Abstract
Mitochondrial DNA (mt-DNA) from Oenothera berteriana tissue culture cells was isolated and characterized with respect to buoyant density in CsCl, melting point, contour length, and restriction fragments.
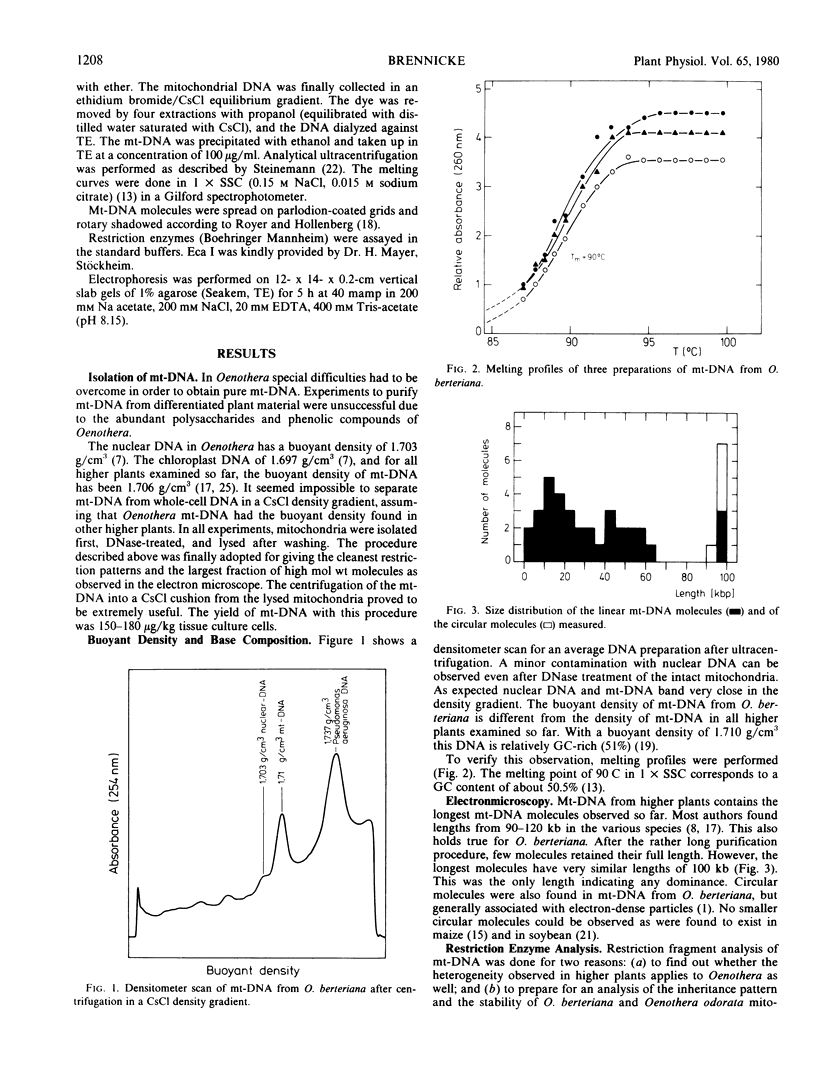
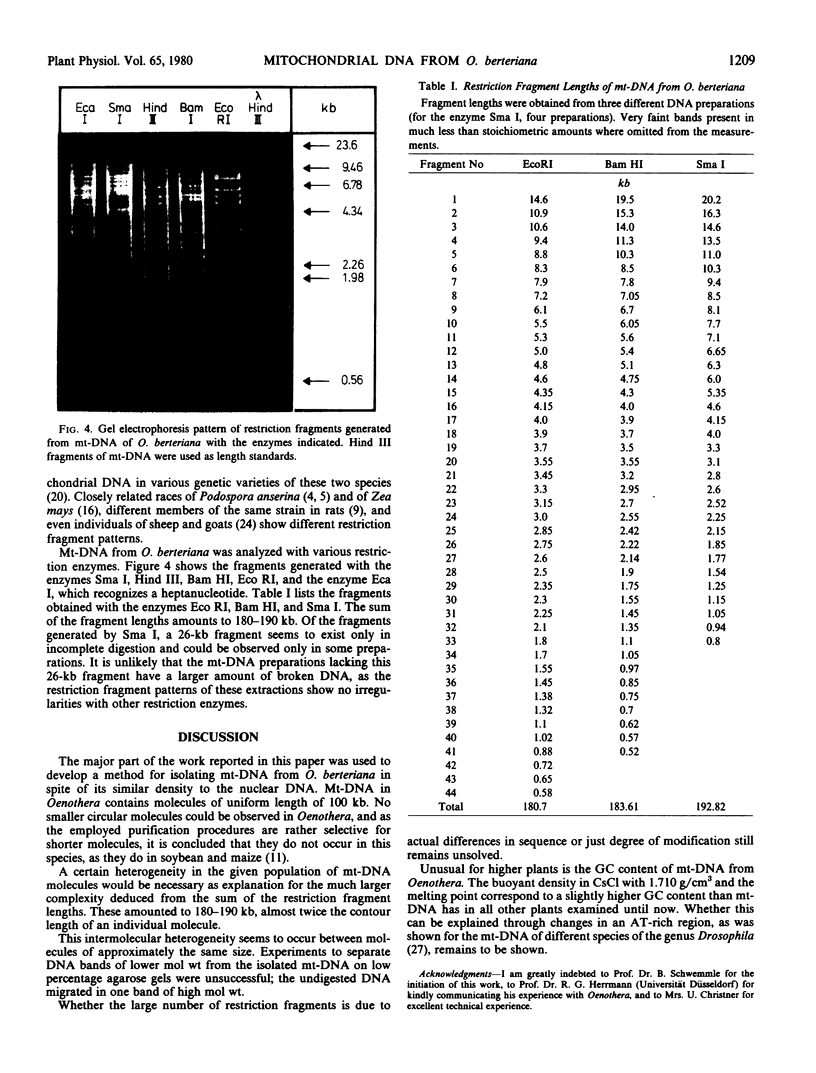
Because of the rather long purification procedure very few molecules retained their circularity. Only one distinct size class of molecules with a length of 100 kilobases was found. Restriction fragments were obtained with the enzymes, restrictionendonuclease I from Serratia marcescens, restrictionendonuclease III from Haemophilus influenzae, restrictionendonuclease I from Bacillus amyloliquefaciens H, and restrictionendonuclease I from Escherichia coli (Bohnert 1977 Exp Cell Res 106: 426-430); the added lengths of these fragments amounted to 180 to 190 kilobases. As in other higher plants, an intermolecular heterogeneity has to be postulated to explain the large number of restriction fragments. Unique to the mt-DNA from Oenothera berteriana, as compared to other higher plants, is the unusual high guanosine + cytosine content with 51% as determined by the buoyant density in CsCl of 1.710 grams/cubic centimeter and the melting point of 90 C.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bohnert H. J. Size and structure of mitochondrial DNA from Physarum polycephalum. Exp Cell Res. 1977 May;106(2):426–430. doi: 10.1016/0014-4827(77)90195-1. [DOI] [PubMed] [Google Scholar]
- Borst P., Hoeijmakers J. H. Kinetoplast DNA. Plasmid. 1979 Jan;2(1):20–40. doi: 10.1016/0147-619x(79)90003-9. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Belcour L., Grandchamp C. Mitochondrial DNA from Podospora anserina. I. Isolation and characterization. Mol Gen Genet. 1979 Mar 27;171(3):229–238. doi: 10.1007/BF00267577. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Belcour L., Grandchamp C. Mitochondrial DNA from Podospora anserina. II. Properties of mutant DNA and multimeric circular DNA from senescent cultures. Mol Gen Genet. 1979 Mar 27;171(3):239–250. doi: 10.1007/BF00267578. [DOI] [PubMed] [Google Scholar]
- Herrmann R. G., Bohnert H. J., Kowallik K. V., Schmitt J. M. Size, conformation and purity of chloroplast DNA of some higher plants. Biochim Biophys Acta. 1975 Jan 20;378(2):305–317. doi: 10.1016/0005-2787(75)90118-5. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Physicochemical characterization of mitochondrial DNA from pea leaves. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1830–1834. doi: 10.1073/pnas.69.7.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon A. M., de Vos W. M., Bakker H. The heterogeneity of rat-liver mitochondrial DNA. Biochim Biophys Acta. 1978 Jun 22;519(1):269–273. doi: 10.1016/0005-2787(78)90079-5. [DOI] [PubMed] [Google Scholar]
- Levings C. S., 3rd, Pring D. R. Restriction endonuclease analysis of mitochondrial DNA from normal and Texas cytoplasmic male-sterile maize. Science. 1976 Jul 9;193(4248):158–160. doi: 10.1126/science.193.4248.158. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Nitsch J. P., Nitsch C. Haploid plants from pollen grains. Science. 1969 Jan 3;163(3862):85–87. doi: 10.1126/science.163.3862.85. [DOI] [PubMed] [Google Scholar]
- Pring D. R., Levings C. S. Heterogeneity of Maize Cytoplasmic Genomes among Male-Sterile Cytoplasms. Genetics. 1978 May;89(1):121–136. doi: 10.1093/genetics/89.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring D. R., Levings C. S., Hu W. W., Timothy D. H. Unique DNA associated with mitochondria in the "S"-type cytoplasm of male-sterile maize. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2904–2908. doi: 10.1073/pnas.74.7.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer H. D., Hollenberg C. P. Saccharomyces cerevisiae 2-mum DNA. An analysis of the monomer and its multimers by electron microscopy. Mol Gen Genet. 1977 Feb 15;150(3):271–284. doi: 10.1007/BF00268126. [DOI] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- Steinemann M. Co-replication of satellite DNA of Chironomus melanotus with mainband DNA during polytenization. Chromosoma. 1978 Mar 31;66(2):127–139. doi: 10.1007/BF00295135. [DOI] [PubMed] [Google Scholar]
- Suyama Y., Bonner W. D. DNA from plant mitochondria. Plant Physiol. 1966 Mar;41(3):383–388. doi: 10.1104/pp.41.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synenki R. M., Levings C. S., Shah D. M. Physicochemical characterization of mitochondrial DNA from soybean. Plant Physiol. 1978 Mar;61(3):460–464. doi: 10.1104/pp.61.3.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upholt W. B., Dawid I. B. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977 Jul;11(3):571–583. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- Wells R., Ingle J. The constancy of the buoyant density of chloroplast and mitochondrial deoxyribonucleic acids in a range of higher plants. Plant Physiol. 1970 Jul;46(1):178–179. doi: 10.1104/pp.46.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfeld P. R., Herrmann R. G., Bottomley W. Mapping of the ribosomal RNA genes on spinach chloroplast DNA. Nucleic Acids Res. 1978 Jun;5(6):1741–1751. doi: 10.1093/nar/5.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]