Abstract
A bacterial community analysis, using a culture-independent method (polymerase chain reaction-denaturing gradient gel electrophoresis), detected 17 species of bacteria including species of the genera Tetragenococcus, Lactobacillus, Pediococcus, Weissella Halanaerobium, Clostridium, and Sphingomonas in a traditional salty-fermented fish paste known as pla-ra or pa-daek in Thailand and Laos, which is used as a storage-stable multi-purpose seasoning. The representative genus of lactic acid bacteria seemed to vary in the 10 products collected from Thailand and Laos. Tetragenococci were common in products from central Thailand and Vientiane in Laos which had salinities of not less than 11% and pH values ranging from 5.6 to 6.1. However, lactobacilli were common in products from northern Thailand which had the lowest salinities (8.3–8.6%) and pH values (4.5–4.8) of all the samples examined. Two Lactobacillus and one Tetragenococcus species were detected in one product from northeastern Thailand containing 10% salt. These results suggest that salinity in pla-ra/pa-daek is an important determinant of the representative genus of lactic acid bacteria such as, Tetragenococcus or Lactobacillus. Additionally, differences in the acidity between these two groups seemed to be related to the production of d-/l-lactic acid in the lactic acid bacteria in each product. This is the first study to report a correlation between bacterial community structure and taste components in pla-ra/pa-daek products from various regions. This scientific work on a traditional fermented food will be useful in helping local producers meet differing consumer preferences in various regions.
Keywords: fermented fish, pla-ra, pa-daek, lactic acid bacteria, bacterial community, PCR-DGGE
INTRODUCTION
The salty-fermented fish paste known as pla-ra or pa-daek in the Thai and Lao languages is popularly used as an all-purpose seasoning in Thai and Laotian cooking. It is produced using freshwater fish in central, northern, and northeastern Thailand, and Laos. For the preparation of pla-ra/pa-daek, the raw fish is de-scaled, eviscerated, and cleaned well with water, then mixed with salt and pickled with rice bran or roasted rice powder. The fermentation generally takes place at ambient temperatures in the tropical climate for at least 6 months [1]. As the use of a starter culture is not common for pla-ra/pa-daek production, fermentation occurs spontaneously with microbes in the raw materials and the production environment. The fermented products are usually stored without refrigeration while being used and can last for a year or longer. It is empirically believed that a longer fermentation makes the product taste better. This shows the benefits of a traditional fermentation technique that furnishes a seasonally available indigenous food resource with additional values such as palatability and a long shelf life. Currently, eating uncooked freshwater fish products including pla-ra/pa-daek is not advisable because of the health risks from parasites [2, 3], therefore it is generally cooked before being eaten.
Pla-ra/pa-daek is often made and consumed at home, particularly in rural areas, or by relatively small-scale manufacturers for local consumption [1]. Consumers often think that the taste and flavor of pla-ra/pa-daek products vary from place to place. Factors producing such differences between products are of scientific interest to those who produce fermented products to meet consumer preferences as well as to those who wish to preserve the diversity of a regional food culture. Seventeen species of lactic acid bacteria, including species of the genera Lactobacillus (7 species), Weissella (3 species), Pediococcus (2 species), Enterococcus (2 species), Tetragenococcus (1 species), Aerococcus (1 species), and Leuconostoc (1 species) have been isolated by culture-based methods from Thai pla-ra products [4,5,6,7]. This suggests that these bacteria have an important role in determining the palatability and keeping quality. However, less attention has been paid to possible variations in bacterial community structures between varieties of pla-ra/pa-daek products from various regions, and how this is related to product qualities.
During the last decade, polymerase chain reaction-denaturant gradient gel electrophoresis (PCR-DGGE) has been extensively used as a culture-independent approach for studying microbial communities in a wide variety of fermented foods around the world [8]. Most recently, the time-dependent development of a bacterial community consisting of six species of lactic acid bacteria, including species of the genera Lactococcus (2 species), Weissella (2 species), Pediococcus (1 species) and Enterococcus (1 species), and other bacteria including species of the genera Staphylococcus (3 species), Macrococcus (1 species), Clostridium (1species), Flavobacterium (1 species) and Plesiomonas (1 spcies) were shown during the production of pa-som, a short-term (up to 4 days) fermented, freshwater fish product from Laos [9]. By using this technique, microbial communities in food samples of interest can be visualized and compared with each other in a single gel image and assessed at the species level regardless of the ability of each bacterium to be cultured in synthetic growth media.
In the present study, the bacterial communities in pla-ra/pa-daek products collected from central, northern, northeastern Thailand, and Vientiane in Laos were investigated using PCR-DGGE. Additionally, the relationship between the lactic acid bacteria community profiles and the product properties, salinity and pH, and content of d-/l-lactic acid was also examined and discussed.
MATERIALS AND METHODS
Sample collection
Pla-ra products made in the Suphanburi, Singburi, and Sukhothai provinces of central, northern, and northeastern Thailand, were obtained from markets in Bangkok, Nakhon Ratchasima, and Khon Kaen in Thailand, respectively. Pa-daek products made in the Vientiane province of Laos were obtained from local markets in the Vientiane capital area. The information about production region, fish materials, and fermentation period for each product was provided by the sellers (Table 1). The scientific name of each fish was assigned, if available, by referring to FishBase (http://www.fishbase.org/search.php). The samples were collected from May 29th to June 4th in 2012. After purchase at the markets, the samples were cold-stored in a refrigerated container and refrigerators to minimize microbial activity. All the samples were shipped by air in the same refrigerated container to the laboratory in Tsukuba, Japan on June 7th, 2012, following the storage in a freezer at −20°C. The samples were stored in the freezer until analysis.
Table 1. Production region, fish material, and fermentaion period of products used in this study.
| Sample number | Province | Region | Local name of fish materials used with academic names in parenthesis* | fermentation period |
| 1 | Suphanburi | Central Thailand | Pla chon (Channa striata) | 1 year |
| 2 | Singburi | Central Thailand | Pla ka di (Trichopodus trichopterus) and other small-sized fish | 1 year |
| 3 | Sukhothai | Northern Thailand | Mixture of small-sized fish | 1 year |
| 4 | Sukhothai | Northern Thailand | Mixture of small-sized fish | 1 year |
| 5 | Sukhothai | Northern Thailand | Pla ka di (Trichopodus trichopterus) | 1 year |
| 6 | Khon Kaen | Northeastern Thailand | Mixture of small-sized fish | 1 year |
| 7 | Vientiane | Laos | Mixture of small-sized fish | 10 months |
| 8 | Vientiane | Laos | Pa keng (Cirrhinus molitorella) | 1 year |
| 9 | Vientiane | Laos | Mixture of small-sized fish | 1 year |
| 10 | Vientiane | Laos | Mixture of small-sized fish | 1 year |
* “small-sized” approximately 10 cm or smaller.
Bacterial community analysis
The bacterial 16S rRNA gene for DGGE analysis was amplified by nested PCR as described by Kim et al [10]. DNA for the PCR template was extracted from 1 g of each pla-ra/pa-daek sample using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). The nearly complete 16S rRNA gene was first amplified using 5 µL of DNA extract by PCR using Takara Ex Taq (Takara Bio Inc., Shiga, Japan) with the pair of primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGCTACCTTGTTACGACTT-3′) in a 50-μl reaction volume. The resulting product was diluted 100-fold, and 2 µL of the diluent was subjected to a second PCR using the pair of primers 338fGC (5′-CGCCCGCCGCGCGCGGC GGGCGGGGCGGGGGCACGGGGG GACTCCTACGGGAGGCAGCAG-3′) and 518r (5′-ATTACCGCGGCTGCTGG-3′) to amplify the V3 region of the 16S rRNA gene. The PCR products (10 µL) were electrophoresed using a DCode Universal Mutation Detection System (Bio-Rad Laboratories, Hercules, CA, USA) at a constant voltage of 50 V in 1×Tris-acetate-EDTA buffer (40 mM Tris-acetate, 1 mM EDTA) for 18 h at 58°C using a polyacrylamide gel consisting of 7% (w/v) acrylamide-bisacrylamide (37.5:1) with a denaturing gradient in the range of 40–70% denaturant (100% denaturation corresponds to 7 M urea and 40% (v/v) formamide). After electrophoresis, the gels were stained with GelRed (Biotium, Hayward, CA, USA) and the DNA fragments were visualized by 254-nm ultraviolet light and photographed. The brightness and contrast of the DGGE images were optimized by Photoscape v3.6.4 software (http://www.photoscape.org/). The reproducibility of the band patterns of PCR-DGGE analysis observed in the present study was confirmed by a repeat experiment starting from PCR (data not shown). The DNA bands selected for nucleotide sequence analysis were excised from the DGGE gel under a 500-nm light-emitting diode. The DNA in each excised gel was eluted in sterilized water and subjected to PCR using the pair of primers, 338fGC and 518r. The purity of the PCR products was confirmed by DGGE as described above. The PCR products were purified using a FastGene Gel/PCR Extraction Kit (Nippon Genetics Co., Ltd., Tokyo, Japan). The nucleotide sequences of the purified PCR products were determined by the laboratory of Eurofins Genomics Co., Ltd. (Tokyo, Japan) using the primer, 338f (5′-ACTCCTACGGGAGGCAGCAG-3′). The nucleotide sequence homology search of each purified PCR product was performed using the blastn search tool against the 16S ribosomal RNA sequences database in the NCBI (National Center for Biotechnology Information (http://www.ncbi.nlm.gov/).
Measurements of salt concentration, pH, and d-/l-lactic acid contents
One gram of each sample was mixed vigorously in 10 mL of sterilized water, followed by centrifugation at 15,000 × g for 10 min at 4°C. The supernatant was collected and used for the analysis. The salt concentration and pH were measured three times with LAQUA twin compact salt and pH meters, respectively (Horiba Ltd., Kyoto, Japan). The lactic acid content was measured three times using a d-/l-lactic acid enzymatic test kit (R-Biopharm AG, Darmstadt, Germany).
RESULTS
Bacterial community analysis by PCR-DGGE
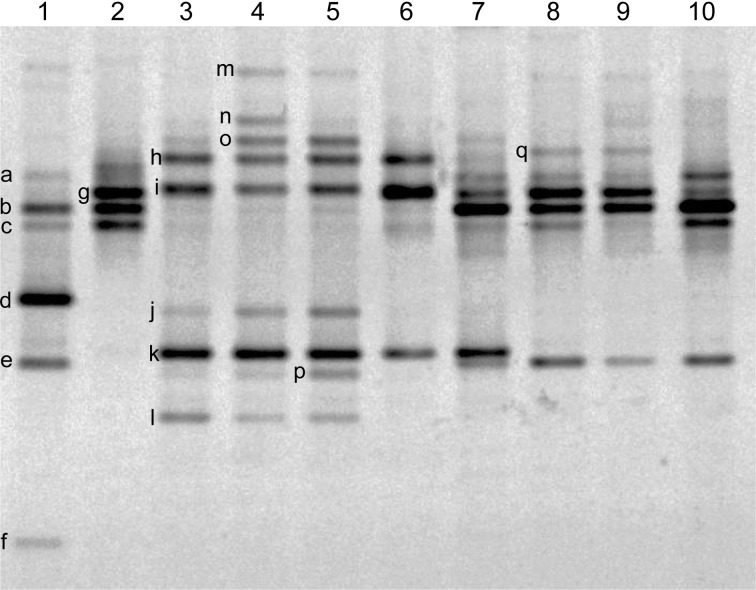
Because the mobility of each DNA band in the DGGE gel is determined by the sequence-dependent partial denaturation of the double strand at the specific denaturant concentration in the gel, the DNA bands exhibiting identical mobility between samples in the gel can be deduced to have been derived from genetically identical or closely related bacterial species. Moreover, in the present study, some DNA bands in the same mobility group were additionally isolated and subjected to nucleotide sequence analysis to confirm their identities (Table 3). As shown in Fig. 1, and Tables 2 and 3, each product appeared to contain two to eight types of lactic acid bacteria. In particular, two types of Tetragenococcus muriaticus (bands “a” and “b”) were detected in a product made in central Thailand (lane 1). In addition, band “a” was detected in products from Laos (lanes 7, 8, and 10). Band “b” was also detected in the other product from central Thailand (lane 2) as well as those from Laos (lanes 7 to 10). In the two bands of Tetragenococcus halophilus detected (bands “g” and “q”), band “g” was common in the same product areas (central Thailand and Laos) as the T. muriaticus species (band “b”), except for the sample 1. Band “g” was also detected in the product made in northeastern Thailand (lane 6). Band “q” appeared relatively weak and was specific to products from Laos (lanes 8 and 9). Bands “m” and “n” corresponded to Weissella paramesenteroides and Pediococcus argentinicus, respectively. It was obvious that Lactobacillus species such as Lactobacillus futsaii (band “o”), Lactobacillus acidipiscis (bands “h” and “i”), and Lactobacillus rennini (bands “j”, “k”, “l” and “p”) were common to the products made in northern Thailand (lanes 3, 4, and 5). Variants of the L. acidipiscis and L. rennini, represented by bands “h” and “k”, respectively, were also detected in the product made in northeastern Thailand (lane 6). Additionally, a product from Laos (lane 7) contained a L. rennini species (band “k”).
Table 3. Salinity, pH, and d-/l-lactic acid content.
| Sample number | Salinity (%) | pH | d-lactic acid | l-lactic acid | Total lactic acid | DGGE bands detetcted |
| 1 | 12.3 | 5.7 | 0.02 | 0.86 | 0.88 | a*, b*, c*, d*, e*, f* |
| 2 | 11.3 | 5.6 | 0.18 | 0.29 | 0.47 | b*, c*, g* |
| 3 | 8.3 | 4.8 | 1.22 | 1.22 | 2.44 | h, i, j, k*, l |
| 4 | 8.4 | 4.6 | 1.58 | 1.49 | 3.08 | h*, i, j, k, l, m*, n*, o* |
| 5 | 8.6 | 4.5 | 1.13 | 1.46 | 2.58 | h, i*, j*, k*, l*, m, o, p* |
| 6 | 10.3 | 4.9 | 0.47 | 1.22 | 1.69 | h*, g*, k |
| 7 | 15.3 | 5.7 | 0.04 | 0.58 | 0.62 | a*, b*, e, g*, k* |
| 8 | 20.3 | 5.7 | 0.16 | 0.37 | 0.53 | a*, b, c*, e, g*, q* |
| 9 | 16.7 | 5.9 | 0.20 | 0.55 | 0.75 | b, e*, g*, q |
| 10 | 18.3 | 6.1 | 0.18 | 0.22 | 0.40 | a, b, c, e, g |
Values shown are the means of three measurements. pH was measured in one tenth diluted samples. d-/l-lactic acid contents are showned as g/100 g sample. The bands that were isolated and subjected to nucleotide sequence analysis are indicated by asterisks (*).
Fig. 1.
PCR-DGGE profiles of the bacterial communities in pla-ra and pa-daek. The PCR products from samples 1 to 10 presented in Table 1 are shown in lanes 1 to 10, respectively. The representative DGGE band in each mobility group is indicated by lower-case letters. The nucleotide sequence of the bands indicated in Table 3 was subjected to a homology search against a 16S rRNA sequence database.
Table 2. Homology search results for the DNA fragments from selected DGGE bands.
| Band | Highest match(es) | Accession number | Identity (%) |
| a | Tetragenococcus muriaticus strain NBRC 100499 | NR113915.1 | 99 |
| Tetragenococcus muriaticus strain LMG 18498 | NR116418.1 | 99 | |
| Tetragenococcus muriaticus strain X-1 | NR025887.1 | 99 | |
| b | Tetragenococcus muriaticus strain NBRC 100499 | NR113915.1 | 100 |
| Tetragenococcus muriaticus strain LMG 18498 | NR116418.1 | 100 | |
| Tetragenococcus muriaticus strain X-1 | NR025887.1 | 100 | |
| c | Halanaerobium fermentans strain R-9 | NR024715.1 | 100 |
| d | Clostridium haemolyticum strain JCM 1402 | NR113381.1 | 100 |
| Clostridium haemolyticum strain DSM 5565 | NR117767.1 | 100 | |
| Clostridium haemolyticum strain ATCC 9650 | NR024749.1 | 100 | |
| Clostridium haemolyticum strain DSM 5565 | NR119281.1 | 100 | |
| e* | Sphingomonas roseiflava strain MK341 | NR117716.1 | 100 |
| f | Clostridium drakei strain FP | NR114863.1 | 100 |
| Clostridium drakei strain SL1 | NR044942.1 | 100 | |
| Clostridium carboxidivorans strain P7 | NR104768.1 | 100 | |
| Clostridium vincentii strain DSM 10228 | NR026336.1 | 100 | |
| Clostridium scatologenes strain ATCC 25775 | NR118727.1 | 100 | |
| g | Tetragenococcus halophilus subsp. flandriensis strain T5 | NR108208.1 | 100 |
| h | Lactobacillus acidipiscis strain NBRC 102163 | NR112693.1 | 99 |
| Lactobacillus acidipiscis strain FS60-1 | NR024718.1 | 99 | |
| i | Lactobacillus acidipiscis strain NBRC 102163 | NR112693.1 | 100 |
| j | Lactobacillus rennini strain CECT 5922 | NR042195.1 | 99 |
| k | Lactobacillus rennini strain CECT 5922 | NR042195.1 | 100 |
| l | Lactobacillus rennini strain CECT 5922 | NR042195.1 | 99 |
| m | Weissella paramesenteroides | NR044759.1 | 100 |
| Weissella paramesenteroides | NR040815.1 | 100 | |
| Weissella hellenica strain NBRC 15553 | NR113775.1 | 100 | |
| Weissella hellenica strain NCFB 2973 | NR119224.1 | 100 | |
| n | Pediococcus pentosaceus ATCC 25745 strain | NR075052.1 | 100 |
| Pediococcus pentosaceus strain DSM 20336 | NR042058.1 | 100 | |
| Pediococcus argentinicus strain CRL 776 | NR042623.1 | 100 | |
| o | Lactobacillus futsaii strain YM 0097 | NR117973.1 | 100 |
| Lactobacillus farciminis strain BCRC 14043 | NR114398.1 | 100 | |
| Lactobacillus heilongjiangensis strain S4-3 | NR109370.1 | 100 | |
| p | Lactobacillus rennini strain CECT 5922 | NR042195.1 | 99 |
| q | Tetragenococcus halophilus strain JCM 5888 | NR115655.1 | 100 |
*Another 33 Sphingomonas, 15 Sphingobium, 9 Sphingopyxis, 2 Erythrobacter, 1 Altererythrobacter, 1 Novosphingobium and 1 Stakelama species were matched with 100% identity (data not shown).
Besides the lactic acid bacteria mentioned above, two Clostridium species (bands “d” and “f”) were found only in a product from central Thailand (lane 1). A band corresponding to Sphingobium or a related species (band “e”) was detected in a product from central Thailand (lane 1) as well as from Laos (lanes 7 to 10). The nucleotide sequence of this DNA band matched to the 16S rRNA genes of more than 60 types of bacteria deposited in the database with 100% identity. Band “c”, corresponding to Haloanaerobium fermentans, was common to products from central Thailand and Laos (lanes 1, 2, 8, and 10).
Salt concentration, pH, and d-/l-lactic acid content
The salinity, pH, and d-/l-lactic acid content of the pla-ra/pa-daek samples are presented in Table 3. Among the samples examined, the products from northern Thailand (samples 3, 4 and 5) contained the lowest levels of salt (8.3–8.6%), while the products from the other regions contained more than 10% salt. The salinity in products from Laos (samples 7 to 10) was relatively higher (15–20%) than those from the other places. The pH value of the products from northern (samples 3, 4, and 5) and northeastern (sample 6) Thailand was less than 5.0, the former exhibiting the lowest pH levels (4.5–4.8). The products from the other regions exhibited pH values of 5.6–6.1.
In the present study, the d-/l-lactic acid content in the samples was measured, because the d-/l ratio in each sample should be related to the characteristics of the lactic acid bacteria community. The highest levels of total lactic acid content (approximately 2.4–3.0 g/100 g of sample) were observed in the products from northern Thailand (samples 3, 4, and 5) and the contents of d- and l-lactic acids were nearly identical in each product. The product from northeastern Thailand (sample 6) contained l-lactic acid at a similar level to the northern Thailand products, but the d-lactic acid content was less than half. In samples from central Thailand (samples 1 and 2) and Laos (samples 7 to 10), the total lactic acid contents varied from 0.40 to 0.88 g/100 g of sample. The d-lactic acid content in these products were noticeably lower than those in products from northern Thailand.
DISCUSSION
In the present study, we first used a culture-independent PCR-DGGE technique to assess the bacteria community structure of pla-ra/pa-daek, which is one of the most frequently used fermented seasonings, particularly in northeastern Thailand and Laos. Through examining the differences in their 16S rRNA gene sequences, 13 species of lactic acid bacteria, including species of the genera Tetragenococcus, Pediococcus, Weissella, and Lactobacillus were detected in the 10 pla-ra/pa-daek products collected, showing the variety and importance of lactic acid fermentation for the production of pla-ra/pa-daek. Other than lactic acid bacteria, 4 species of bacteria including the species of genera Halanaerobium, Clostridium, and Sphingomonas were detected. The H. fermentas, and Sphingomonas or related species were commonly detected in the products exhibiting relatively high salinity. On the other hand, clostridia were detected in only one sample in the present study, suggesting this species might not be common in pla-ra/pa-daek products, although further comprehensive and quantitative analysis will be necessary to fully confirm this. Compared with previous culture-based isolations from Thai pla-ra, in this study, T. muriaticus and L. rennini were newly found by PCR-DGGE analysis in multiple samples, showing the benefit of this culture-independent method in the exploration of the diversity of lactic acid bacteria profiles and the comparison of pla-ra/pa-daek products collected from various regions. Additionally, two to four sequence variations were found in the DGGE bands identified as T. muriaticus (Fig.1, bands “a” and “b”), T. halophilus (bands “g” and “q”), L. acidipiscis (bands “h” and “I”), and L. rennini (bands “j”, “k”, “l”, and “p”). Such variations might suggest that the lactic acid bacteria profiles in pla-ra/pa-daek could be characterized at the strain level and conserved between products in some cases, although further assessment at the genomic level is necessary to fully confirm the species diversity.
It is noteworthy that the pla-ra/pa-daek samples examined in the present study could be classified into two groups by the representative genus of lactic acid bacteria, such as Lactobacillus, or Tetragenococcus, regardless of the fish material used. A salt concentration of approximately 10% might be the dividing point between these two groups, because both Lactobacillus and Tetragenococcus species were detected in the product from northeastern Thailand containing 10% salt. Additionally, the lactic acid bacteria profile appeared to determine the d-/l-lactic acid ratio in each product: lactobacilli are known to produce d- and/or l-lactic acid, while tetragenococci produce only the l-form [11]. The small amount of d-lactic acid detected in the samples where tetragenococci were mainly detected by PCR-DGGE might have been produced by bacteria other than lactic acid bacteria. Alternatively, the population of such d-lactic acid-producing bacteria in these products might be below the detection limit of the PCR-DGGE used in this study, or detectable only at the beginning and/or the middle phase of fermentation.
Tetragenococci appeared to be the representative lactic acid bacteria in products containing more than 11% salt. In such cases, T. muriaticus and halophilus coexisted except in the product from central Thailand (Fig. 1, lane 1). Both species are known to be associated with habitats rich in salt and protein, and play an important role in the halophilic fermentation process [11]. It has been reported that the growth rate and lactic acid production of T. muriaticus in high salinity (23% NaCl) conditions were higher than those in T. halophilus, but they were severely affected by low pH (5.8) conditions compared with T. halophilus [12]. Thus, the coexistence of these two halophilic species might be a reasonable assumption for starting and continuing the lactic acid fermentation during the pla-ra/pa-daek production in adapting to the high salinity as well as to changing pH conditions over time. A strictly anaerobic and moderately halophilic bacterium, H. fermentas, previously reported to coexist with T. muriaticus and T. halophilus in salt-preserved food, such as fermented puffer fish ovaries [13,14,15], was also detected in the present study. The possible contribution of this bacterium to the production of salty-fermented foods is therefore of interest. Sphingomonas or related species was also commonly detected in samples exhibiting relatively high salinity. Because the nucleotide sequence of the corresponding band (e) matched the 16S rRNA genes of more than 60 types of bacteria, isolation of this bacterium and its detailed characterization will be required to understand the role of this bacterium in the fermentation.
Although the salinity of the products from northern Thailand was lower than those from the other regions, the acidic conditions (pH 4.5–4.8) presumably created by the markedly higher levels of d- and l-lactic acid may be advantageous for preventing the growth of food-spoilage bacteria. The growth of lactobacilli in these samples might occur selectively depending on the fermentation methods, particularly under the favorable conditions of relatively low salinity. Indeed, according to previous reports, the type strains of lactobacilli, such as L. acidipiscis FS60-1T [16], L. rennini CECT 5922T [17], and L. futsaii YM0097T [18], are viable in the ranges of salinity (8.3–8.6%) and pH (4.5–4.8) observed in the products from northern Thailand.
For the manufacture of cheddar cheese, it has been proposed that the glutamate dehydrogenase activity of lactobacilli should be considered for use as adjuncts in the ripening process, because the α-keto-glutarate formed by glutamate dehydrogenase activity would enhance the amino acid catabolism of the lactobacilli, thereby generating amino acid-related aroma compounds which would improve flavor development during maturation [19]. The occurrence and degree of glutamate dehydrogenase activity has been shown to be dependent on the strain of lactobacilli [20]. Its viability during cheese ripening is also important for the intensification and acceleration of aroma and flavor formation [20]. Thus, it should be interesting to assess the possible involvement of glutamate dehydrogenase activity of varieties of lactic acid bacteria, such as lactobacilli and tetragenococci in the formation of aroma compounds in pla-ra/pa-daek products, as it might be related to the regional characteristics of flavor and taste.
Since ancient times, fermentation methods for pla-ra/pa-daek have been developed to preserve seasonally available freshwater fish resources in the production areas without any microbial knowledge. Fermentation can give value to small-sized or juvenile fish that are often worth little in a local fresh fish market. A precise elucidation of such traditional techniques using recent molecular microbiology and analytical techniques should be of use in helping local producers to improve and/or maintain their product quality. For example, product improvement could be achieved by monitoring the initial salinity, or possibly using the representative lactic acid bacteria as a fermentation starter so that the characteristic lactic acid bacteria communities develop steadily, leading to the creation of tastes and flavors that can meet the differing consumer preferences of various regions. Further microbial profiling of various pla-ra/pa-daek products in Thailand and Laos, using a combination of culture-independent analysis and a statistical approach, could reveal the regional characteristics and their link to product quality as in the cases of the production of traditional water-buffalo mozzarella cheese [21], and the fermented dairy products of Mongolia [22].
The results of the present study are encouraging for the use of PCR-DGGE analysis as a monitoring technique for the time-dependent development of bacteria community structure during pla-ra/pa-daek fermentation with the different initial salinities used in various regions. This should offer local producers and consumers important scientific evidence for understanding the traditionally based production method and fermentation period. It will also be useful for improving the quality of the traditional fermented products and increasing manufacturing productivity. A laboratory fermentation trial based on local traditional recipes is now in progress.
Acknowledgments
We would like to thank Dr. Gassinee Trakoontivakorn and Ms. Plernchai Tangkanakul of IFRPD (Kasetsart University, Bangkok, Thailand), Mr. Bounsong Vongvichith and Ms. Vonhnida Bounlaphan of the Living Aquatic Resources Research Center (LARReC) (Vientiane, Lao PDR) and Dr. Shinsuke Morioka of JIRCAS (Tsukuba, Japan) for their support in collecting the pla-ra and pa-daek samples.
REFERENCES
- 1.Ishige N, Ruddle K. 1987. Gyosho in Southeast Asia— a study of fermented aquatic products (5). Bull Nat Mus Ethol 12: 235–312. [Google Scholar]
- 2.Wongba N, Thaewnongiew K, Phathee K, Laithavewat L, Duangsong R, Promthet S, Tangsawad S. 2011. Liver fluke prevention and control in the northeast of Thailand through action research. Asian Pac J Cancer Prev 12: 1367–1370. [PubMed] [Google Scholar]
- 3.Xayaseng V, Phongluxa K, van Eeuwijk P, Akkhavong K, Odermatt P. 2013. Raw fish consumption in liver fluke endemic areas in rural southern Laos. Acta Trop 127: 105–111. [DOI] [PubMed] [Google Scholar]
- 4.Miyashita M, Yukphan P, Chaipitakchonlatarn W, Malimas T, Sugimoto M, Yoshino M, Potacharoen W, Tanasupawat S, Nakagawa Y, Kirtikara K, Tanticharoen M, Suzuki K. 2012. 16S rRNA gene sequence analysis of lactic acid bacteria isolated from fermented foods in Thailand. Microbiol Cult Collect 28: 1–9. [Google Scholar]
- 5.Tanasupawat S, Komagata K. 1995. Lactic acid bacteria in fermented foods in Thailand. World J Microbiol Biotechnol 11: 253–256. [DOI] [PubMed] [Google Scholar]
- 6.Tanasupawat S, Okada S, Komagata K. 1998. Lactic acid bacteria found in fermented fish in Thailand. J Gen Appl Microbiol 44: 193–200. [DOI] [PubMed] [Google Scholar]
- 7.Tanasupawat S. 2009. Thai lactic acid bacteria: diversity and applications. SWU Sci. J. 25: 1–13. [Google Scholar]
- 8.Cocolin L, Alessandria V, Dolci P, Gorra R, Rantsiou K. 2013. Culture independent methods to assess the diversity and dynamics of microbiota during food fermentation. Int J Food Microbiol 167: 29–43. [DOI] [PubMed] [Google Scholar]
- 9.Marui J, Boulom S, Panthavee W, Momma M, Kusumoto K, Nakahara K, Saito M. 2014. Culture-independent analysis of the bacterial community during fermentation of pa-som, a traditional fermented fish product in Laos. Fish Sci (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TW, Lee JH, Kim SE, Park MH, Chang HC, Kim HY. 2009. Analysis of microbial communities in doenjang, a Korean fermented soybean paste, using nested PCR-denaturing gradient gel electrophoresis. Int J Food Microbiol 131: 265–271. [DOI] [PubMed] [Google Scholar]
- 11.Lahtinen S, Ouwehand AC, Salminen S, Von Wright A. (eds). 2012. Lactic Acid Bacteria, Microbiological and Functional Aspects, 4th ed. CRC Press, Boca Raton. [Google Scholar]
- 12.Kobayashi T, Kajiwara M, Wahyuni M, Hamada-Sato N, Imada C, Watanabe E. 2004. Effect of culture conditions on lactic acid production of Tetragenococcus species. J Appl Microbiol 96: 1215–1221. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi T, Okuzumi M, Fujii T. 1995. Micoroflora of fermented puffer fish ovaries in rice-bran “Fugunoko Nukazuke”. Fish Sci 61: 291–295. [Google Scholar]
- 14.Kobayashi T, Kimura B, Fujii T. 2000. Differentiation of Tetragenococcus populations occurring in products and manufacturing processes of puffer fish ovaries fermented with rice-bran. Int J Food Microbiol 56: 211–218. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi T, Kimura B, Fujii T. 2000. Haloanaerobium fermentans sp. nov., a strictly anaerobic, fermentative halophile isolated from fermented puffer fish ovaries. Int J Syst Evol Microbiol 50: 1621–1627. [DOI] [PubMed] [Google Scholar]
- 16.Tanasupawat S, Shida O, Okada S, Komagata K. 2000. Lactobacillus acidipiscis sp. nov. and Weissella thailandensis sp. nov., isolated from fermented fish in Thailand. Int J Syst Evol Microbiol 50: 1479–1485. [DOI] [PubMed] [Google Scholar]
- 17.Chenoll E, Macián MC, Aznar R. 2006. Lactobacillus rennini sp. nov., isolated from rennin and associated with cheese spoilage. Int J Syst Evol Microbiol 56: 449–452. [DOI] [PubMed] [Google Scholar]
- 18.Chao SH, Kudo Y, Tsai YC, Watanabe K. 2012. Lactobacillus futsaii sp. nov., isolated from fu-tsai and suan-tsai, traditional Taiwanese fermented mustard products. Int J Syst Evol Microbiol 62: 489–494. [DOI] [PubMed] [Google Scholar]
- 19.Tanous C, Kieronczyk A, Helinck S, Chambellon E, Yvon M. 2002. Glutamate dehydrogenase activity: a major criterion for the selection of flavour-producing lactic acid bacteria strains. Antonie van Leeuwenhoek 82: 271–278. [PubMed] [Google Scholar]
- 20.Williams AG, Withers SE, Brechany EY, Banks JM. 2006. Glutamate dehydrogenase activity in lactobacilli and the use of glutamate dehydrogenase-producing adjunct Lactobacillus spp. cultures in the manufacture of cheddar cheese. J Appl Microbiol 101: 1062–1075. [DOI] [PubMed] [Google Scholar]
- 21.Mauriello G, Moio L, Genovese A, Ercolini D. 2003. Relationships between flavoring capabilities, bacterial composition, and geographical origin of natural whey cultures used for traditional water-buffalo mozzarella cheese manufacture. J Dairy Sci 86: 486–497. [DOI] [PubMed] [Google Scholar]
- 22.Oki K, Dugersuren J, Demberel S, Watanabe K. 2014. Pyrosequencing analysis of the microbial diversity of airag, khoormog and tarag, traditional fermented dairy products of mongolia. Biosci Microbiota Food Health 33: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]