Abstract
Transcription factor GATA2 plays critical roles in hematopoietic stem cell survival and proliferation, GMP differentiation, and basophil and mast cell differentiation. However, precise roles of GATA2 in basophil and mast cell differentiation and maintenance have not been delineated. We have identified GATA2 as an essential transcription factor in differentiation of newly identified common basophil and mast cell progenitors into basophils and mast cells. We observed Gata2 haploinsufficiency for mast cell differentiation but not for basophil differentiation. We examined the precise role of GATA2 in maintaining the expression of a wide range of genes that are important for performing basophil or mast cell functions. The effects of GATA2 on gene expression were broadly based. We demonstrated that GATA2 was required for maintaining Fcer1a mRNA and FcεRIα protein expression on both basophils and mast cells as well as for maintaining Kit mRNA and c-Kit protein expression on mast cells. GATA2 was required for histamine synthesis and was also critical for Il4 mRNA expression in basophils and Il13 mRNA expression in mast cells. We demonstrate a STAT5-GATA2 connection, showing that the STAT5 transcription factor directly bound to the promoter and an intronic region of the Gata2 gene. Overexpression of the Gata2 gene was sufficient to direct basophil and mast cell differentiation in the absence of the Stat5 gene. Our study reveals that the STAT5-GATA2 pathway is critical for basophil and mast cell differentiation and maintenance.
Basophils and mast cells are minor leukocyte populations, constituting less than 1% of peripheral blood and bone marrow cells. Both basophils and mast cells express the high affinity receptor for Immunoglobulin E (IgE), FcεRI. Upon re-exposure to allergens, basophils and mast cells are activated through the binding of allergen-loaded IgE via FcεRI. Activated basophils and mast cells release both overlapping and unique sets of inflammatory mediators, including histamine, proteoglycans, lipid mediators, proteases, chemokines, and cytokines (1–3). Basophils and mast cells are important components of type 2 immune responses that protect against parasitic infection and cause allergic inflammation (4–7). Recent evidence supports non-redundant roles of basophils and mast cells in causing allergic inflammation and in expelling worms (4).
The processes of basophil and mast cell differentiation have received increased attention in recent years. Immature basophils differentiate and undergo maturation in the bone marrow. Mature basophils circulate in the blood stream and enter inflamed tissues. In contrast, immature mast cells develop in the bone marrow prior to taking residence in tissues, where they undergo further maturation (2). The nature of precursors of these cells is a subject of intense debate. Galli and colleagues identified mast cell lineage-restricted progenitors (MCPs) in the bone marrow and proposed that MCPs are derived from multiple potential progenitors (MPPs), but not from common myeloid progenitors (CMPs) or granulocyte-monocyte progenitors (GMPs) (8–9). On the other hand, Akashi and colleagues determined that both basophils and mast cells are derived from CMPs and GMPs (10). Additionally, they described a subset of cells in the spleen, but not in the bone marrow, termed basophil/mast cell progenitors (BMCPs). These cells are suggested to give rise to both basophils and mast cells (10). However, whether or not BMCPs are authentic bipotential basophil/mast cell progenitors was challenged by a recent study (11) and our data (12), which indicate that BMCPs mainly gave rise to mast cells. Furthermore, data from proliferation-tracking experiments support the conclusion that most new basophils are generated in the bone marrow, rather than in the spleen (13).
We have identified a novel population of common basophil/mast cell progenitors in the bone marrow (12). These progenitors were highly enriched in the capacity to differentiate into basophils and mast cells while retaining a limited capacity to differentiate into myeloid cells. Because it was determined that the common basophil/mast cell progenitors were more mature than GMPs and because they possessed great potential to differentiate into basophils and mast cells but had not yet fully committed into bipotential basophil-mast cell potential progenitors, we have designated these progenitor cells “pre-basophil and mast cell progenitors” (pre-BMPs). We showed that pre-BMPs differentiated into basophils and mast cells at the clonal level in vitro and at the population level in vivo (12). We also demonstrated that STAT5 signaling was required for the differentiation of pre-BMPs into both basophils and mast cells and was critical for inducing two downstream transcription factors CCAAT/Enhancer Binding Protein, alpha (C/EBPα) and Microphthalmia-Associated Transcription Factor (MITF). We identified C/EBPα as the critical transcription factor for specifying basophil cell fate and MITF as the crucial transcription factor for specifying mast cell fate. We demonstrated that C/EBPα and MITF silenced each other’s transcription in a directly antagonistic fashion (12).
GATA Binding Protein 2 (GATA2) is a member of the GATA family of zinc finger transcription factors. GATA2 plays critical roles in survival and proliferation of hematopoietic stem cells (HSCs) (14–15). It has been implicated to play a role in GMP differentiation (16). GATA2 has been shown to be critical in both basophil and mast cell differentiation (17–18). The order of GATA2 and C/EBPα expression has been suggested to be crucial in determining basophil cell fate. When GATA2 expression preceded C/EBPα expression at the GMP stage, GATA2 together with C/EBPα drove basophil differentiation. Conversely, when C/EBPα expression preceded GATA2 expression, C/EBPα together with GATA2 drove eosinophil differentiation (18). However, it remains unknown whether GATA2 plays a role in the differentiation of pre-BMPs into basophils and mast cells and in the maintenance of basophil and mast cell identities.
In this study, we demonstrated that GATA2 was essential for the differentiation of pre-BMPs into basophils and mast cells and for the maintenance of basophil and mast cell identities. GATA2 haploinsufficiency was observed for mast cell differentiation but not for basophil differentiation. We further demonstrated that the STAT5 transcription factor directly regulated Gata2 gene expression and that overexpression of the Gata2 gene was sufficient to direct basophil and mast cell differentiation in the absence of the Stat5a/b genes.
Materials and Methods
Mice
C57BL/6J (B6) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Stat5a/bf/−RosaYfp/CreErt2 mice and Stat5a/bf/+RosaYfp/CreErt2 mice were generated as described previously (12). Gata2f/fRosaYfp/YfpTgCreErt2hemi mice were generated by crossing Gata2f/f mice [Gata2tm1Sac (19), the mutant mouse regional resource center at University of California-Davis, Davis, CA] to the mice with an YFP Cre activity reporter gene knocked in the Rosa locus [B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J (20); short name: RosaYfp/Yfp; the Jackson Lab, Bar Harbor, ME] and to the transgenic mice with an inducible Cre enzyme [TgCreErt2hemi (21), the Jackson Lab, Bar Harbor, ME]. Gata2f/+RosaYfp/YfpTgCreErt2hemi mice were generated by crossing Gata2f/fRosaYfp/YfpTgCreErt2hemi mice to Gata2+/+RosaYfp/YfpTgCreErt2hemi mice, which were generated by crossing RosaYfp/Yfp to TgCreErt2hemi mice. All animal experiments were approved by the National Jewish Health Institutional Animal Care and Use Committee.
FACS analysis and sorting
For fluorescence activated cell sorting (FACS) analysis of basophils, mast cells, T cells, B cells, dendritic cells (DCs), neutrophils, and macrophages, cells obtained from various tissues or cell cultures were stained with the following antibodies: basophils and mast cells were stained with Allophycocyanin-CY7-conjugated anti-c-Kit (2B8) and PE-CY7-conjugated anti-FcεRIα (MAR-1) antibodies; T cells and B cells were stained with PE-CY5-conjugated anti-CD3e (145-2C11) and PE-CY7-conjugated anti-CD19 (1D3) antibodies; dendritic cells (DCs) were stained with Allophycocyanin-conjugated anti-MHC Class II (MHC II) (M5/114.15.2) and PE-conjugated anti-CD11c (N418) antibodies; neutrophils and macrophages were stained with PE-conjugated anti-Gr-1 (RB6-8C5) and Allophycocyanin-conjugated anti-CD11b (M1/70) antibodies. Dead cells were stained with propidium iodide (PI) and excluded in all FACS plots. Stained cells were acquired by using CyAN (DakoCytomation, Glostrup, Denmark) and analyzed using the FlowJo software (Tree Star, Ashland, OR). The absolute number of positive cells was calculated by multiplying the total number of cells with the percentages of positive cells. Mean fluorescence intensities (MFIs) were calculated by using the FlowJo software. The percentage of reduction was calculated by the following formula: percent of reduction=[MFI(Control)-MFI(inducible knockout)]/MFI(Control)×100%.
Regular GMPs (FcεRIα− GMPs) and pre-BMPs (FcεRIα+ GMPs) were stained and FACS-sorted according to the published protocol (12). Briefly, Regular GMPs (GMPs) were FACS-sorted as Lin−IL-7Rα−Sca-1−c-kit+CD34+FcγRII/IIIhiFcεRIα − cells. Pre-BMPs were FACS-sorted as Lin−IL-7Rα −Sca-1−c-kit+CD34+FcγRII/IIIhiFcεRIα+ cells. The stained cells were FACS-sorted using a Moflo machine (DakoCytomation, Glostrup, Denmark). All antibodies used for FACS analysis and sorting were purchased from BD PharMingen (San Diego, CA) or eBioscience (San Diego, CA).
In vitro gene deletion
The floxed genes in the cultured cells were deleted by the 4-hydroxytamoxifen (4HT; Calbiochem, Billerica, MA) treatment using a concentration of 25 nM. For the methylcellulose culture, 4HT was included in the culture for 9 days without washing (the 25 nM concentration was determined to generate maximum floxed gene deletion with the least amount of toxicity). For the liquid cultures, 4HT was washed after 3 days of treatment. The washed cells were cultured under the original conditions without 4HT until they were subjected to analysis. The deletion of the floxed Gata2 gene in the FACS-sorted YFP+ basophils or YFP+ mast cells was determined to be near 100%. The deletion of the floxed Stat5a/b genes in the FACS-sorted YFP+ basophils or YFP+ BMMCs was also highly effective (12).
In vitro differentiation of progenitors
To analyze the differentiation of pre-BMPs into basophils and mast cells, pre-BMPs were FACS-sorted from Gata2+/+RosaYfp/YfpTgCreErt2hemi mice and Gata2f/fRosaYfp/YfpTgCreErt2hemi mice. And the sorted pre-BMPs were seeded in a 35 mm Nunc dish (Thermo Fisher Scientific, Rochester, NY) at a density of 1000 cells/dish in 1 ml of 1% methylcellulose containing complete Iscove’s modification of Dullbecco’s modified eagle medium (IDMEM) supplemented with 50 μM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), 20 ng/ml IL-3, and 25 nM 4HT for 9 days (8, 22). Cells were then collected and analyzed by FACS.
Bone marrow-derived mast cell culture
Bone marrow-derived mast cells (BMMCs) were prepared by culturing bone marrow cells from Gata2+/+RosaYfp/YfpTgCreErt2hemi mice, Gata2f/fRosaYfp/YfpTgCreErt2hemi mice, Stat5a/bf/+RosaYfp/CreErt2 mice, or Stat5a/bf/−RosaYfp/CreErt2 mice in complete IDMEM supplemented with 50μM β-mercaptoethanol and 20 ng/ml IL-3 for 4 weeks (23). Greater than 99% of BMMCs were mast cells (FcεRIα+ c-Kit+) as determined by FACS analysis.
Basophil culture
To obtain committed basophils, IL-3 (10 μg) and anti-IL-3 antibody (5 μg; MP2–8F8, BDPharMingen) were mixed together at room temperature for 1 min according to published methods (24). IL-3 and anti-IL-3 antibody complex (IL-3C) was intraperitoneally injected into Gata2+/+RosaYfp/YfpTgCreErt2hemi mice, Gata2f/fRosaYfp/YfpTgCreErt2hemi mice, Stat5a/bf/+RosaYfp/CreErt2 mice, or Stat5a/bf/−RosaYfp/CreErt2 mice 3 days prior to bone marrow harvest. Bone marrow cells from the treated mice were cultured in complete IDMEM containing 50μM β-mercaptoethanol and 20 ng/ml IL-3 for an additional 3 days. CD34 expression on the cultured basophils was no longer detectable and these cells expressed lineage markers Gr-1 and CD11b, indicating that they exhibited a mature phenotype.
In vivo treatment of mice
Gata2+/+RosaYfp/YfpTgCreErt2hemi mice, Gata2f/+RosaYfp/YfpTgCreErt2hemi mice, or Gata2f/fRosaYfp/YfpTgCreErt2hemi mice were injected intraperitoneally with100 μl of tamoxifen (Sigma-Aldrich, St. Louis, MO) mixed with sunflower seed oil (2 mg/100 μl, Spectrum Chemical MFG. Corp, Gardena, CA) once daily for five consecutive days. Two weeks after the last injection, cells and tissues from the treated mice were collected and subjected to FACS and histological analysis.
Retroviral infection
Full-length Stat5a and Gata2 cDNAs were cloned into the retroviral expression vector MSCV2.2-Ires-Thy1a through BglII and NotI sites. The retroviral particles were prepared as described previously (25). For retroviral infection, bone marrow cells from the control (Stat5a/bf/+RosaYfp/CreErt2, Stat5a/bf/+) mice and inducible Stat5 knockout (Stat5a/bf/−RosaYfp/CreErt2, Stat5a/bf/−) mice were stimulated with stem cell factor (50 ng/ml), IL-6 (50 ng/ml), and IL-3 (20 ng/ml) overnight. The stimulated cells (2 × 106) were centrifuged at 1800 rpm for 90 min at room temperature with 1 ml viral supernatant containing recombinant retrovirus containing the Stat5a, Gata2, or Thy1a (CTRL) genes in the presence of polybrene (8 μg/ml; Millipore, Billerica, MA). Infected cells were cultured in complete IDMEM with 20 ng/ml of IL-3 and treated with 25 nM 4HT for three days to delete the Stat5a/b gene. Ten days after the initial treatment of 4HT, cells were collected and analyzed by FACS. Infected cells were identified by the expression of Thy1.1 using Allophycocyanin-labeled anti-Thy1.1 antibody (clone HIS51, eBioscience).
Quantitative PCR and ELISA
Total RNA from FACS-sorted GMPs and pre-BMPs, basophils, or BMMCs was isolated with an RNAeasy Mini Kit (Qiagen, Valencia, CA) according to manufacturer’s instructions. For cytokine gene mRNA analysis, the cells were stimulated with PMA (50 ng/ml, Sigma-Aldrich, St. Louis, MO) and ionomycin (1μM, Sigma-Aldrich) for 6 hours or IgE crosslinking [1 μg/ml IgE (clone D8406, Sigma-Aldrich) and 1 μg/ml anti-IgE antibody (clone R35-72, BD Pharmingen)] overnight, and then total RNA was prepared from the treated cells. cDNA was synthesized by reverse transcription. Quantitative PCR (qPCR) was performed in an ABI PRISMTM 7700 Sequence Detection System. Primer sequences are listed in Supplemental Table I. Relative mRNA amounts were calculated as follows: relative mRNA amount = 2− [Ct(Sample) −Ct(Hprt)]. Percentage of reduction was calculated using the following formula: percentage of reduction = [Relative amount (Control)-Relative amount (inducible knockout)]/Relative amount (Control) × 100%.
For ELISA analysis, basophils or BMMCs were stimulated with PMA (50 ng/ml) and ionomycin (1μM) for 6 hours or IgE cross-linking (1 μg/ml IgE and 1 μg/ml anti-IgE antibody) overnight. IL-4 and IL-13 protein in the supernatants was measured by ELISA (BD PharMingen).
Measurement of histamine content and release in basophils and mast cells
For histamine content, basophils or BMMCs were frozen and thawed three times. Histamine in the cell lysates was measured by using a histamine enzyme immunoassay kit (Beckman Coulter, Fullerton, CA) following manufacturer’s instructions. For histamine release, basophils or BMMCs were stimulated with PMA (50 ng/ml) and ionomycin (1 μM) for 6 hours or IgE cross-linking (1 μg/ml IgE and 1 μg/ml anti-IgE antibody) overnight. Histamine in the supernatants was measured by using the histamine enzyme immunoassay kit following manufacturer’s instructions.
Histology
Ear sections from the tamoxifen-treated mice were fixed in 4% paraformaldehyde and stained with toluidine blue. Histological images were captured on a Nikon E800 microscope (Nikon, Melville, NY).
Chromatin Immunoprecipitation (ChIP) Assay
Anti-STAT5 antibody was purchased from Cell Signaling Technology (Beverly, MA). ChIP experiments were performed using an EpiTect Chip One-Day Kit (Qiagen, Valencia, CA) as described previously (12). The quantity of DNA precipitated by anti-STAT5 antibody was calculated as fold of enrichment using the following formula: Fold Enrichment = 2[ΔCt(IgG)−ΔCt(IP)], where ΔCt(IgG) = Ct(IgG)−[Ct(Input)-Log2(Input Dilution Factor)], ΔCt(IP) = Ct(IP)-[Ct(Input)-Log2(Input Dilution Factor)] (12).
Statistical Analysis
All of the error bars in this report represent the SD. For ELISA and qPCR analyses, means ± SDs were derived from triplicate measurements. Pooled data are indicated in the figure legends. The difference between two samples was analyzed with Student’s t test.
Results
GATA2 is essential for the differentiation of pre-BMPs into basophils and mast cells
To analyze the role of GATA2 in the differentiation of pre-BMPs into basophils and mast cells, we first examined Gata2 mRNA expression in the FACS-sorted pre-BMPs and found that the levels of Gata2 mRNA in the pre-BMPs were 60-fold higher than those in regular GMPs (FcεR1α − GMPs) albeit that expression was still lower compared with that detected in basophils or mast cells (Fig. 1A). We then tested whether GATA2 was necessary for the differentiation of pre-BMPs into basophils and mast cells. We took advantage of the inducible gene deletion system, in which mice with a floxed Gata2 gene were crossed to mice with an inducible Cre enzyme and to mice with a YFP Cre activity reporter (Gata2f/fRosaYfp/YfpTgCreErt2hemi mice). We refer these mice to as the inducible Gata2 knockout mice and Gata2+/+RosaYfp/YfpTgCreErt2hemi mice to as the control mice hereafter. We deleted the Gata2 gene in the FACS-sorted pre-BMPs of the inducible Gata2 knockout mice with 4-hydroxytamoxifen (4HT). As a control, we treated the FACS-sorted pre-BMPs of the control mice with 4HT. YFP+ pre-BMPs of the inducible Gata2 knockout mice (Gata2−/−) failed to differentiate into basophils and mast cells but differentiated into neutrophils and macrophages normally, while YFP+ pre-BMPs of the control mice (Gata2+/+) differentiated into basophils and mast cells (Fig. 1B, throughout this report, YFP+ cells represent YFP+ PI− live cells at the time of analysis). These results indicate that GATA2 plays an essential role in the differentiation of pre-BMPs into basophils and mast cells.
FIGURE 1.
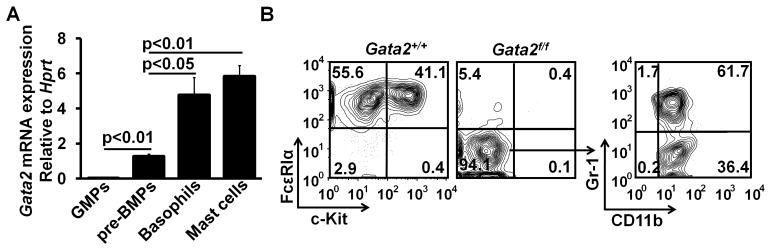
GATA2 is essential for the differentiation of pre-BMPs into both basophils and mast cells. (A) Regular GMPs (FcεRIα − GMPs) and pre-BMPs were FACS-sorted from bone marrow cells of B6 mice. Basophils were purified from bone marrow cells of IL-3C-injected B6 mice. Mast cells were cultured from bone marrow cells of B6 mice in the presence of IL-3 for four weeks. Gata2 mRNA expression in GMPs, pre-BMPs, basophils, and mast cells was measured by qPCR (mean ± SD, triplicates). Data represent two independent experiments with similar results. (B) The FACS-sorted pre-BMPs from Gata2f/fRosaYfp/YfpTgCreErt2hemi (Gata2f/f) mice and Gata2+/+RosaYfp/YfpTgCreErt2hemi (Gata2+/+) mice were cultured in 1% methylcellulose containing medium in the presence of IL-3 with 25nM 4HT for 9 days. Then, the treated cells were collected and analyzed by FACS. YFP+ cells are shown. Data represent two independent experiments with similar results.
GATA2 haploinsufficiency is observed for mast cell differentiation but not for basophil differentiation
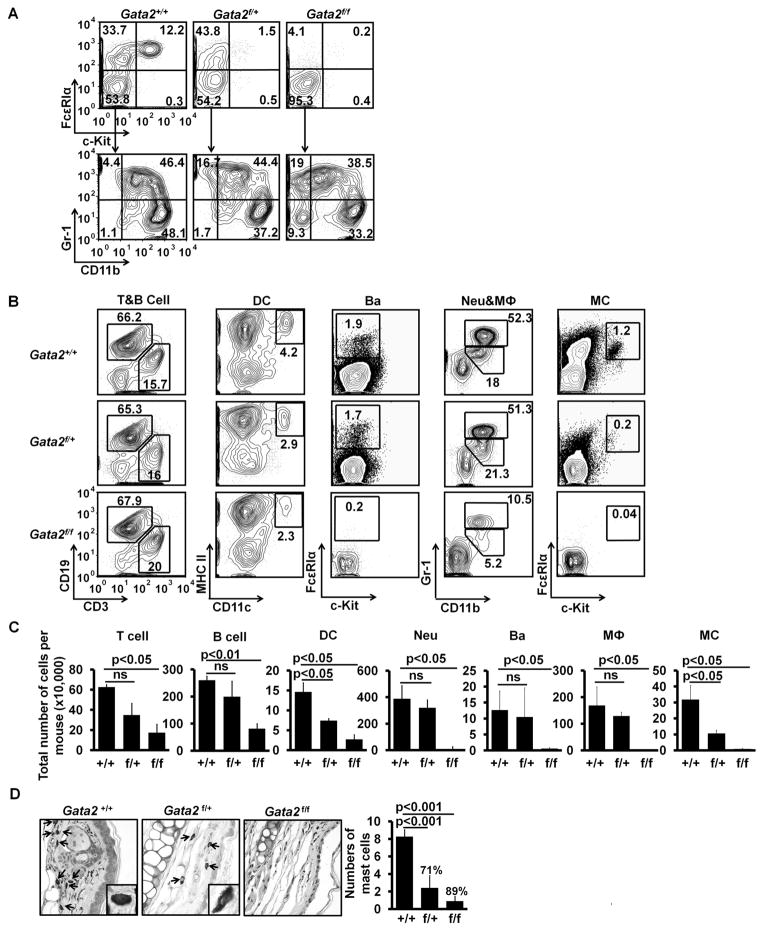
Gata2 haploinsufficiency (one copy of a gene is not sufficient to carry out the gene’s biological functions) has been reported in both mice and humans. In mice, Gata2 haploinsufficiency leads to reduced production and expansion of hematopoietic stem cells in the aorta-gonad-mesonephros region (14). In humans, Gata2 haploinsufficiency results in immune deficiency (26). Although FcεRIα − GMPs failed to differentiate into basophils, these progenitors differentiated into mast cells (data not shown), suggesting that there might exist multiple mast cell progenitors (27). Here, we wished to examine whether there is a Gata2 haploinsufficiency for basophil and mast cell development using experimental systems that assess basophil and mast cell developmental potential of all possible basophil and mast cell progenitors. In vitro, we used semi-solid 1% methylcellulose culture conditions, in which only progenitors can grow colonies, to assess collectively the capacity of mixed bone marrow basophil and mast cell progenitors, including basophil lineage-restricted progenitors (BaPs), MCPs, uncharacterized mast cell progenitors, and pre-BMPs, to differentiate into basophils or mast cells. Whole bone marrow cells of homozygous or heterozygous inducible Gata2 knockout mice, or control mice were cultured in 1% methylcellulose-containing medium with 4HT. We found that Gata2+/− basophil and mast cell progenitors differentiated into basophils but not mast cells, while Gata2+/− myeloid progenitors differentiated into neutrophils and macrophages comparable to Gata2+/+myeloid progenitors (Fig. 2A). To verify this finding in vivo, we treated heterozygous inducible Gata2 knockout mice with tamoxifen (one copy of the Gata2 gene was deleted in all cells). Directly ex vivo analysis of cells prepared from the treated mice revealed that Gata2 haploinsufficiency was indeed detected in the differentiation of mast cells but not in basophils. We did not observe a Gata2 haploinsufficiency in the differentiation of other cell lineages with the exception of dendritic cells (Fig. 2B, 2C). Histological analysis showed that the number of mast cells in the ears of heterozygous inducible Gata2 knockout mice treated with tamoxifen was reduced greatly to a level similar to that of tamoxifen-treated homozygous mice (Fig. 2D). Together, these data demonstrate a Gata2 haploinsufficiency in mast cell differentiation.
FIGURE 2.
GATA2 haploid insufficiency is observed for mast cell differentiation but not for basophil differentiation. (A) Bone marrow cells (not FACS-sorted) of Gata2+/+, Gata2f/+ mice or Gata2f/f mice were cultured in methylcellulose containing medium in the presence of IL-3 and 25 nM 4HT for 9 days. YFP+ cells are shown in the FACS plots. (B) FACS analysis of cells from the tamoxifen-treated mice. YFP+ cells are shown. T cells, B cells, and dendritic cells (DC) were prepared from spleen; basophils (Ba), neutrophils (Neu), and macrophages (MΦ) from bone marrow cells; and mast cells (MC) from peritoneal cavity of the treated mice. Data represent two independent experiments with similar results. (C) Total numbers of YFP+ cells (mean ± SD, n=6). (D) Toluidine blue staining of ear sections (40×; insert, 100×). Mast cells are indicated by arrows. The right panel shows the average number of mast cells in ten different fields (40×) randomly selected from the sections of ears (mean ± SD, n=3). The percentages indicate the percentages of reduction in mast cell numbers in the ear sections.
GATA2 is critical for maintaining FcεRIα expression on basophils and FcεRIα and c-Kit expression on mast cells
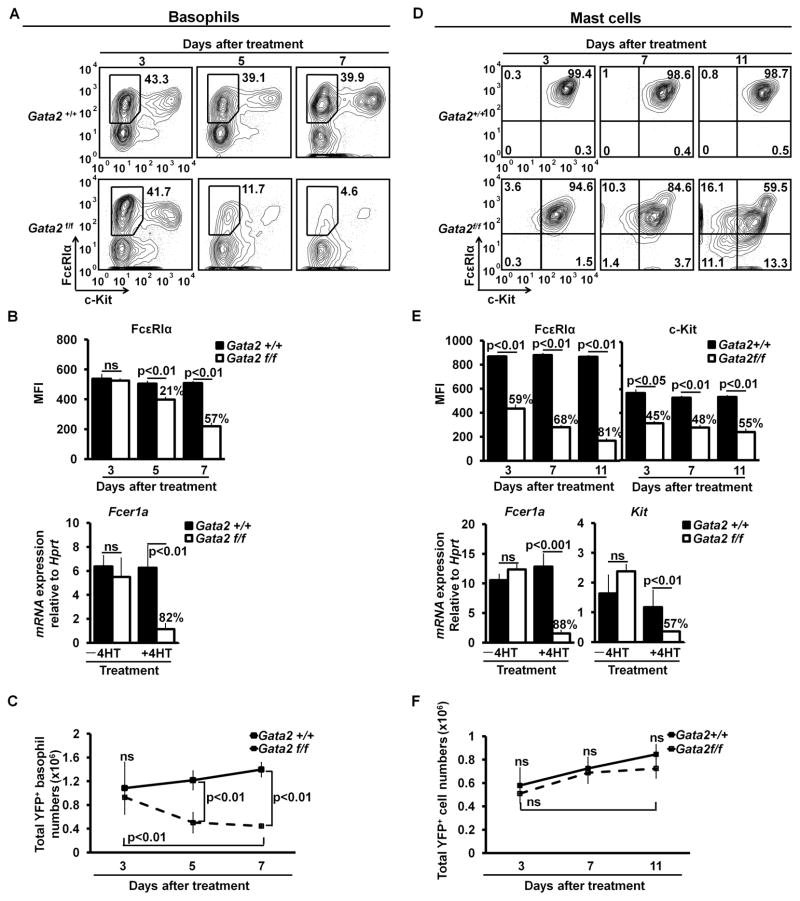
Transcription factors that regulate developmental process leading to cell lineage commitment might not always be needed for maintaining the committed molecular signatures once progenitor cells have differentiated into specific cell types. To determine whether GATA2 is required for the maintenance of basophil and mast cell identities, we deleted the Gata2 gene after progenitors were committed to basophils or mast cells. For the basophil maintenance study, we prepared basophil-enriched bone marrow cells from the inducible Gata2 knockout or control mice injected with IL-3 and anti-IL-3 antibody complex (IL-3C), which rapidly expanded BaPs and basophils in vivo (28–29). The IL-3C-expanded BaPs and basophils were cultured in the presence of IL-3 for an additional 3 days to ensure that basophils were committed. We then deleted the Gata2 gene in the committed basophils with the 4HT treatment. We first examined mRNA and protein expression of FcεRIα. We found that in the absence of the Gata2 gene, FcεRIα protein expression on YFP+ Gata2−/− basophils began to decrease at day 5 after the initial 4HT treatment, reaching the lowest levels at day 7 after the initial 4HT treatment (Fig. 3A, upper panel of 3B). Directly ex vivo Fcer1a mRNA expression in the FACS-sorted YFP+ Gata2−/− basophils decreased even more dramatically when compared with FcεRIα protein expression on YFP+ Gata2−/− basophils at day 5 after the initial 4HT treatment (Fig. 3A, lower panel of Fig. 3B). The number of YFP+ Gata2−/− basophils in the culture was significantly reduced compared with that of YFP+ Gata2+/+ basophils, indicating a pivotal role of GATA2 in basophil survival (Fig. 3C).
FIGURE 3.
GATA2 is critical for maintaining FcεRIα expression on basophils and FcεRIα and c-Kit expression on mast cells. (A) Bone marrow cells from IL-3C-injected Gata2+/+or Gata2f/f mice were cultured with IL-3 for three days. Then, the cells were treated with 25nM 4HT. Three, 5, and 7 days after the initial 4HT treatment, the cells were analyzed by FACS. (B) MFIs of FcεRIα expression on YFP+ Gata2+/+or Gata2−/− basophils (mean ± SD, n=6) (upper panel). The lower panel shows qPCR analysis of Fcer1a mRNA expression in the FACS-sorted YFP+ Gata2+/+or Gata2−/− basophils at 5 days after the initial 4HT treatment (mean ± SD, triplicates). Data represent three independent experiments with similar results. (C) Total numbers of YFP+ Gata2+/+ or Gata2−/− basophils (mean ± SD, n=6). (D) FACS analysis of 4-week BMMCs treated with 25nM 4HT. (E) MFIs of FcεRIα and c-Kit expression on YFP+ Gata2+/+ mast cells (FcεRIα+ c-Kit+) or “Gata2−/− mast cells” (including FcεRIα+/− and c-Kit+/− cells) (mean ± SD, n=4, upper panel). The lower panel shows qPCR analysis of Fcer1a and c-Kit mRNA expression in the FACS-sorted YFP+ Gata2+/+ mast cells or “Gata2−/− mast cells” at 11 days after the initial 4HT treatment (mean ± SD, triplicates). Data represent three independent experiments with similar results. (F) Total numbers of YFP+ Gata2+/+ mast cells or “Gata2−/− mast cells” (mean ± SD, n=4). YFP+ cells are shown (A and D). The percentages of reduction are indicated (B and E).
For the mast cell maintenance study, we prepared 4-week BMMCs from the Gata2 inducible knockout or control mice and used them as committed mast cells [although BMMCs are often considered as immature mast cells, they are committed since they cannot differentiate into other cell lineages even under culture conditions that are appropriate for other cell lineage differentiation (data not shown)]. We found that in the absence of the Gata2 gene, FcεRIα and c-Kit protein expression on YFP+ Gata2−/− mast cells (FcεRIα+ and c-Kit+) decreased beginning at day 3 (Fig. 3D, upper panel of 3E). At day 7 after the initial 4HT treatment, one portion of mast cells completely lost c-Kit expression, while a smaller percentage of mast cells completely lost FcεRIα expression. By day 11, in addition to greater percentages of mast cells that lost either c-Kit or FcεRIα expression, significant percentages of mast cells lost both c-Kit and FcεRIα expression (Fig. 3D). Although Gata2−/− mast cells that have lost FcεRIα, c-Kit, or both expressions no longer met the phenotypical definition of mast cells, they could still maintain mast cell molecular signatures. On the other hand, Gata2−/− mast cells that expressed c-Kit and FcεRIα expression could still lose mast cell molecular signatures. Thus, we analyzed YFP+ FcεRIα+/− and/or c-Kit+/− cells collectively and refer to these cells to as “Gata2−/− mast cells” hereafter. MFIs of c-Kit and FcεRIα expression on YFP+ “Gata2−/− mast cells” reached the lowest level at day 11 after the initial 4HT treatment (Fig. 3E, upper panel). Fcer1a and Kit mRNA expression in the FACS-sorted YFP+ “Gata2−/− mast cells” at day 11 after the initial 4HT treatment were reduced significantly (Fig. 3E, lower panel). In regarding to GATA2 dosage requirement for maintaining FcεRIα and c-Kit expression, committed mast cells, unlike differentiating mast cells, did not exhibit a Gata2 haploinsufficiency (Supplemental Fig. 1).
Interestingly, in contrast to its role in basophil survival, GATA2 did not appear to affect mast cell survival. Although mast cells lost much of FcεRIα and c-Kit protein expression in the absence of the Gata2 gene, the number of YFP+ “Gata2−/− mast cells” at day 11 after the initial 4HT treatment was comparable to that of YFP+ Gata2+/+ mast cells (Fig. 3F), suggesting that GATA2 is not a survival factor for mast cells.
GATA2 is crucial for maintaining the expression of genes that are important in performing basophil or mast cell functions and in histamine synthesis
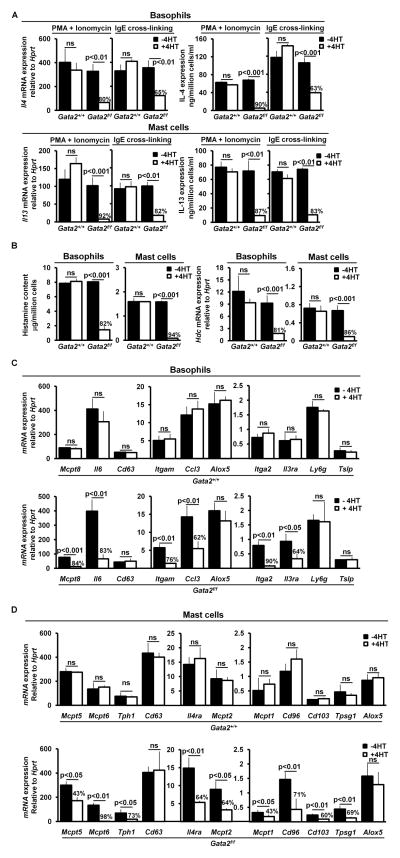
We analyzed further whether deletion of the Gata2 gene affects the expression of genes known to carry out basophil or mast cell functions, which could be expressed in both basophils and mast cells, genes that we previously identified those that are highly expressed in basophils but not in mast cells, or genes that are highly expressed in mast cells but not in basophils (12). Genes that are highly expressed in basophils but not in mast cells include genes encoding protease (Mcpt8), chemokine (Ccl3), receptors (Itgam, Itga2, and Il3ra). Genes that are highly expressed in mast cells but not in basophils include those that encode proteases (Mcpt1, Mcpt2, Mcpt5, Mcpt6, Tpsg1, and Tph1), receptors (Cd96, Cd103, and Il4ra). Genes that are important in performing basophil and/or mast cell functions included those encoding cytokines (Il4, Il6, Il13 and Tslp) and a gene encoding for an enzyme required for histamine synthesis. As a control, we included Cd63 and Alox5, which are commonly expressed in both basophils and mast cells. For cytokine gene expression analysis, we analyzed Il4 gene expression in basophils and Il13 gene expression in mast cells because the cytokines encoded by these two genes perform major basophil and mast cell functions (30–31). We activated basophils and mast cells with IgE cross-linking to determine the effect of reduced FcεRIα expression on cytokine gene expression (to assess the indirect effects of Gata2 gene deletion on cytokine expression) and with PKC stimulator PMA and Ca2+ influx stimulator ionomycin (activate basophils and mast cells bypassing FcεRI) to assess more directly the effects of GATA2 on cytokine gene expression. Fig. 4A (upper panel) shows that Il4 mRNA and IL-4 protein expression were greatly diminished in the FACS-sorted YFP+ Gata2−/− basophils stimulated either with PMA and ionomycin or with IgE cross-linking. Similarly, Il13 mRNA and IL-13 protein expression in the FACS-sorted YFP+ “Gata2−/− mast cells” were nearly abolished (Fig. 4A, lower panel). Cytokine gene expression in WT or Gata2−/− basophils and mast cells was low without stimulation. These results demonstrate that GATA2 plays a critical role in maintaining the expression of the Il4 gene in basophils and the Il13 gene in mast cells.
FIGURE 4.
GATA2 is crucial for maintaining the expression of genes that are important in carrying out basophil or mast cell functions and for histamine synthesis. (A) qPCR and ELISA analyses of Il4 mRNA and IL-4 protein in basophils or Il13 mRNA and IL-13 protein in mast cells not treated or treated with 4HT. YFP+ Gata2+/+ or Gata2−/− basophils at day 5 after the initial 4HT treatment and YFP+ Gata2+/+ mast cells or “Gata2−/− mast cells” at day 11 after the initial 4HT treatment were FACS-sorted and stimulated with PMA and ionomycin for 6 hours or stimulated with IgE cross-linking overnight. (B) ELISA measurement of histamine content and qPCR analysis of Hdc mRNA expression in the FACS-sorted YFP+ Gata2−/− basophils and “Gata2−/− mast cells”. (C) qPCR analysis of mRNA expression of basophil genes in the FACS-sorted YFP+ Gata2+/+ or Gata2−/− basophils at day 5 after the initial 4HT treatment. Different scales were used to present data generated in the same experiment. (D) qPCR analysis of mRNA expression of mast cell genes in the FACS-sorted YFP+ Gata2+/+ mast cells or “Gata2−/− mast cells” at day 11 after the initial 4HT treatment. The percentages of reduction are indicated (A through D). Data represent mean ± SD (triplicates) and two independent experiments with similar results (A through D).
In addition to de novo cytokine synthesis, basophils and mast cells synthesize histamine and store the synthesized histamine in preformed granules. Activated basophils and mast cells then secrete the preformed granules through a cellular process known as degranulation. Histamine is synthesized from the decarboxylation of the amino acid histidine, a reaction catalyzed by the enzyme called histidine decarboxylase (32). Mice deficient in the histidine decarboxylase gene (Hdc) fail to synthesize histamine. IgE-mediated anaphylactic reactions are absent in Hdc-deficient mice (33–34). To determine whether GATA2 affects the histamine synthesis or histamine release in basophils and mast cells, we measured histamine content in the FACS-sorted YFP+ Gata2−/− basophils and “Gata2−/− mast cells” and found that the histamine contents were greatly reduced those cells (Fig. 4B, left two panels). Similar levels of reduction in histamine release by the FACS-sorted YFP+ Gata2−/− basophils and “Gata2−/− mast cells” in response to IgE cross-linking were also observed (not shown). Consistent with a critical role of GATA2 in histamine synthesis, we showed that Hdc mRNA expression in FACS-sorted YFP+ Gata2−/− basophils and “Gata2−/− mast cells” was greatly reduced (Fig. 4B, right two panels). These results demonstrate that GATA2 is required for histamine synthesis in basophils and mast cells.
For expression analysis of the remaining genes, the FACS-sorted basophils and “Gata2−/− mast cells” were not stimulated because expression of those genes generally does not require stimulation. We showed that the majority of basophil- or mast cell-specific genes (except Ly6g and Tslp) depended on GATA2 for their expression, whereas the two commonly expressed genes did not (Fig. 4C, 4D). We did not detect re-expression of the Cebpa gene in “Gata2−/− mast cells” and the Mitf gene in Gata2−/− basophils. We also did not detect re-expression of cell surface markers unique to T cells (CD3), B cells (CD19), dendritic cells (MHC Class II and CD11c), and eosinophils (CCR3 and Siglec-F) or genes that are expressed in macrophages (Mmp12, Mpg-1, and Msr1), or neutrophils (Ela2, Prtn3, and Lactoferrin) in “Gata2−/− mast cells”, indicating that GATA2 is not a cell fate altering factor (Supplemental Fig. 2). Taken together, our data revealed that GATA2 is a crucial transcription factor in the molecular programs that regulate gene expression necessary for maintaining basophil and mast cell identities and for carrying out basophil and mast cell functions. Our data also demonstrate that GATA2 is imperative in basophil survival but not in mast cell survival.
STAT5 is required for maintaining FcεRIα expression on basophils and FcεRIα and c-Kit expression on mast cells
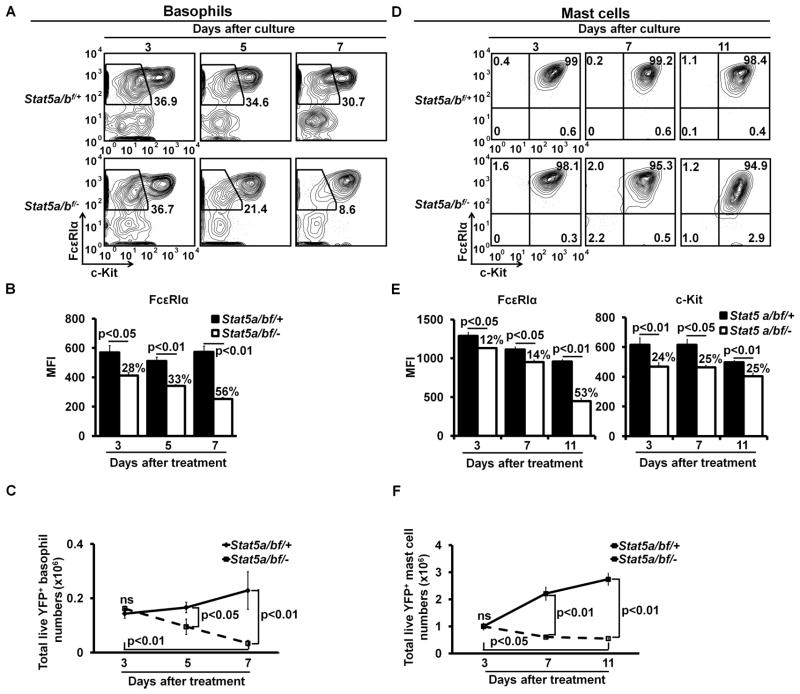
Our and other’s previous studies demonstrated that STAT5 is critical in basophil and mast cell development (12, 28, 35). However, it remains to be determined whether STAT5 is needed for the maintenance of basophil and mast cell identities. We showed that in the absence of the Stat5a/b gene, FcεRIα protein expression on YFP+ Stat5a/b−/− basophils begun to decrease at day 3 after the initial 4HT treatment, reaching the lowest levels at day 7 after the initial 4HT treatment (Fig. 5A and 5B). The number of YFP+ Stat5a/b−/− basophils in the culture after the 4HT treatment was significantly reduced compared with that of YFP+ Stat5a/b+/− basophils, indicating a pivotal role of STAT5 in basophil survival (Fig. 5C). Similarly, we found that in the absence of Stat5a/b gene, FcεRIα and c-Kit expression on YFP+ Stat5a/b−/− mast cells decreased beginning at day 3, reaching the lowest levels at day 11 after the initial 4HT treatment (Fig. 5D, 5E). The absence of the Stat5a/b gene had a less severe effect on FcεRIα and c-Kit expression than the absence of the Gata2 gene since we did not observe a complete loss of FcεRIα and c-Kit expression on YFP+ Stat5a/b−/− mast cells. The number of YFP+ Stat5a/b−/− mast cells decreased at day 7 after the initial 4HT treatment, reaching the lowest numbers at day 11 after the initial 4HT treatment (Fig. 5F), suggesting that STAT5 is a survival factor for mast cells. Thus, our data demonstrate that STAT5 signaling is essential for maintaining FcεRIα expression on basophils and FcεRIα and c-Kit expression on mast cells and for basophil and mast cell survival.
FIGURE 5.
STAT5 is required for maintaining FcεRIα expression on basophils and FcεRIα and c-Kit expression on mast cells. (A) Bone marrow cells prepared from the IL-3C-injected Stat5a/bf/+RosaCreErt2/Yfp (Stat5a/bf/+) or Stat5a/bf/−RosaCreErt2/Yfp (Stat5a/bf/−) mice were cultured in the presence of IL-3 for 3 days. The resulting cells were treated with 25 nM 4HT for an additional 3 days. Three, 5, or 7 days after the initial 4HT treatment, the cells were analyzed by FACS. (B) MFIs of FcεRIα expression on YFP+ Stat5a/b+/− or Stat5a/b−/− basophils (mean ± SD, n=6). (C) Total numbers of YFP+ Stat5a/b+/− or Stat5a/b−/− basophils (mean ± SD, n=6). (D) FACS analysis of BMMCs treated with 4HT. (E) MFIs of FcεRIα and c-Kit expression on YFP+ Stat5a/b+/− or Stat5a/b−/− mast cells (mean ± SD, n=6). (F) Total numbers of YFP+ Stat5a/b+/− or Stat5a/b−/− mast cells (mean ± SD, n=6). YFP+ cells are shown (A and D). The percentages of reduction are indicated (B and E).
STAT5 directly regulates the Gata2 gene and overexpression of the Gata2 gene is sufficient to direct basophil and mast cell differentiation in the absence of the Stat5 gene
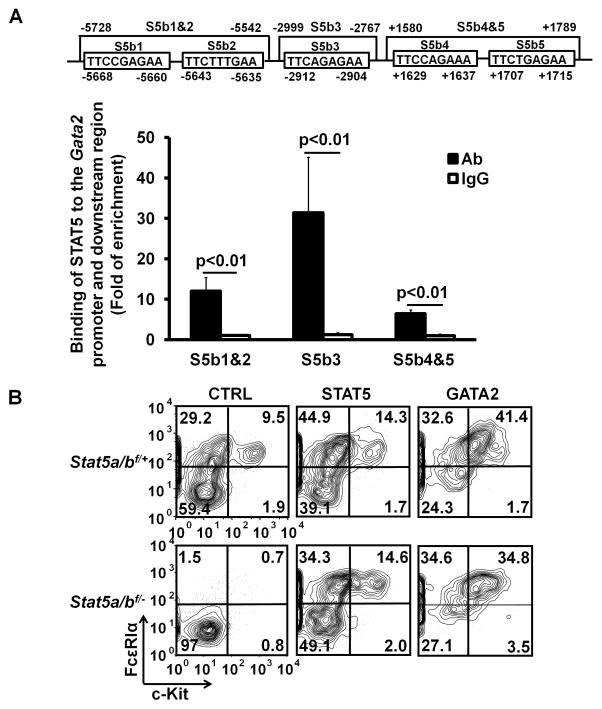
Stat5 deficiency and Gata2 deficiency resulted in the same developmental phenotype—a failure of pre-BMPs to differentiate into both basophils and mast cells. These results suggested that STAT5 and GATA2 might operate in the same pathway to regulate basophil and mast cell development. To define the relationship of STAT5 and GATA2 in basophil and mast cell differentiation, we searched the STAT5-binding sites (TTCNNNGAA, N means any nucleotide) in the +/− 30 kb from the transcription start site of the Gata2 gene and found three STAT5-binding sites in the Gata2 promoter (S5b1, S5b2, and S5b3) and two STAT5-binding sites in the +2 kb intronic region of the Gata2 gene (S5b4 and S5b5) (Fig. 6A, upper panel). Using ChIP assay, we found that STAT5 was recruited into the promoter and the +2 kb intronic region of the Gata2 gene (Fig. 6A, lower panel). Overexpression of the Gata2 gene in the Stat5a/b−/− bone marrow basophil and mast cell progenitors rescued basophil and mast cell differentiation to the levels that were comparable to those driven by Stat5a gene overexpression (Fig. 6B). We also noticed that the Stat5a virus-infected or Gata2 virus-infected Stat5a/b−/− or Stat5a/b+/+ bone marrow progenitors consistently differentiated into higher percentages of basophils and mast cells than the control bone marrow progenitors with vector virus transduction, presumably due to higher than WT levels of STAT5A and GATA2 protein expression in the infected progenitors. Compared to STAT5A, GATA2, when overexpressed, appeared to possess a higher capacity to drive mast cell differentiation (Fig. 6B). These results demonstrate that the Gata2 gene is a direct STAT5 target gene in the signaling pathway that drives the differentiation of bone marrow basophil and mast cell progenitors into basophils and mast cells.
FIGURE 6.
STAT5 directly regulates the Gata2 gene and overexpression of the Gata2 gene is sufficient to direct basophil and mast cell differentiation in the absence of the Stat5 gene. (A) STAT5-binding sites (S5bs1-5, upper panel) and ChIP analysis of STAT5 binding to the Gata2 promoter and downstream region (lower panel). BMMCs without stimulation were used for the ChIP analysis. (B) Bone marrow cells of Stat5a/bf/+ and Stat5a/bf/− mice were infected with retrovirus containing Stat5a, Gata2, or Thy1a (CTRL) gene. Twenty-four hours post infection, the infected cells were treated with 25nM 4HT for three days. Ten days after the initial 4HTtreatment, the cells were analyzed by FACS. YFP+ Thy1.1+ cell populations are shown. Data represent two independent experiments with similar results.
Discussion
In this study, we examined the role of GATA2 in the differentiation of pre-BMPs into basophils and mast cells. GATA2 has been shown to be important for mast cell differentiation (17). Enforced expression of GATA2 together with PU.1 drives the differentiation of mast cells from myeloid progenitors (36). GATA2 has also been implicated to play a role in basophil development. Akashi and colleagues showed that the order of GATA2 and C/EBPα expression is a deciding factor in driving GMPs into basophils (18). However, the previous approaches of deleting the Gata2 gene in the germ cells or overexpressing the Gata2 gene in GMPs failed to determine the precise role of GATA2 in basophil and mast cell development. Because GATA2 exerts its functions at the multiple developmental stages of hematopoiesis, it is pivotal to analyze the precise role of GATA2 in the defined progenitors of basophils and mast cells. We employed an approach combining prospective FACS sorting with an inducible gene deletion system to analyze the precise role of GATA2 in the differentiation of pre-BMPs into basophils and mast cells. Our analyses demonstrate that GATA2 is required for the differentiation of pre-BMPs into basophils and mast cells.
We document a Gata2 haploinsufficiency for mast cell differentiation but not for basophil differentiation in vitro and in vivo. Gata2haploinsufficiency has been reported for production and expansion of HSCs in the aorta-gonad-mesonephros region during embryonic development (14). In adult Gata2+/− mice, hematopoiesis appears to be normal. Only when a more robust test known as serial or competitive transplantation assay is used, defects in HSC self-renewal (14) and GMP functions in Gata2+/− mice (16) are revealed. Our in vitro results that Gata2+/− basophil and mast cell progenitors failed to differentiate into mast cells prompted us to further examine if there is a Gata2 haploinsufficiency for mast cells in vivo. We demonstrate a selective Gata2 haploinsufficiency in mast cell differentiation. However, it is still unknown why a full dose of GATA2 is required for mast cell differentiation but not for basophil development or mast cell maintenance. It is likely that GATA2 inefficiently transcribes genes whose protein products are required for the mast cell progenitors to pass mast cell developmental checkpoint(s). Once mast cell progenitors pass the checkpoints, one copy of the Gata2 gene is sufficient to maintain the expression of those important developmental genes. Alternatively, GATA2 downstream genes that maintain the mast cell identities differ from those that direct mast cell differentiation. Thus, the mast cell molecular program that directs mast cell differentiation and the mast cell molecular program that maintains mast cell identities require a different dose of the Gata2 gene. Identification of GATA2 target genes that are important in mast cell differentiation and maintenance is needed in order to test these possibilities.
If a gene is required for basophil or mast cell differentiation, then deletion of the gene will result in loss of basophils or mast cells, making it difficult to study gene functions in these cells. For example, STAT5 (35), GATA1 (37), GATA2 (17), and MITF (38–39) have each been demonstrated to be critical for mast cell differentiation, while STAT5 (28), Runx1 (11), GATA2, and C/EBPα (12, 18) have been shown as crucial transcription factors for basophil differentiation. Deletion of these transcription factors all resulted in loss or abnormal development of basophils and mast cells. Our approach has overcome this problem. We developed an experimental system, in which the Gata2 gene was deleted after basophils and mast cells become committed, to examine the precise roles of GATA2 in maintaining the expression of a wide range of genes that are important in carrying out basophil or mast cell functions, including genes encoding FcεRIα, c-Kit, and cytokines, genes that are expressed highly in basophils or mast cells, and genes that are commonly expressed in basophils and mast cells. We found that the effects of GATA2 on those gene expressions were broadly based. We demonstrate that GATA2 was required for maintaining Fcer1a mRNA and FcεRIα protein expression on basophils as well as for maintaining Fcer1a mRNA, FcεRIα protein expression, Kit mRNA and c-Kit protein expression on mast cells. We showed that GATA2 was also required for histamine synthesis by upregulating Hdc gene expression. Our data suggest that GATA2 regulates Il4 mRNA expression in basophils and Il13 mRNA expression in mast cells through both indirect and direct mechanisms. In the indirect mechanism, GATA2 regulates Il4 and Il13 gene transcription by regulating Fcer1a mRNA expression or critical signaling proteins. However, the reduced FcεRIα protein expression caused by the lack of the Gata2 gene could not fully account for the reduction in Il4 and Il13 mRNA expression in Gata2−/− basophils and “Gata2−/− mast cells” when those cells were activated by PMA and ionomycin, which bypass the FcεRI receptor stimulation to induce Il4 and Il13 gene transcription. We propose that GATA2 might regulate Il4 and Il13 gene transcription by either binding to the Il4 and Il13 regulatory regions or by inducing transcription factors that bind to Il4 and Il13 regulatory regions.
Unlike the Cebpa or Mitf genes, deletion of which resulted in re-expression of Mitf mRNA in Cebpa−/− basophils and re-expression of Cebpa mRNA in Mitf mutant mast cells and thus resulted in a basophil-mast cell fate conversion (12), deletion of the Gata2 gene did not lead to re-expression of neither Mitf mRNA in Gata2−/− basophils nor re-expression of Cebpa mRNA in Gata2−/− mast cells (Supplemental Fig. 2A) and thus did not lead to a basophil-mast cell fate conversion. We also did not detect re-expression of genes that are important for T cell, B cell, dendritic cell, eosinophil, neutrophil or macrophage molecular programming. Moreover, deletion of the Gata2 gene in mast cells does not appear to affect mast cell survival. We found a significant number of live Gata2−/− cells that had lost FcεRIα and c-Kit expression at the end of culture. The identities of those Gata2−/− cells remain unknown and further extensive analysis of gene expression profiles will be required.
Our experimental data also make a STAT5-GATA2 connection. Previous work has established that STAT5 plays critical roles in basophil and mast cell development (28, 35). Our work advances the understanding of STAT5 downstream transcription factor that exerts the STAT5-mediated biological functions. We demonstrate that STAT5 binds to the promoter and an intronic regulatory region of the Gata2 gene and overexpression of the Gata2 gene is sufficient to direct basophil and mast cell differentiation in the absence of the Stat5 gene. The finding that STAT5 was required for mast cell survival is consistent with a published work that STAT5 is required for mast cell survival by maintaining the expression of pro-survival molecules, such as Bcl-2 and Bcl-x(L) (35). Together, our analyses of the roles of GATA2 using an improved approach reveal a novel STAT5-GATA2 pathway in the differentiation and maintenance of basophils and mast cells.
Supplementary Material
Acknowledgments
Grant support:
This work is supported by a grant from the National Institutes of Health (RO1AI083986).
We thank D. Tracy for animal husbandry; S. Sobus and J. Looms for FACS analysis and cell sorting; H. Chu for assisting with microscopy.
Abbreviations
- 4HT
4-hydroxytamoxifen
- BaPs
basophil lineage-restricted progenitors
- BMCPs
basophil/mast cell progenitors
- BMMCs
bone marrow-derived mast cells
- C/EBPα
CCAAT/Enhancer Binding Protein, alpha
- CMPs
common myeloid progenitors
- FACS
fluorescence activated cell sorting
- GATA2
GATA Binding Protein 2
- GMPs
granulocyte-monocyte progenitors
- IDMEM
Iscove’s modification of Dullbecco’s modified eagle medium
- IgE
Immunoglobulin E
- HSCs
hematopoietic stem cells
- MCPs
mast cell lineage-restricted progenitors
- MITF
Microphthalmia-Associated Transcription Factor
- MPPs
multiple potential progenitors
- pre-BMPs
pre-basophil and mast cell progenitors
- YFP
yellow fluorescent protein
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Stone KD, Prussin C, Metcalfe DD. IgE, mast cells, basophils, and eosinophils. J Allergy Clin Immunol. 2010;125:S73–80. doi: 10.1016/j.jaci.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CM, Tsai M. Mast cells as “tunable” effector and immunoregulatory cells: recent advances. Annu Rev Immunol. 2005;23:749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- 3.Siracusa MC, Kim BS, Spergel JM, Artis D. Basophils and allergic inflammation. The Journal of allergy and clinical immunology. 2013;132:789–801. doi: 10.1016/j.jaci.2013.07.046. quiz 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karasuyama H, Mukai K, Obata K, Tsujimura Y, Wada T. Nonredundant roles of basophils in immunity. Annu Rev Immunol. 2011;29:45–69. doi: 10.1146/annurev-immunol-031210-101257. [DOI] [PubMed] [Google Scholar]
- 5.Kim HY, DeKruyff RH, Umetsu DT. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat Immunol. 2010;11:577–584. doi: 10.1038/ni.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voehringer D. Protective and pathological roles of mast cells and basophils. Nat Rev Immunol. 2013;13:362–375. doi: 10.1038/nri3427. [DOI] [PubMed] [Google Scholar]
- 7.Wedemeyer J, Tsai M, Galli SJ. Roles of mast cells and basophils in innate and acquired immunity. Current opinion in immunology. 2000;12:624–631. doi: 10.1016/s0952-7915(00)00154-0. [DOI] [PubMed] [Google Scholar]
- 8.Chen CC, Grimbaldeston MA, Tsai M, Weissman IL, Galli SJ. Identification of mast cell progenitors in adult mice. Proc Natl Acad Sci U S A. 2005;102:11408–11413. doi: 10.1073/pnas.0504197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franco CB, Chen CC, Drukker M, Weissman IL, Galli SJ. Distinguishing mast cell and granulocyte differentiation at the single-cell level. Cell stem cell. 2010;6:361–368. doi: 10.1016/j.stem.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, Tenen DG, Austen KF, Akashi K. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci U S A. 2005;102:18105–18110. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukai K, BenBarak MJ, Tachibana M, Nishida K, Karasuyama H, Taniuchi I, Galli SJ. Critical role of P1-Runx1 in mouse basophil development. Blood. 2012;120:76–85. doi: 10.1182/blood-2011-12-399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi X, Hong J, Chaves L, Zhuang Y, Chen Y, Wang D, Chabon J, Graham B, Ohmori K, Li Y, Huang H. Antagonistic regulation by the transcription factors C/EBPalpha and MITF specifies basophil and mast cell fates. Immunity. 2013;39:97–110. doi: 10.1016/j.immuni.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohnmacht C, Voehringer D. Basophil effector function and homeostasis during helminth infection. Blood. 2009;113:2816–2825. doi: 10.1182/blood-2008-05-154773. [DOI] [PubMed] [Google Scholar]
- 14.Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim KC, Hosoya T, Brandt W, Ku CJ, Hosoya-Ohmura S, Camper SA, Yamamoto M, Engel JD. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J Clin Invest. 2012;122:3705–3717. doi: 10.1172/JCI61619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues NP, Boyd AS, Fugazza C, May GE, Guo Y, Tipping AJ, Scadden DT, Vyas P, Enver T. GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood. 2008;112:4862–4873. doi: 10.1182/blood-2008-01-136564. [DOI] [PubMed] [Google Scholar]
- 17.Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- 18.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charles MA, Saunders TL, Wood WM, Owens K, Parlow AF, Camper SA, Ridgway EC, Gordon DF. Pituitary-specific Gata2 knockout: effects on gonadotrope and thyrotrope function. Mol Endocrinol. 2006;20:1366–1377. doi: 10.1210/me.2005-0378. [DOI] [PubMed] [Google Scholar]
- 20.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Developmental biology. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Chen L, Huang Z, Alkan S, Bunting KD, Wen R, Wang D, Huang H. Cutting edge: IL-5 primes Th2 cytokine-producing capacity in eosinophils through a STAT5-dependent mechanism. J Immunol. 2004;173:2918–2922. doi: 10.4049/jimmunol.173.5.2918. [DOI] [PubMed] [Google Scholar]
- 23.Razin E, Ihle JN, Seldin D, Mencia-Huerta JM, Katz HR, LeBlanc PA, Hein A, Caulfield JP, Austen KF, Stevens RL. Interleukin 3: A differentiation and growth factor for the mouse mast cell that contains chondroitin sulfate E proteoglycan. J Immunol. 1984;132:1479–1486. [PubMed] [Google Scholar]
- 24.Finkelman FD, Madden KB, Morris SC, Holmes JM, Boiani N, Katona IM, Maliszewski CR. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J Immunol. 1993;151:1235–1244. [PubMed] [Google Scholar]
- 25.Zhuang Y, Huang Z, Nishida J, Brown M, Zhang L, Huang H. A continuous T-bet expression is required to silence the interleukin-4-producing potential in T helper type 1 cells. Immunology. 2009;128:34–42. doi: 10.1111/j.1365-2567.2009.03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostergaard P, Simpson MA, Connell FC, Steward CG, Brice G, Woollard WJ, Dafou D, Kilo T, Smithson S, Lunt P, Murday VA, Hodgson S, Keenan R, Pilz DT, Martinez-Corral I, Makinen T, Mortimer PS, Jeffery S, Trembath RC, Mansour S. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome) Nat Genet. 2011;43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- 27.Huang H, Li Y. Mechanisms controlling mast cell and basophil lineage decisions. Curr Allergy Asthma Rep. 2014;14:457. doi: 10.1007/s11882-014-0457-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohmori K, Luo Y, Jia Y, Nishida J, Wang Z, Bunting KD, Wang D, Huang H. IL-3 induces basophil expansion in vivo by directing granulocyte-monocyte progenitors to differentiate into basophil lineage-restricted progenitors in the bone marrow and by increasing the number of basophil/mast cell progenitors in the spleen. J Immunol. 2009;182:2835–2841. doi: 10.4049/jimmunol.0802870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anthony RM, Kobayashi T, Wermeling F, Ravetch JV. Intravenous gammaglobulin suppresses inflammation through a novel T(H)2 pathway. Nature. 2011;475:110–113. doi: 10.1038/nature10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motomura Y, Morita H, Moro K, Nakae S, Artis D, Endo TA, Kuroki Y, Ohara O, Koyasu S, Kubo M. Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity. 2014;40:758–771. doi: 10.1016/j.immuni.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Wang M, Takeda K, Shiraishi Y, Okamoto M, Dakhama A, Joetham A, Gelfand EW. Peanut-induced intestinal allergy is mediated through a mast cell-IgE-FcepsilonRI-IL-13 pathway. The Journal of allergy and clinical immunology. 2010;126:306–316. 316 e301–312. doi: 10.1016/j.jaci.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichikawa A, Sugimoto Y, Tanaka S. Molecular biology of histidine decarboxylase and prostaglandin receptors. Proceedings of the Japan Academy. Series B, Physical and biological sciences. 2010;86:848–866. doi: 10.2183/pjab.86.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtsu H, Tanaka S, Terui T, Hori Y, Makabe-Kobayashi Y, Pejler G, Tchougounova E, Hellman L, Gertsenstein M, Hirasawa N, Sakurai E, Buzas E, Kovacs P, Csaba G, Kittel A, Okada M, Hara M, Mar L, Numayama-Tsuruta K, Ishigaki-Suzuki S, Ohuchi K, Ichikawa A, Falus A, Watanabe T, Nagy A. Mice lacking histidine decarboxylase exhibit abnormal mast cells. FEBS letters. 2001;502:53–56. doi: 10.1016/s0014-5793(01)02663-1. [DOI] [PubMed] [Google Scholar]
- 34.Nakazawa S, Sakanaka M, Furuta K, Natsuhara M, Takano H, Tsuchiya S, Okuno Y, Ohtsu H, Nishibori M, Thurmond RL, Hirasawa N, Nakayama K, Ichikawa A, Sugimoto Y, Tanaka S. Histamine synthesis is required for granule maturation in murine mast cells. European journal of immunology. 2014;44:204–214. doi: 10.1002/eji.201343838. [DOI] [PubMed] [Google Scholar]
- 35.Shelburne CP, McCoy ME, Piekorz R, Sexl V, Roh KH, Jacobs-Helber SM, Gillespie SR, Bailey DP, Mirmonsef P, Mann MN, Kashyap M, Wright HV, Chong HJ, Bouton LA, Barnstein B, Ramirez CD, Bunting KD, Sawyer S, Lantz CS, Ryan JJ. Stat5 expression is critical for mast cell development and survival. Blood. 2003;102:1290–1297. doi: 10.1182/blood-2002-11-3490. [DOI] [PubMed] [Google Scholar]
- 36.Walsh JC, DeKoter RP, Lee HJ, Smith ED, Lancki DW, Gurish MF, Friend DS, Stevens RL, Anastasi J, Singh H. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 2002;17:665–676. doi: 10.1016/s1074-7613(02)00452-1. [DOI] [PubMed] [Google Scholar]
- 37.Migliaccio AR, Rana RA, Sanchez M, Lorenzini R, Centurione L, Bianchi L, Vannucchi AM, Migliaccio G, Orkin SH. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. The Journal of experimental medicine. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takemoto CM, Lee Y-N, Jegga AG, Zablocki D, Brandal S, Shahlaee A, Huang S, Ye Y, Gowrisankar S, Huynh J, McDevitt MA. Mast cell transcriptional networks. Blood Cells, Molecules, and Diseases. 2008;41:82–90. doi: 10.1016/j.bcmd.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitamura Y, Morii E, Jippo T, Ito A. Regulation of mast cell phenotype by MITF. Int Arch Allergy Immunol. 2002;127:106–109. doi: 10.1159/000048178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.