Abstract
Regular exercise improves learning and memory, including during aging process. Interestingly, the imbalance of epigenetic mechanisms has been linked to age-related cognitive deficits. However, studies about epigenetic alterations after exercise during the aging process are rare. In this preliminary study we investigated the effect of aging and exercise on DNA methyltransferases (DNMT1 and DNMT3b) and H3-K9 methylation levels in hippocampus from 3 and 20-months aged Wistar rats. The animals were submitted to two exercise protocols: single session or chronic treadmill protocol. DNMT1 and H3-K9 methylation levels were decreased in hippocampus from aged rats. The single exercise session decreased both DNMT3b and DNMT1 levels in young adult rats, without any effect in the aged group. Both exercise protocols reduced H3-K9 methylation levels in young adult rats, while the single session reversed the changes on H3-K9 methylation levels induced by aging. Together, these results suggest that an imbalance on DNMTs and H3-K9 methylation levels might be linked to the brain aging process and that the outcome to exercise seems to vary through lifespan.
Keywords: aging, exercise, rat hippocampus, epigenetic mechanisms, methylation markers
1. Introduction
Recently, epigenetic mechanisms have been linked to normal aging-related changes in the brain, as well as neuropsychiatric and neurodegenerative diseases (Saha and Pahan, 2006). Epigenetic typically involve modifications in the micro and macrostructure of chromatin, DNA and nuclear proteins, particularly histones, which can modulate the transcriptional machinery and allow long lasting modifications in the genome. DNA and histone methylation, in addition to histone acetylation, are the most extensively studied post-translational modifications, which can influence gene transcription (Kouzarides, 2007).
The histones can be methylated on either lysine (K) or arginine (R)-residues by histone methyltransferases. Site-specific methylation of amino acid residues can condensate or relax the chromatin structure, such as mono-methylation of histone H3 at K9 (H3-K9) is associated to transcriptional activation, whereas transcriptionally silent regions contain di- and tri-methylation of H3-K9 (Gupta et al., 2010). The impact of aging process and exercise on H3-K9 methylation is poorly exploited. While, DNA methylation is catalyzed by a group of enzymes called DNA methyltransferases (DNMTs), DNMT1, DNMT2, DNMT3a, and DNMT3b that transfer the methyl group from the donor S-adenosylmethionine (SAM) to 5′ position of the cytosine pyramidal ring. This process usually represses the gene transcription. DNMT1 is primarily involved in maintenance of DNA methylation after replication, while DNMT3a and DNMT3b are particularly important for de novo methylation (Reik et al., 1999). It has been described a genome-wide tendency to DNA hypomethylation in multiple vertebrate organs during aging process (Wilson and Jones, 1983; Richardson, 2002). In addition, the age-related global hypomethylation is related to DNMT1 deficits in senescent human fibroblasts (Lopatina et al., 2002). However, studies reporting DNMTs content in the brain during aging process are lack.
Interestingly, epigenetic mechanisms have been linked to the age-related cognitive decline, since histone deacetylases (HDAC) inhibitors have been shown to improve memory in aged rodents (Levenson and Sweatt, 2005; Reolon et al., 2011). Accordantly, some evidences demonstrated that exercise ameliorates aging-related cognitive function in rodents (Pietrelli et al., 2012; Radak et al., 2001), in addition, recent findings have demonstrated that the exercise was able to modulate the histone acetylation status, enhancing transcription of genes related to brain function (Elsner et al., 2011; Gomez-Pinilla et al., 2011).
Considering that exercise restores the age-related memory deficits and epigenetic mechanisms may be related to protective effects of exercise, it is crucial access the modulation of exercise on epigenetic parameters in the normal aging process. Therefore, the aim of this investigation was to study the effect of aging and two treadmill exercise protocols, single session of treadmill or chronic treadmill protocol on methylation parameters, specifically, DNA methyltransferases 1 and 3b (DNMT1and DNMT3b) and histone H3 lysine 9 (H3-K9) methylation levels.
2. Material and Methods
2.1 Animals and Training
Male Wistar rats of different ages, 3 and 20-months-old were used. The animals were maintained under standard conditions (12-h light/ dark, 22±2 °C) with food and water ad libitum. The NIH “Guide for the Care and Use of Laboratory Animals” (No. 80-23, revised 1996) was followed in all experiments. The Local Ethics Committee (CEUA/ UFRGS) approved all handling and experimental conditions (nr. 21449).
Rats were randomly divided into sedentary (SED) or exercised group (EXE). The exercise training consisted of running sessions in an adapted motorized rodent treadmill (INBRAMED TK 01, Porto Alegre, Brazil) at 60% of the animals maximal oxygen uptake (Brooks and White, 1987). Peak oxygen uptake (VO2) was measured indirectly in all animals before training (Arida et al., 1999; Brooks and White, 1987; Elsner et al., 2011). Two exercise protocols were employed: a single session of treadmill exercise (20 min) and chronic treadmill protocol (20 min running session each day for 2 weeks). SED was handled exactly as the experimental animals and was left on the treadmill for 5 min without any stimulus to run. All the procedures took place between 14:00 and 17:00 h.
2.2 Preparation of samples
In order to verify the acute and delayed effects of exercise, rats were decapitated 1h and 18h after a single session or after the last training session of chronic treadmill exercise. The whole hippocampi were quickly dissected out and immediately snap-frozen in liquid nitrogen, and stored at -80°C. In the day of the assay, the whole hippocampi were homogenized in 1:3 volumes of specific ice-cold lysis buffer.
2.3 Determination of DNMT1, DNMT3b and methylation of histone H3 lysine 9 (H3-K9) levels
Specific ELISA Assay Kits (Colorimetric Detection, catalog # P-3011, #P-3013, # P-3018, respectively, EpiQuik®) were used. All the procedures were done according to the manufacturer's instructions. The Pierce BCA Protein Assay kit ® was used to determine the protein concentration.
2.4 Statistical analysis
All results were expressed as the percent of control (mean ± S.E.M.). The results were analyzed by Three-Way Analysis of Variance (ANOVA) with age, exercise and time points after the exercise as factors followed by post hoc Tukey's multiple range test when appropriate. The influence of age on epigenetic markers was evaluated by Student's T-Test. In all tests, p < 0.05 was considered.
3. Results
3.1 Aging process reduces epigenetic markers in rat hippocampus
Firstly, we evaluated the effect of the aging process on histone methylation levels. 20-months-old rats hippocampi displayed lowers histone H3-K9 methylation levels (about 50%) compared to the young adult group (Fig. 1; p=0.0002). It was also observed that DNMT1 levels were significantly diminished (about 25%) in the aged group (Fig. 1; p=0.009). The level of DNMT3b enzyme was not modified by age.
Fig. 1.
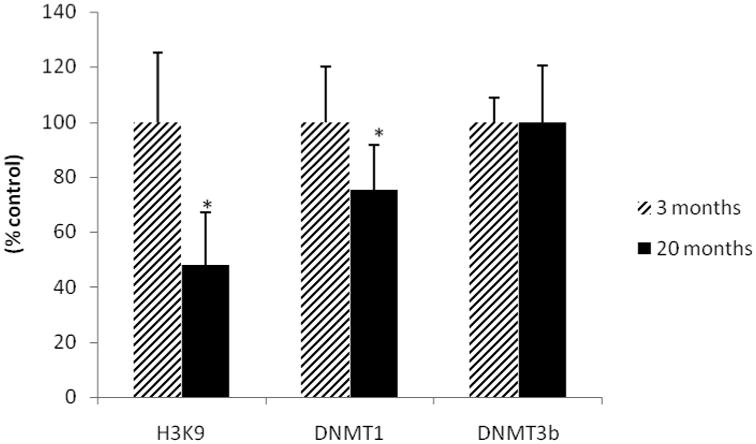
Effects of aging process on methylation parameters in rat hippocampus. Results are expressed as percentage of young adult group and columns represent mean ± S.D. (n=9-12). Student t-Test, * = different as compared to control group.
3.2. Exercise decreased DNMT3b and DNMT1 levels in hippocampi from young adult rats
The DNMT3b levels after single session of exercise are illustrated in Figure 2A. Three-way ANOVA revealed significant effect of the exercise factor (F(1,39) = 5.845; p=0.021), without any effect of age. When measured 1 h after session ended, young adult exercised rats exhibited lowers levels of DNMT3b (about 30%) when compared to its sedentary group (p=0.042), while no delayed (18h) effects of exercise were observed. The DNMT3b levels were not modified by the chronic exercise regimen in all groups (data not shown). Three-way ANOVA showed the effects of both factors, single session of exercise (F(1,39)= 29.505; p<0.001) and age (F(1,39)=10.073; p=0.003), on DNMT1 levels (Fig 2B). In addition, there was a significant interaction between age and exercise factors (F(1,39)= 7.302; p=0.011). This exercise protocol diminished acutely DNMT1 levels in hippocampi from 3 months-aged rats (approximately 45%; p<0.001), without any change on this parameter 18 h after exercise. There was no significant effect of chronic exercise protocol on DNMT1 levels in both young adult and aged groups (data not shown).
Fig. 2.
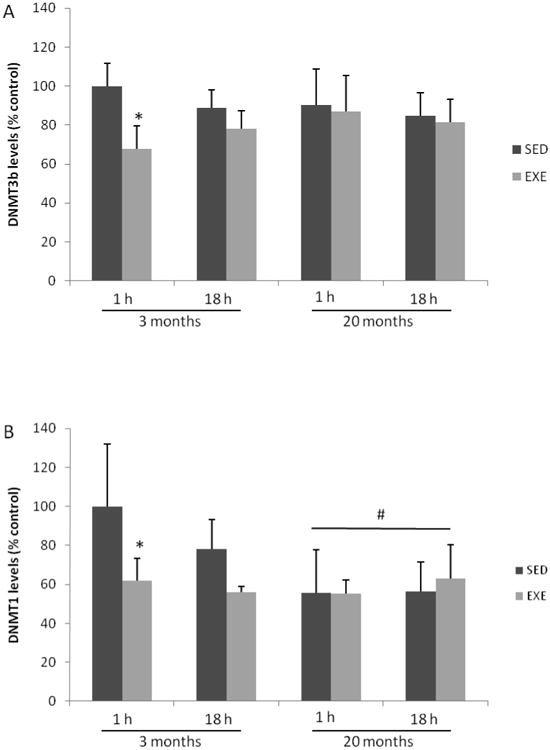
Effect of single exercise session on DNMT3b (A) and DNMT1 (B) levels in rat hippocampus. Results are expressed as percentage of the young SED group (1h) and columns represent mean ± S.D. (n=5-7). Three-way ANOVA followed by Tukey's test, *= different from its respective SED control group (p<0.05); # = different from the young adult groups (p<0.05).
3.3 Exercise affects differently hippocampal H3-K9 methylation levels in young adult and aged rats
Fig. 3 shows H3-K9 methylation levels in both 3 and 20-months-aged rat hippocampus at different time-points. Three-way ANOVA indicated effect of age (F(1,39) = 11.818; p=0.002) and a significant interaction between age and exercise factors (Fig. 3A; F(1,39)=42.165; p<0.0001). This exercise protocol diminished acutely and delayed H3-K9 methylation levels in young adult hippocampi (about 50%, p=0.006 and 60%, p<0.0001, respectively). Differently, hippocampi from 20-months-old rats have higher H3-K9 methylation levels 1 h and 18 h after single exercise session (respectively, about 30 and 100%; p=0.024 and p<0.001).
Fig. 3.
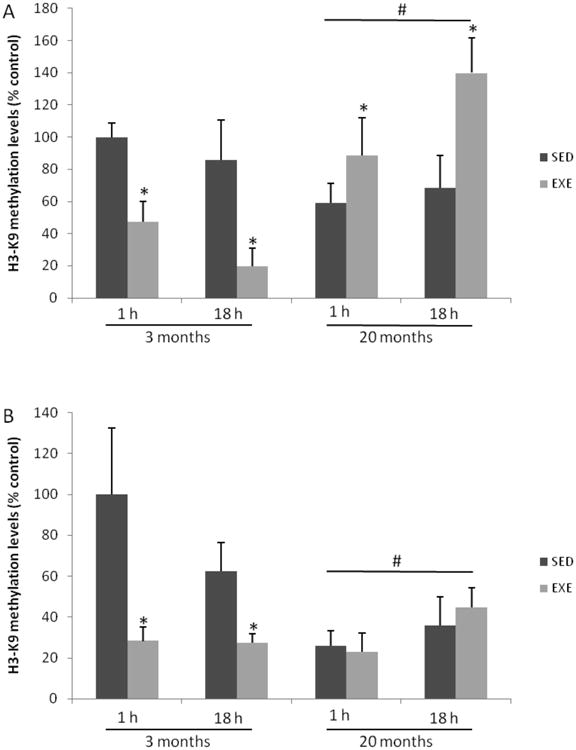
Effect of single exercise session (A) and chronic protocol (B) on H3-K9 methylation levels in rat hippocampus. Results are expressed as percentage of the young SED group (1h) and columns represent mean ± S.D. (n=5-7). Three-way ANOVA followed by Tukey's test, *= different from the respective SED control group; # = different from the young adult groups (p<0.05).
Additionally, it was observed the effect of both factors, chronic exercise (Fig. 3B; F(1,39)=7.431; p=0.011), and age (F(1,39)= 10.709, p=0.003), in addition to an interaction between the factors (F(1,39) = 13.072; p=0.001). The chronic exercise induced an acute and delayed decrease (about 70%; p=0.008 and 50%, p=0.015, respectively) on H3-K9 methylation levels in hippocampi from young adult rats, without any effect on aged group (Fig 3B).
4. Discussion
We described here that aged hippocampus has diminished DNMT1 and H3-K9 methylation levels. Our data corroborates to those obtained by Lopatina and colleagues (2002), demonstrating decreases on DNMT1 activity in senescent human fibroblasts. These findings can be related to genome-wide tendency to DNA hypomethylation during aging process (Wilson and Jones, 1983; Richardson, 2002). The relevance of our finding is not currently known, although we can suggest that might reflect age-related global patterns of genomic destabilization. Additionally, we could suggest that the age-related folate deficiency (Keyes et al., 2007) might potentiate the dysregulation of DNA methylation levels in the aged brain, since folate exerts the main role in the supply of one-carbon moieties for DNA synthesis and the synthesis of SAM, the most important methyl donor in the DNA methylation process (Selhub, 2002). Furthermore, low folate levels has been linked to neuropsychiatric and neurodegenerative diseases, such as Alzheimer disease (Querfurth and LaFerla 2010), what could be related to hypomethylation status.
Our result support the idea that dysregulation of histone methylation is also related to aging process, since aged hippocampi have lowers H3-K9 methylation levels. It is known that histone methylation can display opposite effects, resulting in either gene activation or repression, but it is impossible at this moment to establish if aging is able to alter mono-, di- or tri-methylation. We can suggest that the reduced levels of H3-K9 in hippocampi from 20-months aged rats may be related to mono-methylation of H3-K9, contributing to the age-related reduction of gene transcription. Consistent with our finding, the expression of genes with important role on memory and synaptic plasticity, including Arc (activity-regulated cytoskeletal gene), zif268 (nerve growth factor inducible-A) and BDNF (Brain-derived neurotrophic factor) are decreased in normal aging process, as well in many models of memory disorders (Kohman et al., 2011). In accordance, lowers levels of global histone H4 acetylation were observed in hippocampi from aged rats (Lovatel et al., 2012; submitted). Considering that mono-methylation of H3-K9 and histone acetylation are typically associated with transcriptional activation, our findings suggest that both epigenetic markers may be related to aging-associated abnormal brain gene expression patterns.
A major finding that emerged from our study involves a potential interaction between aging and exercise on DNA and histone methylation markers. To our knowledge, we provide here, for the first time, evidence of age- dependent exercise modulation on epigenetic mechanisms. The single exercise session significantly decreased both DNMT1 and DNMT3b levels in young adult rats, which may reduce DNA methylation, affecting gene expression. Our findings are in agreement with those recently obtained by Gomez-Pinilla and colleagues (2011), where exercise reduced DNA methylation in the BDNF gene promoter IV in the hippocampi of 3 months-aged rats, augmenting BDNF gene expression. Together, these data are consistent with the idea that exercise may increase the transcriptional activity of genes involved in neuronal plasticity and memory formation through the modulation of DNA methylation in hippocampi from young adult rats.
In contrast to the young adult rat group, our exercise protocol did not modify DNMT1 nor DNMT3b levels in hippocampi from 20 months-aged rats, suggesting that exercise might not influence the transcription activity through DNMT activity in aging brain. The differences between young adult and aged samples are not surprising, since it is well documented that aged animals can respond differently to same stimulus in both behavioral tasks and biochemical assays. Buchanan and colleagues (2008) observed that a stress model increased hippocampal IL-1β expression associated with a cognitive impairment in aged mice, without any effect in adult mice. Therefore, these findings demonstrated that exercise can impact methylation parameters in an age-dependent manner.
Another remarkable result is the opposite effects of exercise on H3-K9 methylation levels between young adult and old groups. Both exercise protocols reduced H3-K9 methylation levels in the young group. We might hypothesize that exercise reduced di- and tri-methylation of H3-K9 levels in hippocampus from young rats, which may enhance the expression of specific gene expression involved to neuronal survival, plasticity and cognitive functions, since di- and tri-methylation of histone H3-K9 are associated to transcriptional repression (Gupta et al., 2010). Interestingly, it was observed that the single exercise session reversed the diminished H3-K9 methylation levels in aged group, therefore we could suppose that exercise can impact positively the transcriptional activity through mono-methylation of H3-K9. Taken together, it is plausible to suppose that histone lysines might be methylated in response to exercise with different patterns (mono-, di- or tri-methylation) depending on the age.
The single exercise session had transiently effects on DNMT1 and DNMT3b enzyme levels, reducing acutely their levels (1 h after exercise), without any delayed effect (18 h after). This finding corroborate our previous results, since the same exercise protocol modulates HAT and HDAC in rat hippocampus, enzymes involved in the acetylation status, immediately and 1 hour after exercise, but had no delayed effects (Elsner et al., 2011). Similarly, Chandramohan and colleagues (2008) demonstrated that forced swimming increased phosphoacetylation histone levels in rat dentate gyrus up to 4 h, but not 24 h after training. These findings led us to hypothesize that exercise can alter enzymatic activity/levels up to the first hours after training, without any delayed influence. Although it is impossible to establish at this moment why exercise-induced acetylation and methylation changes did not persist after training, we might suggest this short-lived effect as a protective mechanism to maintain the homeostasis of the transcriptional machinery, avoiding maladaptive conditions.
5. Conclusions
The present findings bring new insights into the effects of aging process and exercise on epigenetic modulation in rat hippocampus. Summarizing, our results support the hypothesis that an imbalance on epigenetic mechanisms, specifically DNMTs and H3-K9 methylation levels, are linked to brain aging process that might ultimately lead to age-related dysfunctions. Furthermore, exercise induced age-dependent changes on methylation markers, providing evidence that the epigenetic mechanisms in response to exercise might vary through lifespan. Additional work will be required to investigate the molecular mechanism behind this phenomenon. Further studies are necessary to support our preliminary findings. (1) We will use specific antibodies to distinguish between the three types of H3K9 methylation (H3K9me1, H3K9me2 and H3k9me3), (2) we need to evaluate the roles played by H3K9 methyl-transferases (G9a, GLP, etc) and (3) we will evaluate the effects of exercise in the DNA methylation levels itself.
Acknowledgments
This work was supported by the Brazilian funding agencies: Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (Dr. I.R. Siqueira; V.R. Elsner; C.F. Spindler); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES (K. Bertoldi; G.A. Lovatel; F. Moysés); Programa Institucional de Bolsas de Iniciação Cientifica – PIBIC CNPq-UFRGS (L.R. Cechinel). Dr. A.R.Muotri was supported from the National Institutes of Health through the NIH Director's New Innovator Award Program, 1-DP2-OD006495-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arida RM, Scorza FA, dos Santos NF, Peres CA, Cavalheiro EA. Effect of physical exercise on seizure occurrence in a model of temporal lobe epilepsy in rats. Epilepsy Res. 1999;37:45–52. doi: 10.1016/s0920-1211(99)00032-7. [DOI] [PubMed] [Google Scholar]
- Brooks GA, White TP. Determination of metabolic and heart rate responses of rats to treadmill exercise. J Appl Physiol. 1978;45:1009–1015. doi: 10.1152/jappl.1978.45.6.1009. [DOI] [PubMed] [Google Scholar]
- Buchanan JB, Sparkman NL, Chen J, Johnson RW. Cognitive and neuroinflammatory consequences of mild repeated stress are exacerbated in aged mice. Psychoneuroendocrinol. 2008;33:755–765. doi: 10.1016/j.psyneuen.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramohan Y, Droste SK, Arthur JS, Reul JM. The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-D aspartate/extracellular signal-regulated kinase/mitogen- and stress-activated kinase signalling pathway. Eur J Neurosci. 2008;27:2701–2713. doi: 10.1111/j.1460-9568.2008.06230.x. [DOI] [PubMed] [Google Scholar]
- Elsner VR, Lovatel GA, Bertoldi K, Vanzella C, Santos FM, Spindler C, de Almeida EF, Nardin P, Siqueira IR. Effect of different exercise protocols on histone acetyltransferases and histone deacetylases activities in rat hippocampus. Neurosci. 2011;29:580–587. doi: 10.1016/j.neuroscience.2011.06.066. [DOI] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Zhuang Y, Feng J, Ying Z, Fan G. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur J Neurosci. 2011;33:383–90. doi: 10.1111/j.1460-9568.2010.07508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohman RA, Rodriguez-Zas SL, Southey BR, Kelley KW, Dantzer R, Rhodes JS. Voluntary Wheel Running Reverses Age-Induced Changes in Hippocampal Gene Expression. PLoS One. 2011;6:8. doi: 10.1371/journal.pone.0022654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;4:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt J. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Lopatina N, Haskell JF, Andrews LG, Poole JC, Saldanha S, Tollefsbol T. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J Cell Biochem. 2002;84:324–334. doi: 10.1002/jcb.10015. [DOI] [PubMed] [Google Scholar]
- Pietrelli A, Lopez-Costa J, Goni R, Brusco A, Basso N. Aerobic exercise prevents age-dependent cognitive decline and reduces anxiety-related behaviors in middle-aged and old rats. Neurosci. 2012;202:252–266. doi: 10.1016/j.neuroscience.2011.11.054. [DOI] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM. Alzheimer's disease. NEJM. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- Radak Z, Kaneko T, Tahara S, Nakamoto H, Pucsok J, Sasvari M, Nyakas C, Goto S. Regular exercise improves cognitive function and decreases oxidative damage in rat brain. Neurochem Int. 2001;38:17–3. doi: 10.1016/s0197-0186(00)00063-2. [DOI] [PubMed] [Google Scholar]
- Reik W, Kelsey G, Walter J. Dissecting de novo methylation. Nat Genet. 1999;23:380–382. doi: 10.1038/70476. [DOI] [PubMed] [Google Scholar]
- Reolon GK, Maurmann N, Werenicz A, Garcia VA, Schröder N, Wood MA, Roesler R. Posttraining systemic administration of the histone deacetylase inhibitor sodium butyrate ameliorates aging-related memory decline in rats. Behav Brain Res. 2011;221:329–332. doi: 10.1016/j.bbr.2011.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr. 2002;132:2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selhub J. Folate, vitamin B12 and vitamin B6 and one carbon metabolism. J Nutr Health Aging. 2002;6:39–42. [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyl-deoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]


