Abstract
Intrinsically disordered proteins (IDPs) are important components of the cellular signaling machinery, allowing the same polypeptide to undertake different interactions with different consequences. IDPs are subject to combinatorial post-translational modifications and alternative splicing, adding complexity to regulatory networks and providing a mechanism for tissue-specific signaling. IDPs participate in assembly of signaling complexes and in the dynamic self-assembly of membrane-less nuclear and cytoplasmic organelles. Experimental, computational and bioinformatic analysis combine to identify and characterize disordered regions of proteins, leading to a greater appreciation of their widespread role in biological processes.
The abundance and functional significance of protein disorder in eukaryotes was largely unrecognized before the mid-1990s. Around that time, experimental studies on regulatory proteins and parallel bioinformatics interrogation of the complete genome sequences that were just beginning to emerge revealed that regions of disorder are very common in eukaryotic proteins, especially those involved in cellular regulation and signaling (reviewed in 1,2). Intrinsically disordered proteins (IDPs) are characterized by their biased amino acid composition and low sequence complexity and by their low content of bulky hydrophobic amino acids. Such protein sequences are unable to fold spontaneously into stable, well-defined globular three-dimensional structures but are dynamically disordered and fluctuate rapidly over an ensemble of conformations that cover a continuum of conformational space ranging from extended statistical coils to collapsed globules3. Some proteins are predicted to be entirely disordered, while others contain disordered sequences, referred to as intrinsically disordered regions (IDRs), in combination with structured globular domains. The majority of proteins in eukaryotic proteomes contain both intrinsically disordered and structured regions. In this review, we use IDP as a generic term to denote a protein that contains extensive disorder that is important for function. A discussion of the classification of IDPs and IDRs was recently published4.
Intrinsically disordered proteins frequently interact with or function as hubs in protein interaction networks5,6. They perform a central role in regulation of signaling pathways and crucial cellular processes, including regulation of transcription, translation and the cell cycle1,7–9. The abundance of IDPs in the cell is tightly regulated to ensure precise signaling in time and space, and mutations in IDPs or changes in their cellular abundance are associated with disease10–12. In addition to their regulatory functions, IDPs play a central role in the ordered assembly of macromolecular machines such as the ribosome, in organization of chromatin, in assembly and disassembly of microfilaments and microtubules, in transport through the nuclear pore, in binding and transport of small molecules, in the functioning of protein and RNA chaperones, and as flexible “entropic” linkers that separate functional protein domains13–16.
An exciting recent finding is that many proteins containing low-complexity or prion-like sequences can promote phase separation to form membrane-less organelles within the cytoplasm or nucleoplasm, thus contributing to their compartmentalization in a regulated manner17,18. The broader roles of IDPs in biology have been discussed in many excellent recent reviews and will not be revisited here.
The physical characteristics of IDPs allow an exquisite level of control of cellular signaling processes. Their favorable characteristics include: the presence of small recognition elements that fold upon partner binding; a degree of flexibility that enables IDPs to interact promiscuously with different targets on different occasions; accessible sites for post-translational modification; efficient utilization of conserved sequence motifs to mediate binding interactions; the ability to bind partners with high specificity but modest affinity, leading to rapid and spontaneous dissociation and termination of the signal1,3,19; kinetic advantages in signaling20 as their extremely fast association rates allow signals to be rapidly turned on. In performing their signaling functions, IDPs bind transiently to multiple interaction partners in dynamic regulatory networks21 that respond precisely and quantitatively to cellular signals and have the potential for complex information processing. The molecular interactions are transient and dynamic: IDPs exchange binding partners and compete for binding to central hub proteins, which are often present in limiting amounts. These interactions are fine-tuned by post-translational modifications that enable them to function as switches and rheostats3,22–26.
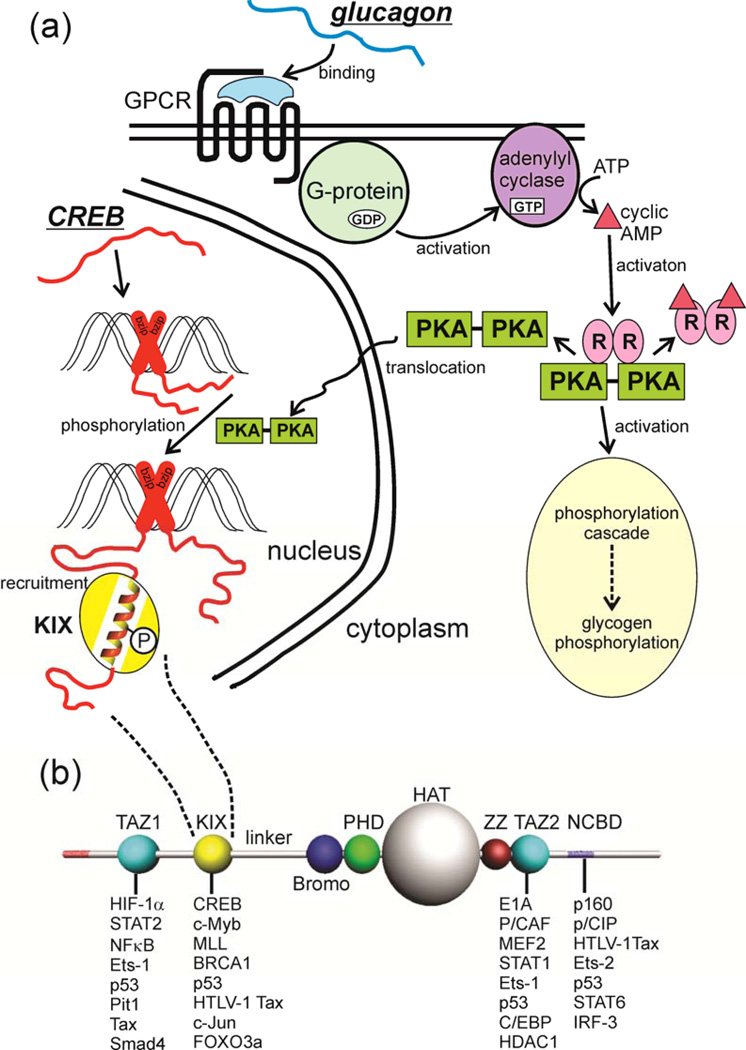
In this Review, we focus on the well-documented roles of IDPs in the regulation of intracellular signaling, where the role of proteins with flexible structures and extensive dynamics is well understood. We discuss how intrinsic disorder functions to enhance and propagate signaling. The multiplicity of protein interaction motifs in IDPs and their capacity for regulation through post-translational modification brings important advantages to the signaling process. For example, the same amino acid sequence can be used in different contexts and in response to different signals to turn on or turn off different signaling pathways and hence cause different cellular responses. These attributes also contribute to pathway crosstalk and to the operation of positive and negative feedback circuits. An illustration of the involvement of disordered proteins in a canonical signaling pathway is shown in Figure 1.
Figure 1. Intrinsic Disorder in Signaling.
(a) The metabolic hormone glucagon (an early example of intrinsic disorder in a functional molecule 144) binds to a structured, membrane-bound cell surface receptor, a G-protein-coupled receptor (GPCR) causing the translocation of the α subunit of the coupled G protein to the membrane-bound adenylyl cyclase, with concomitant formation of GTP from GDP. Cyclic AMP is generated, and activates protein kinase A (PKA), which has two downstream effects, firstly initiating the phosphorylation cascade that results in the phosphorylation of glycogen and the mobilization of stored glucose. The second effect is that activated PKA is translocated to the nucleus, where it phosphorylates the transcription factor cyclic-AMP response element binding protein (CREB), an intrinsically disordered protein. It appears that CREB is constitutively bound to the CRE DNA sequence through dimerization of the C-terminal basic leucine zipper domain (bzip, red cross). Phosphorylation of the kinase-inducible (KID) domain causes this domain to fold into a helical structure on the KIX domain of the transcriptional coactivator CREB-binding protein (CBP) (yellow), recruiting it to the promoter and promoting the transcription of downstream signal-response genes (reviewed in 145). In this case, intrinsically disordered proteins function both in the original reception of the signal and in the promotion of gene transcription in response to the signal. (b) Domain organization of CBP, showing a subset of the IDPs that bind to each of the four main interaction domains, the folded domains TAZ1, KIX and TAZ2, and the disordered (probably molten globular) NCBD.
Characterization of Protein Disorder
Several web servers have been developed for prediction of protein disorder on the basis of sequence analysis. A database has been established that contains consensus disorder predictions for all proteins coded by the human genome27 (Box 1). Major advances have been made in the development of experimental and computational tools to characterize disorder and to generate structural ensembles of disordered proteins. Modeling of ensembles28–30 is based primarily on nuclear magnetic resonance (NMR) and small angle X-ray scattering (SAXS) data and a public database pE-DB31 (http://pedb.vib.be) has been established for deposition of conformational ensembles. Structures of IDRs in their bound states can be obtained by X-ray crystallography, if well ordered, or by NMR, which has the additional advantage of providing insights into dynamic interactions between IDPs and their targets. Single molecule fluorescence energy transfer (smFRET) and computer simulations based on polymer physics are playing an increasingly important role in characterization of IDPs32–34.
Box 1 Prediction of Intrinsic Disorder from Sequence.
The likelihood that an amino acid sequence will be disordered rather than part of a well-structured three-dimensional fold can be evaluated both experimentally and using bioinformatics. The most convenient method to identify disordered regions in amino acid sequences is using the powerful bioinformatics prediction tools that have been developed over the past 15 years or so. Programs such as DISPROT134, IUPRED135,136 (and the associated program ANCHOR137,138, a predictor of binding sites), PONDR139, PrDOS140, and ESpritz141, analyze local sequence composition, with relatively higher proportions of small hydrophilic amino acids indicating a sequence with greater probability of disorder. These programs are available through web servers. Hidden Markov Models have also been applied to identify disordered and structured domains 142. An extremely convenient means of accessing disorder predictions is through the D2P2 database (http://d2p2.pro/)27. D2P2 is an interactive website that presents a compilation of disorder predictions for all sequences in the human proteome, both as predictions made using each of the above algorithms and also as an overall consensus.
Interaction Motifs
Intracellular signaling is accomplished by dynamic networks of interacting proteins; intrinsic disorder has a prominent role in mediating these interactions. The disordered regions of signaling and regulatory proteins frequently contain multiple conserved sequence motifs that interact with nucleic acids or other proteins3,35. Amphipathic sequence motifs and short linear motifs that mediate binding can readily be identified by bioinformatics analysis36,37.
Recent estimates suggest that the human proteome may contain more than 100,000 short linear binding motifs located within intrinsically disordered regions38. An important feature of the recognition elements in many IDPs is that they exhibit structural polymorphism, adopting different structures on different targets3. An extreme example is the nuclear coactivator binding domain (NCBD) of the CREB-binding protein CBP, which folds into two quite different structures when bound to the activation domain of p160 nuclear receptor coactivators39,40 or the interferon regulatory factor IRF341.
Coupled folding and binding
The kinetics and mechanism by which disordered interaction motifs associate with and fold upon binding to their targets, a process known as coupled folding and binding35,42, have recently received much attention through numerous experimental and theoretical studies that are beyond the scope of this review. The presence of pre-formed secondary structural elements in the conformational ensemble of the IDP has been predicted to favor the binding process43, but experimental evidence so far does not uniformly support this hypothesis.
Binding of the phosphorylated kinase-inducible domain (pKID) of the cyclic-AMP responsive element binding protein (CREB) to the KIX interaction domain of CREB-binding protein (CBP) occurs through an induced folding mechanism, in which pKID binds in a disordered state and folds on the surface of KIX44. Subsequent studies of the kinetics of binding of five different IDPs to KIX45–47 and of the disordered protein PUMA to MCL-148 show that, for each of these IDPs, folding is induced by binding and that pre-formed helical structure does not influence the rate of association with their target proteins. Association is remarkably fast and is diffusion-limited, which is clearly advantageous for a rapid signaling response in the cell. Initial association of the disordered activation domain of the p160 coactivator ACTR with the disordered, molten globular NCBD domain of CBP is also extremely rapid, but is followed by slow conformational transitions that are presumably associated with the folding process49. However, in contrast to the results described above, an increase in the population of a pre-formed ACTR helix led to a modest increase in the association rate50.
The effect that stabilizing a pre-formed helical structure has on partner binding affinity has been investigated through amino acid substitution to insert helix-favoring amino acids, and by covalent modification to “staple” the peptide into a helical conformation51. Stabilization of pre-formed helix in some disordered peptides appears to have a rather small effect in enhancing the binding affinity for the target proteins, and can even destabilize the complex52,53.
Two recent papers suggest that the population of helical structure in the unbound state, which is controlled by the IDP sequence, is an important determinant of biological function. The population of pre-formed helical structure in a linker region of the p27 cyclin-dependent kinase inhibitor directly mediates its ability to regulate the cell cycle, with variants that reduce the intrinsic linker helicity being deficient in promoting cell cycle arrest54. The population of pre-formed helical structure in the intrinsically disordered N-terminal transactivation domain of the p53 tumor suppressor has been implicated as a determinant of Mdm2 binding affinity55. In the absence of a binding partner, wild-type p53 has a very low helical content in the Mdm2 binding motif. The population of helix is tuned by conserved proline residues; substitution of a flanking proline residue (Pro27) with alanine greatly increases the residual helicity and enhances the affinity for binding Mdm2. This enhanced Mdm2 binding affinity, however, upsets the delicate balance of protein-protein interactions in p53 signaling pathways and is deleterious to p53 function. Substitution of mutant p53 for endogenous p53 in cells alters the dynamics of p53 accumulation, impairs target gene expression, and culminates in failure to induce cell cycle arrest upon DNA damage55.
Overall, much remains to be understood about the functional importance of preformed structures, which seems to depend strongly on the particular system under study. Nevertheless, for p53 and p27 at least, it is already clear that the intrinsic helical propensity is finely tuned by the protein sequence and is an important determinant of signaling fidelity54,55.
Fuzzy complexes
Not all IDPs undergo folding transitions in performing their biological functions; some disordered regions appear to function as flexible unstructured linkers between globular or disordered interaction domains3,56,57, while other IDPs remain disordered even after binding to their targets58,59, forming so-called “fuzzy” complexes60. IDPs often form dynamically heterogeneous complexes with their targets, interacting through well-ordered and fully structured “static” interfaces plus additional disordered “dynamic” (or “fuzzy”) sites on distinct, non-overlapping surfaces of the target protein61,62. Functionally, such dynamic binding interactions can enhance target binding affinity, mediate pathway crosstalk through formation of ternary complexes with other binding partners, and modulate allosteric interactions63.
IDPs as Signaling Hubs
Because IDPs and IDRs of large multi-functional proteins frequently contain multiple interaction motifs that mediate binding to diverse targets, they commonly function as central hubs in signaling networks5,6. The ability to bind their targets through multiple sites confers on IDPs specific properties that facilitate the dynamic assembly of ternary and higher order complexes and integrate diverse signaling pathways. The existence of multiple binding sites also enables allosteric responses in biological signaling, and the energetics of the binding process are exquisitely tuned by the large variations in entropy between free and bound states64.
Non-homogeneous distribution of binding free energy
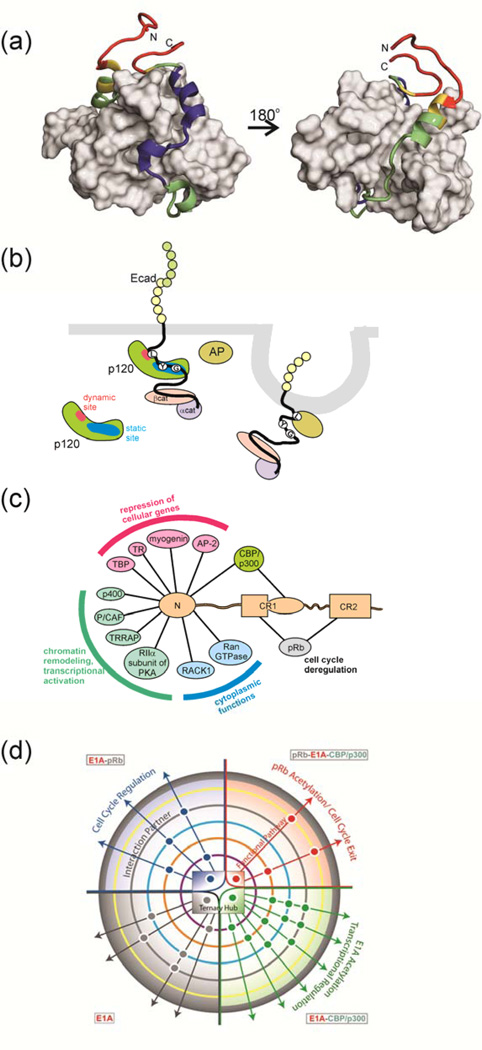
Knowledge of how the binding free energy is distributed over the IDP–target interface is of great importance for understanding the functional interactions of IDPs. The interactions between an IDP containing multiple binding motifs and its target will only rarely be energetically uniform. Binding will frequently be dominated by residues in local “hotspots” that contribute most of the binding free energy, whereas other regions may interact only weakly and contribute to a lesser extent, but still significantly, to the overall binding affinity. An example of this can be seen in the interaction between the transcriptional activation domain of RelA and the TAZ1 domain of CBP (Figure 2a)65. Alanine mutagenesis shows that binding is dominated by three amphipathic motifs in RelA that dock within hydrophobic grooves in the surface of the TAZ1 domain. Additional contacts are made by a transient amphipathic helix in the N-terminal region of the RelA activation domain, but these contribute little to the overall binding energy. NMR experiments show that the interactions are dynamic and that this region of the RelA sequence fluctuates between bound and free states and between helical and extended conformations65. Nevertheless, the transient binding sites contribute non-zero binding free energy, since truncation of these regions leads to a measurable decrease in affinity65.
Figure 2. Variable Binding Affinities of IDPs.
(a) One member of an NMR-derived family of the structure of the complex between the folded CBP TAZ1 domain (grey surface) and the transactivation domain of RelA (NFκB p65)65. Backbone dynamics of RelA in the complex were estimated using 1H-15N NOE measurements, and are mapped onto the RelA backbone in red (most flexible), yellow (less flexible), green (less flexible again) and blue (least flexible). The regions of RelA colored blue coincide with the hydrophobic docking interactions that dominate association with the TAZ1 domain65. The N-terminal helix (green) is only transiently populated, and is dynamically disordered on a nanosecond timescale, yet contributes to binding affinity. The figure was made from coordinates 2LWW and data in 65. (b) Schematic illustration of pathway cross talk mediated by differential binding of an IDP. The disordered cytoplasmic tail of E-cadherin binds to the armadillo repeat region of the p120 catenin through interaction of conserved sequence motifs containing phosphorylated tyrosines (Y) and glycines (G) at a high-affinity static binding site (blue), while the LL motif binds at the dynamic site (red), effectively chaperones this region. The adaptor protein (AP) recognizes the exposed LL motif, leading to clathrin-mediated endocytosis of E-cadherin. Figure adapted from 62. (c) Interactions between adenovirus E1A and cellular proteins. The intrinsically disordered N-terminal region of E1A binds numerous cellular proteins to disrupt cellular regulation. Interactions that function in the repression of cellular genes are shown in pink, in chromatin remodeling and transcriptional activation in green and in the cytoplasm in blue. Interactions with pRb (gray) are essential for deregulation of the cell cycle, while binding to CBP/p300 (green) disrupts cellular transcriptional programs. (d) Schematic summary of the allosteric modulation of the E1A signaling network through complex formation with CBP and pRb. Signaling pathways are modulated allosterically by interactions with various binding partners, as represented by a central phase diagram of the hub, with four states of E1A: free (gray), E1A–pRb (blue), E1A–CBP/p300 (green) and ternary complex (red). Circles outside the hub show additional protein partner interactions that influence regulatory pathways within the cell. Figure reproduced from 63 with permission.
Pathway crosstalk
The presence of a weak dynamic interface has advantages in a signaling network: when IDPs containing multiple interaction motifs bind to their targets through weak, dynamic interfaces as well as well-structured “static” interfaces, conformational fluctuations transiently expose the dynamic interaction motif, facilitating posttranslational modification or interactions with other target proteins. The p120 catenin, for example, regulates the stability of cell-cell adhesion by binding the intrinsically disordered cytoplasmic tail of cadherin through both static and dynamic interfaces62 (Figure 2b). The core region of the cadherin tail imparts specificity to the interaction and binds strongly to p120 through a well-structured “static” interface. In contrast, an N-terminal flanking region interacts only weakly and dynamically with p120, fluctuating between free and bound states. This “dynamic” binding site contributes little to binding affinity but plays an important function in determining the cellular fate of cadherin, by masking a critical Leu-Leu motif to hinder internalization by clathrin-mediated endocytosis.
A similar situation obtains in the intrinsically disordered adenovirus early region 1A oncoprotein (E1A)66, which illustrates how promiscuous interactions through multiple binding motifs can modulate signaling outcomes. E1A uses its N-terminal region and conserved regions CR1 and CR2 to recruit key cellular regulatory proteins to subvert cellular signaling pathways, force entry into S phase of the cell cycle, and activate transcription of viral genes67 (Figure 2c and Box 2). By incorporation of multiple binding motifs within an intrinsically disordered region, E1A functions as a hub that can bind promiscuously to a large number of cellular proteins and organize them combinatorially into higher-order complexes that efficiently disrupt regulatory networks and reprogram gene expression. Indeed, the N-terminal region and CR1 function synergistically to activate CBP/p300-mediated transcription of viral genes from the adenovirus E2 promoter68 and to repress transcription of a subset of cellular genes involved in proliferation and differentiation69.
Box 2 Motif Mimicry and the Role of IDPs in Viral Infection.
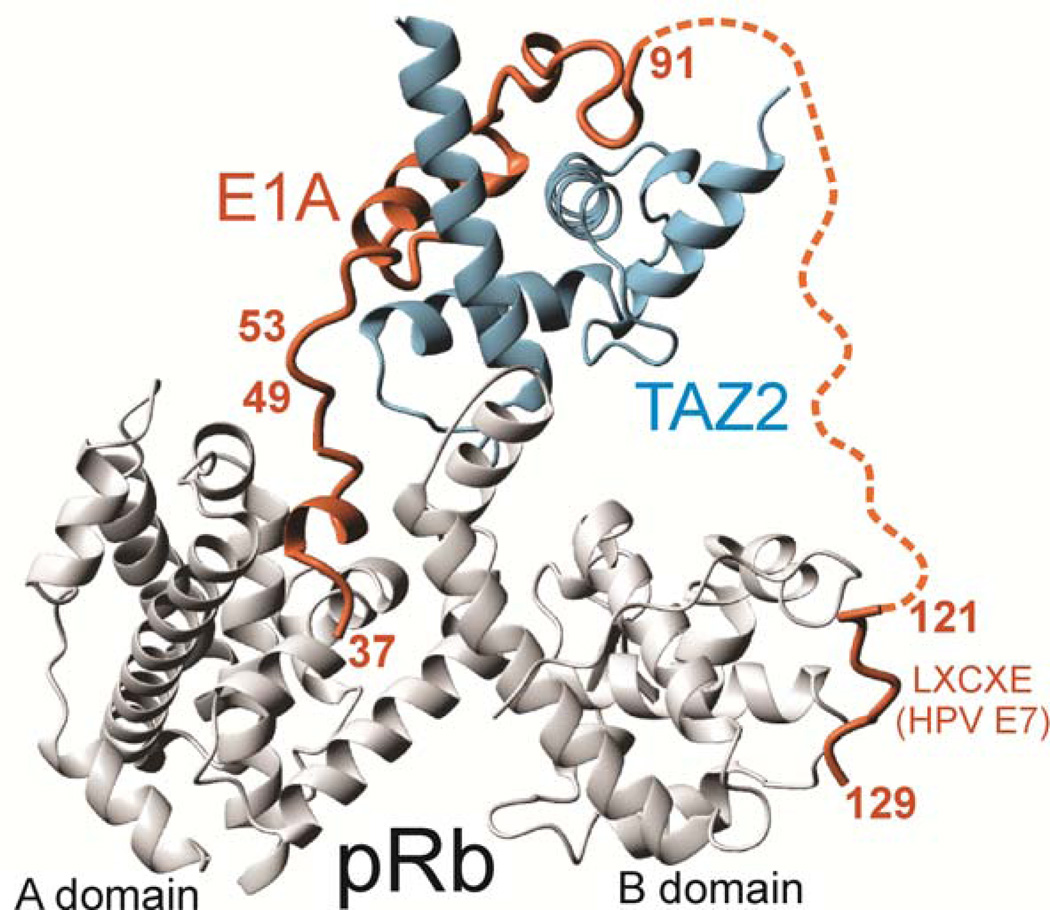
There are numerous examples of cellular signaling cascades that are subverted by viral proteins132. The proteins that viruses use to hijack cellular signaling networks are very often intrinsically disordered, and they frequently have higher affinities for their cellular targets than the natural sequences. For example, the adenovirus early region 1A (E1A) oncoprotein is disordered in solution and binds to the TAZ2 domain of the cellular transcriptional coactivator CBP with a dissociation constant (Kd) of 3 nM63 whereas the Kd values for binding cellular partners such as p53 (26 nM90) or STAT1 (52 nM143) are much weaker, allowing the viral protein to compete successfully for CBP 61 and block cellular transcription programs. The subversion of cellular regulation by viral proteins does not consist solely of out-competing cellular proteins for binding to the scarce CBP and p300 coactivator molecules. Intrinsically disordered proteins such as E1A provide multiple binding sites that allow them to recruit cellular proteins into higher order assemblies. Formation of an E1A–mediated ternary complex between the CBP/p300 TAZ2 domain and the retinoblastoma protein pRb (Figure) facilitates the disruption of the cell cycle by the virus: by bringing the CBP histone acetyl transferase (HAT domain) and the cell cycle regulatory protein pRb into proximity, E1A promotes acetylation and degradation of pRb, forcing S phase entry and uncontrolled proliferation61. By mimicking intrinsically disordered cellular proteins, viruses make very efficient use of their small genomes.
Figure reproduced from 61 with permission.
Weak binding sites function in synergy
Even in the absence of a localized high affinity interaction motif, IDRs can bind tightly to their targets through the synergistic action of multiple weak binding sites. The anaphase-promoting complex/cyclosome (APC/C) controls cell division by promoting ubiquitin-mediated degradation of cyclins and other proteins involved in regulation of the cell cycle. The 1.5 MDa APC/CCDH1 complex is inhibited during interphase by a 143 residue region of the early mitotic inhibitor protein 1 (EMI1), containing a zinc binding domain embedded in an intrinsically disordered region70. The intrinsically disordered region contains multiple interaction motifs that mediate interactions with multiple sites on APC/CCDH1. The individual binding sites in EMI1 interact only weakly with the APC/C but function synergistically to form a high affinity complex through multisite binding. Because binding at individual sites is weak, fluctuations between free and bound states make EMI1 accessible to kinases that can regulate the APC/C interaction through phosphorylation. By using intrinsic disorder, EMI1 is able to bind dynamically through multiple recognition motifs to regulate a large molecular machine 100 times its size.
Allostery in signaling
Allostery plays a central role in the regulation of cellular signaling networks71. Because of their conformational plasticity, their ability to bind multiple targets with high specificity and low affinity, and their propensity for posttranslational modification, IDPs display complex allosteric behavior that can fine-tune their regulatory interactions72. Indeed, theoretical considerations suggest that allosteric coupling is optimal when one or both of the coupled binding sites is intrinsically disordered73. Allosteric regulation by IDPs was first observed experimentally for the phd/doc toxin-antitoxin operon from bacteriophage P174. Allosteric coupling between an intrinsically disordered domain and an unstable folded domain of the protein Phd represses or derepresses transcription, a phenomenon termed conditional cooperativity, in response to changes in the relative concentrations of Phd and Doc. Allosteric coupling can also occur between functionally distinct domains that are intrinsically disordered, for example in the disordered N-terminal region of the glucocorticoid receptor75 and in the adenovirus E1A oncoprotein63. Interactions between E1A and the cellular proteins CBP and pRb can display either positive or negative allostery, depending on the available E1A binding sites63. Modulation of allosteric interactions will likely emerge as a common mechanism by which the signaling functions of intrinsically disordered hub proteins are regulated and by which the ultimate outcome of signaling is determined (Figure 2d).
Allosteric effects associated with binding to IDRs could potentially be exploited in a novel strategy for drug development. It has recently been shown that the small molecule inhibitor MSI-1436 binds to the C-terminal disordered region of the protein tyrosine phosphatase PTP1B and, through an allosteric effect, locks the enzyme in an inactive state76. By inhibiting PTP1B, MSI-1436 inhibits HER2 signaling and limits tumor growth, making it a viable therapeutic candidate for treatment of HER2-positive breast cancer.
Post-translational modifications
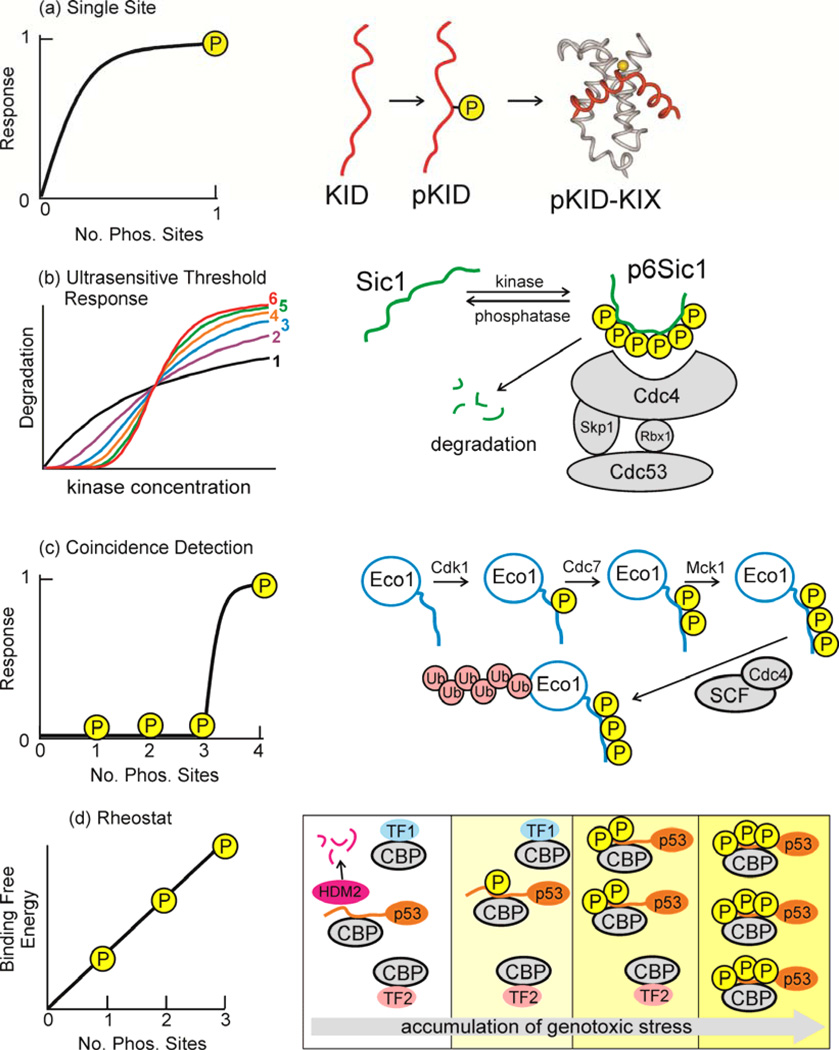
The enhanced flexibility and conformational plasticity of disordered regions of proteins renders them readily accessible for post-translational modification, often resulting in dense clusters of modifications77. It has recently been estimated that, when post-translational modifications are taken into account, there may be as many as one million instances of peptide interaction motifs within intrinsically disordered regions of the human proteome38. This enormous number underscores the central role that IDPs play in cellular signaling and regulation and sheds light on their remarkable functional diversity. Modifications of an intrinsically disordered protein by different kinases, acetylases, methylases, or other modifying enzymes can result in different signaling outputs, adding great complexity to signaling pathways. Phosphorylation sites are located predominantly in intrinsically disordered regions78 and phosphorylation plays a major role in modulating the conformational ensemble and interactions of disordered signaling proteins. Signaling can be regulated by addition (or removal) of a single phosphoryl group, as for example in the activation of the cAMP-regulated transcription factor CREB through phosphorylation at Ser133, in the intrinsically disordered kinase inducible activation domain79,80 (Fig. 3a). However, intrinsically disordered signaling proteins frequently contain multiple phosphorylation sites that can be modified sequentially or combinatorially to exert exquisite control over the signaling output (Figure 3b–d). By changing the bulk electrostatics, multisite phosphorylation can generate protein rheostats or ultrasensitive protein switches that are triggered at a threshold level of phosphorylation.
Figure 3. Response to multisite phosphorylation in IDRs.
Multisite phosphorylation of disordered proteins can give rise to a range of signaling responses. (a) For phosphorylation at a single site, the response takes the form of a simple hyperbolic saturation curve, as would be the case for single-site phosphorylation of CREB at Ser133 and its interaction with the KIX domain of CBP79,80. (b) In the Sic1-SCF ubiquitin ligase system81, the response to phosphorylation at multiple sites takes the form of sigmoidal threshold response curves, with cooperativity increasing as an increasing number of sites (1–6) are phosphorylated. (c) The interaction of Eco1 with the Cdc4 subunit of the SCF ubiquitin ligase85 shows coincidence detection, where a certain threshold level of phosphorylation must be achieved before response is initiated. (d) Under conditions of genotoxic stress the affinity of the p53 transactivation domain for CBP/p30023 increases with each successive phosphorylation event, relative to the affinities of other transcription factors (denoted TF1 and TF2). This is an example of a rheostat response. Figure adapted from 81,85,23 with permission.
Phosphorylation switches and threshold responses
Multisite phosphorylation of the intrinsically disordered Cdk1 inhibitor Sic1 late in the G1 phase of the yeast cell cycle acts as a switch that commits the cell to proceed to S phase81. Progressive phosphorylation of Sic1 by Cdk1 promotes degradation of Sic1 by enhancing binding to the Cdc4 subunit of the SCF ubiquitin ligase. Sic1 contains nine sub-optimal phosphodegron motifs, which individually bind only weakly to Cdc4. High affinity binding to Cdc4 occurs only after phosphorylation of any six of the phosphodegron sites. Progression from G1 phase is delayed until this phosphorylation threshold has been reached, after which Sic1 becomes degraded in a switch-like, ultrasensitive, sigmoidal response (Figure 3b)81. Activation of the switch is entirely dependent upon intrinsic disorder in Sic1 and its complex with Cdc482. Sic1 forms a highly disordered complex with Cdc4, in which individual phosphodegron motifs interact only weakly with Cdc4 and exchange dynamically between free and bound states. Unbound phosphate groups contribute to binding affinity through electrostatics, and the cumulative electrostatic interactions drive an ultrasensitive switch-like response once a threshold level of phosphorylation is reached24.
Coincidence detectors
Sequentially ordered phosphorylation cascades can function as logical AND operations in signaling networks83,84; signaling occurs only when all sites are phosphorylated. Such a cascade functions to degrade the Eco1 protein and prevent excess chromatid adhesion after the S phase of the cell cycle85. Degradation requires sequential phosphorylation of Ser and Thr residues in an intrinsically disordered region of Eco1. Phosphorylation of Ser99 by the cyclin-dependent kinase Cdk1 acts as a priming event that allows stepwise phosphorylation at Ser98 by the cell cycle-regulated kinase Cdc7-Ddf4, then at Thr94 by the Mck1 kinase. Precise spacing between the phosphoryl groups at Ser98 and Thr94 is required to create a binding site for the Cdc4 subunit of the SCF ubiquitin ligase85. This phosphorylation cascade functions as a coincidence detector (Figure 3c), integrating inputs from three distinct kinases and thus imparting exquisite control over the biological outcome, degradation of Eco1 and prevention of sister chromatid adhesion.
Molecular rheostats
Transcriptional pathways regulated by the p53 tumor suppressor are activated by a cascade of posttranslational modifications in the intrinsically disordered N- terminal transactivation and C-terminal regulatory domains of p5386. In unstressed cells, p53 levels are kept low by continual proteasomal degradation mediated by the Mdm2 ubiquitin ligase. Genotoxic stress initiates a phosphorylation and acetylation cascade that leads to stabilization and accumulation of p53, arrest of cell growth, and apoptosis87,88. The stability and transcriptional activity of p53 is tightly regulated through its interactions with Mdm2 and the transcriptional coactivators CBP and p30089, 90. Phosphorylation at Thr18, which requires prior phosphorylation at Ser15, decreases the affinity for binding the E3 ubiquitin ligase Mdm2 and thereby helps to stabilize p53 against proteasomal degradation90–92. Activation of p53-regulated transcriptional programs requires recruitment of CBP or p300 and subsequent acetylation of the C-terminal regulatory domain of p53. Whereas interactions with Mdm2 are primarily regulated by a Thr18 phosphorylation switch, binding of p53 to CBP/p300 is modulated by a phosphorylation rheostat, with successive phosphorylation events in the intrinsically disordered N-terminal transactivation domain enhancing the binding affinity in an additive manner (Figure 3d)23. This graded response to multisite phosphorylation progressively enhances the ability of p53 to recruit CBP/p300 in competition with cellular transcription factors, gradually increasing the efficiency of the p53 response following severe or prolonged cellular stress23.
Molecular clocks
The fractional content of charged residues and their distribution in the sequence has a profound influence on the dimensions and degree of compaction of intrinsically disordered proteins33,34,93. Changes in net charge and charge distribution resulting from multi-site Ser and Thr phosphorylation can thus strongly influence the conformational propensities of an IDP through bulk electrostatic effects. An example of this is seen in the time-delayed regulation of the Neurospora circadian clock by progressive phosphorylation of the protein FREQUENCY (FRQ)94. FRQ is predicted to be disordered over most of its length, with an asymmetric distribution of positive and negative charge. Over the course of a day, FRQ becomes progressively phosphorylated at as many as 113 sites95,96. At low levels of phosphorylation, two isolated amphipathic motifs interact to form a closed state that recruits casein kinase 1a. Slowly progressing phosphorylation of up to 46 non-consensus sites in the disordered N-terminal domain of FRQ leads to a steady accumulation of negative charge that destabilizes the closed state and triggers a transition to an open conformation. This timed change in conformation or compaction of FRQ exposes a PEST signal which results in protein degradation to reset the circadian clock. FRQ functions as a “molecular hourglass”94, in which time is “measured” by the number of phosphorylated residues and not by specific interactions mediated by individual phosphorylation sites. This clock mechanism is driven by the extensive intrinsic disorder in FRQ, which ensures that a large number of phosphorylation sites are accessible within a flexible polypeptide that can readily undergo changes in molecular compaction in response to changes in electrostatic charge.
Temporal regulation of the cell cycle is also achieved through multisite phosphorylation processes, at clustered sites within intrinsically disordered regions97. Timing and coordination of the cell cycle is orchestrated by cyclin-dependent kinases (Cdk), which catalyze phosphorylation of numerous downstream targets. Multisite phosphorylation by the yeast cyclin-Cdk1-Cks1 complex, for example, is controlled by the distance between phosphorylation sites on the substrate protein, the distribution of serine and threonine phosphoacceptors in these sites, and the efficiency of phosphorylation of individual motifs98. Thus, the output signal is determined in part by the sequence and spatial pattern of the multisite phosphorylation cluster embedded in an intrinsically disordered region.
Autoinhibitory sequences
The activities of many signaling proteins are negatively regulated in cis by autoinhibitory sequences99. A recent bioinformatic analysis has shown that the inhibitory modules of autoinhibited proteins are enriched in disorder and contain multiple phosphorylation sites and structural variability that can function combinatorially to ensure tight control of activation100. In the Vav1 guanine nucleotide exchange factor, for example, the catalytic Dbl homology (DH) domain is inhibited in cis by a dynamic helical motif in the adjacent acidic domain101,102 (Figure 4). The ~50 residue acidic domain, which is predicted to be intrinsically disordered in the free state, interacts with the neighboring calponin (CH) and Dbl homology domains in the autoinhibited state to form localized elements of helical structure separated by disordered regions. The CH and DH domains act cooperatively to bind the acidic domain, leading to tight inhibition and finely tuned mechanisms of activation. Dynamic fluctuations between the bound and free (dissociated) states of the acidic domain expose critical tyrosine residues to phosphorylation, resulting in a phosphorylation cascade that triggers dissociation and unfolding of the inhibitory helix and leads to activation of the DH domain101,102. The dynamic and cooperative nature of the interactions between the intrinsically disordered inhibitory module and the core domains of Vav1 provide fine control of Vav1 activity. Alternate splicing (see below) is also common in intrinsically disordered autoinhibitory domains, providing a mechanism for regulation of the activation process in a tissue-specific manner100.
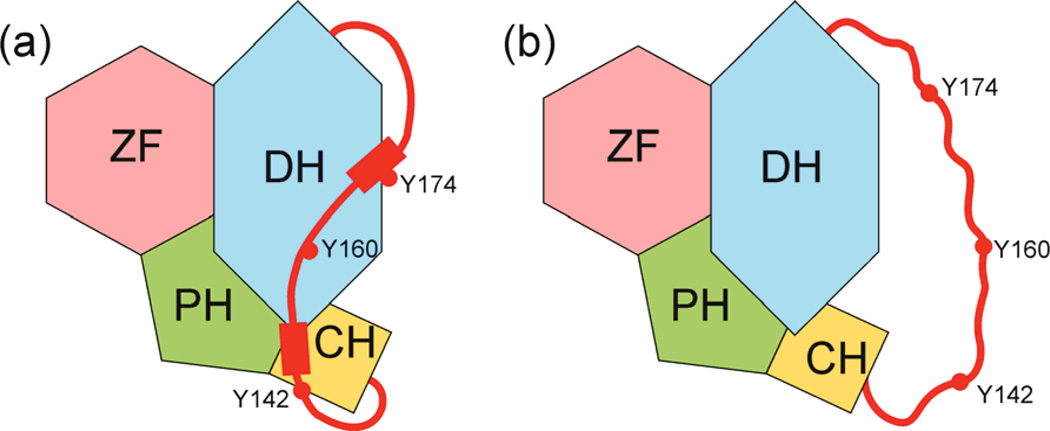
Figure 4. Autoinhibition through interactions with IDRs.
A disordered acidic domain inhibits the Vav1 nucleotide exchange factor by interacting in cis with the catalytic Dbl homology (DH) and calponin (CH) domains, forming localized elements of helical structure (red rectangles) that incorporate tyrosine residues. The acidic domain undergoes dynamic fluctuations, exposing the tyrosines to phosphorylation which results in dissociation of the bound inhibitory domain and activation of Vav1101,102. Figure adapted from 102 with permission.
Higher-order signaling assemblies
Recent studies have shown that regulatory proteins frequently form higher-order assemblies – signalosomes – that amplify signals, reduce noise, promote threshold signaling responses, and provide spatial and temporal control over signaling103. Intrinsically disordered regions play an important role in the assembly of a subset of signaling complexes, both through fully reversible protein-protein interactions that promote formation of reversible cellular assemblies104 and through formation of ultra-stable amyloid scaffolds105. The RIP1 and RIP3 kinases assemble into a signaling complex required for programmed necrosis by formation of a heterodimeric amyloid fibril105. Fibril formation is mediated by short, amyloidogenic RHIM sequences, embedded within intrinsically disordered regions of RIP1 and RIP3. It has been suggested that the disordered RHIM sequences are hidden in the inactive state but become exposed for amyloid/necrosome formation upon kinase activation105. The mechanism by which these ultra-stable amyloid fibrils are disassembled to terminate signaling is not yet understood.
In an exciting recent advance, it has been demonstrated that low affinity, multivalent protein-protein interactions, often mediated by low complexity and prion-like intrinsically disordered sequences, can promote liquid-liquid demixing to form membrane-less cytoplasmic and nuclear granules17,18,106,107. These granules behave like dynamic liquid droplets, rapidly exchanging component proteins and RNA with the cytoplasm or nucleoplasm. By sequestering regulatory proteins under conditions of high macromolecular concentration, IDP-mediated phase separation can have a profound influence on cellular signaling. For example, in response to cellular stress, the mechanistic target of rapamycin complex 1 (mTORC1), a kinase that controls cellular growth and metabolism, becomes sequestered in an inactive form in stress granules. Reactivation of mTORC1 requires stress granule dissolution, a process that is mediated by the dual specificity kinase DYRK3108. The intrinsically disordered N-terminal region of inactive DYRK3 targets it to the stress granules and prevents their dissolution. Activation of the DYRK3 kinase promotes dissolution of the stress granules and, by direct phosphorylation of the mTORC1 inhibitor PRAS40, releases reactivated mTORC1 for signaling108 (Figure 5). Given the abundance of multivalent low complexity sequences and prion-like glutamine/asparagine-rich sequences in RNA-binding proteins and transcription factors17,109, it appears likely that phase separation plays a quite general but poorly understood role in cellular signaling.
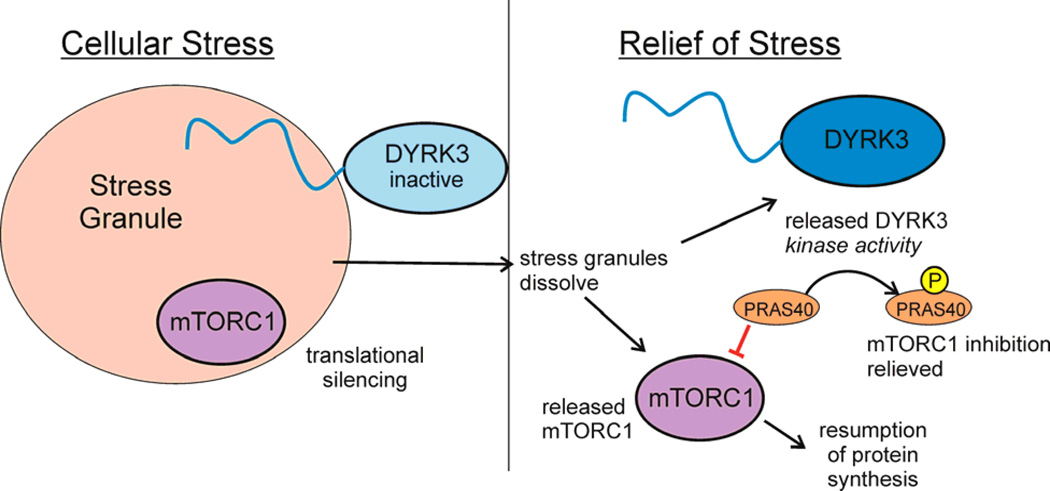
Figure 5. Disorder mediates stress-induced translational silencing.
Stress granules, which contain RNA and protein in a membraneless condensed particle, form and coalesce in response to cellular stress, sequestering proteins, including the dual-specificity kinase DYRK3 and mTORC1, the cellular factor that activates translation108. Translational silencing is mediated by DYRK3 under stress conditions in two ways, through stabilization of stress granules by the interaction of the disordered N-terminal tail, thus prolonging the sequestration of mTORC1. When cellular stress is relieved, DYRK3 and mTORC1 are released from the stress granules. Active DYRK3 acts as a kinase to phosphorylate PRAS40, relieving its inhibition of mTORC1 and allowing the resumption of protein synthesis. Figure adapted from 108 with permission.
Alternative Splicing and Disorder
More than 90% of human genes undergo alternative splicing, which frequently leads to expression of distinct protein isoforms in different cell types and tissues110.
Tissue-specific splicing
Tissue-specific splicing, which results in protein isoforms that make distinct interactions in different tissues, plays a central role in development and cellular differentiation111,112. Protein segments encoded by tissue-specific exons are enriched in disorder, whereas constitutive exons more often encode folded protein domains111,112. These intrinsically disordered regions are rich in interaction motifs and sites for posttranslational modification and tend to occupy central positions in cellular interaction networks. Tissue-specific splicing thus modulates the binding properties of critical regulatory proteins and differentially rewires the signaling networks in different cell types or tissues.
Disorder in the splicing machinery
Intrinsically disordered proteins play a key role in both constitutive pre-mRNA splicing and alternative splicing, processes that are catalyzed by the spliceosome, a large and highly dynamic ribonucleoprotein machine. The protein components of the spliceosome are highly enriched in intrinsic disorder113 as are the alternatively spliced segments of the protein substrates114. Proteins involved in spliceosome assembly and mRNA recognition, for example, the retention-and-splicing complex RES, have a strong propensity for disorder whereas proteins such as the snRNP proteins that comprise the catalytic core of the spliceosome tend to be highly ordered113. Spliceosome assembly and conformational rearrangement is regulated by reversible posttranslational modifications in disordered regions. Splicing of pre-mRNA is regulated through a dynamic cycle of multisite phosphorylation and dephosphorylation of serine residues in intrinsically disordered arginine/serine-rich regions (termed RS or SR domains) of splicing factors115,116. Recent NMR studies show that unphosphorylated RS domains are fully disordered and highly dynamic and are susceptible to efficient phosphorylation by a number of kinases117. Multisite phosphorylation acts as a dynamic switch that favors a more rigid arch-like structure, with well-defined orientations of the arginine and serine side chains in the RS repeats. The extent of ordering of the RS domain depends upon the number of RS repeats and the number of phosphoryl groups. It has been suggested that the interactions of the RS domain with RNA and with other proteins is modulated through entropic changes and increased charge associated with progressive phosphorylation117. Indeed, recent evidence suggests a role for RS domains in regulating the compartmentalization of splicing factors within the nucleus. RS domain proteins first localize to the nucleolus in a hypophosphorylated state, then are distributed to nuclear speckles in response to phosphorylation by CLK1/2 family kinases118.
RNA polymerase and splicing
The carboxy terminal domain (CTD) of RNA polymerase II plays a central role in coupling transcription to pre-mRNA splicing and alternative splicing (reviewed in 119). This domain contains 52 YSPTSPS heptad repeats in vertebrates, is predicted to be intrinsically disordered, and is subject to extensive posttranslational modification120. The CTD functions as a long and flexible tether, regulated by phosphorylation, acetylation, and methylation, that recruits splicing factors and other proteins to the polymerase elongation complex121. The intrinsically disordered CTD may also promote efficient cotranscriptional splicing across introns of highly variable length by tethering the downstream and upstream exons in close proximity122.
Future Perspectives
In the past decade, concepts of protein disorder have penetrated deeply into molecular and cell biology. It is now recognized that intrinsically disordered regions of proteins are involved mechanistically in a staggering array of cellular processes. Recent technical advances give great promise of rich future perspectives in the detection and characterization of IDPs. One of the most exciting developments has been the beginnings of biophysical studies of IDPs within the cell. By introducing isotopically or fluorescently labeled protein into the cell, NMR and fluorescence methods can be used to investigate the structural propensities and interactions of IDPs in their natural cellular environments. While such studies are currently in their infancy, they offer enormous promise for probing the dynamic interactions of IDPs within the cell. Early skepticism about the existence of disorder in proteins in the crowded cellular environment has been refuted by numerous studies that demonstrate that IDPs remain disordered in vivo (reviewed in 123). Many important questions remain. We need to acquire information on the abundance, concentration and subcellular localization of IDPs and proteins containing functional IDRs. Characterization of interactions and structural states of low-abundance proteins at their natural abundance in cells will require a whole new repertoire of high-sensitivity methods. The most promising methods for analysis of signaling IDPs, which are mostly present at extremely low concentration in the cell, are fluorescence-based, with single-molecule fluorescence imaging and single-molecule FRET already producing intriguing insights 124,125. Many questions remain to be answered about the structure, interactions, and localization of IDPs within the environment of the cell. Are signaling IDPs always fully sequestered by target proteins or are there significant populations of unbound and disordered protein inside the cell? How do PTMs modify the conformational ensemble, the sub-cellular localization, and interaction networks of IDPs? Are IDPs distributed homogeneously through the cytoplasm or nucleoplasm or are they concentrated in sub-cellular organelles and puncta? Can we characterize the dynamic interactions between IDPs and their target proteins within living cells?
Another major challenge will be structural characterization of full-length IDPs containing both folded and disordered domains. Until quite recently, experimental approaches have of necessity been reductionist, where large multi-domain proteins have been analyzed as fragments, and their interactions with physiological partners examined in vitro by atomic-scale biophysical approaches such as X-ray crystallography and NMR. Very few full-length IDPs have been characterized structurally, a notable exception being the p53 tetramer126. However, large modular IDPs do not function as isolated domains; instead their component regions, both ordered and disordered, act synergistically in performing their cellular functions. To fully understand these complex systems, it will be important to take a more holistic approach, characterizing the structural ensembles, dynamics, and interactions of the full-length proteins, or at least of large multi-domain fragments. The challenges should not be underestimated. Large proteins with both structured and disordered domains are extremely difficult to prepare intact, even from eukaryotic expression systems, and frequently undergo degradation to form truncated fragments during purification, likely due to the protease-vulnerability of their disordered regions. Such problems even arise during transient expression in mammalian cells. New approaches for expression, purification, and biophysical analysis will be required. The rewards will be great, offering new insights into the mechanism by which structured domains, disordered regions, interaction motifs, and posttranslational modifications function synergistically to control cellular signaling networks.
The discovery that protein phase separation leads to formation of membrane-less compartments that have important functional roles in the cell has also opened up an important new field of study. Many questions remain to be addressed, including the roles that IDPs and IDRs play in the assembly and disassembly of these organelles, and how they function to localize signaling proteins and thereby influence signaling pathways. Can we identify sequence motifs or identify prion-like domains that direct the assembly of phase separated states, and can this knowledge be used to predict the subcellular localization and potential function of IDPs?
Finally, there is growing interest in IDPs as potential targets for drug design. IDPs play a central role in key cellular signaling pathways and are frequently associated with disease127. They usually bind, often with modest affinity, to concave grooves in the surface of their targets through predominantly hydrophobic interactions128, making them very attractive therapeutic targets. This has led to recent efforts to synthesize conformationally-constrained molecules that mimic the bound conformation of IDRs as a new and rational approach to design potent inhibitors of protein-protein interactions in vivo129,130. It may even prove possible to design drugs targeted against the IDP itself, rather than its globular target, and success in this approach has recently been reported76,131. We have much to learn from viruses, which frequently mimic cellular IDR motifs to subvert signaling networks and hijack the cellular regulatory machinery132,133. Despite the promise of exploiting intrinsic disorder therapeutically, critical issues remain to be answered such as whether it is better to target the IDPs themselves, or to direct design efforts towards the grooves and pockets into which IDRs bind. Either way, targeting intrinsic disorder holds promise for a new era in drug discovery.
Online Summary.
Intrinsically disordered proteins (IDPs) and disordered regions (IDRs) of proteins that may also contain structured domains mediate critical signaling processes in eukaryotic cells
Disorder is advantageous in these processes because disordered sequences have the potential to bind to multiple partners, often utilizing different structures
Disordered regions are relatively accessible, often contain multiple binding motifs, and are frequently the sites for post-translational modification, an important mediator of the control of signaling pathways
Disordered proteins have central roles in formation of higher-order signaling assemblies and in the operation of circadian clocks
Acknowledgments
The authors apologize to the many colleagues whose work could not be cited owing to space limitations. This work was supported by grants CA096865 (PEW) and GM71862 (HJD) from the National Institutes of Health and by the Skaggs Institute for Chemical Biology (PEW).
Glossary
- Conformational ensemble
A structural description of proteins that do not have a single well-ordered three-dimensional structure. Conformational ensembles contain a multitude of different structures that, in sum, are consistent with observed parameters such as NMR spectra or SAXS data
- Binding Free Energy
The difference in free energy (AG) between the free and bound states of a complex. If the complex is stable, the binding free energy is negative
- Association Rate and Dissociation Rate
Measured quantities that describe the rate of formation of a complex from the component parts, and the reverse process, dissociation of the components. The binding affinity is determined by the relative magnitude of the association and dissociation rates; for example a fast association rate (“on-rate”) and a slow dissociation rate (“off-rate”) is characteristic of a high-affinity complex
- IDP and IDR
Intrinsically disordered proteins (IDPs), also known as intrinsically unstructured proteins, do not form stable three-dimensional structures under normal conditions, despite being perfectly functional. IDPs and disordered regions (IDRs) within larger proteins that may contain structured domains are characterized by amino acid sequences that have low content of bulky hydrophobic amino acids and high proportions of charged and hydrophilic amino acids
Biography
Peter E. Wright is a Professor at The Scripps Research Institute. His research has focused on applications of NMR and other biophysical methods to characterize the structural properties and interactions of intrinsically disordered proteins, mechanisms of protein folding and mis-folding, the structural basis of protein-protein and protein-nucleic acid interactions in the regulation of gene expression, and the role of dynamics in protein function. H. Jane Dyson is a Professor at the Scripps Research Institute. Her research interests are in the conformation of peptides and intrinsically disordered proteins, protein folding and dynamics, and structure and functional studies of proteins using NMR and other spectroscopic techniques.
Contributor Information
Peter E. Wright, Email: wright@scripps.edu.
H. Jane Dyson, Email: dyson@scripps.edu.
References
- 1.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. Journal of Molecular Biology. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 3.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 4.van der Lee R, et al. Classification of intrinsically disordered regions and proteins. Chemical Reviews. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBSJournal. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 6.Kim PM, Sboner A, Xia Y, Gerstein M. The role of disorder in interaction networks: a structural analysis. Molecular Systems Biology. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. Journal of Molecular Biology. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, et al. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW. Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47:7598–7609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 12.Babu MM, van der Lee R, de Groot NS, Gsponer Jr. Intrinsically disordered proteins: regulation and disease. Current Opinion in Structural Biology. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Letters. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 14.Frey S, Richter RP, Gorlich D. FG-Rich Repeats of Nuclear Pore Proteins Form a Three-Dimensional Meshwork with Hydrogel-Like Properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 15.Tompa P. Structure and Function of Intrinsically Disordered Proteins. Boca Raton: Chapman & Hall; 2010. [Google Scholar]
- 16.Guharoy M, Szabo B, Martos SC, Kosol S, Tompa P. Intrinsic Structural Disorder in Cytoskeletal Proteins. Cytoskeleton. 2013;70:550–571. doi: 10.1002/cm.21118. [DOI] [PubMed] [Google Scholar]
- 17.Kato M, et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber SC, Brangwynne CP. Getting RNA and Protein in Phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 19.Oldfield CJ, et al. Coupled folding and binding with a-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 20.Pontius BW. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends in Biochemical Sciences. 1993;18:181–186. doi: 10.1016/0968-0004(93)90111-y. [DOI] [PubMed] [Google Scholar]
- 21.Stein A, Pache RA, Bernado P, Pons M, Aloy P. Dynamic interactions of proteins in complex networks: a more structured view. FEBSJournal. 2009;276:5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x. [DOI] [PubMed] [Google Scholar]
- 22.Gsponer J, Babu MM. The rules of disorder or why disorder rules. Progress in Biophysics and Molecular Biology. 2009;99:94–103. doi: 10.1016/j.pbiomolbio.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borg M, et al. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Roey K, Dinkel H, Weatheritt RJ, Gibson TJ, Davey NE. The switches.ELM resource: a compendium of conditional regulatory interaction interfaces. Science Signaling. 2013;6 doi: 10.1126/scisignal.2003345. rs7. [DOI] [PubMed] [Google Scholar]
- 26.Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Current Opinion in Structural Biology. 2012;22:378–85. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Oates ME, et al. D2P2: database of disordered protein predictions. Nucleic Acids Research. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsh JA, Forman-Kay JD. Ensemble modeling of protein disordered states: Experimental restraint contributions and validation. Proteins: Structure, Function and Bioinformatics. 2012;80:556–572. doi: 10.1002/prot.23220. [DOI] [PubMed] [Google Scholar]
- 29.Salmon L, et al. NMR Characterization of Long-Range Order in Intrinsically Disordered Proteins. Journal of the American Chemical Society. 2010;132:8407–8418. doi: 10.1021/ja101645g. [DOI] [PubMed] [Google Scholar]
- 30.Sibille N, Bernado P. Structural characterization of intrinsically disordered proteins by the combined use of NMR and SAXS. Biochemical Society Transactions. 2012;40:955–962. doi: 10.1042/BST20120149. [DOI] [PubMed] [Google Scholar]
- 31.Varadi M, et al. pE-DB: a database of structural ensembles of intrinsically disordered and of unfolded proteins. Nucleic Acids Research. 2014;42:D326–D335. doi: 10.1093/nar/gkt960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreon AC, Moran CR, Gambin Y, Deniz AA. Single-molecule fluorescence studies of intrinsically disordered proteins. Methods in Enzymology. 2010;472:179–204. doi: 10.1016/S0076-6879(10)72010-3. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann H, et al. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das RK, Pappu RV. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Current Opinion in Structural Biology. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 36.Mohan A, et al. Analysis of molecular recognition features (MoRFs) Journal of Molecular Biology. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 37.Neduva V, Russell RB. Linear motifs: Evolutionary interaction switches. FEBS Letters. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Tompa P, Davey NE, Gibson TJ, Babu MM. A million peptide motifs for the molecular biologist. Molecular Cell. 2014;55:161–169. doi: 10.1016/j.molcel.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 39.Demarest SJ, et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 40.Waters L, et al. Structural diversity in p160/CREB-binding protein coactivator complexes. Journal of Biological Chemistry. 2006;281:14787–14795. doi: 10.1074/jbc.M600237200. [DOI] [PubMed] [Google Scholar]
- 41.Qin BY, et al. Crystal Structure of IRF-3 in complex with CBP. Structure. 2005;13:1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Spolar RS, Record MT. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 43.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. Journal of Molecular Biology. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 45.Shammas S, Travis AJ, Clarke J. Remarkably fast coupled folding and binding of the intrinsically disordered transactivation domain of cMyb to CBP KIX. Journal of Physical Chemistry B. 2013;117:13346–13356. doi: 10.1021/jp404267e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gianni S, Morrone A, Giri R, Brunori M. A folding-after-binding mechanism describes the recognition between the transactivation domain of c-Myb and the KIX domain of the CREB-binding protein. Biochemical and Biophysical Research Communications. 2012;428:205–209. doi: 10.1016/j.bbrc.2012.09.112. [DOI] [PubMed] [Google Scholar]
- 47.Shammas SL, Travis AJ, Clarke J. Allostery within a transcription coactivator is predominantly mediated through dissociation rate constants. Proceedings of the National Academy of Sciences. 2014;111:12049–12054. doi: 10.1073/pnas.1405815111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogers JM, Wong CT, Clarke J. Coupled Folding and Binding of the Disordered Protein PUMA Does Not Require Particular Residual Structure. Journal of the American Chemical Society. 2014;136:5197–5200. doi: 10.1021/ja4125065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dogan J, Schmidt T, Mu X, Engström Å, Jemth P. Fast association and slow transitions in the interaction between two intrinsically disordered protein domains. Journal of Biological Chemistry. 2012;287:34316–34324. doi: 10.1074/jbc.M112.399436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iesmantavicius V, Dogan J, Jemth P, Teilum K, Kjaergaard M. Helical Propensity in an Intrinsically Disordered Protein Accelerates Ligand Binding. Angewandte Chemie International Edition in English. 2014 doi: 10.1002/anie.201307712. [DOI] [PubMed] [Google Scholar]
- 51.Schafmeister CE, Po J, Verdine GL. An All-Hydrocarbon Cross-Linking System for Enhancing the Helicity and Metabolic Stability of Peptides. Journal of the American Chemical Society. 2000;122:5891–5892. [Google Scholar]
- 52.Bienkiewicz EA, Adkins JN, Lumb KJ. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27Kip1. Biochemistry. 2002;41:752–759. doi: 10.1021/bi015763t. [DOI] [PubMed] [Google Scholar]
- 53.Baek S, et al. Structure of the Stapled p53 Peptide Bound to Mdm2. Journal of the American Chemical Society. 2011;134:103–106. doi: 10.1021/ja2090367. [DOI] [PubMed] [Google Scholar]
- 54.Otieno S, Kriwacki R. Probing the role of nascent helicity in p27 function as a cell cycle regulator. PLoS One. 2012;7:e47177. doi: 10.1371/journal.pone.0047177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borcherds W, et al. Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat Chem Biol. 2014;10:1000–1002. doi: 10.1038/nchembio.1668. [DOI] [PubMed] [Google Scholar]
- 56.Bertagna A, Toptygin D, Brand L, Barrick D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochemical Society Transactions. 2008;36:157–166. doi: 10.1042/BST0360157. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, et al. Intrinsic disorder mediates the diverse regulatory functions of the Cdk inhibitor p21. Nature Chemical Biology. 2011;7:214–221. doi: 10.1038/nchembio.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker JMR, et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nature Structural and Molecular Biology. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mittag T, et al. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends in Biochemical Sciences. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Ferreon JC, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13260–13265. doi: 10.1073/pnas.0906770106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishiyama N, et al. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell. 2010;141:117–128. doi: 10.1016/j.cell.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 63.Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498:390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flock T, Weatheritt RJ, Latysheva NS, Babu MM. Controlling entropy to tune the functions of intrinsically disordered regions. Current Opinion in Structural Biology. 2014;26:62–72. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Mukherjee SP, et al. Analysis of the RelA:CBP/p300 Interaction Reveals Its Involvement in NFkB-Driven Transcription. PLoS Biology. 2013;11:e1001647. doi: 10.1371/journal.pbio.1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pelka P, Ablack JNG, Fonseca GJ, Yousef AF, Mymryk JS. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. Journal of Virology. 2008;82:7252–7263. doi: 10.1128/JVI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berk AJ. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 68.Fax P, Lipinski KS, Esche H, Brockmann D. cAMP-independent activation of the adenovirus type 12 E2 promoter correlates with the recruitment of CREB-1/ATF-1, E1A(12S), and CBP to the E2-CRE. Journal of Biological Chemistry. 2000;275:8911–8920. doi: 10.1074/jbc.275.12.8911. [DOI] [PubMed] [Google Scholar]
- 69.Green M, Panesar NK, Loewenstein PM. The transcription-repression domain of the adenovirus E1A oncoprotein targets p300 at the promoter. Oncogene. 2008;27:4446–4455. doi: 10.1038/onc.2008.85. [DOI] [PubMed] [Google Scholar]
- 70.Frye JJ, et al. Electron microscopy structure of human APC/CCDH1-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nature Structural and Molecular Biology. 2013;20:827–835. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nussinov R, Tsai CJ. Allostery in Disease and in Drug Discovery. Cell. 2013;153:293–305. doi: 10.1016/j.cell.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 72.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garcia-Pino A, et al. Allostery and Intrinsic Disorder Mediate Transcription Regulation by Conditional Cooperativity. Cell. 2010;142:101–111. doi: 10.1016/j.cell.2010.05.039. [DOI] [PubMed] [Google Scholar]
- 75.Li J, Motlagh HN, Chakuroff C, Thompson EB, Hilser VJ. Thermodynamic Dissection of the Intrinsically Disordered N-terminal Domain of Human Glucocorticoid Receptor. Journal of Biological Chemistry. 2012;287:26777–26787. doi: 10.1074/jbc.M112.355651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krishnan N, et al. Targeting the disordered C terminus of PTP1B with an allosteric inhibitor. Nature Chemical Biology. 2014;10:558–566. doi: 10.1038/nchembio.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pejaver V, et al. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Science. 2014;23:1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Research. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chrivia JC, et al. Phosphorylated CREB binds specifically to nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 80.Radhakrishnan I, et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:Coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 81.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 82.Mittag T, et al. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Samaga R, Klamt S. Modeling approaches for qualitative and semiquantitative analysis of cellular signaling networks. Cell Communications and Signaling. 2013;11:43. doi: 10.1186/1478-811X-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferrell JE, Jr, Ha SH. Ultrasensitivity part II: multisite phosphorylation, stoichiometric inhibitors, and positive feedback. Trends in Biochemical Sciences. 2014;39:556–569. doi: 10.1016/j.tibs.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lyons NA, Fonslow BR, Diedrich JK, Yates JR, Morgan DO. Sequential primed kinases create a damage-responsive phosphodegron on Eco1. Nature Structural and Molecular Biology. 2013;20:194–201. doi: 10.1038/nsmb.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meek DW, Anderson CW. Posttranslational Modification of p53: Cooperative Integrators of Function. Cold Spring Harbor Perspectives in Biology. 2009;1 doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 89.Grossman SR, et al. Polyubiquitination of p53 by a Ubiquitin Ligase Activity of p300. Science. 2003;300:342. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 90.Ferreon JC, et al. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6591–6596. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sakaguchi , et al. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase Effect on Mdm2 binding. Journal of Biological Chemistry. 2000;275:9278–9283. doi: 10.1074/jbc.275.13.9278. [DOI] [PubMed] [Google Scholar]
- 92.Schon O, Friedler A, Freund S, Fersht AR. Binding of p53-derived Ligands to MDM2 Induces a Variety of Long Range Conformational Changes. Journal of Molecular Biology. 2004;336:197–202. doi: 10.1016/j.jmb.2003.11.051. [DOI] [PubMed] [Google Scholar]
- 93.Müller-Späth S, et al. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Querfurth C, et al. Circadian Conformational Change of the Neurospora Clock Protein FREQUENCY Triggered by Clustered Hyperphosphorylation of a Basic Domain. Molecular Cell. 2011;43:713–722. doi: 10.1016/j.molcel.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 95.Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell. 2009;34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tang CT, et al. Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10722–10727. doi: 10.1073/pnas.0904898106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holt LJ, et al. Global Analysis of Cdk1 Substrate Phosphorylation Sites Provides Insights into Evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koivomagi M, et al. Multisite phosphorylation networks as signal processors for Cdk1. Nature Structural and Molecular Biology. 2013;20:1415–1424. doi: 10.1038/nsmb.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pufall MA, Graves BJ. AUTOINHIBITORY DOMAINS: Modular Effectors of Cellular Regulation. Annual Review of Cell and Developmental Biology. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 100.Trudeau T, et al. Structure and Intrinsic Disorder in Protein Autoinhibition. Structure. 2013;21:332–341. doi: 10.1016/j.str.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 101.Li P, Martins IRS, Amarasinghe GK, Rosen MK. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nature Structural and Molecular Biology. 2008;15:613–618. doi: 10.1038/nsmb.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu B, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140:246–256. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu H. Higher-Order Assemblies in a New Paradigm of Signal Transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weatheritt RJ, Gibson TJ, Babu MM. Asymmetric mRNA localization contributes to fidelity and sensitivity of spatially localized systems. Nature Structural and Molecular Biology. 2014;21:833–839. doi: 10.1038/nsmb.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li J, et al. The RIP1/RIP3 Necrosome Forms a Functional Amyloid Signaling Complex Required for Programmed Necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends in Biochemical Sciences. 2013 doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wippich F, et al. Dual Specificity Kinase DYRK3 Couples Stress Granule Condensation/Dissolution to mTORC1 Signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 109.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buljan M, et al. Tissue-Specific Splicing of Disordered Segments that Embed Binding Motifs Rewires Protein Interaction Networks. Molecular Cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ellis JD, et al. Tissue-specific alternative splicing remodels protein-protein interaction networks. Molecular Cell. 2012;46:884–892. doi: 10.1016/j.molcel.2012.05.037. [DOI] [PubMed] [Google Scholar]
- 113.Korneta I, Bujnicki JM. Intrinsic disorder in the human spliceosomal proteome. PLoS Computational Biology. 2012;8:e1002641. doi: 10.1371/journal.pcbi.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Romero PR, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mermoud JE, Cohen P, Lamond AI. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Research. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes & Development. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 117.Xiang S, et al. Phosphorylation Drives a Dynamic Switch in Serine/Arginine-Rich Proteins. Structure. 2013;21:2162–2174. doi: 10.1016/j.str.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 118.Kwon I, et al. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Muñoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends in Biochemical Sciences. 2010;35:497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 120.Hsin J-P, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes & Development. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes & Development. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dye MJ, Gromak N, Proudfoot NJ. Exon Tethering in Transcription by RNA Polymerase II. Molecular Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 123.Theillet FX, et al. Physicochemical Properties of Cells and Their Effects on Intrinsically Disordered Proteins (IDPs) Chem Rev. 2014;114:6661–714. doi: 10.1021/cr400695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sakon JJ, Weninger KR. Detecting the conformation of individual proteins in live cells. Nature Methods. 2010;7:203–205. doi: 10.1038/nmeth.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gebhardt JC, et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nature Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wells M, et al. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cheng Y, et al. Rational drug design via intrinsically disordered protein. Trends in Biotechnology. 2006;24:435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 128.Mészáros B, Tompa P, Simon I, Dosztányi Z. Molecular principles of the interactions of disordered proteins. Journal of Molecular Biology. 2007;372:549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 129.Lao BB, et al. Rational Design of Topographical Helix Mimics as Potent Inhibitors of Protein-Protein Interactions. Journal of the American Chemical Society. 2014;136:7877–7888. doi: 10.1021/ja502310r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lao BB, et al. In vivo modulation of hypoxia-inducible signaling by topographical helix mimetics. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7588–7593. doi: 10.1073/pnas.1402393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-myc. Journal of the American Chemical Society. 2009;131:7390–7401. doi: 10.1021/ja900616b. [DOI] [PubMed] [Google Scholar]
- 132.Davey NE, Trave G, Gibson TJ. How viruses hijack cell regulation. Trends in Biochemical Sciences. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 133.Hagai T, Azia A, Babu MM, Andino R. Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Reports. 2014;7:1729–1739. doi: 10.1016/j.celrep.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sickmeier M, et al. DisProt: the database of disordered proteins. Nucleic Acids Research. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. Journal of Molecular Biology. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 136.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 137.Dosztanyi Z, Meszaros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Meszaros B, Simon I, Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Obradovic Z, et al. Predicting intrinsic disorder from amino acid sequence. Proteins: Structure, Function and Bioinformatics. 2003;53(Suppl 6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 140.Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Walsh I, Martin AJ, Di Domenico T, Tosatto SC. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012;28:503–509. doi: 10.1093/bioinformatics/btr682. [DOI] [PubMed] [Google Scholar]
- 142.Fukuchi S, Homma K, Minezaki Y, Gojobori T, Nishikawa K. Development of an accurate classification system of proteins into structured and unstructured regions that uncovers novel structural domains: its application to human transcription factors. BMC Structural Biology. 2009;9 doi: 10.1186/1472-6807-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBOJournal. 2009;28:948–958. doi: 10.1038/emboj.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bösch C, Bundi A, Oppliger M, Wüthrich K. 1H nuclear-magnetic-resonance studies of the molecular conformation of monomeric glucagon in aqueous solution. European Journal of Biochemistry. 1978;91:209–214. doi: 10.1111/j.1432-1033.1978.tb20953.x. [DOI] [PubMed] [Google Scholar]
- 145.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature Reviews Molecular Cell Biology. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]