fitness of the organism. Selective pressure for improvement in the new function can result in two alternative outcomes. The gene encoding the newly bi-functional protein may duplicate and diverge so as to encode two proteins, each of which serves only one function. Alternatively, genetic changes that minimize adaptive conflict between the two functions and/or improve control over the time and place at which each function is served can lead to a moonlighting protein. Importantly, genetic changes that enhance a moonlighting function can occur in the gene encoding the moonlighting protein itself, in a gene that affects the structure of its new partner, or in a gene encoding a transcription factor that controls expression of either partner. The evolutionary history of each moonlighting protein is complex, depending upon the stochastic occurrence of genetic changes such as gene duplication and point mutations, and the effects of those changes on fitness. Population effects, particularly loss of promising individuals due to random genetic drift, also play a role in the emergence of a moonlighting protein. The ultimate outcome is not necessarily the “optimal” solution to the problem of serving two functions, but may be “good enough” that fitness becomes limited by some other function.
In the early days of molecular biology, each gene was expected to encode a single protein that serves a single function[1, 2]. This appealingly simple paradigm has been shattered by numerous examples to the contrary, including identification of “moonlighting” proteins that serve multiple functions, often in different places or at different times. Each case of moonlighting begs a number of interesting evolutionary questions. How did the secondary function arise? Why are different moonlighting functions seen in orthologous proteins in different organisms? And, most interestingly, why has the moonlighting protein not been replaced by two proteins, each of which performs a specialized function?
Acquisition of a new function
Moonlighting functions arise as a result of an adventitious interaction with a new partner, often another protein, but sometimes DNA or RNA. Possibilities for new interactions are rife in the crowded cytoplasm of cells. A simulation of the E. coli cytoplasm that includes the 50 most abundant macromolecules suggests that proteins have about 25 neighbors at any moment, and encounter over 100 different molecules within 15 µsec [3]. The external milieu also offers many opportunities for new interactions that may confer a selective advantage, especially for pathogens and multi-cellular organisms.
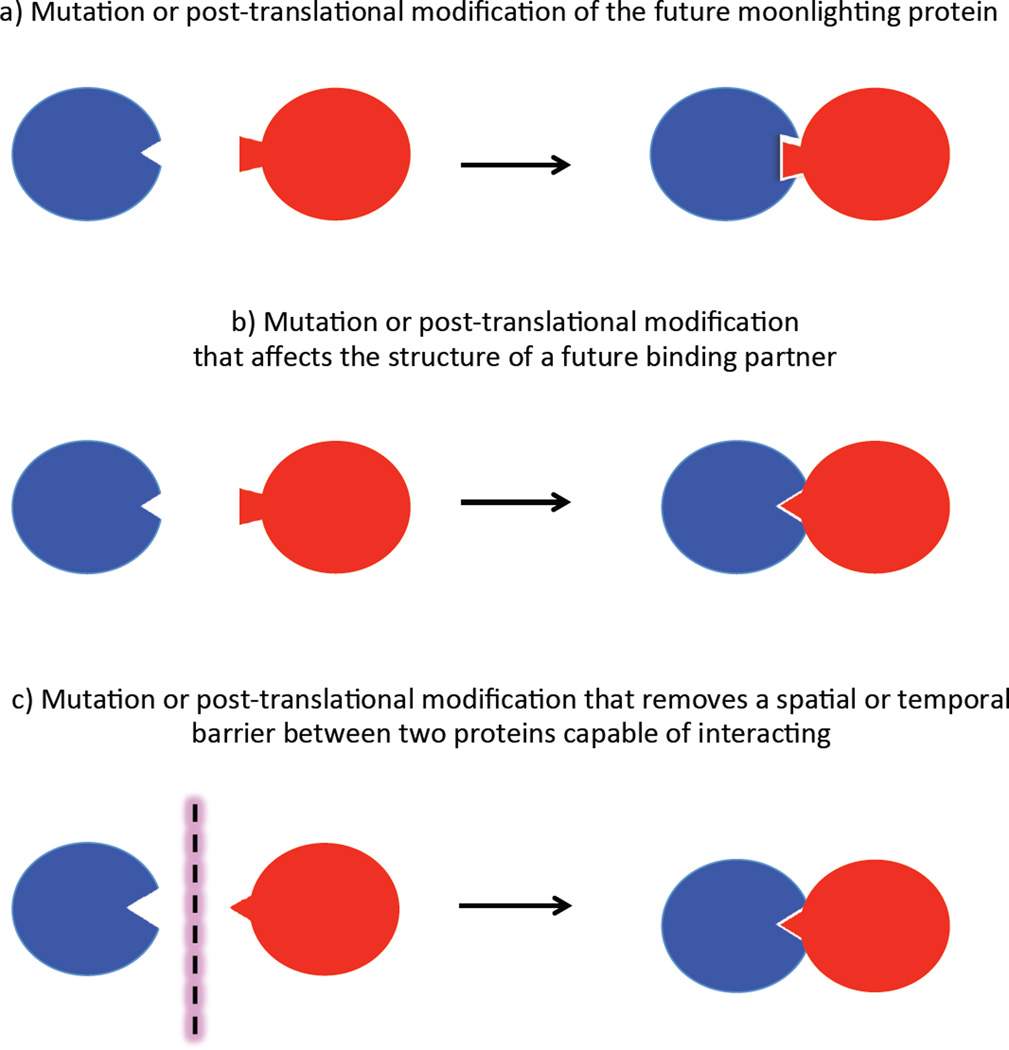
New binding interactions can involve any part of a protein’s surface. Much of a protein’s surface is not involved in its canonical function and thus free to drift via mutations that do not affect the canonical function. If a new interaction is beneficial, natural selection will favor persistence of mutations and/or post-translational modifications of either the moonlighting protein or its new partner that enhance the affinity or orientation of the interaction. Alternatively, new binding interactions can result from mutations that change the time of expression or the location of binding partners, thus bringing together two proteins that are already capable of interacting but that were never before present in the same place at the same time (see Figure 1).
Figure 1.
New interactions can be enabled by either mutations or new post-translational modifications of a future moonlighting protein (blue) and a new partner (red), which may be a protein or another macromolecule.
A study of the affinities of variants of the transcriptional regulator MarA for 64 DNA binding sites illustrates that new binding partners can be acquired as a result of only one or two mutations [4]. In wild-type MarA, Trp42, Gln45 and Arg46 interact with a GCA motif in the target promoter. W42R MarA is specific for TCC, whereas W42T Q45R MarA is specific for GAC. In contrast, W42S Q45A MarA has low specificity and binds to 42 of the 64 binding sites.
The probability that a given protein will acquire a moonlighting function depends upon many factors. The protein must be present under the conditions in which a new physical interaction will improve fitness. Consequently, proteins that are present under nearly all growth conditions may be the most likely to acquire a new function. Proteins that are abundant are more likely to acquire a new function simply because the frequency of encounters between potential interaction partners is proportional to the concentrations of both partners. The abundance of moonlighting functions exhibited by glycolytic enzymes [5–7] and ribosomal proteins [8] may be due to their nearly ubiquitous presence and abundance.
A framework for thinking about the evolutionary fate of a newly bifunctional protein
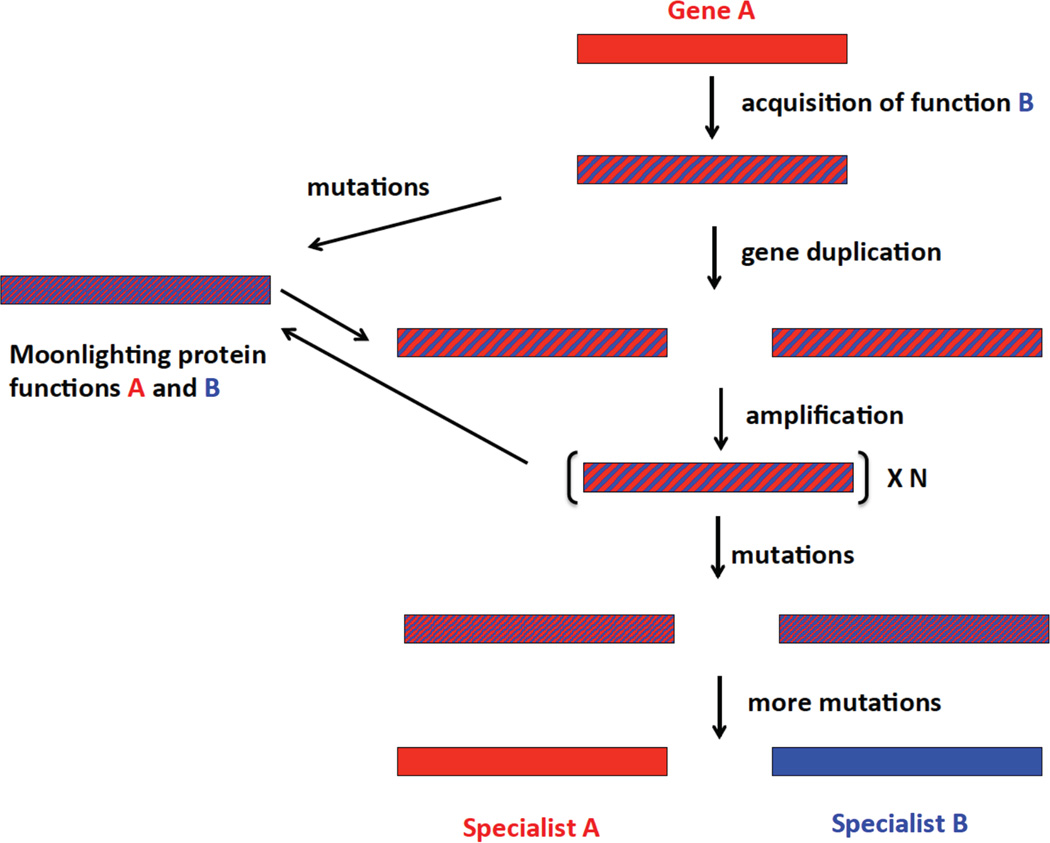
Figure 2 depicts possible evolutionary fates of a protein that has acquired a new moonlighting function that does not yet operate optimally. On the left is a trajectory in which mutations enhance the moonlighting function without significantly damaging the ancestral function. Such mutations might be in the regulatory and/or coding region of the gene encoding the moonlighting protein. A moonlighting function might also be enhanced by mutations in the regulatory and/or coding region of other genes, including the new interaction partner or transcriptional regulators that control expression of either of the new binding partners. Thus, improvement in a moonlighting function need not involve any changes in the protein itself. As a consequence, the gene encoding a moonlighting protein may not carry any traces of the acquisition of a second function. This factor will complicate efforts to use sequence analysis to predict moonlighting functions and to identify the surface regions involved in a moonlighting interaction.
Figure 2.
Possible evolutionary fates of a protein that has acquired a second function. Mutations can occur in the regulatory and/or coding regions of the gene, and in the case of a moonlighting protein, in the regulatory and/or coding regions of other genes.
The right side depicts a trajectory that begins with gene duplication [9, 10]. If fitness is enhanced by an increase in gene dosage, further amplifications may occur, up to the point at which there is no further benefit to increased copy number. Gene duplication/amplification provides an expanded opportunity to search sequence space because there are multiple targets for mutation and because deleterious mutations in an individual allele can be buffered by other alleles. Recombination between alleles carrying different mutations can allow relatively rapid sampling of the effects of various combinations of mutations. Upon emergence of two alleles, each of which encodes a sufficiently effective specialist, additional copies that do not contribute to fitness can be lost. Again, it is important to keep in mind that mutations that allow divergence of function may occur in the promoter of one or both genes and/or a gene encoding a regulatory protein.
The evolutionary trajectories shown in Figure 2 are not uni-directional. Mutations in the context of a gene amplification may lead to a protein that can serve both its ancestral and moonlighting functions well enough to allow loss of extra gene copies. Thus, a moonlighting protein may be the outcome of a process that begins by gene duplication or amplification. Alternatively, a gene encoding a moonlighting protein that has accumulated some mutations may duplicate and the copies may subsequently diverge to two specialist proteins.
The evolutionary trajectory followed by a bi-functional protein depends on the occurrence of stochastic events (gene duplication/amplification and beneficial point mutations and/or indels), as well as the fitness effects realized by each process. These processes will be discussed in the following sections.
The frequencies and fitness effects of gene duplications
The evolutionary fate of a bifunctional protein is influenced by the frequency and mechanism of duplication, as well as the size, copy number and gene content of a duplicated/amplified region. All of these parameters vary between organisms and between sites in the same genome.
It is important to recognize that duplications usually involve segments larger than a single gene. The term “segmental duplication” is a more accurate description in such cases, although “gene duplication” is commonly used when the focus is upon a single gene within a duplicated region. The fitness effects of a segmental duplication depend upon the effects of increasing the dosage of every gene in the duplicated region. Increasing the dosage of some proteins may increase fitness, but beyond a certain point the effect on fitness may level off or become detrimental. The latter is commonly seen when a precise ratio between a protein and other cellular components is critical, or when the protein aggregates at high concentrations [11].
Gene duplication occurs at a frequency of 5×10−6 per gene per generation in S. cereviseae [12], and about 50-fold lower in C. elegans [13]. However, these are average values; duplication frequency vary within genomes. A study of the frequency of gene duplication at 38 sites in S. enterica revealed a 550-fold difference in duplication frequency, between 5.8×10−5 to 3.2×10−2 [14]. In the human and mouse genomes, there are more than 25,000 and 47,000 recombination hot-spots, respectively [15, 16]. In humans, some chromosomes are enriched for segmental duplications [17], and regions near the centromeres and telomeres are enriched for recent duplications [11].
Duplication that occurs by illegitimate or homologous recombination usually results in retention of the regulatory regions upstream of a gene. Although tandem duplications are common in bacteria [14, 18–20], duplicated genes or segments sometimes end up on different replicons. Recent gene duplicate pairs are sprinkled throughout the 9 replisomes of the cyanobacterium Acaryochloris [21], and many duplicate pairs in humans are found on different chromosomes or different regions of the same chromosome [17]. Gene duplication via retrotransposition, which results in integration of an intron-less copy of a gene that lacks its regulatory sequences, often deposits genes in a distant location in the genome. Depending upon the sequence upstream of the new copy, the gene may lose the capacity for expression, or, more interestingly, may acquire a totally different set of regulatory factors.
Once duplication occurs, the evolutionary fate of a duplicated gene or segment depends upon whether an increase in gene dosage increases fitness. If it does, further amplifications of genes or segments can occur. If it does not, loss of one copy by homologous recombination or deleterious mutations that lead to a pseudogene will likely occur before beneficial mutations that increase an inefficient function in one or the other of the copies [22].
The frequency and fitness effects of mutations
Mutations will often be required to improve the performance of a newly acquired moonlighting function, regardless of whether the gene has been duplicated/amplified. Like gene duplication, the frequencies and fitness effects of mutations are highly variable. In the following discussion, it will be assumed that point mutations that improve the function of a moonlighting protein may occur in the gene encoding either partner. This discussion will focus on point mutations, but the principles discussed are applicable to indels, as well.
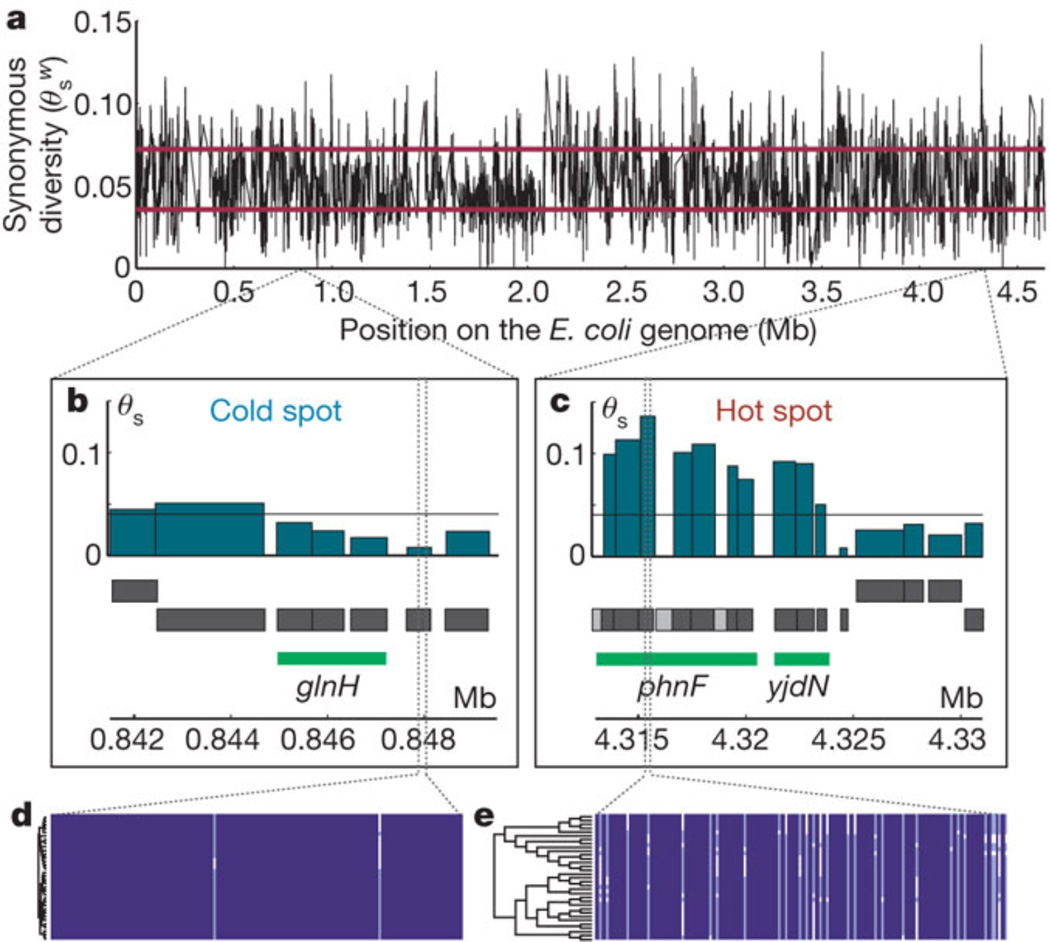
Point mutations in cellular organisms are rare relative to the frequency of gene duplication. The frequency in E. coli is 2.2×10−10 per bp per cell division [23]. Frequencies in eukaryotic nuclear DNA range from about 10−10 to 10−9 per bp per cell division. The mutation rate is often higher in mitochondrial DNA, reaching 12×10−9 per bp per cell division in S. cereviseae mitochondrial DNA [24]. Frequencies in viruses are also variable, ranging from 2×10−8 per nt per strand-copying in the double-stranded DNA phage T2 to 4.4 × 10−5 per nt per strand-copying in the (-)-strand RNA measles virus [25]. However, these average values conceal a striking variation in mutation frequencies within the genome. Mutations occur at CpG sites 10-fold more often than at other sites [24]. In yeast nuclear DNA, the most frequent point mutation (G:C → A:T transition) is 4.5-fold more common than the least frequent mutation (A:T → T:A transversion) [12]. In addition, there are mutation “hot spots” and “cold spots” in which synonymous mutations occur at widely varying frequencies in genomes. In E. coli, these hot and cold spots often span multiple genes (see Figure 3) [26].
Figure 3.
Mutational cold spots and hot spots in the E. coli genome based upon a comparison of 34 fully sequenced genomes. a) Synonymous diversity plotted along the E. coli genome; b) a representative cold spot that spans 3 kb; c) a representative hot spot that spans 10 kb; d and e) multiple sequence alignments used to generate the data. Reprinted by permission from Macmillan Publishers Ltd: Nature 485, 95–98, copyright 2012.
The fitness consequences of point mutations are generally either neutral, due to the structure of the genetic code, or detrimental because there are many ways to destroy the function of a protein, including truncation, destabilization, or loss of critical amino acid residues required for binding or catalysis. Only a small fraction of mutations in coding regions is expected to have beneficial effects on one or both of the functions of a moonlighting protein. Thus, achieving an improvement of an inefficient function may be a long and difficult process.
One of the primary determinants of the consequence of a mutation is a protein’s stability. Since many amino acid changes are destabilizing [27, 28], proteins that are more stable to begin with have a greater capacity to accumulate the changes needed to improve an inefficient function. Further, even in quite stable proteins, a successful evolutionary trajectory may require intermediate stages in which “enabling” mutations restore stability but do not contribute to improvement of function, before further beneficial mutations can be tolerated.
Structural characteristics affect the consequence of point mutations for other reasons, as well. The abundance of proteins that share certain folds suggests that some folds are more evolvable than others. The common (αβ) barrel provides sites at the ends of the barrel at which variation is easily achieved by changes in the loops connecting the α helices and β strands in the context of a stable structure. This versatility can enhance the acquisition and refinement of moonlighting functions. The plasminogen binding site in α-enolase from Streptococcus pneumoniae is located in one of these loops at the surface of a (αβ) barrel [29].
The effect of a point mutation on an inefficient function depends in a surprisingly strong way upon sequence, even among orthologous proteins that share a common structure and function. Mutations can be beneficial in some sequence contexts, but detrimental in others [30–32]. A mutation that changes a Glu near the active site of gamma-glutamyl phosphate reductase to Ala increases an inefficient promiscuous activity by 770-fold in the Yersinia enterocolytica enzyme, but by only 14-fold in the Lactococcus casei enzyme (Khanal, Yu McLoughlin, Kershner and Copley, in press). This strong intra-protein epistasis is perhaps the least predictable of the factors that influence the potential for evolutionary innovation in different organisms.
Given the difficulty of improving an inefficient function via mutations in the coding region of the gene, the most accessible mechanism for improvement in fitness often involves regulatory mutations [33]. Such mutations may alter the strength of a promoter, result in recruitment of a new transcription factor, or alter RNA-based control mechanisms that depend upon a sequence or a secondary structure in the encoded mRNA [34]. Mutations in genes encoding transcription factors can also alter the regulation of expression of a moonlighting protein. Indeed, both cis and trans effects have been found to contribute to regulatory differences between gene duplicates in yeast [35].
Finally, the effects of point mutations depend upon the nature of the moonlighting function. Evolution of a novel catalytic function is more demanding than evolution of a novel binding site, since catalysis requires precise positioning of multiple catalytic groups in addition to residues involved in ligand binding. Thus, moonlighting functions usually involve new binding interactions, and it is rare, although not unprecedented [36], for a moonlighting function to be catalytic.
Which trajectory?
Given the relative frequencies of gene duplication and point mutations, the most likely event to follow acquisition of a new function is gene duplication. Although duplication and divergence is a common outcome, the existence of moonlighting proteins demonstrates that this is not the only fate of a bifunctional protein. When beneficial mutations that alter regulation are more frequent than beneficial mutations in coding regions of the genes involved in the moonlighting function, regulatory mutations may result in loss of some gene copies, and consequently a loss in the ability to search sequence space afforded by the presence of multiple gene copies. Further, when gene duplication does not increase fitness, duplication and divergence is unlikely, and refinement of a new function may have to rely upon mutations in the single copy of the gene encoding the moonlighting function or in genes encoding its new partner or transcriptional regulators.
There is no guarantee that evolution of a new moonlighting protein will lead inexorably to the most “optimal” solution from a design standpoint. Mutations leading to the optimal solution may be rare. Multiple mutations may be required to traverse a valley in fitness space, requiring a decrease in fitness before better solutions can be accessed. In either case, mutants that arise more frequently but cannot access a trajectory toward the optimal solution may take over the population. Further, selective pressure ceases when a solution that is “good enough” emerges and fitness becomes limited by some other component of the cell or organism.
The previous discussion has focused on the stochastic genetic events that lead to either a moonlighting protein or two specialized proteins. However, the evolution of a moonlighting protein takes place in the context of an entire genome that is subject to genetic changes, and within a cell or organism that may be subject to variable selective pressures. The situation is further complicated when something else is more important for fitness than the function of a moonlighting protein. In such cases, particular alleles that contribute to the moonlighting function may persist because they are physically linked with genes that are more critical for fitness, and not because they provide the optimal performance of the moonlighting protein.
An additional layer of complexity is introduced by the characteristics of the population and the environment in which a bifunctional protein has emerged. Random genetic drift, particularly in small populations, and population bottlenecks can result in loss of individuals that carry advantageous genetic traits. Thus, the population may lose promising individuals, and retain individuals that are constrained to a trajectory leading to a sub-optimal solution to the challenge presented by a bifunctional protein. Even in larger populations, a common mutation that confers an increase in fitness may sweep through a population and preclude evolutionary trajectories that are more difficult to find but that have the potential to lead to more optimal solutions.
Why proteins acquire different moonlighting functions in different lineages
One of the intriguing aspects of moonlighting proteins is that a single protein often has different moonlighting functions in different lineages. Conversely, a common moonlighting function is often served by different proteins in different lineages. An obvious reason is that the repertoire of potential interaction partners varies in different organisms, and even in the same organism under different environmental conditions. In multicellular organisms, the repertoire of potential interaction partners also varies in different organelles and different tissues, and at different times during development. Even within a given species, the protein repertoire can differ dramatically. A comparison of 35 serovars of Salmonella enterica revealed 2811 proteins common to all strains. However, each serovar contained between 50 and 504 proteins found in no other serovar [37]. In addition, random drift in surface regions of proteins means that novel binding interactions between orthologous components may be possible in some organisms but not in others.
The evolution of a moonlighting function takes place in the context of a complex metabolic and regulatory network. Consequently, epistatic effects play a strong role in the evolutionary fate of a newly bifunctional protein. For example, compromises in a canonical function due to improvement of a moonlighting function may be better tolerated in organisms in which the canonical function is less critical due to genetic redundancy or a better ability to re-route metabolic flux. Alterations in regulation needed to improve the timing or location of expression of a moonlighting program may be more accessible in some organisms than in others due to a different complement of transcriptional regulators, different “wiring” of the regulatory network, and/or differences in promiscuous binding activities of transcriptional regulators that can jump-start the emergence of a new regulatory interaction.
Moonlighting is favored when gene duplication is detrimental
Small DNA and RNA viruses use multiple strategies to maintain their streamlined genomes. Selection against increases in size is believed to be due to the constraints of the protein capsid [38]. Common strategies include the use of overlapping genes [38, 39], alternative splicing [40], antisense transcription [41], programmed translational frameshifting [42] and moonlighting [43, 44], all of which allow a virus to squeeze greater functionality out of a small genome.
Ebola virus protein VP40 is an example of a moonlighting viral protein [43, 44]. The (-)-strand RNA genome of Ebola virus encodes only 7 proteins. VP40 is the matrix protein that connects the nucleocapsid to the viral membrane. It also moonlights as a transcriptional regulator earlier in the infectious process. The N-terminal domain of the protein forms an octamer that binds RNA and down-regulates transcription of the genome. This dual functionality makes sense; high levels of VP40 serve as a signal to stop transcription and start packaging progeny viruses.
Moonlighting may be favored when there is no adaptive conflict between the functions of a bi-functional protein
Many moonlighting proteins serve different functions in different places. In such cases, a single protein may be able to perform more than one function because only one function is required in a certain place. For example, the glycolytic enzyme enolase converts 2-phosphoglycerate to phosphoenolpyruvate in the cytoplasm, and also binds plasminogen on the surface of many cells, including bacteria [29, 45, 46], tumor cells [47], leukocytes [48],[49], neurons [50], and muscle precursor cells [51]. Cleavage of plasminogen to the enzymatically active plasmin form contributes to degradation of proteins in the extracellular matrix and allows motile cells to move through tissues. This contributes to the spread of pathogens and cancer cells [47]. On a more positive note, the same moonlighting function contributes to the migration of peripheral blood monocytes into infected tissue [52] and the ability of muscle precursor cells to repair damaged muscle tissue [51].
Moonlighting may be favored when the two functions of a bi-functional protein are complementary
When two functions are mutually exclusive, combining them a single protein may benefit the organism by allowing efficient switching between functions depending on the environmental conditions. An example is aconitase [53], whose canonical function is isomerization of citrate and isocitrate in the TCA cycle. The active site of aconitase contains an iron-sulfur cluster that is required for catalysis. When iron levels are low, the iron-sulfur cluster cannot be assembled, and the protein adopts a conformation that binds stem-loop structures known as iron-regulatory elements (IREs) in the 5’ or 3’ UTR of certain mRNAs, many of which are involved in iron uptake or storage. Binding to the 5’-UTR decreases transcription of genes; binding to the 3’-UTR stabilizes transcripts and increases the levels of translation. Since aconitase cannot perform its enzymatic role in the absence of sufficient iron, its use as an iron sensor allows a simple direct readout of cytoplasmic iron levels, as well as an important function for a protein that would otherwise be useless.
Duplication and divergence is favored when the adaptive conflict between two functions cannot be easily solved in the context of a single protein
The existence of large superfamilies of enzymes, transcriptional regulators and ligand receptors attests to the increase in fitness permitted by the process of duplication and divergence. In these cases, fitness is clearly increased by the emergence of specialist proteins with high specificity for their substrates or ligands. In the case of transcriptional regulators and receptors, high specificity enables precise responses to environmental clues. In the case of enzymes, specialization allows increased catalytic efficiency as well as independent control over fluxes in metabolic pathways.
Genes encoding members of the enolase superfamily have duplicated and diverged many times, leading to families of enzymes that catalyze more than 20 different reactions [54], while retaining the ancestral protein fold as well as the active site architecture that promotes abstraction of a hydrogen from a position alpha to a carboxylate. After this initial step, the reactions differ depending upon the disposition of catalytic groups in the active site. The benefit of duplication and divergence in this superfamily is evident, as specific catalytic groups are required in different positions [55].
Genes encoding G-protein coupled receptors have also duplicated and diverged. The human genome encodes an astonishing 800 G-protein receptors that are involved in a wide variety of signal transduction functions [56]. The ligand binding domains of these receptors have diverged to bind a range of small molecules, peptides, and proteins with high specificity. No doubt the ability to sense and respond precisely to extracellular stimuli has provided selective pressure to allow the enormous expansion of this protein family.
Moonlighting and duplication and divergence are not necessarily mutually exclusive solutions
The stochastic processes that affect the fate of a bifunctional protein may not follow a simple evolutionary trajectory toward only one of the two possibilities shown in Figure 2. For example, in metazoa, the gene for aconitase has duplicated. One copy encodes a moonlighting protein, while the other encodes a protein that is 60% identical but has lost enzymatic activity and serves only as an iron-regulatory protein [53, 57].
Conclusion
Studies of the growing number of characterized moonlighting proteins provide a fascinating view of the marvelous functional versatility of moonlighting proteins. However, we have limited insight into the process by which these intriguing multi-tasking abilities arose in the distant past. The evolutionary history of each moonlighting protein is an individual story that played out in the context of a set of genetic, environmental and population conditions that have been obscured by the ensuing millions of years of evolution. We can, however, hope to reveal some of the critical events in the evolution of moonlighting proteins. Identifying the binding partners of moonlighting proteins and the regions of each macromolecule that are involved in the moonlighting interaction will focus our attention on the genetic sequences that have undergone changes. Likewise, identifying the proteins and processes that regulate the temporal and spatial control of the individual functions of moonlighting proteins will point us toward additional genes in which mutations may have contributed to the refinement of a moonlighting function.
References
- 1.Horowitz NH. One-gene-one-enzyme: remembering biochemical genetics. Protein Sci. 1995;4:1017–1019. doi: 10.1002/pro.5560040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beadle GW, Tatum EL. Genetic control of biochemical reactions in Neurospora. Proc. Natl. Acad. Sci. USA !941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shultzaberger RK, et al. Probing the informational and regulatory plasticity of a transcription factor DNA-binding domain. PLoS Genet. 2012;8:e1002614. doi: 10.1371/journal.pgen.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sirover MA. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim. Biophys. Acta. 2011;1810:741–751. doi: 10.1016/j.bbagen.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Gomez-Arreaza A, et al. Extracellular functions of glycolytic enzymes of parasites: unpredicted use of ancient proteins. Mol. Biochem. Parasitol. 2014;193:75–81. doi: 10.1016/j.molbiopara.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Tunio SA, et al. The moonlighting protein fructose-1, 6-bisphosphate aldolase of Neisseria meningitidis: surface localization and role in host cell adhesion. Mol. Microbio. 2010;7:605–615. doi: 10.1111/j.1365-2958.2010.07098.x. [DOI] [PubMed] [Google Scholar]
- 8.Aseev LV, Boni IV. Extraribosomal functions of bacterial ribosomal proteins. Mol. Biol. 2011;45:739–750. [PubMed] [Google Scholar]
- 9.Bergthorsson U, Andersson DI, Roth JR. Ohno's dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. U S A. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. B. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 11.Conrad B, Antonarakis SE. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- 12.Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. U S A. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipinski KJ, et al. High spontaneous rate of gene duplication in Caenorhabditis elegans . Curr. Biol. 2011;21:306–310. doi: 10.1016/j.cub.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson P, Roth J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc. Natl. Acad. Sci. U S A. 1981;78:3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunschwig H, et al. Fine-scale maps of recombination rates and hotspots in the mouse genome. Genetics. 2012;191:757–764. doi: 10.1534/genetics.112.141036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers S, et al. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 17.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 18.Reams AB, Neidle EL. Gene amplification involves site-specific short homology-independent illegitimate recombination in Acinetobacter sp. strain ADP1. J. Mol. Biol. 2004;338:643–656. doi: 10.1016/j.jmb.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 19.Devers M, Rouard N, Martin-Laurent F. Fitness drift of an atrazine-degrading population under atrazine selection pressure. Environ. Microbiol. 2008;10:676–684. doi: 10.1111/j.1462-2920.2007.01490.x. [DOI] [PubMed] [Google Scholar]
- 20.Nasvall J, et al. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338:384–387. doi: 10.1126/science.1226521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller SR, et al. Dynamics of gene duplication in the genomes of chlorophyll d-producing cyanobacteria: implications for the ecological niche. Genome Biol. Evol. 2011;3:601–613. doi: 10.1093/gbe/evr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 23.Lee H, et al. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. U S A. 2012;109:E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanjuan R, et al. Viral mutation rates. J. Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martincorena I, Seshasayee AS, Luscombe NM. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature. 2012;485:95–98. doi: 10.1038/nature10995. [DOI] [PubMed] [Google Scholar]
- 27.Tokuriki N, et al. The stability effects of protein mutations appear to be universally distributed. J. Mol. Biol. 2007;369:1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 28.Bloom JD, et al. Thermodynamic prediction of protein neutrality. Proc. Natl. Acad. Sci. U S A. 2005;102:606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehinger S, et al. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 2004;343:997–1005. doi: 10.1016/j.jmb.2004.08.088. [DOI] [PubMed] [Google Scholar]
- 30.Parera M, Martinez MA. Strong epistatic interactions within a single protein. Mol. Biol. Evol. 2014;31:1546–1553. doi: 10.1093/molbev/msu113. [DOI] [PubMed] [Google Scholar]
- 31.Weinreich DM, et al. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 32.Parera M, et al. Epistasis among deleterious mutations in the HIV-1 protease. J. Mol. Biol. 2009;392:243–250. doi: 10.1016/j.jmb.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 33.Shultzaberger RK, et al. The fitness landscapes of cis-acting binding sites in different promoter and environmental contexts. PLoS Genet. 2010;6:e1001042. doi: 10.1371/journal.pgen.1001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Nunez MA, et al. New insights into the regulatory networks of paralogous genes in bacteria. Microbiology. 2010;156:14–22. doi: 10.1099/mic.0.033266-0. [DOI] [PubMed] [Google Scholar]
- 35.Dong D, Yuan Z, Zhang Z. Evidences for increased expression variation of duplicate genes in budding yeast: from cis- to trans-regulation effects. Nucleic Acids Res. 2011;39:837–847. doi: 10.1093/nar/gkq874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, et al. Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site. J. Biol. Chem. 2009;284:36711–36719. doi: 10.1074/jbc.M109.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobsen A, et al. The Salmonella enterica pan-genome. Microb. Ecol. 2011;62:487–504. doi: 10.1007/s00248-011-9880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chirico N, Vianelli A, Belshaw R. Why genes overlap in viruses. Proc. Biol. Sci. 2010;277:3809–3817. doi: 10.1098/rspb.2010.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabath N, Wagner A, Karlin D. Evolution of viral proteins originated de novo by overprinting. Mol. Biol. Evol. 2012;29:3767–3780. doi: 10.1093/molbev/mss179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoltzfus CM. Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv. Virus Res. 2009;74:1–40. doi: 10.1016/S0065-3527(09)74001-1. [DOI] [PubMed] [Google Scholar]
- 41.Barbeau B, Mesnard J-M. Making sense out of anti-sense transcription in human T-cell lymphotrophic viruses (HTLVs) Viruses. 2011;3:456–468. doi: 10.3390/v3050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacks T, et al. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1998;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 43.Gomis-Ruth FX, et al. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure. 2003;11:423–433. doi: 10.1016/S0969-2126(03)00050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bornholdt ZA, et al. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 2013;154:763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agarwal S, et al. alpha-Enolase binds to human plasminogen on the surface of Bacillus anthracis . Biochim. Biophys. Acta. 2008;1784:986–994. doi: 10.1016/j.bbapap.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 46.Knaust A, et al. Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis . J. Bacteriol. 2007;189:3246–3255. doi: 10.1128/JB.01966-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capello M, et al. alpha-Enolase: a promising therapeutic and diagnostic tumor target. FEBS J. 2011;278:1064–1074. doi: 10.1111/j.1742-4658.2011.08025.x. [DOI] [PubMed] [Google Scholar]
- 48.Lopez-Alemany R, et al. Inhibition of cell surface mediated plasminogen activation by a monoclonal antibody against alpha-Enolase. Am. J. Hematol. 2003;7:234–242. doi: 10.1002/ajh.10299. [DOI] [PubMed] [Google Scholar]
- 49.Miles LA, et al. Role of cell-surface lysines in plasminogen binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry. 1991;30:1682–1691. doi: 10.1021/bi00220a034. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima K, et al. Plasminogen binds specifically to alpha-enolase on rat neuronal plasma membrane. J. Neurochem. 1994;63:2048–2057. doi: 10.1046/j.1471-4159.1994.63062048.x. [DOI] [PubMed] [Google Scholar]
- 51.Diaz-Ramos A, et al. Requirement of plasminogen binding to its cell-surface receptor alpha-enolase for efficient regeneration of normal and dystrophic skeletal muscle. PLoS One. 2012;7:e50477. doi: 10.1371/journal.pone.0050477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wygrecka M, et al. Enolase-1 promotes plasminogen-mediated recruitment of monocytes to the acutely inflamed lung. Blood. 2009;113:5588–5598. doi: 10.1182/blood-2008-08-170837. [DOI] [PubMed] [Google Scholar]
- 53.Volz K. The functional duality of iron regulatory protein 1. Curr. Opin. Struct. Biol. 2008;18:106–111. doi: 10.1016/j.sbi.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerlt JA, et al. Divergent evolution in enolase superfamily: strategies for assigning functions. J. Biol. Chem. 2012;287:29–34. doi: 10.1074/jbc.R111.240945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch. Biochem. Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 56.Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo B, Yu Y, Leibold EA. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J. Biol. Chem. 1994;269:24252–24260. [PubMed] [Google Scholar]