Abstract
The BAF (mammalian SWI/SNF) complexes are a family of multi-subunit ATP-dependent chromatin remodelers that use ATP hydrolysis to alter chromatin structure. Distinct BAF complex compositions are possible through combinatorial assembly of homologous subunit families and can serve non-redundant functions. In mammalian neural development, developmental stage-specific BAF assemblies are found in ES cells, neural progenitors and postmitotic neurons. In particular, the neural progenitor-specific BAF complexes are essential for controlling the kinetics and mode of neural progenitor cell division, while neuronal BAF function is necessary for the maturation of postmitotic neuronal phenotypes as well as long-term memory formation. The microRNA-mediated mechanism for transitioning from npBAF to nBAF complexes is instructive for the neuronal fate and can even convert fibroblasts into neurons. The high frequency of BAF subunit mutations in neurological disorders underscores the rate-determining role of BAF complexes in neural development, homeostasis and plasticity.
Keywords: BAF, mammalian SWI/SNF, chromatin remodeling, neural development, microRNA, intellectual disability, Coffin-Siris syndrome, Nicolaides-Baraitser syndrome, autism, schizophrenia
INTRODUCTION
Putting together a stable yet versatile network such as the mammalian central nervous system poses a formidable developmental challenge. While all neurons must acquire a common set of properties, such as the ability to form synapses and fire action potentials, there is a tremendous degree of additional specialization in each of the thousands of neuronal subtypes differing in size, shape, location and neurotransmitter phenotypes (Killackey et al., 1989; Wise and Jones, 1977). Moreover, remarkable architectural integrity is required both at the cellular and system-wide level over the course of the animal’s lifetime; yet the system must retain the ability to reshape some of its connections based on experience so that learning can occur (Kandel, 2012; Engert and Bonhoeffer, 1999).
How is our genetic information utilized to achieve this feat? In part, it involves the hierarchical regulation of gene expression programs in which a small number of lineage-specifying master transcription factors activate or repress a large number of downstream targets (Weintraub, 1993; Nutt et al., 1999; Stoykova et al., 2000; Boyer et al., 2005). However, these concerted changes in gene expression profile must occur within the physical dimensions of the cell’s 5-micron nucleus that necessitates extraordinary compaction of DNA. The two billion base pairs of the human genome would total two meters in length if extended, but in the nucleus they are packaged around histone octamers and further compacted to form chromatin (Kornberg, 1974). The degree of compaction is not uniform across all chromatin loci, and while ‘open’ (euchromatic) regions are accessible to nuclear factors, ‘closed’ or repressed (heterochromatic) regions are unable to be activated. Thus, there must be dynamic interplay between chromatin regulation and the actions of other nuclear factors throughout development.
Three main epigenetic mechanisms cooperate to regulate the state of the chromatin (Bernstein et al., 2007; Hargreaves and Crabtree, 2011): DNA methylation on the cytosine residues of CpG dinucleotides; covalent modifications of histone tails; and ATP-dependent chromatin remodeling which utilizes the energy of ATP hydrolysis to move or exchange nucleosomes. In this review, we focus on a family of multimeric ATP-dependent chromatin remodeling complexes called BAF (BRG1/BRM-associated factors), also known as mammalian SWI/SNF (mSWI/SNF) for their limited similarity to the yeast SWI/SNF complex. BAF complexes have emerged at the forefront of human neurological diseases following recent discoveries which implicate BAF subunit mutations in syndromic and non-syndromic intellectual disability (Santen et al., 2013, 2012; Tsurusaki et al., 2013, 2012; Hoyer et al., 2012; Halgren et al., 2012; Van Houdt et al., 2012; Backx et al., 2011; Wieczorek et al., 2013), sporadic autism (Neale et al., 2012; O’Roak et al., 2012), schizophrenia (Loe-Mie et al., 2010; Koga et al., 2009) and amyotrophic lateral sclerosis (Chesi et al., 2013). These important findings from human genetics indicate that BAF complexes make rate-limiting contributions to the establishment of the diversity, stability and plasticity found in our nervous system.
We begin by describing the emergence of mammalian BAF complexes, then look at specific BAF assemblies found during neural development, starting with pluripotent stem cells from which cells of all three embryonic germ layers are derived. Finally, we will examine recent evidence for the role of BAF complexes in human neurological disorders.
Evolution from the yeast SWI/SNF complex to mammalian BAF complexes [caps]
The subunits of the yeast SWI/SNF complex were first discovered in genetic screens for their requirement for mating type switching and the use of sucrose as a nutrient source (Neigeborn and Carlson, 1984; Stern et al., 1984). In addition to the ATPase subunit, SWI2/SNF2, the yeast complex includes SWI1/ADR6, SWI3, SNF5 and SNF6 (Cairns et al., 1994), and has an activating role in transcription through chromatin regulation (Table I) (Peterson and Herskowitz, 1992; Hirschhorn et al., 1992; Laurent et al., 1991; Laurent and Carlson, 1992). A homologous complex in Drosophila was also discovered from screens for suppressors of Polycomb mutations that cause homeotic transformations (Tamkun et al., 1992; Elfring et al., 1994). In flies, the core ATPase subunit is a homolog of the yeast SWI2/SNF2, called Brahma (BRM), and the complex is termed the Brahma-associated protein (BAP) complex (Table 1).
Table 1. BAF complex subunit nomenclature and conservation in yeast and fly.
The subunits are organized into families and presented in the order of decreasing protein size. For the mammalian subunits, the names used in this review are highlighted in bold. Adapted from Tang, Yoo & Crabtree, Curr Opinion Genet & Dev (2013).
| BAF | Aliases | SMARC nomenclature | SWI/SNF (S. cerevisiae) | BAP (D. Melanogaster) |
|---|---|---|---|---|
| BAF250a | ARID1A | - | Osa/eyelid | |
| BAF250b | ARID1B | - | ||
| BAF200 | ARID2 | - | BAP170 | |
| BAF190a | BRG1/SNF2β | SMARCA4 | SWI2 | |
| BAF190b | BRM/SNF2α | SMARCA2 | Brahma | |
| BAF180 | PBRM1 | - | BAP180 | |
| BAF170 | SMARCC2 | SWI3 | Moira | |
| BAF155 | SMARCC1 | |||
| BAF100a | Bcl11a/Ctip1 | - | CG9650 | |
| BAF100b | Bcl11b/Ctip2 | - | ||
| BAF60a | SMARCD1 | SWP73 | BAP60 | |
| BAF60b | SMARCD2 | |||
| BAF60c | SMARCD3 | |||
| BAF57 | SMARCE1 | - | Dalao/BAP111 | |
| BAF55a | SS18 | - | - | |
| BAF55b | CREST/SS18L | - | - | |
| β-actin | - | Actin 5C | ||
| BAF53a | ACTL6A | ARP7, APR9 | BAP55 | |
| BAF53b | ACTL6B | |||
| BAF47 | hSNF5/INI1 | SMARCB1 | SNF5 | SNR1 |
| BAF45a | PHF10 | - | SAYP | |
| BAF45b | DPF1 | - | D4 | |
| BAF45c | DPF3 | - | ||
| BAF45d | DPF2 | - | ||
| BAF40a | Bcl7a | - | Bcl7-like | |
| BAF40b | Bcl7b | - | ||
| BAF40c | Bcl7c | - | ||
| Brd7 | - | CG7154 | ||
| Brd9 | - |
In human, the DEAD/H helicase family of 29 genes codes for homologs of the yeast SWI2/SNF2, and are involved in a diverse range of functions that pertain to the chromatin including DNA replication and repair, transcriptional initiation and elongation, mRNA splicing and gene silencing (Hargreaves and Crabtree, 2011; Yoo and Crabtree, 2009). Several members also serve as core subunits in ATP-dependent chromatin remodeling complexes of which there are 4 families: BAF (which uses BRG1 or hBRM), INO80/SWR1 (which uses hINO80, hDomino or SRCAP), ISWI (which uses hSNF2H or hSNF2L) and CHD (which uses CHD1 through CHD9).
The 2-megadalton mammalian BAF complexes consist of at least 15 subunits, five of which have homologs in the yeast SWI/SNF (BRG1/BRM, BAF155/170, BAF60, BAF53, BAF47) (Figure 1, Table I). However, the mamm alian BAF has diverged significantly from the yeast counterpart, losing some subunits while gaining novel core subunit families including BAF250a/b (ARID1a/b), BAF45a/b/c/d, BAF57, β-actin, BRD7, BRD9 and SS18/CREST; of note, BAF250a/b are the most frequently mutated BAF subunits in human malignancy and neurological disorders (Ho et al., 2009b; Kadoch et al., 2013). Furthermore, while the yeast SWI/SNF has an exclusively activating role, BAF complexes can both activate or repress their genomic targets (Boyer et al., 2005; Ho et al., 2009a, 2011). Thus, the mammalian BAF complexes only have limited similarities with the yeast SWI/SNF complex. The changes in subunit composition correspond to the emergence of multicellularity, and even later changes correspond to the development of complex nervous systems; the later evolutionary point is reflected in their frequent implication in neurologic disease.
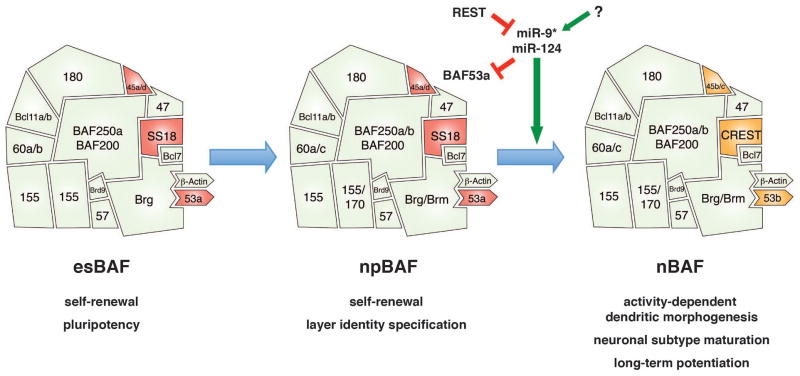
Figure 1. BAF complex assemblies in neural development.
Distinct BAF compositions exist in ES cells (esBAF), neural progenitors (npBAF) and postmitotic neurons (nBAF). Each complex serves critical context-dependent functions by interacting with a unique set of cellular factors. During mitotic exit, miR-9* and miR-124 mediate switching at three subunit positions (highlighted in red and orange in esBAF/npBAF and nBAF, respectively).
Another distinct feature of BAF complexes is the existence of alternative subunits for many of the positions within the complex. Only one of the homologous subunits can be incorporated at each of the polymorphic positions, such that there are hundreds of theoretically possible assemblies of BAF. The subunits are not freely exchanged and are stably associated with the complex even in highly denaturing conditions in vitro (Zhao et al., 1998; Kadoch et al., 2013). As we will discuss, each unique composition can translate to a specific biological meaning which can be instructive for a particular cell fate. Distinct BAF assemblies have been found in a number of tissues (de la Serna et al., 2001; Ohkawa et al., 2006; Cvekl and Duncan, 2007; Xiong et al., 2013; Li et al., 2013; Yu et al., 2013; Lickert et al., 2004; Lessard et al., 2007; Olave et al., 2002) and it is possible that many more cell types have chromatin remodeling mechanisms tailored to their specific needs.
ES cell-specific BAF (esBAF) complexes (caps)
Embryonic stem (ES) cells are derived from the inner cell mass (ICM) of the pre-implantation embryo and maintain unlimited self-renewal capacity and multi-lineage differentiation potential. A suite of transcription factors and chromatin regulators are critical for achieving the chromatin landscape in ES cells which remains poised for differentiation without losing pluripotency. An ES cell-specific assembly of BAF, called esBAF, is characterized by the presence of BRG1, BAF250a, BAF60a/b and BAF155, while excluding BRM, BAF250b, BAF60c and BAF170 at the variable positions (Ho et al., 2009b; Kaeser et al., 2008; Kidder et al., 2009) (Figure 1).
esBAF in stem cell self-renewal and pluripotency
The peri-implantation lethal phenotype of mice lacking Brg1, BAF155 or BAF47 indicates that esBAF has an essential role in early embryonic development (Bultman et al., 2000; Kim et al., 2001; Klochendler-Yeivin et al., 2000). Cre-mediated deletion of Brg1 impairs self-renewal of cultured ES cells but not mouse embryonic fibroblasts (MEFs) (Ho et al., 2009b) or glial cells (Lessard et al., 2007). Brg1-deleted ES cells first lose proliferative capacity and, after several passages, lose the expression of ES cell markers Oct4, Sox2 and Nanog; they are also defective in forming ectodermal and mesodermal lineages during spontaneous differentiation, indicating a loss of pluripotency.
The specific configuration of subunits in esBAF is crucial for the ES cell state. Brm, the alternative ATPase subunit, is not found in esBAF and does not compensate for the absence of Brg1 (Bultman et al., 2000); moreover, Brm−/− mice are viable despite being slightly larger than normal (Reyes et al., 1998). Similarly, the esBAF component BAF155 and its homolog BAF170 do not have redundant roles in ES cells. Knockdown of BAF155 in ES cells causes defects similar to Brg1 knockdown, with decreased proliferation followed by loss of Oct4 and increased cell death (Ho et al., 2009b). This phenotype is rescued by overexpressing BAF155 but not BAF170. In addition, BAF170-expressing ES cells do not contribute efficiently to teratoma formation in vivo, indicating that forced assembly of BAF170-containing BAF complexes in ES cells is detrimental to pluripotency.
esBAF, LIF/STAT3 signaling and opposition of Polycomb
Murine ES cells require leukemia inhibitory factor (LIF) signaling, which leads to phosphorylation and nuclear localization of the transcription factor STAT3 (Matsuda et al., 1999; Niwa et al., 1998). The genome-wide binding sites of STAT3 overlap extensively with Brg1 occupancy (Ho et al., 2009a), and the transcriptional changes caused by LIF withdrawal and acute Brg1 deletion are highly similar (Ho et al., 2011). This is in contrast to Oct4 (Loh et al., 2006; Masui et al., 2007) and Sox2 (Shao et al., 1999) whose targets also overlap with Brg1 binding sites but do not show consistent transcriptional co-regulation. Although there is no detectable physical interaction between STAT3 and Brg1, Brg1 knockout abolishes STAT3 binding at over 80% of sites; these sites become more resistant to DNase I digestion (Ho et al., 2011) indicative of a more ‘closed’ chromatin state (Dorschner et al., 2004). Creating accessibility at genomic sites may be a key function of the esBAF complex, and 80% of BRG1 target sites in ES cells are indeed DNase I hypersensitive (Schnetz et al., 2010).
One histone modification which is significantly altered in Brg1-knockout ES cells is histone H3 lysine-27 trimethylation (H3K27me3) (Ho et al., 2011), which is a repressive mark deposited by Polycomb repressive complex 2 (PRC2). Many Brg1-dependent STAT3 targets, as well as genes that are co-activated by Brg1 and Oct4 or Sox2, gain aberrant H3K27me3 upon Brg1 deletion, even though PRC2 components are not upregulated and the overall level of H3K27me3 remains constant. The ectopic H3K27me3 domains are localized near promoters and transcription start sites (TSSs) where Brg1 peaks are normally located, suggesting a direct opposition between BAF and PRC2. Consistent with their functional antagonism, the genome-wide binding of Brg1 is largely mutually exclusive with Suz12, a PRC2 component (Ho et al., 2009a), and the ectopic H3K27me3 marks in Brg1-deleted cells can be suppressed by knockdown of Suz12 (Ho et al., 2011). This antagonistic relationship is conserved in flies, where homeotic transformations caused by PcG mutations can be suppressed by compensating trithorax mutations (Tamkun et al., 1992; Elfring et al., 1994; Kennison and Tamkun, 1988). However, the interaction seems more complex in mammals: importantly, BAF synergizes with Polycomb at all four Hox loci in ES cells to prevent precocious differentiation (Ho et al., 2011). Nevertheless, one of the key functions of esBAF is to maintain the accessibility of actively transcribed genes by preventing the formation of Polycomb repressive domains in the ES cell genome.
Neural progenitor BAF (npBAF) complexes
The development of the brain follows a stereotyped pattern beginning with the expansion of neuroepithelial cells, which go on to form distinct pools of progenitors in a number of anatomical locations (Chenn and McConnell, 1995). In the cortex, most γ-aminobutyric acid (GABA)-producing local inhibitory neurons arise in the ventral telencephalon and migrate long distances to the dorsal cortex (Wonders and Anderson, 2006). On the other hand, excitatory projection neurons, which are glutamatergic and project to other cortical areas as well as subcortical targets, originate from the dorsal telencephalon in the germinal zone lining the ventricles, called the ventricular zone (VZ) (Hendelman, 1991). Radial glial cells (RGs) are bipolar neural precursors whose cell body resides in the VZ; they are bona fide neural stem cells (NSCs) in that they can divide symmetrically to self-renew or asymmetrically to produce more differentiated progeny (Götz and Huttner, 2005; Kriegstein et al., 2006). Initially, RGs generate neurons directly (‘direct neurogenesis’), but begin producing a second population of neural precursors called intermediate progenitors (IPs) around E13.5 (Hendelman, 1991). The late phase of corticogenesis is dominated by an expansion of the pool of RG-derived IPs in the subventricular zone (SVZ) which undergo limited rounds of division to produce neurons (‘indirect neurogenesis’).
Since the number of precursors and their mode of division critically influence final cortical size and complexity, the neural precursor state is a focal point for regulation in brain development. Neural precursors must downregulate the pluripotency gene network promoted in ES cells while remaining proliferative; they must also suppress the neuronal differentiation program until mitotic exit. The chromatin landscape associated with this particular state requires the neural progenitor-specific BAF (npBAF) complex (Lessard et al., 2007). In contrast to esBAF, npBAF can have alternative subunits at a number of positions, including either Brg1 or Brm as the core ATPase; either BAF250a or BAF250b; and either a homodimer of BAF155 or a heterodimer of BAF155 and BAF170 (Figure 1). Three of the subunits which are unchanged during neural induction – BAF53a, SS18 and BAF45a/d – are also important for specifying the undifferentiated, proliferative state of the progenitors; as we will discuss, these three positions later undergo switching around the time of mitotic exit, such that the postmitotic neuronal BAF (nBAF) contains BAF53b, CREST and BAF45b/c, at those positions, respectively.
npBAF in neural progenitor self-renewal
Similar to the role of esBAF in ES cells, components of the nBAF complex have a critical role in the self-renewal of neural progenitors. Mice heterozygous for Brg1 or BAF155 have defects in neural tube closure, indicative of insufficient cell numbers (Bultman et al., 2000; Kim et al., 2001). The rate-limiting role of BAF in progenitor division is evolutionarily conserved: in C. elegans, the asymmetric division of NSCs requires psa-1 and psa-4 (homologs of BRG1 and BAF155, respectively) (Sawa et al., 2000). Deleting Brg1 specifically in Nestin+ murine NSCs causes reduced proliferation and subsequent depletion of the neural progenitor pool; these mice die perinatally and exhibit thinning of the cortex and midbrain, as well as near-complete absence of the cerebellum (Matsumoto et al., 2006; Lessard et al., 2007; Zhan et al., 2011). Interestingly, even though the total number of GFAP+ astrocytes is reduced in the Nestin::Brg1−/− animals, the astrocytes themselves are not impaired in their ability to divide, indicating that the effect on proliferation is specific to the multipotent progenitors. At the transcriptional level, Brg1-containing npBAF complexes activate Notch signaling pathway components to promote NSC proliferation, while repressing genes involved in Sonig Hedgehog (Shh) signaling which plays a vital role in neural patterning (Lessard et al., 2007; Rowitch et al., 1999; Chiang et al., 1996).
As would be expected, genetic perturbations of the three npBAF-specific subunits, BAF53a, BAF45a/d and SS18, also cause proliferation defects in NSCs. Knockdown of either BAF45a or BAF53a, or both, in E13.5 cortical cultures reduces the rate of BrdU incorporation without affecting cell survival or terminal differentiation (Lessard et al., 2007). Interestingly, while ES cells tolerate knockdown of SS18 to 20% of wild-type protein level, NSCs fail to self-renew when SS18 is decreased by only 20% (Staahl et al., 2013), indicating their heightened sensitivity to the levels of this subunit. Consistent with this, SS18+/− embryos are able to implant but die between E8.5 and E9.5 (de Bruijn et al., 2006). Importantly, perturbing neuron-specific subunits which are highly homologous to the npBAF subunits does not cause the same phenotype in NSCs. While overexpression of BAF45a causes a 2- to 4-fold increase in the number of cycling cortical and cerebellar progenitors at E14.5, ectopic BAF45b expression has no discernable effect (Lessard et al., 2007). Conversely, knockdown of BAF45a, but not BAF45b, leads to a reduced number of proliferative neural progenitors in the same neurogenic regions of the brain. This functional discrepancy stems from the N-terminal domain and the Krüpple zinc finger domain which are highly divergent between the two homologs: deleting either domain from BAF45a prevents its enhancement of progenitor proliferation, but deleting the C-terminal double PHD domain, which is more conserved between BAF45a and BAF45b, does not have a significant effect.
BAF170 in the specification of layer identity
The timing of neurogenic division is indicative of the laminar identity of the newborn cortical neuron. Neurons born in the dorsal VZ migrate in an inside-out fashion starting around E12.5, first to deep cortical layers (in the order: layers VI, V, and IV) on successive days and finally to the upper layers (II/III) around E15.5 (Arlotta et al., 2005). While direct neurogenesis from RGs produces deep-layer neurons during early corticogenesis, indirect neurogenesis via production of IPs contributes most to the neurons in the upper layer (Pontious et al., 2007). Pax6 is a master regulator of dorsal telencephalic fate (Muzio et al., 2002; Schuurmans et al., 2004; Stoykova et al., 2000; Toresson et al., 2000; Yun et al., 2001), but also has a role in regulating the kinetics of progenitor cell division (Quinn et al., 2007); in particular, Pax6-null mice display a specific thinning of the upper cortical layers due to premature asymmetrical cell division of the progenitors (Estivill-Torrus et al., 2002). BAF has been shown to interact with Pax6 to regulate the decision between direct and indirect neurogenesis (Tuoc et al., 2013). Specifically, BAF170-containing npBAF complex restricts indirect neurogenesis during the early phase by recruiting the transcriptional repressor REST to Pax6 targets (Tuoc et al., 2013; Battaglioli et al., 2002). Notably, the amino acid sequences of BAF170 and BAF155 diverge the most in the C-terminal region, and it is the C-terminus of BAF170 that is required for its interaction with REST in vitro. The loss of BAF170 in Emx1+ cortical progenitors has the opposite effect from Pax6 deletion and causes an abnormal expansion of IPs and a bias toward upper-layer identities, while BAF170 overexpression causes a phenotype similar to the Pax6-null with a depletion of upper-layer neurons (Tuoc et al., 2013). Thus, specific assemblies of npBAF determine the size as well as composition of the cortex.
Neuronal BAF (nBAF) complexes
At the final neurogenic cell division, a shift in BAF composition occurs at three subunit positions: BAF53a is switched for BAF53b, SS18 for CREST, and BAF45a/d for BAF45b/c (Wu et al., 2007; Olave et al., 2002) (Figure 1). Immunostaining in the developing embryonic spinal cord reveals a clear boundary between the Ki67+ proliferative zone which expresses npBAF subunits, and the β-Tubulin-III+ postmitotic zone which expresses the homologous neuronal BAF (nBAF) subunits (Lessard et al., 2007; Staahl et al., 2013). The nBAF-specific subunits are not found in any other cell type examined to date and are thus exclusively involved in remodeling the postmitotic neuronal chromatin. Deletion of the nBAF-specific subunits affects differentiated neurons, in contrast to npBAF subunits whose disruption affects the progenitor population.
nBAF in dendritic morphogenesis
Functional nBAF complexes have an essential role in dendritic morphogenesis, a process which is critical for the proper integration of postmitotic neurons into their appropriate circuitry (Jan and Jan, 2003; Whitford et al., 2002). This is conserved in flies, where perturbation of BAP55, BRM, BAF60 or SNR1 (fly homologs of BAF53a/b, BRM, BAF60c and BAF47) results in PNS dendritic morphogenesis defects (Parrish et al., 2006); in addition, the loss of BAP55 produces a highly specific, fully penetrant retargeting phenotype in Drosophila olfactory projection neurons and can be rescued by expressing the human BAF53A or BAF53B (Tea and Luo, 2011). Deletion of ham-3 (homolog of BAF60) in C. elegans, on the other hand, disrupts axon pathfinding in a specific subset of serotonergic neurons (Weinberg et al., 2013).
In mice, BAF53b deletion results in perinatal lethality owing to a failure to nurse, and only one in ten survive to adulthood with hyperactivity phenotypes (Lessard et al., 2007; Wu et al., 2007). Interestingly, the brain of BAF53b−/− mice at postnatal day 0 (P0) does not differ appreciatively from wild-type in its size and complexity, indicating that intrinsic, activity-independent dendritogenesis is not affected (Wu et al., 2007). However, ex vivo cultures of BAF53b−/− hippocampal and cortical neurons reveal a striking deficit in the ability to elaborate dendritic processes following KCl stimulation. This phenotype can be rescued by the introduction of wild-type BAF53b, but not its homolog, BAF53a. BAF53a and BAF53b belong to the family of actin-related proteins (Arps) and contain actin folds with four distinct subdomains, the most divergent of which is subdomain 2. Remarkably, chimeric BAF53a containing the actin fold subdomain 2 of BAF53b was able to rescue the dendritic outgrowth phenotype of BAF53b-null, demonstrating that functional specificity is conferred by the structural divergence between homologous subunits.
Refinement of dendrites occurs in response to stimulation which causes Ca2+ influx through voltage-sensitive Ca2+ channels and activates downstream signaling pathways. As a result, local cytoskeletal rearrangements are triggered, as well as transcription-dependent global dendritic remodeling. The latter mechanism involves a number of transcriptional factors including cAMP-responsive element binding protein (CREB), CREB-binding protein (CBP) and NeuroD (Redmond et al., 2002). The nBAF component CREST is a calcium-sensitive transcriptional coactivator which itself does not bind DNA but interacts with CREB to actuate transcriptional outcome (Aizawa et al., 2004). While CREST-null mice are viable, they exhibit dendritic defects in the cortex and the hippocampus similar to a BAF53b-null (Aizawa et al., 2004). The proper localization of the CREST-containing nBAF complex at key target promoters, including Ephexin1, requires BAF53b, although BAF53b is not necessary for the assembly of nBAF (Wu et al., 2007).
Bcl11b in neuronal subtype maturation
A recent proteomic study has uncovered Bcl11b (also known as Ctip2) to be a novel, dedicated subunit of BAF which stably associates with the complex even in denaturing conditions (Kadoch et al., 2013). Bcl11b is a zing-finger domain transcription factor critical in several developmental paradigms, including T cell development (Wakabayashi et al., 2003; Albu et al., 2007, 2011; Ikawa et al., 2010) and odontogenesis (Golonzhka et al., 2009). During neural development, Bcl11b has critical roles in consolidating the subtype identities of postmitotic neurons in multiple localized regions.
In the developing neocortex, Bcl11b is strongly expressed across layer V in subcerebral projection neurons as well as weakly in layer VI corticothalamic neurons (Arlotta et al., 2005), and specifies their respective laminar identities. Layer V subcerebral neurons are early-born neurons (E12.5-E14.5) which project to targets caudal to the cerebrum, including the brainstem and spinal cord (Killackey et al., 1989; Glickstein et al., 1985; Molnár and Cheung, 2006; Wise and Jones, 1977). Bcl11b deletion affects the later stages of subcerebral axonal outgrowth and refinement, including fasciculation, pathfinding and pruning. In particular, Bcl11b is haploinsufficient for axonal pruning in corticospinal neurons, a subtype of subcerebral neurons that project to the spinal cord (Arlotta et al., 2005). Similarly, in the striatum, Bcl11b is exclusively expressed in postmitotic medium spiny neurons (MSNs) and is required for their maturation, including the expression of MSN markers and organization into patches (Arlotta et al., 2008).
Bcl11b deletion also affects the maturation of vomeronasal sensory neurons (VSNs) which detect pheromones and project to the olfactory bulb (OB) (Dulac and Axel, 1995; Herrada and Dulac, 1997; Matsunami and Buck, 1997; Ryba and Tirindelli, 1997). In Bcl11b−/− mice, VSN precursors are produced normally and terminal differentiation is induced, but the immature VSNs display increased apoptosis, reduced axonal outgrowth and mistargeting to incorrect regions in the OB, leading to abnormal layer structure in the anterior olfactory bulb (AOB) (Enomoto et al., 2011). Transcriptionally, Bcl11b is necessary for activating the vomeronasal receptor genes V1r and V2r, as well as Big2/Contactin4, an axonal guidance molecule in the olfactory system. Bcl11b also has a role in finer subtype specification of VSNs, regulating the fate decision between basal and apical VSNs.
During pre- and postnatal hippocampal development, Bcl11b expression is again restricted to postmitotic granule neurons and excluded from the dividing progenitors in the subgranular zone (SGZ) of the dentate gyrus (Simon et al., 2012). In the Bcl11b-null brain, newborn neurons have reduced calbindin 1 (Calb1) expression, are no longer confined to the innermost compartment of the granule cell layer (GCL) and have shorter dendrites than wild-type, reminiscent of the dendritic outgrowth phenotype seen in BAF53b−/− hippocampal neurons in vitro (Wu et al., 2007). Intriguingly, there is a severe reduction in the size and cell number of the dentate gyrus in the absence of Bcl11b due to a decrease in BrdU+ progenitors without increased apoptosis (Simon et al., 2012); given the exclusion of Bcl11b from the progenitors in the SGZ in wild-type animals, this suggests indirect mechanisms are at work. It will be of interest to examine whether Bcl11b-containing BAF complexes cooperate with subtype-specific transcription factors to target distinct chromatin loci for activation or repression in each neuronal type.
BAF53b in learning and memory
Formation of long-term memory is the basis for learned behavior in higher organisms. The cellular correlate of long-term memory is long-term potentiation (LTP), which requires transcription and cytoskeletal rearrangements downstream of activity-triggered Ca2+ signaling (Kandel, 2012). Epigenetic mechanisms have important roles in LTP, the most well-studied examples being histone modifications and DNA methylation (Alarcón et al., 2004; Cohen-Armon et al., 2004). The contribution of chromatin remodeling to memory formation remains largely unexplored, but a recent study provides evidence that the nBAF complexes have a role in this process (Vogel-Ciernia et al., 2013). Mice heterozygous for BAF53b, as well as mice expressing dominant negative form of BAF53b (BAF53ΔHD) in Camk2a+ forebrain excitatory neurons, have abnormal spine morphology and impaired synaptic plasticity. While the short-term memory of the mice is normal, they have deficits in various facets of long-term memory including object location, object recognition and contextual fear. Consistent with this, hippocampal slices from BAF53ΔHD-expressing animals exhibit short-term potentiation (STP) but are unable to consolidate LTP. RNA-seq analysis of the dorsal hippocampus reveals that BAF53b+/− mice misregulate a variety of genes following object location memory training as compared to wild-type, including genes involved in transcriptional regulation, neurogenesis and chromatin modification, as well as those involved in cytoskeletal rearrangements and postsynaptic density. The role of nBAF in the adult CNS may be the basis for the pathogenicity of BAF subunit mutations in psychiatric disorders (Loe-Mie et al., 2010; Koga et al., 2009; Neale et al., 2012; O’Roak et al., 2012; Nord et al., 2011).
Regulation of BAF complex switching during neural development and reprogramming (caps)
The switching of npBAF subunits with nBAF subunits at mitotic exit is mediated by a triple-negative genetic circuitry involving non-coding micro-RNAs (miRNAs) (Figure 1) (Yoo et al., 2009). miRNAs achieve repression of their target genes by binding to transcripts and inhibiting translation or causing mRNA degradation (Lewis et al., 2005). The 3′ untranslated region (UTR) of BAF53a contains binding sites for miR-9* and miR-124, two neurogenic miRNAs which are abundant in neural tissue (Krichevsky et al., 2006; Lagos-Quintana et al., 2002; Cao et al., 2007; Makeyev et al., 2007; Visvanathan et al., 2007). The miRNAs are in turn subject to regulation by REST (Conaco et al., 2006), the transcriptional repressor which prevents neuronal gene expression in non-neuronal cells by binding to repressive element 1 (RE-1) on target genes and recruiting other repressors including CoREST and mSin3A (Grimes et al., 2000; Lunyak et al., 2002; Ballas et al., 2005; Chen et al., 1998; Chong et al., 1995).
In neural progenitors, REST-mediated repression of miR-9* and miR-124 maintains BAF53a expression (Yoo et al., 2009). Forced expression of the miRNAs in Nestin+ neural progenitors leads to loss of BAF53a and impaired proliferation, consistent with the requirement for the BAF53a subunit in progenitor cell division (Lessard et al., 2007). On the other hand, REST is repressed in postmitotic neurons by the unliganded retinoic acid receptor (RAR) repressor complex (Ballas et al., 2005); this allows miR-9* and miR-124 to be expressed and as a result, BAF53a is targeted for repression by the miRNAs (Yoo et al., 2009). Mutating the miRNA binding sites on the BAF53a 3′ UTR leads to persistent BAF53a expression and reciprocal downregulation of BAF53b in β-Tubulin-III+ neurons, indicating that BAF53a antagonizes BAF53b gene activation. Prolonged expression of npBAF subunits, including SS18, is inhibitory to dendritogenesis, albeit being insufficient to reactivate cell cycle in postmitotic neurons (Staahl et al., 2013; Yoo et al., 2009). The SS18 gene also contains putative miR-9* and miR-124 binding sites, but it is unclear whether the same regulatory mechanism governs the switch from SS18 to CREST at this subunit position.
This conserved mechanism of neuronal differentiation is not only necessary but instructive for the neuronal fate, allowing reprogramming from an unrelated lineage to postmitotic induced neurons (iNs) (Yoo et al., 2011). Expressing miR-9* and miR-124 in fibroblasts induces a cell fate conversion to functional neurons. The miRNA-overexpressing fibroblasts exit the cell cycle within one week, and while the starting cells express BAF53a, the resulting iNs robustly express the nBAF-specific subunits, BAF53b, BAF45b/c and CREST (Yoo et al., 2011; Staahl et al., 2013), indicating that the nBAF complex is operational. The efficiency of conversion is enhanced when the neurogenic transcription factor NEUROD2 is expressed in combination with the miRNAs, demonstrating their functional synergy. Unlikely many other lineages which are specified by master transcription factors (Weintraub, 1993; Davis et al., 1987; Nutt et al., 1999), a master regulator transcription factor for the diverse neuronal fates has not yet emerged. Intriguingly, the two neurogenic miRNAs as well as the nBAF-specific subunits are found throughout the CNS and are exclusive to the neuronal lineage (Yoo et al., 2009; Wu et al., 2007). Given the instructive capacity, it is tempting to speculate that the miRNA-nBAF regulatory network defines a universal ground state of the neuronal chromatin, which then requires patterning by subtype-specific transcription factors for maturation.
BAF complexes in adult neurogenesis (caps)
After embryonic development, neurogenesis ceases in most regions of the CNS. The subependymal zone (SEZ) in the lateral walls of the lateral ventricles is one of the two neurogenic niches in the adult brain, and supplies new neurons to the olfactory bulb (OB) (Luskin, 1993; Lois and Alvarez-Buylla, 1994) as well as glia to the cortex and corpus callosum (Menn et al., 2006) throughout life. Adult NSCs have similar properties to embryonic NSCs and are thought to arise from the latter (Ahlenius et al., 2009). The precise composition of BAF complexes in adult NSCs is unknown, but Brg1 has been shown to suppress non-neuronal fates in this context while, intriguingly, not affecting self-renewal. The interaction between Pax6 and the Brg1-containing BAF complex is essential for specifying the neurogenic fate of adult NSCs in the SEZ (Ninkovic et al., 2013): ablation of either Pax6 or Brg1 diverts adult NSCs from the neuronal lineage to the ependymal or glial lineages. Gene expression profiling of Brg1-deleted SEZ and OB shows misregulation of genes involved in cell cycle, neurogenesis, axonogenesis and synaptic transmission. Surprisingly, Brm is unable to compensate for Brg1 in adult NSCs despite being expressed at high levels. The interaction of BAF with Pax6 seems functionally specific: forced neurogenesis by Pax6 expression in SEZ-derived neurospheres requires a catalytically active form of Brg1, whereas overexpression of another neurogenic transcription factor, Ngn2, efficiently generates neurons in the absence of Brg1 (Ninkovic et al., 2013). Therefore, the interaction between Pax6 and BAF, which also regulates embryonic neural progenitor division (Tuoc et al., 2013), may have been repurposed for adult neurogenesis in the SEZ.
BAF complexes in neurological disorders
Given the far-reaching contributions of BAF complexes during neural development and beyond, it is not surprising that mutations targeting their subunits perturb normal brain function. With the advent of next-generation sequencing, a host of BAF mutations have been discovered in human neurological disorders ranging from syndromic and non-syndromic intellectual disability to psychiatric conditions and neurodegenerative disease (Figure 2).
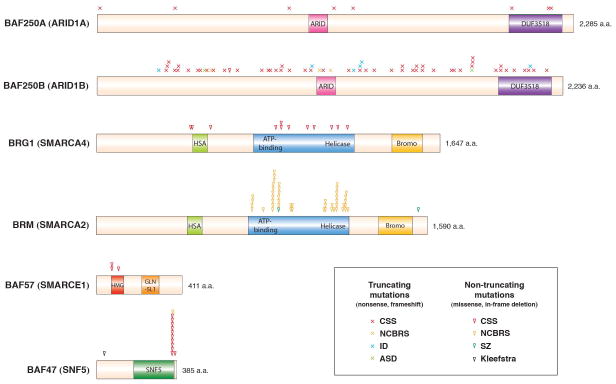
Figure 2. Most commonly mutated BAF subunits in human cognitive disorders.
Dominant de novo BAF subunit mutations have been found in Coffin-Siris syndrome (CSS), Nicolaides-Baraitser syndrome (NCBRS), sporadic intellectual disability (ID), autism spectrum disorder (ASD), schizophrenia (SZ) and Kleefstra syndrome. Mutations in BAF250A and BAF250B lead to truncated proteins, indicating that haploinsufficiency of these subunits causes disease. Mutations in the BRG1 and BRM ATPases as well as BAF57 and BAF47 are missense mutations or in-frame deletions, suggesting gain-of-function or dominant negative mechanisms. Microdeletions or translocations are not included. Data taken from the following studies: Hoyer et al. (2012); Kleefstra T. Am J Hum Genet (2012); Koga et al. (2009); Nord et al. (2011); Santen et al. (2012, 2013); Tsurusaki et al. (2012, 2013); Van Houdt et al. (2012); Wieczorek et al. (2013); Wolff et al. (2011).
Intellectual disability
It is estimated that between 2–3% of the general population have a form of intellectual disability (ID), and the vast majority of those are thought to have genetic causes (Ropers, 2010). Interestingly, mutations in BAF250B, the largest BAF subunit, have emerged as an important factor in non-familial ID. It has been known that rare de novo deletions or translocations involving the BAF250B (ARID1B) gene in chromosomal region 6q are associated with microcephaly, growth delay and frequent agenesis of the corpus callosum (AgCC) (Pirola et al., 1998; Narahara et al., 1991; Sukumar et al., 1999; Hopkin et al., 1997; Oliveira-Duarte et al., 1990; Rivas et al., 1986; Meng et al., 1992; Valtat et al., 1992; Nagamani et al., 2009; Backx et al., 2011). More recently, high-resolution molecular karyotyping and next-generation trio sequencing studies have identified additional chromosomal legions involving the BAF250B gene as well as truncating BAF250B mutations in sporadic ID patients with similar clinical features as above (Hoyer et al., 2012; Halgren et al., 2012); in one of the studies, inactivating BAF250B deletions were found in 8/887 (0.9%) of the patient cohort (Hoyer et al., 2012), making it the single most frequently mutated gene in idiopathic ID. All BAF250B legions are predicted to abolish wild-type protein expression; as outlined below, this is a recurrent theme in neurologic disorders and indicates a dosage-sensitive role of BAF250B in the CNS.
Coffin-Siris syndrome
A series of exome sequencing studies have uncovered dominant de novo BAF mutations in patients with Coffin-Siris syndrome (CSS), a rare congenital disorder with microcephaly, AgCC, moderate-to-profound ID, developmental delay, coarse facial features, hypoplasia of the fifth fingernails and/or toenails and multiple organ abnormalities (Tsurusaki et al., 2013, 2012; Santen et al., 2012, 2013; Wieczorek et al., 2013). In each of the large-scale studies, between 41–87% of the subjects were positive for BAF mutations. A combined total of 106 lesions have been reported in five subunits: BAF250A (8/106, 7.5%), BAF250B (72/106, 6.8%), BRG1 (12/106, 11.3%), BAF47 (12/106, 11.3%) and BAF57 (3/106, 2.8%)
Similar to in sporadic ID, all but one BAF250B mutations in CSS lead to truncation of the polypeptide (Figure 2); it is likely that unknown genetic factors contribute to the exact phenotypic outcome in patients with BAF250B mutations. All mutations in the homologous BAF250A gene are also nonsense or frameshift mutations, and are found in a smaller number of CSS patients who appear to present with greater disease severity than do BAF250B patients. The two homologous subunits, also known as ARID1A/B, contain the DNA-binding AT-rich interaction domains (ARID domains) and can have opposing roles in cell cycle through specific interactions with the E2F family of transcription factors (Nagl et al., 2007). Both homologs are found mutated in a wide variety of human malignancies, with BAF250A being more highly mutated than BAF250B (Kadoch et al., 2013; Ronan et al., 2013). Interestingly, a Drosophila RNAi screen for regulators of the neuroblast fate revealed that knockdown of Brahma, Moira and Osa (homologs of BRM, BAF47 and BAF250, respectively) results in the generation of extra neuroblasts at the expense of differentiated neurons (Neumüller et al., 2011); Osa was subsequently shown to control the progression of neural differentiation program and prevent tumorigenesis by regulating the number of progenitor divisions (Eroglu et al., 2014). The prevalence of BAF250A/B mutations prompts a greater understanding of how they function in the mammalian CNS.
CSS mutations in BRG1, the core ATPase, are either missense or in-frame deletions. They are located near the HSA domain and in the ATPase domain that contains an ATP-binding region and a helicase region (Figure 2). The absence of truncating mutations indicates that these mutations do not lead to complete loss of function similar to the null allele in Brg1 heterozygous knockout mice which display severe neural tube closure defects (Lessard et al., 2007). Rather, these CSS mutations may produce BRG1 that can be incorporated into BAF complexes which could then exhibit impaired or altered activity and act dominantly over the wild-type BRG1-containing complexes.
BAF47, also known as INI1 and SNF5, is a known tumor suppressor whose biallelic inactivation is responsible for ~100% of the cases of malignant rhabdoid tumor (MRT), an aggressive childhood tumor in soft tissues (Biegel et al., 2002). While the cancer mutations in BAF47 are inactivating and lead to loss of heterozygosity, de novo BAF47 mutations found in CSS patients are not truncating and act in a genetically dominant manner. The CSS mutations are exclusively located in the positively charged lysine or arginine residues in the hydrophilic C-terminus of BAF47 (Figure 2) (Tsurusaki et al., 2013, 2012; Santen et al., 2013). One recurrent in-frame deletion (p.Lys364del) is found in 9 unrelated patients, suggesting a critical role for this residue in the function of BAF47 within the CNS.
Most recently, de novo SOX11 mutations have been found in two CSS patients with mild phenotypes (Tsurusaki et al., 2014). SOX11 is a transcription factor that is targeted by the Pax6-BAF interaction in adult NSCs (Ninkovic et al., 2013). This suggests that a functional network emanating from BAF governs aspects of CSS pathology. It will be interesting to see if the host of CSS mutations can be experimentally linked to specific molecular and cellular phenotypes downstream of BAF malfunction, such as insufficient progenitor expansion (which could cause microcephaly), defects in the maturation of callosal projection neurons (which could cause AgCC) and misregulation of the Hedgehog signaling pathway (which could cause digit malformations).
Nicolaides-Baraitser syndrome
De novo missense mutations in the alternative ATPase subunit BRM are causal for Nicolaides-Baraitser syndrome (NCBRS), a developmental disorder closely related to CSS. NCBRS patients have severe ID, microcephaly, seizures, stunted growth and sparse hair (Nicolaides and Baraitser, 1993); they exhibit enlarged interphalangeal joints rather than hypoplasia of fifth fingernails seen in CSS patients (Sousa et al., 2009). Van Houdt and colleagues found BRM missense mutations in 36/44 (82%) patients with NCBRS (Van Houdt et al., 2012), and additional missense and in-frame deletion mutations were discovered by other groups (Figure 2) (Wolff et al., 2012; Wieczorek et al., 2013; Santen et al., 2013). Interestingly, Wieczorek and colleagues also found 3 truncating mutations in BAF250B and a C-terminal mutation in BAF47 (p.Arg366Cys) (Wieczorek et al., 2013). Similar to the BRG1 mutations in CSS, most BRM mutations are located in the conserved ATPase domain, particularly in the ATP-binding region and the helicase region; this suggests that these missense mutations alter the activity of the ATPase and lead to gain-of-function or dominant negative effects. Given the overall patterns of subunit mutations in CSS and NCBRS, the BAF47 mutation in one NCBRS patient is unusual and may reflect the extensive overlap of clinical features between the two disorders.
Autism spectrum disorder and schizophrenia
Autism spectrum disorder (ASD) is a developmental condition found in as much as 1% of all children. It is characterized by impaired social and language skills as well as repetitive behavior and narrow range of interests. The genetic etiology of ASD is highly heterogenous, with copy-number variations (CNVs) playing an important role (Szatmari et al., 2007; Glessner et al., 2009; Guilmatre et al., 2009; Pinto et al., 2008; Sebat et al., 2007; Weiss et al., 2008). Exome sequencing studies have begun to uncover a highly interconnected network of ASD risk factors centered around DYRK1A, CHD8 (a chromatin remodeling enzyme) and Wnt/β-catenin signaling (Neale et al., 2012; O’Roak et al., 2012). A small but significant number of BAF-related mutations have been found in non-familial ASD, including a splice site mutation in BAF170; one missense mutation each in BAF155, BAF180 and REST; and a truncating mutation and a microdeletion in BAF250B (Figure 2) (Nord et al., 2011; O’Roak et al., 2012; Neale et al., 2012).
Schizophrenia (SZ) is a mental disorder affecting 0.3–0.7% of the population during their lifetime (van Os and Kapur, 2009). Typically having a post-adolescent onset, SZ is characterized by paranoia, confused speech and thought, delusions, hallucinations and social withdrawal. In a study of single-nucleotide polymorphisms (SNPs) in SZ, Koga and colleagues identified a missense mutation and two intronic mutations in BRM (Figure 2) (Koga et al., 2009). The missense mutation (p.Asp1546Glu) lowers the nuclear localization efficiency of BRM. Brm−/− mice display impaired social interaction and prepulse inhibition, and the gene expression changes in their prefrontal cortex is correlated with those seen in the postmortem prefrontal cortices of SZ patients. In addition, a BRM-centered interaction network involves 8 other SZ-associated genes (Loe-Mie et al., 2010), supporting the role of BRM perturbation in SZ.
The BAF components implicated in ASD and SZ play vital roles in npBAF/nBAF complexes and their switching at mitotic exit; thus, the above mutations may adversely impact multiple stages of development. Notably, synaptic dysfunction is a hallmark of many psychiatric disorders (Zoghbi, 2003) and ASD/SZ-associated BAF mutations are consistent with the important role of the nBAF complexes in the maturation of neurites and synaptic plasticity (Wu et al., 2007; Vogel-Ciernia et al., 2013).
Amyotrophic lateral sclerosis
De novo mutations in the nBAF subunit CREST has been found in a trio sequencing study of sporadic amyotrophic lateral sclerosis (ALS) (Chesi et al., 2013). ALS is a fatal late-onset neurodegenerative disease in which specific death of motor neurons causes progressive paralysis and death (Rowland and Shneider, 2001). From 47 ALS patients, one missense mutation (p.Ile123Met) and one nonsense mutation (p.Gln388*) in CREST have been found. These mutations may warrant further investigation since CREST function is important for neuronal maturation (Aizawa et al., 2004; Wu et al., 2007; Staahl et al., 2013); intriguingly, the nonsense mutation lies in the C-terminal end of the protein required for binding CBP (Aizawa et al., 2004).
BAF-Polycomb opposition in human disease
Oncogenic transformation can be caused by the imbalance between the opposing functions of BAF and Polycomb, for instance, at the Ink4a locus in rhabdoid tumor (Wilson et al., 2010) and at the Sox2 locus in synovial sarcoma (Kadoch and Crabtree, 2013). It is possible that a direct mechanistic link also exists between the two chromatin regulators in human neurological disorders. To this end, it is worth noting that mutations in EZH2 cause Weaver’s syndrome (Gibson et al., 2012; Tatton-Brown et al., 2013; Tatton-brown et al., 2011), a developmental disorder which presents with macrocephaly, in contrast to the microcephalic phenotype seen in BAF-related neurodevelopmental disorders. Polycomb inhibits Wnt signaling to antagonize the neurogenic fate of neural precursors (Hirabayashi et al., 2009), while BAF complexes have been shown to interact directly with β-catenin to activate Wnt target genes in a colon carcinoma cell line (Barker et al., 2001). Thus, one of the rate-limiting regulatory mechanisms in neural development and homeostasis may be the opposition between BAF and Polycomb complexes.
Summary and outlook
Once thought to be simply permissive, mammalian BAF complexes have critical functions in multiple aspects of neural development. The specific BAF assemblies found in ES cells, neural progenitors and postmitotic neurons have non-redundant functions which stem from structural divergence within closely related homologous subunit families. The npBAF complex is essential for controlling the kinetics and mode of neural progenitor cell division; in particular, it regulates the generation of upper-layer cortical neurons which have co-emerged with the expansion of the SVZ in recent mammalian brain evolution (Aboitiz et al., 2003; Marin-Padilla, 1992). On the other hand, nBAF function is necessary for the mature phenotypes of postmitotic neurons such as the refinement of axons and dendrites, and for determining the finer subtypes of several classes of neurons including deep-layer cortical neurons and hippocampal neurons. Surprisingly, the conserved mechanism for transitioning from npBAF to nBAF is instructive for the neuronal fate even in unrelated lineages, and may be a universal determinant of the core neuronal identity. Furthermore, BAF is essential for the plasticity of the adult brain and contributes to hippocampal LTP as well as adult neurogenesis; thus, BAF complexes are critical not only for the initial wiring of neural circuitry but also in maintaining and reshaping the connections based on the experience of the organism. The large number of BAF mutations in cognitive disorders prompts a deeper inquiry into the role BAF in brain development and function, particularly with regard to subunits with frequent or recurrent patterns of mutation, such as BAF250B and BAF47.
The precise biophysical mechanism of ATP-dependent chromatin remodelers is not well understood; however, loosening heterochromatic loci across the genome appears to be a critical function of BAF. This is achieved at least in part by antagonizing the repressive Polycomb complexes and facilitating the removal of H3K27me3 – a conserved regulatory interaction which co-emerged with multicellularity. The accessibility thus created by BAF allows other nuclear factors that interact with BAF, such as STAT3, Pax6 and REST, to bind their genomic targets and regulate downstream genes in a context-dependent manner. The composite surfaces created by the assembly of cell type-specific subunit combinations may dictate the versatility of BAF interactions. Of note, BAF also cooperates with lineage-specifying transcription factors during the differentiation of oligodendrocytes and Schwann cells (Limpert et al., 2013; Marathe et al., 2013), which are the glial cells responsible for myelinating axons in the CNS and PNS, respectively.
While rodent studies have informed much of what we know about human neural development, the human brain has undergone tremendous evolution particularly in the neocortex, with a concomitant increase in the complexity of precursor-progeny relationships. For instance, a small subset of human GABAergic interneurons originates from precursors whose final mitotic division takes place in the dorsal telencephalon (Letinic et al., 2002). An even more striking difference in the human is a massive expansion of a novel population of RGs, termed outer RGs (oRGs), which lack the apical process of ventricular RGs (vRGs) and reside in the outser SVZ (oSVZ) (Hansen et al., 2010; Fietz et al., 2010; Wang et al., 2011). Although oRGs do not undergo the highly stereotyped interkinetic nuclear movement (INM) of vRGs during mitosis (Noctor et al., 2004), instead exhibiting mitotic somal translocation (MST) (LaMonica et al., 2013), the two RG populations display similar cycling kinetics. Understanding how BAF contributes to the regulation of this novel population of progenitors may shed light on the emergence of human-specific cognitive traits and their disruption in disease states.
Acknowledgments
The authors thank Miriam Y. Son for help with the figures, and members of the Crabtree lab for helpful discussions on this topic. E.Y.S. is a Walter V. and Idun Berry Postdoctoral Fellow. G.R.C. is funded by grants from the National Institutes of Health and the Howard Hughes Medical Institute.
Biographies
Esther Y. Son is a postdoctoral fellow in the Department of Pathology at Stanford University studying the role of chromatin regulation in neural development and disease.
Gerald R. Crabtree is a Professor of Pathology and Developmental Biology at Stanford University with a long-standing interest in gene regulation. His current research focuses on chromatin regulation in development and cancer.
Footnotes
The authors have no conflict of interest to declare.
References
- Aboitiz F, Morales D, Montiel J. The evolutionary origin of the mammalian isocortex: towards an integrated developmental and functional approach. Behav Brain Sci. 2003;26:535–552. doi: 10.1017/s0140525x03000128. discussion 552–585. [DOI] [PubMed] [Google Scholar]
- Ahlenius H, Visan V, Kokaia M, Lindvall O, Kokaia Z. Neural stem and progenitor cells retain their potential for proliferation and differentiation into functional neurons despite lower number in aged brain. J Neurosci. 2009;29:4408–4419. doi: 10.1523/JNEUROSCI.6003-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Hu S-C, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albu DI, VanValkenburgh J, Morin N, Califano D, Jenkins NA, Copeland NG, Liu P, Avram D. Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc Natl Acad Sci U S A. 2011;108:6211–6216. doi: 10.1073/pnas.1014304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, MacKlis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backx L, Seuntjens E, Devriendt K, Vermeesch J, Van Esch H. A balanced translocation t(6;14)(q25.3;q13.2) leading to reciprocal fusion transcripts in a patient with intellectual disability and agenesis of corpus callosum. Cytogenet Genome Res. 2011;132:135–143. doi: 10.1159/000321577. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglioli E, Andrés ME, Rose DW, Chenoweth JG, Rosenfeld MG, Anderson ME, Mandel G. REST repression of neuronal genes requires components of the hSWI. SNF complex J Biol Chem. 2002;277:41038–41045. doi: 10.1074/jbc.M205691200. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Biegel JA, Kalpana G, Knudsen ES, Tumors TR, Kalpana G, Packer RJ, Roberts CWM, Thiele CJ, Weissman B, Smith M. The Role of INI1 and the SWI/SNF Complex in the Development of Rhabdoid Tumors: Meeting Summary from the Workshop on Childhood Atypical Teratoid/Rhabdoid Tumors The Role of INI1 and the SWI/SNF Complex in the Development of Rhabdoid Tumors: Meetin; 2002. pp. 323–328. [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruijn DRH, Allander SV, van Dijk AHA, Willemse MP, Thijssen J, van Groningen JJM, Meltzer PS, van Kessel AG. The synovial-sarcoma-associated SS18-SSX2 fusion protein induces epigenetic gene (de)regulation. Cancer Res. 2006;66:9474–9482. doi: 10.1158/0008-5472.CAN-05-3726. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci U S A. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Chesi A, Staahl BT, Jovičić A, Couthouis J, Fasolino M, Raphael AR, Yamazaki T, Elias L, Polak M, Kelly C, Williams KL, Fifita JA, Maragakis NJ, Nicholson GA, King OD, Reed R, Crabtree GR, Blair IP, Glass JD, Gitler AD. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat Neurosci. 2013;16:851–5. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Katzoff A, Levitan D, Susswein AJ, Klein R, Valbrun M, Schwartz JH. Long-term memory requires polyADP-ribosylation. Science. 2004;304:1820–1822. doi: 10.1126/science.1096775. [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han J-J, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Duncan MK. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar aB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dorschner MO, Hawrylycz M, Humbert R, Wallace JC, Shafer A, Kawamoto J, Mack J, Hall R, Goldy J, Sabo PJ, Kohli A, Li Q, McArthur M, Stamatoyannopoulos JA. High-throughput localization of functional elements by quantitative chromatin profiling. Nat Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Enomoto T, Ohmoto M, Iwata T, Uno A, Saitou M, Yamaguchi T, Kominami R, Matsumoto I, Hirota J. Bcl11b/Ctip2 controls the differentiation of vomeronasal sensory neurons in mice. J Neurosci. 2011;31:10159–10173. doi: 10.1523/JNEUROSCI.1245-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu E, Burkard TR, Jiang Y, Saini N, Homem CCF, Reichert H, Knoblich JA. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell. 2014;156:1259–1273. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- Estivill-Torrus G, Pearson H, van Heyningen V, Price DJ, Rashbass P. Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development. 2002;129:455–466. doi: 10.1242/dev.129.2.455. [DOI] [PubMed] [Google Scholar]
- Fietz Sa, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, Huttner WB. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–9. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, Mungall AJ, Eydoux P, Babul-Hirji R, An J, Marra Ma, Chitayat D, Boycott KM, Weaver DD, Jones SJM. Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2012;90:110–8. doi: 10.1016/j.ajhg.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PMA, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SFA, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickstein M, May JG, Mercier BE. Corticopontine projection in the macaque: the distribution of labelled cortical cells after large injections of horseradish peroxidase in the pontine nuclei. J Comp Neurol. 1985;235:343–359. doi: 10.1002/cne.902350306. [DOI] [PubMed] [Google Scholar]
- Golonzhka O, Metzger D, Bornert J-M, Bay BK, Gross MK, Kioussi C, Leid M. Ctip2/Bcl11b controls ameloblast formation during mammalian odontogenesis. Proc Natl Acad Sci U S A. 2009;106:4278–4283. doi: 10.1073/pnas.0900568106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–88. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, Atouf F, Holdener BC, Mandel G, Kouzarides T. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca A-L, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit J-M, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frébourg T, Saugier Veber P, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren C, Kjaergaard S, Bak M, Hansen C, El-Schich Z, Anderson CM, Henriksen KF, Hjalgrim H, Kirchhoff M, Bijlsma EK, Nielsen M, den Hollander NS, Ruivenkamp CaL, Isidor B, Le Caignec C, Zannolli R, Mucciolo M, Renieri a, Mari F, Anderlid B-M, Andrieux J, Dieux a, Tommerup N, Bache I. Corpus callosum abnormalities, intellectual disability, speech impairment, and autism in patients with haploinsufficiency of ARID1B. Clin Genet. 2012;82:248–55. doi: 10.1111/j.1399-0004.2011.01755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendelman WJ. Neocortical Development. J Psychiatry Neurosci. 1991;16:233. [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb Limits the Neurogenic Competence of Neural Precursor Cells to Promote Astrogenic Fate Transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009a;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–13. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009b;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin RJ, Schorry E, Bofinger M, Milatovich A, Stern HJ, Jayne C, Saal HM. New insights into the phenotypes of 6q deletions. Am J Med Genet. 1997;70:377–386. [PubMed] [Google Scholar]
- Van Houdt JKJ, Nowakowska BA, Sousa SB, van Schaik BDC, Seuntjens E, Avonce N, Sifrim A, Abdul-Rahman Oa, van den Boogaard M-JH, Bottani A, Castori M, Cormier-Daire V, Deardorff Ma, Filges I, Fryer A, Fryns J-P, Gana S, Garavelli L, Gillessen-Kaesbach G, Hall BD, Horn D, Huylebroeck D, Klapecki J, Krajewska-Walasek M, Kuechler A, Lines Ma, Maas S, Macdermot KD, McKee S, Magee A, de Man Sa, Moreau Y, Morice-Picard F, Obersztyn E, Pilch J, Rosser E, Shannon N, Stolte-Dijkstra I, Van Dijck P, Vilain C, Vogels A, Wakeling E, Wieczorek D, Wilson L, Zuffardi O, van Kampen AHC, Devriendt K, Hennekam R, Vermeesch JR. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–9. S1. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- Hoyer J, Ekici AB, Endele S, Popp B, Zweier C, Wiesener A, Wohlleber E, Dufke A, Rossier E, Petsch C, Zweier M, Göhring I, Zink AM, Rappold G, Schröck E, Wieczorek D, Riess O, Engels H, Rauch A, Reis A. Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet. 2012;90:565–72. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. The Control of Dendrite Development. Neuron. 2003;40:229–242. doi: 10.1016/s0896-6273(03)00631-7. [DOI] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, Crabtree GR. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser MD, Aslanian A, Dong M-Q, Yates JR, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Killackey HP, Koralek KA, Chiaia NL, Rhodes RW. Laminar and areal differences in the origin of the subcortical projection neurons of the rat somatosensory cortex. J Comp Neurol. 1989;282:428–445. doi: 10.1002/cne.902820309. [DOI] [PubMed] [Google Scholar]
- Kim JK, Huh S, Choi H, Lee K, Shin D, Lee C, Nam J, Kim H, Chung H, Lee HANW, Park SD, Seong RHOH. Srg3, a Mouse Homolog of Yeast SWI3. Is Essential for Early Embryogenesis and Involved in Brain Development. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K, Iritani S, Itokawa M, Inada T, Iwata N, Ozaki N, Ujike H, Kunugi H, Sasaki T, Takahashi M, Watanabe Y, Someya T, Kakita A, Takahashi H, Nawa H, Muchardt C, Yaniv M, Arinami T. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet. 2009;18:2483–94. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag K-C, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- De la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific MicroRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- LaMonica BE, Lui JH, Hansen DV, Kriegstein AR. Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nat Commun. 2013;4:1665. doi: 10.1038/ncomms2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent BC, Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992;6:1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- Laurent BC, Treitel MA, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci U S A. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish Ja, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef Ia, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–15. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li W, Xiong Y, Shang C, Twu KY, Hang CT, Yang J, Han P, Lin C-Y, Lin C-J, Tsai F-C, Stankunas K, Meyer T, Bernstein D, Pan M, Chang CP. Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proc Natl Acad Sci U S A. 2013;110:1738–43. doi: 10.1073/pnas.1218072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Limpert AS, Bai S, Narayan M, Wu J, Yoon SO, Carter BD, Lu QR. NF-κB forms a complex with the chromatin remodeler BRG1 to regulate Schwann cell differentiation. J Neurosci. 2013;33:2388–97. doi: 10.1523/JNEUROSCI.3223-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loe-Mie Y, Lepagnol-Bestel A-M, Maussion G, Doron-Faigenboim A, Imbeaud S, Delacroix H, Aggerbeck L, Pupko T, Gorwood P, Simonneau M, Moalic JM. SMARCA2 and other genome-wide supported schizophrenia-associated genes: regulation by REST/NRSF, network organization and primate-specific evolution. Hum Mol Genet. 2010;19:2841–57. doi: 10.1093/hmg/ddq184. [DOI] [PubMed] [Google Scholar]
- Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong K-Y, Sung KW, Lee CWH, Zhao X-D, Chiu K-P, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei C-L, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze S-H, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 Promotes Neuronal Differentiation by Triggering Brain-Specific Alternative Pre-mRNA Splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe HG, Mehta G, Zhang X, Datar I, Mehrotra A, Yeung KC, de la Serna IL. SWI/SNF Enzymes Promote SOX10- Mediated Activation of Myelin Gene Expression. PLoS One. 2013:8. doi: 10.1371/journal.pone.0069037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Padilla M. Ontogenesis of the pyramidal cell of the mammalian neocortex and developmental cytoarchitectonics: A unifying theory. J Comp Neurol. 1992;321:223–240. doi: 10.1002/cne.903210205. [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MSH, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- Meng J, Fujita H, Nagahara N, Kashiwai A, Yoshioka Y, Funato M. Two patients with chromosome 6q terminal deletions with breakpoints at q24.3 and q25.3. Am J Med Genet. 1992;43:747–750. doi: 10.1002/ajmg.1320430419. [DOI] [PubMed] [Google Scholar]
- Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár Z, Cheung AFP. Towards the classification of subpopulations of layer V pyramidal projection neurons. Neurosci Res. 2006;55:105–115. doi: 10.1016/j.neures.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Muzio L, DiBenedetto B, Stoykova A, Boncinelli E, Gruss P, Mallamaci A. Conversion of cerebral cortex into basal ganglia in Emx2(−/−) Pax6(Sey/Sey) double-mutant mice. Nat Neurosci. 2002;5:737–745. doi: 10.1038/nn892. [DOI] [PubMed] [Google Scholar]
- Nagamani SCS, Erez A, Eng C, Ou Z, Chinault C, Workman L, Coldwell J, Stankiewicz P, Patel A, Lupski JR, Cheung SW. Interstitial deletion of 6q25.2-q25.3: a novel microdeletion syndrome associated with microcephaly, developmental delay, dysmorphic features and hearing loss. 2009 doi: 10.1038/ejhg.2008.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl NG, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narahara K, Tsuji K, Yokoyama Y, Namba H, Murakami M, Matsubara T, Kasai R, Fukushima Y, Seki T, Wakui K. Specification of small distal 6q deletions in two patients by gene dosage and in situ hybridization study of plasminogen and alpha-L-fucosidase 2. 1991. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin C-F, Stevens C, Wang L-S, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser R, Jabado O, Peralta Z, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y, Lewis L, Han Y, Voight BF, Lim E, Rossin E, Kirby A, Flannick J, Fromer M, Shakir K, Fennell T, Garimella K, Banks E, Poplin R, Gabriel S, DePristo M, Wimbish JR, Boone BE, Levy SE, Betancur C, Sunyaev S, Boerwinkle E, Buxbaum JD, Cook EH, Devlin B, Gibbs Ra, Roeder K, Schellenberg GD, Sutcliffe JS, Daly MJ. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller RA, Richter C, Fischer A, Novatchkova M, Neumüller KG, Knoblich JA. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 2011;8:580–593. doi: 10.1016/j.stem.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides P, Baraitser M. An unusual syndrome with mental retardation and sparse hair. 1993. [PubMed] [Google Scholar]
- Ninkovic J, Steiner-Mezzadri A, Jawerka M, Akinci U, Masserdotti G, Petricca S, Fischer J, Von Holst A, Beckers J, Lie CD, Petrik D, Miller E, Tang J, Wu J, Lefebvre V, Demmers J, Eisch A, Metzger D, Crabtree G, Irmler M, Poot R, Götz M. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13:403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Nord AS, Roeb W, Dickel DE, Walsh T, Kusenda M, O’Connor KL, Malhotra D, McCarthy SE, Stray SM, Taylor SM, Sebat J, King B, King M-C, McClellan JM. Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. Eur J Hum Genet. 2011;19:727–731. doi: 10.1038/ejhg.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink aG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–62. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson Da, Bernier R, Shendure J, Eichler EE. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CGA, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Duarte MH, Martelli-Soares LR, Sarquis-Cintra T, Machado ML, Lison MP. Distal monosomy of the long arm of chromosome 6 (6q25----6qter) inherited by maternal translocation t(6q;17q) 1990. [PubMed] [Google Scholar]
- Van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Pinto L, Mader MT, Irmler M, Gentilini M, Santoni F, Drechsel D, Blum R, Stahl R, Bulfone A, Malatesta P, Beckers J, Götz M. Prospective isolation of functionally distinct radial glial subtypes-Lineage and transcriptome analysis. Mol Cell Neurosci. 2008;38:15–42. doi: 10.1016/j.mcn.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Pirola B, Bortotto L, Giglio S, Piovan E, Janes A, Guerrini R, Zuffardi O. Agenesis of the corpus callosum with Probst bundles owing to haploinsufficiency for a gene in an 8 cM region of 6q25. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2007;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Martynoga BS, Zaki PA, Faedo A, Bulfone A, Hevner RF, West JD, Price DJ. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev Biol. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas F, Ruiz C, Rivera H, Möller M, Serrano-Lucas JI, Cantú JM. De novo del(6)(q25) associated with macular degeneration. 1986. [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–59. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, S-Jacques B, Lee SM, Flax JD, Snyder EY, McMahon AP. Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J Neurosci. 1999;19:8954–8965. doi: 10.1523/JNEUROSCI.19-20-08954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland LP, Shneider NA. Amyotrophic Lateral Sclerosis. N Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- Ryba NJP, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Santen GWE, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, Kant SG, Snoeck IN, Peeters EaJ, Hilhorst-Hofstee Y, Wessels MW, den Hollander NS, Ruivenkamp CaL, van Ommen G-JB, Breuning MH, den Dunnen JT, van Haeringen A, Kriek M. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012;44:379–80. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- Santen GWE, Aten E, Vulto-van Silfhout AT, Pottinger C, van Bon BWM, van Minderhout IJHM, Snowdowne R, van der Lans CaC, Boogaard M, Linssen MML, Vijfhuizen L, van der Wielen M, JR, Vollebregt MJE, Breuning MH, Kriek M, van Haeringen A, den Dunnen JT, Hoischen A, Clayton-Smith J, de Vries BBa, Hennekam RCM, van Belzen MJ. Coffin-Siris syndrome and the BAF complex: genotype-phenotype study in 63 patients. Hum Mutat. 2013;34:1519–28. doi: 10.1002/humu.22394. [DOI] [PubMed] [Google Scholar]
- Sawa H, Kouike H, Okano H. Components of the SWI/SNF complex are required for asymmetric cell division in C. elegans. Mol Cell. 2000;6:617–624. doi: 10.1016/s1097-2765(00)00060-5. [DOI] [PubMed] [Google Scholar]
- Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Filipe Pereira C, Fisher AG, Adams DJ, Flicek P, Crawford GE, LaFramboise T, Tesar P, Wei CL, Scacheri PC. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:1–15. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin L-M, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee Y-H, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King M-C, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Simon R, Brylka H, Schwegler H, Venkataramanappa S, Andratschke J, Wiegreffe C, Liu P, Fuchs E, Jenkins NA, Copeland NG, Birchmeier C, Britsch S. A dual function of Bcl11b/Ctip2 in hippocampal neurogenesis. EMBO J. 2012;31:2922–2936. doi: 10.1038/emboj.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SB, Abdul-Rahman OA, Bottani A, Cormier-Daire V, Fryer A, Gillessen-Kaesbach G, Horn D, Josifova D, Kuechler A, Lees M, MacDermot K, Magee A, Morice-Picard F, Rosser E, Sarkar A, Shannon N, Stolte-Dijkstra I, Verloes A, Wakeling E, Wilson L, Hennekam RCM. Nicolaides-Baraitser syndrome: Delineation of the phenotype. Am J Med Genet Part A. 2009;149:1628–1640. doi: 10.1002/ajmg.a.32956. [DOI] [PubMed] [Google Scholar]
- Staahl BT, Tang J, Wu W, Sun A, Gitler AD, Yoo AS, Crabtree GR. Kinetic analysis of npBAF to nBAF switching reveals exchange of SS18 with CREST and integration with neural developmental pathways. J Neurosci. 2013;33:10348–61. doi: 10.1523/JNEUROSCI.1258-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Stoykova A, Treichel D, Hallonet M, Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J Neurosci. 2000;20:8042–8050. doi: 10.1523/JNEUROSCI.20-21-08042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar S, Wang S, Hoang K, Vanchiere CM, England K, Fick R, Pagon B, Reddy KS. Subtle overlapping deletions in the terminal region of chromosome 6q24.2-q26: Three cases studied using FISH. Am J Med Genet. 1999;87:17–22. [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu X-Q, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu C-E, Rogé B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bölte S, Feineis-Matthews S, Herbrecht E, Schmötzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci aM, Kaufman TC, Kennison Ja. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–72. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tatton-brown K, Hanks S, Ruark E, Zachariou A, Duarte DV, Ramsay E, Snape K, Murray A, Elizabeth R. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127–1133. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Murray A, Hanks S, Douglas J, Armstrong R, Banka S, Bird LM, Clericuzio CL, Cormier-Daire V, Cushing T, Flinter F, Jacquemont ML, Joss S, Kinning E, Lynch SA, Magee A, Mcconnell V, Medeira A, Ozono K, Patton M, Rankin J, Shears D, Simon M, Splitt M, Strenger V, Stuurman K, Taylor C, Titheradge H, Van Maldergem L, Temple IK, Cole T, Seal S, Rahman N. Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am J Med Genet Part A. 2013;161:2972–2980. doi: 10.1002/ajmg.a.36229. [DOI] [PubMed] [Google Scholar]
- Tea JS, Luo L. The chromatin remodeling factor Bap55 functions through the TIP60 complex to regulate olfactory projection neuron dendrite targeting. Neural Dev. 2011;6:5. doi: 10.1186/1749-8104-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toresson H, Potter SS, Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 2000;127:4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Koshimizu E, Ohashi H, Phadke S, Kou I, Shiina M, Suzuki T, Okamoto N, Imamura S, Yamashita M, Watanabe S, Yoshiura K, Kodera H, Miyatake S, Nakashima M, Saitsu H, Ogata K, Ikegawa S, Miyake N, Matsumoto N. De novo SOX11 mutations cause Coffin-Siris syndrome. Nat Commun. 2014;5:4011. doi: 10.1038/ncomms5011. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Okamoto N, Ohashi H, Kosho T. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Okamoto N, Ohashi H, Mizuno S, Matsumoto N, Makita Y, Fukuda M, Isidor B, Perrier J, Aggarwal S, Dalal A, Al-Kindy a, Liebelt J, Mowat D, Nakashima M, Saitsu H, Miyake N. Coffin-Siris syndrome is a SWI/SNF complex disorder. Clin Genet. 2013:1–7. doi: 10.1111/cge.12225. [DOI] [PubMed] [Google Scholar]
- Tuoc T, Boretius S, Sansom S, Pitulescu ME, Frahm J, Livesey F, Stoykova A. Chromatin Regulation by BAF170 Controls Cerebral Cortical Size and Thickness. Dev Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Valtat C, Galliano D, Mettey R, Toutain A, Moraine C. Monosomy 6q: report on four new cases. 1992. [DOI] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel-Ciernia A, Matheos DP, Barrett RM, Kramár Ea, Azzawi S, Chen Y, Magnan CN, Zeller M, Sylvain A, Haettig J, Jia Y, Tran A, Dang R, Post RJ, Chabrier M, Babayan AH, Wu JI, Crabtree GR, Baldi P, Baram TZ, Lynch G, Wood Ma. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci. 2013;16:552–61. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai J-W, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–61. doi: 10.1038/nn.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg P, Flames N, Sawa H, Garriga G, Hobert O. The SWI/SNF chromatin remodeling complex selectively affects multiple aspects of serotonergic neuron differentiation. Genetics. 2013;194:189–98. doi: 10.1534/genetics.112.148742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The MyoD Family and Myogenesis: Redundancy, Networks, and Thresholds Minireview. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MAR, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu B-L, Daly MJ. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Whitford KL, Dijkhuizen P, Polleux F, Ghosh A. Molecular control of cortical dendrite development. Annu Rev Neurosci. 2002;25:127–149. doi: 10.1146/annurev.neuro.25.112701.142932. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Bögershausen N, Beleggia F, Steiner-Haldenstätt S, Pohl E, Li Y, Milz E, Martin M, Thiele H, Altmüller J, Alanay Y, Kayserili H, Klein-Hitpass L, Böhringer S, Wollstein A, Albrecht B, Boduroglu K, Caliebe A, Chrzanowska K, Cogulu O, Cristofoli F, Czeschik JC, Devriendt K, Dotti MT, Elcioglu N, Gener B, Goecke TO, Krajewska-Walasek M, Guillén-Navarro E, Hayek J, Houge G, Kilic E, Simsek-Kiper PÖ, López-González V, Kuechler A, Lyonnet S, Mari F, Marozza A, Mathieu Dramard M, Mikat B, Morin G, Morice-Picard F, Ozkinay F, Rauch A, Renieri A, Tinschert S, Utine GE, Vilain C, Vivarelli R, Zweier C, Nürnberg P, Rahmann S, Vermeesch J, Lüdecke H-J, Zeschnigk M, Wollnik B. A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum Mol Genet. 2013;22:5121–35. doi: 10.1093/hmg/ddt366. [DOI] [PubMed] [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho Y-J, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CWM. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–28. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise SP, Jones EG. Cells of origin and terminal distribution of descending projections of the rat somatic sensory cortex. J Comp Neurol. 1977;175:129–157. doi: 10.1002/cne.901750202. [DOI] [PubMed] [Google Scholar]
- Wolff D, Endele S, Azzarello-Burri S, Hoyer J, Zweier M, Schanze I, Schmitt B, Rauch A, Reis A, Zweier C. In-frame deletion and missense mutations of the c-terminal helicase domain of SMARCA2 in three patients with nicolaides-baraitser syndrome. Mol Syndromol. 2012;2:237–244. doi: 10.1159/000337323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave Ia, Qiu Z, Ghosh A, Graef Ia, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Li W, Shang C, Chen R, Han P, Yang J, Stankunas K, Wu B, Pan M, Zhou B, Longaker M, Chang CP. Brg1 Governs a Positive Feedback Circuit in the Hair Follicle for Tissue Regeneration and Repair. Dev Cell. 2013;25:169–181. doi: 10.1016/j.devcel.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19:120–6. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin- remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–31. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LMN, Mao M, Chan JR, Wu J, Lu QR. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–61. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- Zhan X, Shi X, Zhang Z, Chen Y, Wu JI. Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc Natl Acad Sci U S A. 2011;108:12758–12763. doi: 10.1073/pnas.1018510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–30. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]