Abstract
Clines in life history traits, presumably driven by spatially varying selection, are widespread. Major latitudinal clines have been observed, for example, in Drosophila melanogaster, an ancestrally tropical insect from Africa that has colonized temperate habitats on multiple continents. Yet, how geographic factors other than latitude, such as altitude or longitude, affect life history in this species remains poorly understood. Moreover, most previous work has been performed on derived European, American and Australian populations, but whether life history also varies predictably with geography in the ancestral Afro-tropical range has not been investigated systematically. Here, we have examined life history variation among populations of D. melanogaster from sub-Saharan Africa. Viability and reproductive diapause did not vary with geography, but body size increased with altitude, latitude and longitude. Early fecundity covaried positively with altitude and latitude, whereas lifespan showed the opposite trend. Examination of genetic variance-covariance matrices revealed geographic differentiation also in trade-off structure, and QST-FST analysis showed that life history differentiation among populations is likely shaped by selection. Together, our results suggest that geographic and/or climatic factors drive adaptive phenotypic differentiation among ancestral African populations and confirm the widely held notion that latitude and altitude represent parallel gradients.
Keywords: Clines, climate, geography, adaptation, life history, spatially varying selection
Introduction
Understanding how spatially varying selection causes adaptation to heterogeneous environments is a long-standing issue in evolutionary biology (Endler 1986). A powerful approach towards this end is to study clines, i.e. changes in phenotypes or allele frequencies across environmental gradients (Huxley 1938; Slatkin 1973; Endler 1977; Barton 1999). The study of clines is especially informative with regard to adaptation when fitness-related phenotypes are associated with environmental gradients. Although clines can also result from population structure and demography, they are typically thought to be caused by spatially varying selection since clines are often consistently replicated across geographical regions, populations and species (Dobzhansky 1970; Endler 1977, 1986).
One of the premier models for investigating the adaptive role of clinality is the fruit fly Drosophila melanogaster (David & Bocquet 1975; De Jong & Bochdanovits 2003; Hoffmann & Weeks 2007). This species originated in sub-Saharan Africa and has subsequently migrated out of Africa, colonizing Eurasia, America and Australia (David & Capy 1988). As a result, clines have been formed on several continents, spanning temperate to subtropical/tropical regions, thus providing an excellent opportunity for studying adaptation to spatial and climatic heterogeneity in a naturally replicated system. The large body of work on clines in D. melanogaster makes this species a unique system for assessing the generality of clinal adaptation.
Latitudinal clines, presumably driven by gradients in temperature and/or seasonality, have been documented for many fitness-related and morphological traits in D. melanogaster, including pigmentation, developmental time, body size, ovariole number, fecundity, egg size, stress resistance, lifespan, diapause and overwintering ability, with many of them showing parallel differentiation on multiple continents (Capy et al. 1993; James & Partridge 1995; Azevedo et al. 1996; James et al. 1997; Zwaan et al. 2000; Mitrovski & Hoffmann 2001; Hoffmann et al. 2002; Schmidt et al. 2005a, b; Pool & Aquadro 2007; Schmidt & Paaby 2008). Genetic signatures of latitudinal clinality have also been identified, including on a genome-wide scale at the level of single nucleotide polymorphisms (SNPs) (Fabian et al. 2012; Reinhardt et al. 2014). Evidence supports the notion that temperature may be the most important factor underlying latitudinal clines (Partridge et al. 1994; Umina et al. 2005; Santos et al. 2006). Thus, latitudinal clines have been extensively studied, and ongoing efforts focus on understanding the genetic causes and consequences of this clinality (Schmidt et al. 2008; Paaby et al. 2010; Fabian et al. 2012; Kapun et al. 2014; Cogni et al. 2014). In contrast, our knowledge of altitudinal and longitudinal clines is much more limited.
While elevational gradients have major effects on phenotypic differentiation in numerous organisms (Hodkinson 2005; Körner 2007; Yi et al. 2010; Bresson et al. 2011; Keller et al. 2013; Hille & Cooper 2014), considerably less is known about altitudinal clines in Drosophila. Some notable exceptions include effects of altitude on pigmentation, developmental time, size-related traits, ovariole number, fecundity, desiccation resistance and cold tolerance in D. melanogaster (Louis et al. 1982; Munjal et al. 1997; Collinge et al. 2006; Pool & Aquadro 2007; Parkash et al. 2008; Rajpurohit et al. 2008; Pitchers et al. 2013; Klepsatel et al. 2014).
Depending on geographical region, population or species, some studies have found that altitudinal clines exhibit opposite directionality (starvation resistance in D. buzzatii: Sørensen et al. 2005 versus Sarup & Loeschcke 2010; fecundity in D. yakuba versus D. teissieri: Devaux & Lachaise 1987). Others have failed to find clear altitudinal clines altogether: Bubliy & Loeschcke (2005) measured eight traits across an elevational transect in D. buzzatii and D. simulans but did not find clinality. Similarly, since temperature decreases with increasing altitude and latitude, the effects of altitude might mirror those of latitude (Lencioni 2004). Yet, this prediction has not always been confirmed. ‘Bergmann’s rule’, for instance, posits that body size is larger at both higher altitudes and latitudes (Partridge & Coyne 1997; Blanckenhorn & Demont 2004) but evidence for this pattern in insects is mixed (Shelomi 2012). Although many studies in Drosophila support ‘Bergmann’s rule’, others have failed to do so (Norry et al. 2001; Bubliy & Loeschcke 2005; Dillon et al. 2006; Pitchers et al. 2013, Klepsatel et al. 2014).
Some of these discrepancies might be due to the fact that temperature is not the only variable correlated with elevation. Other factors related to altitude but not latitude include decreased atmospheric pressure (entailing reduced pressure of oxygen); increased incoming and outgoing radiation (including higher UV-B radiation); absence of seasonality in tropical high-altitude environments as compared to temperate high-latitude habitats; altitudinal gradients in humidity and vapor pressure; and relatively high levels of gene flow across relatively short distances (Blanckenhorn 1997; Körner 2000; Hodkinson 2005; Körner 2007). Altitudinal gradients are thus not as well understood as latitudinal clines, and this is especially true for Drosophila.
Even less is known about longitudinal clines. Some studies have identified longitudinal clines in planktonic foraminifers, plants and humans (Samis et al. 2008, 2012; Ramachandran & Rosenberg 2011; Ujiié et al. 2012; Kooyers et al. 2014), and Wittkopp et al. (2011) have found a longitudinal cline for pigmentation in Drosophila americana. In D. melanogaster, longitudinal clines have been observed for allozymes and chromosomal inversions (Smith et al. 1978; Knibb 1982; Aulard et al. 2002), but phenotypic longitudinal clines are largely unknown. Factors that might vary predictably with longitude include gradients with respect to humidity, ‘continentality’, or the seasonal phenology of plants (Wittkopp et al. 2011; Samis et al. 2008, 2012.
Another aspect of clinal variation that is understudied is the extent of clinality in the tropics (~23°N to 23°S). A few studies have examined clinal variation in- and outside the Australian tropics (Mitrovski & Hoffmann 2001; Sgrò et al. 2013), and others have investigated clinality on the Indian subcontinent, which includes both tropical and subtropical regions (Rajpurohit & Nedved 2013). In addition, a handful of studies have investigated altitudinal clines for size-related traits in Afro-tropical populations (Louis et al. 1982; Pitchers et al. 2013, Klepsatel et al. 2014). However, whether life history traits other than size vary predictably with geographic or climatic factors within the ancestral Afro-tropical range of D. melanogaster remains unclear.
Clinality in the tropics is an important issue since the effects of climatic factors might differ from those seen in subtropical or temperate regions. For example, within the narrow band of the tropics one might expect to find weaker latitudinal gradients as compared to the much steeper ones that span across temperate and subtropical/tropical regions, as is the case for the North American and Australian clines in D. melanogaster. In contrast, elevational gradients might be ‘physiologically’ (not topographically) steeper for tropical organisms, which are adapted to constantly warm ambient conditions, than for temperate species (Janzen 1967; Ghalambor et al. 2006). More generally, since tropical terrestrial ectotherms live in relatively stable aseasonal climates, they tend to have more narrow thermal tolerance than higher-latitude species (Tewksbury et al. 2008). Consistent with this notion, Australian D. melanogaster populations from higher altitudes have greater cold tolerance at temperate but not tropical latitudes (Collinge et al. 2006).
Here, we have examined variation in life history (viability, body size, early fecundity, lifespan, reproductive diapause) among a total of 14 populations of D. melanogaster from sub-Saharan Africa under common garden conditions in the laboratory. Our study represents the first systematic attempt of characterizing geographic and clinal patterns of life history variation within the ancestral subtropical/tropical range of this species, thus extending previous efforts investigating derived European, American or Australian populations. We had four specific objectives. First, we aimed to examine geographic differentiation and clinality of African populations for life history. Second, we examined reproductive diapause, a plastic life history syndrome induced by low temperature and short photoperiod and known to be clinal across latitude in North American and Europe (Saunders et al. 1989; Schmidt & Paaby 2008). Preliminary data indicate that cold-induced diapause might be absent or at low frequency in ancestral African populations (Schmidt 2011), suggesting that it represents a ‘derived’ temperate adaptation, but Afro-tropical high-elevation populations have not yet been examined. Third, we investigated whether clinality shapes trade-off structure by examining genetic variance-covariance (G) matrices, which might be expected if constraints on trait evolution depend upon the environment. Finally, we aimed to test whether geographic life history divergence is likely caused by selection or drift by contrasting phenotypic and genetic differentiation using QST-FST analysis (Leinonen et al. 2013). This approach might indicate whether clinally varying traits are – as expected – shaped by clinal selection due to environmental gradients or rather shaped by drift. Similarly, for non-clinally varying traits, such an analysis might reveal if differences among populations are likely shaped by spatially varying selection due local adaptation or by drift.
Materials and Methods
Fly populations and maintenance
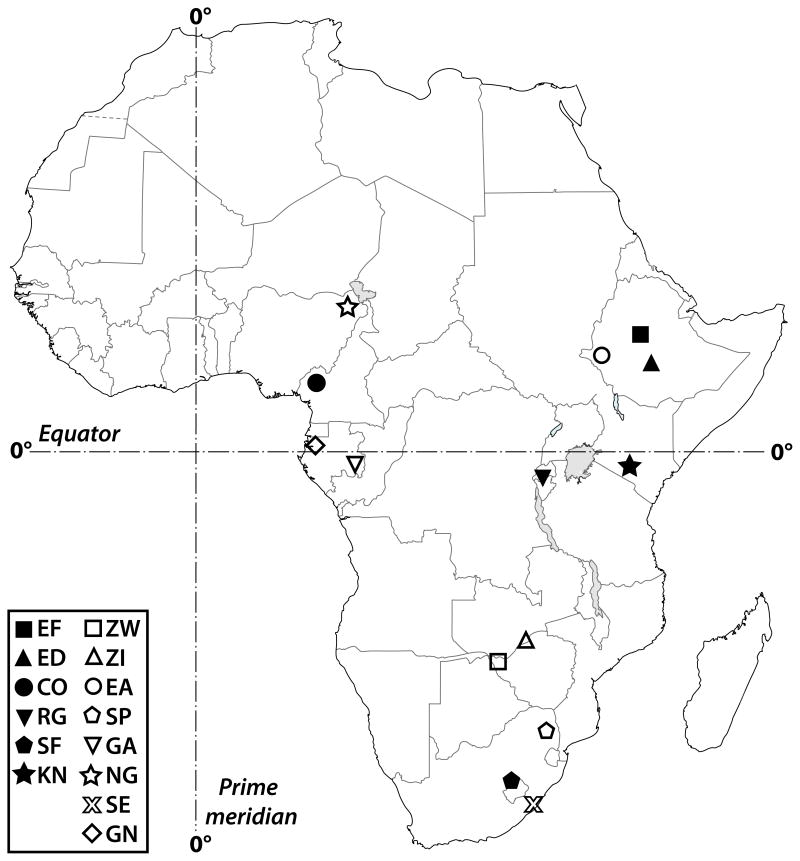
We used multiple wild-derived isofemale lines from each of 14 sub-Saharan populations of D. melanogaster (Fig. 1, Table S1). For practical reasons, we could not measure all traits for all populations; for each trait we measured a total of 62 to 119 isofemale lines from a total of 8 to 10 populations (Tables S1, S2). For most populations individual-line whole-genome and admixture data are available (Pool et al. 2012). The coordinates of the populations ranged from 11.85°N to 31.06°S latitude, 9.75° to 39.18°E longitude, and 17 m to 3070 m above mean sea level (AMSL), covering a major portion of tropical sub-Saharan Africa and also including three subtropical South African populations (Fig. 1). Isofemale lines were maintained and measured under standard laboratory conditions at 25°C (12:12 L:D) and 60% relative air humidity in uncrowded vials on standard cornmeal-agar-yeast medium with dry yeast sprinkled on top.
Figure 1.
Sampling locations of sub-Saharan African populations examined in this study. Open symbols: low-altitude populations (17–900 m AMSL); filled symbols: high-altitude populations (1661–3070 m AMSL). Dashed horizontal line: equator (0° latitude); vertical dashed line: meridian (0° longitude).
Life history measurements
We measured viability (proportion egg-to-adult survival); thorax length (a proxy of body size); early fecundity; lifespan; and reproductive diapause under common garden conditions in the laboratory. Tables S1 and S2 give details of sample sizes; raw data are available from Dryad (doi:10.5061/dryad.738bp).
For assays of viability and thorax length, we placed ~10–20 adults from each line into vials, containing apple juice-agar medium with dry yeast sprinkled on top, and allowed females to oviposit overnight. The next day, eggs from each line were gently brushed off the surface of the medium and allocated to vials (15–22 eggs per vial) containing standard medium with yeast sprinkled on top. Vials were checked every 12 h for eclosing adults until all flies had emerged. Thorax length of 1–3 day old adult females was measured, as described by French et al. (1998), with a stereo dissecting microscope (Leica M205 FA; Leica Microsystems GmbH, Wetzlar, Germany) connected to a digital camera (DFC 300 FX) and using the Leica Application Software (LAS v.3.8).
To measure early fecundity and lifespan, we collected flies within 12 h upon eclosion and allocated virgin females individually to vials together with two males, following brief CO2 anesthesia. Within the first 10 days of adulthood, flies were transferred to new vials with fresh food every 24 h and dead males were replaced. We counted the number of eggs laid by each female in each 24-h period over the first 10 days of adulthood and, from these egg counts, estimated daily per capita fecundity between days 1–5, 6–10, and 1–10. After the first 10 days, flies were transferred to fresh vials every second day. Throughout the assay, female age death was recorded when flies were transferred to new vials.
We also examined reproductive diapause, following the method of Saunders et al. (1989). Reproductive diapause is a plastic state of female ovarian arrest, triggered by low temperature and short photoperiod, associated with increased stress resistance and improved survival, and known to be clinal in North America and Europe (Schmidt & Paaby 2008; Schmidt 2011). Females were collected from each isofemale line within 2 h post-eclosion, transferred to fresh vials containing standard cornmeal-molasses medium, and placed in photoperiodic chambers on a photoperiod of 10L:14D at 11°C and >50% relative air humidity. Four weeks later, we dissected flies in PBS and determined the stages of ovarian development according to King (1970). A given line was classified as expressing diapause if the majority of individuals exhibited reproductive quiescence, as determined by the most advanced stage being ≤ stage 7.
Analysis of single traits
Analyses were performed using JMP v.10 (SAS, Raleigh, NC, USA), unless stated otherwise. We did not analyze data on diapause since the majority of populations did not exhibit diapause (see Results; Supporting Information File, Table S3). To examine among-population variation in viability, thorax length and fecundity we performed two-way mixed effects ANOVA, with ‘population’ as a fixed factor and ‘line’ nested in ‘population’ as a random factor. The random effect was modeled using restricted maximum likelihood (REML); see Table S4 for variance component estimates of the ‘line’ effect. Data for viability were arcsine transformed to achieve normality of residuals (as tested with Shapiro-Wilk test; not shown).
Next, we examined whether traits are clinally differentiated using multiple linear mixed model regression by fitting the intercept (β0), regression coefficients for altitude (β1Alt), latitude (β2 Lat) and longitude (β3 Long) as continuous predictors, and ‘line’ as a random effect using REML (see Table S4 for variance component estimates). Simultaneously fitting the effects of altitude, latitude and longitude allows to model population structure without the need for specifying a population term while at the same time reducing potential collinearity between these individual factors and population (Pitchers et al. 2012). Geographic parameters were fitted by using meters (m) as a unit for altitude, absolute degrees for latitude, and degrees east (°E) for longitude. Given the geographical distribution of our populations, we did not have sufficient power for estimating two- and three-way interactions among altitude, latitude and longitude. Degrees of freedom were calculated using Satterthwaite’s approximation, implemented in JMP.
To examine among-population variation in lifespan we used Cox (proportional hazards) regression with ‘population’ as a categorical predictor and ‘line’ nested in ‘population’ as random effect, using the package ‘coxme’ in R (Therneau 2012). Similarly, to investigate clinality of lifespan, we performed Cox regression using altitude, latitude and longitude as continuous predictors and modeling ‘line’ as random effect. To visualize the effects of these predictors, we plotted range risk ratios showing the change in hazard ratio over the range of a given regressor from Xmin to Xmax (exp[estimate(Xmax - Xmin)]).
Finally, to determine which climatic factors covary with altitude, latitude and longitude, we performed multiple regression (fitting all three predictors as main effects) of 6 climatic variables (mean annual air temperature [°C]; diurnal temperature range [°C]; relative humidity [%]; precipitation [P50, mm]; number of days with rainfall; and wind velocity [m/s]; data from ‘International Water Management Institute’, http://www.iwmi.cgiar.org/). Only one climatic factor, mean annual air temperature, covaried with altitude and latitude; none of the factors covaried with longitude. We therefore repeated all above-mentioned analyses of trait variation using temperature as a predictor variable (i.e., instead of fitting geographic parameters). To further identify if trait variation can be explained by other climatic variables we also fit a ‘full model’ containing all six climatic variables as predictors.
Analysis of trait relationships
To survive and reproduce organisms must function as phenotypically integrated wholes, not as single traits assumed to be separable from others (Stearns 1984, 1989). Indeed, life history traits are often tightly integrated through developmental, physiological and genetic mechanisms, resulting in life history correlations and trade-offs (Stearns 1992; Flatt & Heyland 2011). We therefore examined how geography affects (1) overall multi-trait life history and (2) genetic variance-covariance (G) matrices.
To investigate whether geography/clinality affect variation in multi-trait life history we used MANOVA, which accounts for phenotypic correlations among traits. MANOVA was performed on the combination of dependent variables (isofemale line means for viability, thorax length, early fecundity, lifespan), using ‘geographic effect’ and ‘population’ nested in ‘geographic effect’ as factors. This analysis was done for each geographic effect (altitude, latitude, longitude) separately, classifying data as ‘low’ or ‘high’ for altitude; as ‘above’, ‘at’ or ‘below’ equator for latitude; or as ‘east’ or ‘west’ for longitude (Table S5).
To estimate pairwise genetic correlations among traits we calculated Pearson’s product-moment correlation coefficients using isofemale line means, separately for each level within altitude (low, high), latitude (above, at, below equator), and longitude (east, west). Genetic correlations based on line means often provide similar estimates to those based on variances and covariances (Via 1984; Geber 1990; Donovan & Ehleringer 1994; Schmidt et al. 2005b; David et al. 2005). Our analysis indicated that several genetic correlations might differ between levels of particular geographic factors (Table S6); to verify whether line mean genetic variance-covariance (G) matrices differ significantly between levels of altitude, latitude, or longitude, we performed MANOVA on G pseudo-estimates obtained from the jackknife method (Roff 2002; Schmidt et al. 2005b; Roff et al. 2012), using ‘geographic effect’ and ‘population’ nested in ‘geographic effect’ as factors. Jackknife pseudo-values were created in R v.3.0.0.
QST-FST analysis
We also aimed to test whether among-population life history differentiation is likely due to selection by using QST-FST comparisons. QST quantifies the between-population proportion of quantitative trait variation (Lande 1992; Spitze 1993). For individuals raised in the same environment, QST can be estimated directly from within- and between-population phenotypic differences (Kohn et al. 2008). QST is often compared against neutral estimates of population genetic differentiation (FST) from multi-locus or genomic data to determine whether phenotypic differences among populations exceed that expected from genetic drift (Merilä & Crnokrak 2001; Leinonen et al. 2013). Values of QST that fall in the upper tail of the genomic distribution of FST values between two populations indicate that divergent or directional selection is likely to be acting on the trait (or on another trait pleiotropically correlated with it), whereas QST values in the lower tail suggest the possibility of stabilizing selection.
We used previously published genomes studied by Pool et al. (2012) and Lack et al. (2015) to generate genome-wide distributions of FST between all pairs of populations. The assembly of these genomes is detailed in the above-cited papers, and all sequences used here are available as part of the Drosophila Genome Nexus (http://www.johnpool.net/genomes.html). While these sequences were not generated from the same lines examined for life history characteristics in this study, they are representative samples of genetic diversity in the focal populations. The resulting genome-wide FST distributions were contrasted with pairwise QST values estimated from means of isofemale lines.
A critical factor in QST-FST comparisons is to ensure that the neutral variance of QST is no larger than that of FST (Miller et al. 2008). We sought to fulfill this requirement in two ways. First, the number of (homozygous) genomes analyzed for FST did not exceed the number of strains phenotyped from each population (hence the sampling variance of QST will not exceed that of FST). Second, the genomic distribution of FST between each pair of populations was obtained from short windows, defined by 10 non-singleton segregating sites from the full Rwanda RG population sample (Pool et al. 2012). These windows, each containing 10 SNPs, have an average length of 500 bp while scaling with the diversity along chromosomes (i.e., window-sizes are larger in regions of low diversity); they should have a lower neutral variance than a quantitative trait with a mutational target size greater than the mean FST window length, which seems plausible for the complex traits studied here. Also note that FST between African populations shows little broad-scale genomic variation and is similar for sites with higher and lower levels of constraint (Pool et al. 2012). P-values were estimated using an outlier approach by determining the position of a given value of QST in the FST distribution (i.e., a QST value falling into the upper or lower 0.5% of the FST distribution corresponds to P < 0.01 in a two-tailed comparison). Diapause was omitted from analysis due to negligible geographic variation.
Results
Geographic and clinal variation in life history
We first asked whether populations vary in life history and detected substantial phenotypic differentiation among populations for 3 out of 5 fitness-related traits. While viability did not vary among populations (ANOVA; ‘population’: F9, 67.54 = 1.98, P = 0.06), we found strong geographic differentiation for thorax length (F7, 54.15 = 15.18, P < 0.0001), early fecundity (daily per-capita fecundity between days 1–5: F9, 76.71 = 5.93, P < 0.0001; days 6–10: F9,68.81 = 2.45, P < 0.05; and days 1–10: F9,71.27 = 2.95, P < 0.01), and lifespan (Cox regression; ‘population’: likelihood ratio [LR] χ2 = 60.39, df = 11, P < 0.0001). We also investigated the incidence of diapause within and among populations (Table S3). Only two populations showed weak evidence for a diapause response: 1 line from the South African SP population (diapause frequency among lines: 1/5 lines = 0.2; incidence within the single diapausing line: 0.6) and 1 line from the Zambian ZI population (1/37 lines ≈ 0.03; incidence within the line: 0.57). Hence, among the 119 lines examined across populations, only 1.7% (2/119) showed a diapause response (Table S3). Thus, when measured using the standard assay (Saunders et al. 1989), cold-induced diapause is rare among ancestral sub-Saharan populations and, when present, exhibits a very low frequency within populations.
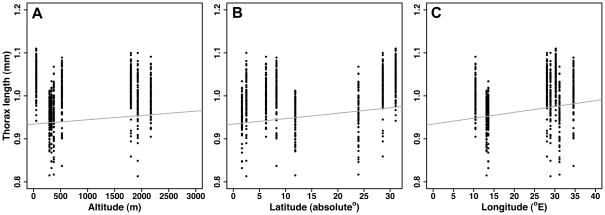
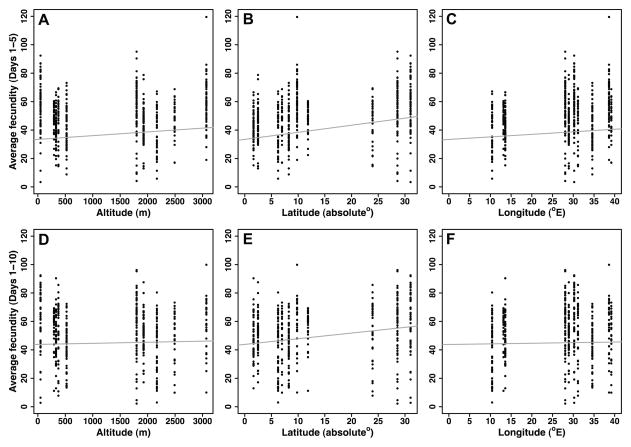
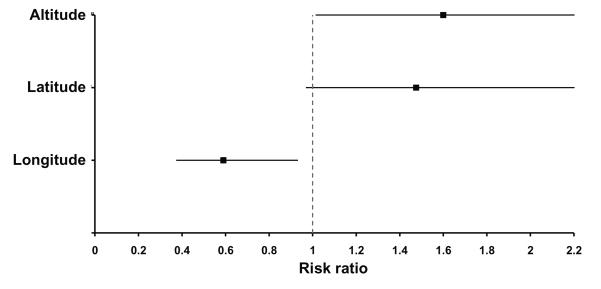
Second, to examine whether life history traits vary clinally, we analyzed the effects of altitude, latitude and longitude (Table 1). Viability did not vary with geography, but thorax length increased with altitude, latitude and longitude (Table 1, PFig. 2). Early fecundity over the first 5 days of adulthood covaried positively with altitude and latitude, and early fecundity over the first 10 days increased with latitude (Table 1, Fig. 3). In contrast, early fecundity between days 6 to 10 did not exhibit clinality (Table 1, Fig. 3). This suggests that early fecundity, especially within 5 days post-eclosion, is most strongly influenced by latitude and more weakly by altitude. Longitude did not measurably affect early fecundity estimates. Opposite to the pattern for fecundity, lifespan tended to decrease with altitude and latitude (multiple Cox regression; ‘altitude’: z = 2.02, < 0.05; ‘latitude’: z = 1.81, P = 0.07, marginally non-significant); moreover, lifespan covaried positively with longitude (z = −2.26, P < 0.05) (Fig. 4). Thus, geography has significant effects on life history differentiation among sub-Saharan African populations.
Table 1.
Clinal variation in life history traits. Summary of multiple regression mixed models for viability, thorax length, and three estimators of early daily per capita fecundity.
| Trait | Effect | F ratio | βo | βi | Adjusted R2 |
|---|---|---|---|---|---|
| Egg to adult survival (viability) | Alt | F1,73.65 = 2.19 | 0.71 (±0.08) | 4x10−5 (±3x10−5) | 0.72 |
| Lat | F1,72.56 = 0.13 | −9x10−4 (±3x10−3) | |||
| Long | F1,74.14 = 2.98 | −5x10−3 (±3x10−3) | |||
|
| |||||
| Body size (thorax length) | Alt | F1,57.5 = 4.26* | 0.93 (±0.01) | 1x10−5 (±5x10−6) | 0.50 |
| Lat | F1,59.47 = 11.09** | 1x10−3 (±4x10−4) | |||
| Long | F1,57.39 = 7.58** | 1x10−3 (±5x10−4) | |||
|
| |||||
| Fecundity 1–5 Days | Alt | F1,82.89 = 4.49* | 33.40 (±3.49) | 3x10−3 (±1x10−3) | 0.50 |
| Lat | F1,81.35 = 17.64*** | 0.50 (±0.12) | |||
| Long | F1,84.44 = 1.87 | 0.18 (±0.13) | |||
|
| |||||
| Fecundity 6–10 Days | Alt | F1,76.33 = 0.01 | 53.21 (±4.94) | −2x10−4 (±2x10−3) | 0.32 |
| Lat | F1,71.07 = 3.38 | 0.31 (±0.17) | |||
| Long | F1,74.65 = 0.29 | −0.10 (±0.19) | |||
|
| |||||
| Fecundity 10 Days | Alt | F1,78.7 = 0.30 | 43.82 (±3.95) | 8x10−4 (±1x10−3) | 0.39 |
| Lat | F1,73.78 = 8.60** | 0.40 (±0.14) | |||
| Long | F1,76.75 = 0.07 | 0.04 (±0.15) | |||
Abbreviations: Alt, altitude; Lat, latitude; and Long, longitude (all modeled as fixed effects). βo denotes the intercept, and βi denotes the slopes of fixed effects (i.e., β1Alt; or β2 Lat; or β3 Long). Note that the full mixed models include ‘line’ as a random effect; variance components of the ‘line’ effect are shown in Table S4. Standard errors are shown in parentheses.
P<0.05,
P<0.01,
P<0.0001.
Figure 2.
Clinal variation in female thorax length (mm), a major proxy of body size. The plots show regression of size against (A) altitude, (B) latitude, and (C) longitude. The grey line represents the regression line for each geographic factor, obtained from the multiple regression mixed model.
Figure 3.
Clinal variation for two different estimators of early fecundity. Upper panel (A–C): early daily per capita fecundity between days 1–5. Lower panel (D–F): early daily per capita fecundity between days 1–10. The plots show regression of fecundity against (A, D) altitude, (B, E) latitude, and (C, F) longitude. The grey line represents the regression line for each geographic factor as obtained from the multiple regression mixed model.
Figure 4.
Clinal variation in adult mortality. The figure shows the range risk ratio between the lowest and highest value of each of the geographic factors (altitude, latitude, longitude) as estimated from Cox regression. Risk ratios < 1 indicate a decrease in the hazard ratio with increasing values of the geographic factor (i.e., a negative slope), whereas risk ratios > 1 correspond to an increase in the hazard ratio (i.e., a positive slope). Error bars represent 95% confidence intervals.
Climatic effects on life history variation
Next, we tested whether clinality might be explained by geographic differences in climatic factors. As predicted, mean annual air temperature decreased with altitude (F1,6 = 37.39, P < 0.001) and latitude (F1,6 = 8.36, P < 0.05), whereas the other five climatic variables did not covary with geography (not shown). Notably, we did not find any climatic component to be significantly correlated with longitude. We therefore first asked whether variation in temperature explains clinal life history differences. Consistent with our clinal analyses above, thorax length (F1,58.93 =16.61, P < 0.001) and fecundity between 1–5 days (F1,85.07 = 12.26, P < 0.001) decreased with temperature (Fig. S1), while lifespan increased with temperature, although this trend was not significant (Cox regression; z = −1.68, P = 0.09) (Fig. S1). Clinality of these traits is thus likely driven by variation in temperature or by factors that covary with temperature.
To identify (also non-clinal) climatic factors potentially important for local adaptation, we analyzed trait variation by fitting all six climatic variables as predictors. Using this full model, variation in fecundity between 1–5 days was best explained by temperature, while lifespan tended to be affected by temperature and diurnal temperature range, thus confirming our above analysis of temperature effects (not shown). In contrast, thorax length showed a significant increase with wind velocity (F1,54.56 = 6.15, P < 0.01), whereas the effect of temperature became non-significant in the full model. Fecundity between 6–10 days, a trait that did not exhibit any clinality, decreased with the amount of rainfall (P50 mm) (F1,72.69 = 5.2, P < 0.01). Thus, wind velocity and the amount of rainfall (or correlated factors) might represent climatic variables responsible for local adaptation independent of climatic (clinal) gradients.
Variation in multi-trait life history and trade-off structure
We also asked whether geography affects multi-trait life history by using MANOVA, which accounts for trait interrelationships. Both latitude and longitude, but not altitude, influenced multivariate life history (Table S7). This indicates that latitudinal and longitudinal effects separate out distinct geographic combinations of trait values; it also suggests that geography might structure correlations between life history traits.
Overall, altitude, latitude or longitude did not affect G matrices for the multivariate combination of all traits (not shown), but MANOVA on G matrices for specific trait pairs revealed two cases where geography structures genetic correlations. First, early fecundity between days 1–5 and lifespan were genetically negatively correlated at low elevation (r = −0.53, P < 0.001) but uncorrelated at high elevation (r = 0.03, P > 0.05), a difference confirmed by MANOVA (‘altitude’: exact F3,75 = 3.69, P < 0.05; ‘population’ nested in altitude not significant). Second, while viability and early fecundity between days 6–10 were genetically positively correlated among West African populations (r = 0.50, P < 0.05), they were uncorrelated among East African populations (r = 0.08, P > 0.05), with MANOVA confirming an effect of longitude (‘longitude’: exact F3,65 = 3.27, P < 0.05; ‘population’ nested in longitude non-significant). Hence, geography and/or climate shape genetic correlation structure, with different local conditions favoring distinct combinations of trait values.
QST-FST comparisons
The above results imply the existence of major clinal differentiation in fitness-related traits and correlations among them, but they do not conclusively demonstrate that these patterns are driven by selection. We therefore aimed to distinguish between the effects of selection and drift in shaping life history divergence by using QST-FST analysis. QST-FST analysis supports an important role for selection in promoting life history divergence among populations (Table S8). We detected numerous QST outliers in the upper tail of the FST distribution (range of FST values: 0.01 to 0.16, average: 0.09) (Table S8). Even those traits with the fewest outlier QST values, viability (QST range: 0–0.92; mean: 0.17) and early fecundity between days 6–10 (range: 0–0.91; mean: 0.18), still had 18 and 23 pairwise QST values (out of 45 population comparisons), respectively, in the upper 2.5% tail of the FST distribution. For viability, this suggests that – even though we were unable to find geographic/clinal variation – this character could be shaped by geographically variable selection. For the other two fecundity estimates (days 1–5, range: 0–0.92; mean: 0.20; and days 1–10, range: 0–0.86; mean: 0.20) and for lifespan (range: 0.01–0.82; mean: 0.16), we identified even more outlier QST values: 26, 26, and 27 out of 45 pairwise comparisons, respectively; and for thorax length (range: 0–0.98; mean: 0.18) we found 22 significant QST outliers out of 28 comparisons. Across all traits, a majority of QST values with P < 0.05 also gave P < 0.01, with extreme QST values around 0.9 or higher observed in some cases (Table S8). Some populations contributed more heavily than others to outlier QST values for certain traits (e.g., EA for viability and fecundity between days 6–10), but in general QST outliers came from diverse geographic comparisons, thus potentially indicating that multiple selection pressures act on life history traits across geography.
Discussion
Despite decades of work on clinality in D. melanogaster (De Jong & Bochdanovits 2003), whether life history varies predictably with geography within the ancestral African range remains largely unknown. Here, we have characterized life history differentiation among 14 populations of D. melanogaster from sub-Saharan Africa. We found that three out of five life history traits (body size, early fecundity, lifespan) vary clinally across altitude, latitude or longitude, suggesting that they might be shaped by spatially varying (clinal) selection. In contrast, viability and reproductive diapause did not exhibit major geographic differentiation or clinality.
Viability and diapause are not clinal in African populations
The absence of clinal patterns for viability is in agreement with the findings of Van’t Land et al. (1999) for populations from the South American west coast, but inconsistent with reports demonstrating that viability increases with altitude and/or decreases with latitude in populations from Cameroon (Louis et al. 1982) and Argentina (Folguera et al. 2008). Since we failed to detect variation for viability among populations, one explanation for our inability to detect clinality may simply be a lack of geographic differentiation. However, QST-FST comparisons revealed that populations have experienced differential selection affecting viability, so insufficient statistical power is a more likely explanation. Alternatively, viability may be truly non-clinal among African populations and shaped by local adaptation independent of altitude, latitude or longitude.
For reproductive diapause, we also failed to detect clinality: in fact, the majority of populations and lines were unable to enter diapause under cold temperature and short photoperiod. Our results parallel those of a preliminary survey of African lines (Schmidt 2011): among isofemale lines from Zimbawe, Kenya, Gabon, and Malawi none exhibited diapause. However, these lines were from low-altitude localities, thus leaving open the possibility that African high-altitude flies might be able to undergo diapause. However, we found that African flies, even those from high elevations, do not typically enter diapause in response to cold temperature and short day length. Overall, only 2 lines from 2 out of 10 populations exhibited diapause (2/10 = 20% of all populations; 2/119 = 1.7% of all lines). Our results thus support the hypothesis that winter diapause in D. melanogaster is a ‘derived’ trait of recent evolutionary origin, which evolved as an adaptation to temperate habitats after the out-of-Africa expansion of this species (Saunders & Gilbert 1990; Schmidt 2011; Flatt et al. 2013). The results further suggest a limited phenotypic influence of admixture from non-African temperate populations, which are known to have diapause ability (Saunders et al. 1989; Tauber et al. 2007; Schmidt & Paaby 2008). However, even though we believe this to be unlikely, we cannot exclude that diapause is ancestral: it might be an exaptation segregating at very low frequencies in African populations, or it might be deleterious and thus have been purged from these populations. Similarly, we cannot rule out that diapause in Afro-tropical populations might be regulated by environmental cues other than low temperature and short photoperiod, for example by humidity/aridity.
Body size clines are consistent with Bergmann’s rule
While there are many reports of clines for size-related traits in D. melanogaster, few have examined clinality of such traits within the ancestral African range. Our analysis of thorax length revealed clear patterns of altitudinal and latitudinal clinality among African populations, and QST-FST analysis indicated that among-population differentiation is likely shaped by selection. Our data confirm recent findings of Pitchers et al. (2012) and Klepsatel et al. (2013) who have analyzed clinality of wing size and thorax length among sub-Saharan African populations, including a subset of populations we have studied here. Together with these observations, our results are consistent with ‘Bergmann’s rule’ (Partridge & Coyne 1997) and suggest that altitude and latitude represent parallel gradients (Lencioni 2004).
Our analyses further show that simple linear regression with mean temperature explains size variation as well as multiple regression with altitude, latitude and longitude (R2 = 0.5 for both analyses). Temperature is therefore the most parsimonious candidate for the selective agent underlying latitudinal and altitudinal clines for size-related traits, a notion that is supported by experimental evolution studies of thermal adaptation (Partridge et al. 1994). However, the mechanisms underlying evolutionary divergence in body size in ectotherms are generally not well understood (Partridge & Coyne 1997); one explanation for the latitude-size correlation in Drosophila might be that, due to increased competition, larval food resources might be more ephemeral in the tropics, causing selection to favor rapid development, whereas temperate habitats might favor longer developmental time and thus larger adult size (James & Partridge 1995).
Early fecundity and lifespan show opposite clinality
Early fecundity and lifespan showed opposite clinal patterns among Afro-tropical populations: fecundity increased with latitude and altitude, whereas lifespan covaried negatively with both predictors (with the effect of latitude being marginally not significant). Thus, as with body size, altitude and latitude had parallel effects on each trait. Moreover, QST-FST analysis showed that for both traits population divergence is likely driven by spatially varying selection and/or local adaptation.
The clinal pattern of fecundity is consistent with previous studies, even though African populations have not yet been systematically examined. For example, early fecundity increases with altitude in D. buzzatii (Norry et al. 2006), and in terms of latitude, temperate populations of D. melanogaster are often more fecund than tropical ones (Boulétreau-Merle et al. 1982; Klepsatel et al. 2013). Similarly, fecundity increases linearly with latitude-of-origin in Australian populations when flies are measured under tropical winter conditions (Hoffmann et al. 2003), and is also positively correlated with latitude on the Indian subcontinent (Rajpurohit & Nedved 2013). In contrast to fecundity, lifespan decreased with altitude and – more weakly – with latitude in our study. Similar results with respect to altitude have been reported for Californian Melanoplus grasshoppers (Tatar et al. 1997) and for Argentinian D. buzzatii populations (Norry et al. 2006). Latitudinal clines have also been documented for lifespan (see below). Together, these results for fecundity and lifespan are compatible with the ‘slow-fast continuum’ or ‘pace-of-life’ framework, which posits that high extrinsic mortality selects for a fast pace of life, i.e. high early reproductive effort and accelerated senescence (Promislow & Harvey 1990).
While our results agree with several previous reports, clinal patterns for fecundity and lifespan are often inconsistent across studies. For fecundity, no relationship with elevation has been found for Indian populations (Rajpurohit & Nedved 2013), and flies from Florida have higher fecundity than flies from Maine, with flies from the mid-Atlantic region being intermediate (Schmidt et al. 2005a; Schmidt & Paaby 2008). Moreover, when Australian flies are assayed in field cages under temperate conditions, the relationship between fecundity and latitude is curvilinear and opposite to that seen under tropical winter conditions (Mitrovski & Hoffmann 2001). For lifespan, clinal patterns also appear to be complex. For the Australian cline, the association between latitude and longevity is concave-upward when flies are assayed under temperate conditions (Mitrovski & Hoffmann 2001) but concave-downward when flies are assayed under laboratory conditions (Sgrò et al. 2013). For the North American cline, in contrast, lifespan increases linearly with latitude (Schmidt & Paaby 2008). A caveat is that here we have measured these (and other) traits under common garden conditions at a specific temperature in the laboratory; our results therefore capture real differences among populations, but we do not know whether they reflect general patterns across other environments.
Despite the fact that for many traits qualitatively identical clines have been observed on multiple continents (De Jong & Bochdanovits 2003), discrepancies are not uncommon, in particular for complex traits (Introduction; Sgrò et al. 2013). Several factors might account for such differences, but their precise causes are unknown. For clinal patterns of fecundity and lifespan in Australia, inconsistencies might be due to differences in assay conditions between studies, e.g. seasonality or genotype by assay environment interactions (Hoffmann et al. 2003; Sgrò et al. 2013). Furthermore, the few studies that have identified clinality of fecundity and lifespan in D. melanogaster vary substantially in the latitudinal ranges examined: ~16°S to 43°S for Australia (Mitrovski & Hoffmann 2001; Hoffmann et al. 2003; Sgrò et al. 2013); ~25°N to 44°N for North America (Schmidt & Paaby 2008); and ~31°S to 12°N for the African cline reported here. Thus, existing studies differ markedly in their relative coverage of temperate, subtropical and tropical regions. A related issue is that, even across comparable latitudinal ranges, climatic conditions can be very distinct across continents: for North American high-latitude populations, for example, winter conditions are likely much more extreme than those experienced by southeastern Australian populations. In addition, clines might also differ between ancestral African and derived populations in unknown ways.
Temperature might be the major factor underlying altitudinal and latitudinal clines, but how thermal gradients affect fecundity and lifespan is not well understood (Klepsatel et al. 2013; Rajpurohit & Nedved 2013; Sgrò et al. 2013). In our study, simple linear regression with average temperature explained clinality of early fecundity between 1 and 5 days post-eclosion and lifespan (R2 = 0.51 and 0.28, respectively) equally well as more complex multiple regression fitting effects of altitude, latitude and longitude (R2 = 0.50 and 0.28, respectively). The significant effects of temperature on these traits were confirmed when simultaneously fitting all six climatic predictors. Moreover, while fecundity between days 6–10 did not vary clinally with our geographic variables, this trait was negatively correlated with the amount of rainfall. We conclude that, at least in our dataset, clinal effects of altitude and latitude on fecundity and lifespan are most parsimoniously explained by temperature. In addition to temperature, the amount of rainfall might also represent an important (but still poorly understood) selective factor that may be relevant for non-clinal local adaptation, especially in terms of its effects on fecundity (this study; see above) but potentially also by influencing morphology (Klepsatel et al. 2013).
It is noteworthy that fecundity and longevity varied inversely across altitude, latitude and temperature in our study. Although for lifespan the effects of latitude and temperature were not nominally significant, likely due to a lack of statistical power, both effects showed a clear trend in the same direction. Importantly, our observed pattern qualitatively matches results from the North American cline where fecundity and lifespan also covary inversely across geography (Schmidt & Paaby 2008). Although in our study the directionality of this association is reversed as compared to North America, the spatially tight negative coupling of these traits is consistent with a genetic trade-off between reproduction and survival (Stearns & Partridge 2001; Flatt 2011). Thus, while we do not yet understand differences in directionality among distinct clines, the available data suggest that fecundity and lifespan co-evolve antagonistically in response to spatially varying selection, leading to a trade-off across geography (Paaby & Schmidt 2009; Flatt et al. 2013). The notion that clinal gradients structure trade-offs is also underscored by our finding that early fecundity and lifespan are negatively genetically correlated for low-altitude populations but uncorrelated for high-elevation populations. A likely reason is that the genetic basis of adaptation to low versus high altitude is different, so that different loci might be under selection in both environments; this in turn, might differentially affect patterns of pleiotropy and thus genetic covariances. Yet, due to the scarcity of studies describing such phenomena, it remains presently difficult to explain why certain genetic correlations change across environmental gradients.
Longitudinal life history clines are poorly understood
Interestingly, we also found evidence that body size increases with longitude and that longitude modulates the sign of the genetic correlation between viability and early fecundity. To date, unfortunately, extremely little is known about longitudinal clines: some examples have been reported in foraminifers, plants and humans (Ramachandran & Rosenberg 2011; Ujiié et al. 2012; Kooyers et al. 2014), for instance longitudinal clines for flowering time in Eurasian and North American Arabidopsis thaliana (Samis et al. 2008, 2012). In D. melanogaster, longitudinal clines have been found for chromosomal inversions (Knibb 1982; Aulard et al. 2002), but whether such clines are associated with phenotypes is unclear. Known phenotypic gradients in Drosophila include longitudinal clines for pigmentation in D. americana (Wittkopp et al. 2011) and for wing shape among African D. melanogaster (Pitchers et al. 2012).
However, the climatic variables that covary with longitude are poorly understood. In fact, our analyses failed to reveal any climatic factor clearly associated with longitude. Nevertheless, several other factors might underlie longitudinal gradients, including spatial variation in ‘continentality’, or the seasonal phenology of plants (Wittkopp et al. 2011; Samis et al. 2008, 2012). Moreover, since fruit flies are commensal with humans, longitudinal differences among African D. melanogaster populations might potentially be due to well-known east-west differences in human land use, agriculture, and so forth (Gomez et al. 2014). Indeed, eastern and western African populations of D. melanogaster are genetically distinct groups (Pool & Aquadro 2006), which is in line with the phenotypic differences across longitude. Clearly, the effects and causes of longitudinal gradients deserve much more attention; they should be relatively easy to investigate in well-studied models such as Arabidopsis or Drosophila.
Conclusions
Here we have provided evidence for major patterns of clinality in several important fitness-related traits among sub-Saharan African populations of D. melanogaster. Our main findings are that (1) body size, early fecundity and lifespan exhibit strong clinality among subtropical and tropical African populations, especially with respect to altitude and latitude; (2) altitudinal and latitudinal clines are best explained by temperature gradients, and for most traits altitude and latitude represent parallel gradients; (3) in contrast to derived, temperate populations, winter diapause is absent – or at least extremely rare – among ancestral African populations; (4) clinality structures the genetic trade-off between early fecundity and lifespan and the genetic correlation between viability and early fecundity, suggesting that the genetic basis of life history adaptation varies across tropical clines; and (5) QST-FST analysis supports an important role for selection in shaping life history differentiation among African populations.
A potential drawback of our isofemale line approach is that we cannot rule out that the measured populations, which have been kept under laboratory conditions for different periods of time, vary in levels of inbreeding and/or laboratory adaptation. We believe this to be unlikely for the following reasons (also see discussion in David et al. 2005). First, trait means and variances obtained from isofemale lines have previously been shown to remain stable across generations (Gibert et al. 1998). Second, our results on size-related traits and Bergmann’s rule are entirely consistent with those obtained by Klepsatel et al. (2014) who measured several outbred populations constructed from a subset of the isofemale lines we have measured here. Third, if present at all, we would expect laboratory adaptation and/or variation in inbreeding levels to act against (or even erase) the clinal signals we have detected here.
A major implication of our study is that life history clinality, presumably driven by spatial differences in climatic factors, can be very pronounced in the tropics, not just across continental gradients that span temperate, subtropical and tropical regions, as might often be assumed. While this has to some extent been appreciated for the effects of tropical elevational gradients, the effects of latitude and longitude within tropical environments have received much less attention. Importantly, our findings from African populations suggest that even within tropical environments thermal gradients can have a major selective influence on life history differentiation.
Supplementary Material
Acknowledgments
We thank V. Zehetbauer, S. Bauer, A. Suvorov and E. Versace for help with experiments and A. J. Betancourt for discussion. We are indebted to the people who have collected the fly strains used here (Table S1). Our research was supported by the Austrian Science Fund (FWF) (P21498-B11 to T.F.; and W1225 ‘Doktoratskolleg Populationsgenetik’); the Swiss National Science Foundation (SNF) (PP00P3_133641 to T.F.); the National Institutes of Health (NIH) (5R01GM100366 to P.S.S.; 1R01GM111797 to J.E.P; Kirschstein–National Research Service Award Postdoctoral Fellowship, 1F32GM106594, to J.B.L.); and the National Science Foundation (NSF) (DEB 0921307 to P.S.S.).
Footnotes
Data accessibility
Our raw data have been deposited at Dryad (doi:10.5061/dryad.738bp).
References
- Aulard S, David JR, Lemeunier F. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet Res. 2002;79:49–63. doi: 10.1017/s0016672301005407. [DOI] [PubMed] [Google Scholar]
- Azevedo RBR, French V, Partridge L. Thermal evolution of egg size in Drosophila melanogaster. Evolution. 1996;50:2338–2345. doi: 10.1111/j.1558-5646.1996.tb03621.x. [DOI] [PubMed] [Google Scholar]
- Barton NH. Clines in polygenic traits. Genet Res. 1999;74:223–236. doi: 10.1017/s001667239900422x. [DOI] [PubMed] [Google Scholar]
- Blanckenhorn WU. Altitudinal life history variation in the dung flies Scathophaga stercoraria and Sepsis cynipsea. Oecologia. 1997;109:342–352. doi: 10.1007/s004420050092. [DOI] [PubMed] [Google Scholar]
- Blanckenhorn WU, Demont M. Bergmann and converse Bergmann latitudinal clines in arthropods: Two ends of a continuum? Integr Comp Biol. 2004;44:413–424. doi: 10.1093/icb/44.6.413. [DOI] [PubMed] [Google Scholar]
- Boulétreau-Merle J, Allemand R, Cohet Y, David JR. Reproductive Strategy in Drosophila melanogaster: Significance of a Genetic Divergence between Temperate and Tropical Populations. Oecologia. 1982;53:323–329. doi: 10.1007/BF00389008. [DOI] [PubMed] [Google Scholar]
- Bresson CC, Vitasse Y, Kremer A, Delzon S. To what extent is altitudinal variation of functional traits driven by genetic adaptation in European oak and beech? Tree Physiol. 2011;31:1164–1174. doi: 10.1093/treephys/tpr084. [DOI] [PubMed] [Google Scholar]
- Bubliy OA, Loeschcke V. Variation of life-history and morphometrical traits in Drosophila buzzatii and Drosophila simulans collected along an altitudinal gradient from a Canary island. Biol J Linn Soc. 2005;84:119–136. [Google Scholar]
- Capy P, Pla E, David JR. Phenotypic and genetic variability of morphometrical traits in natural populations of Drosophila melanogaster and Drosophila simulans 1 Geographic variations. Genet Sel Evol. 1993;25:517–536. [Google Scholar]
- Cogni R, Kuczynski C, Koury S, Lavington E, Behrman EL, O’Brien KR, et al. The intensity of selection acting on the couch potato gene--spatial-temporal variation in a diapause cline. Evolution. 2014;68:538–548. doi: 10.1111/evo.12291. [DOI] [PubMed] [Google Scholar]
- Collinge JE, Hoffmann AA, McKechnie SW. Altitudinal patterns for latitudinally varying traits and polymorphic markers in Drosophila melanogaster from eastern Australia. J Evol Biol. 2006;19:473–482. doi: 10.1111/j.1420-9101.2005.01016.x. [DOI] [PubMed] [Google Scholar]
- David JR, Bocquet C. Evolution in a cosmopolitan species: genetic latitudinal clines in Drosophila melanogaster wild populations. Experientia. 1975;31:164–166. doi: 10.1007/BF01990682. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- David JR, Gibert P, Legout H, Pétavy G, Capy P, Moreteau B. Isofemale lines in Drosophila: an empirical approach to quantitative trait analysis in natural populations. Heredity. 2005;94:3–12. doi: 10.1038/sj.hdy.6800562. [DOI] [PubMed] [Google Scholar]
- De Jong G, Bochdanovits Z. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J Genet. 2003;82:207–223. doi: 10.1007/BF02715819. [DOI] [PubMed] [Google Scholar]
- Devaux J, Lachaise D. Alternative smooth or stepped altitudinal cline of fecundity in Drosophila teissieri and D. yakuba in the Ivory Coast. Japan J Genet. 1987;63:43–50. [Google Scholar]
- Dillon ME, Frazier MR, Dudley R. Into thin air: Physiology and evolution of alpine insects. Integr Comp Biol. 2006;46:49–61. doi: 10.1093/icb/icj007. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of the Evolutionary Process. Columbia Univ. Press; New York, NY: 1970. [Google Scholar]
- Donovan LA, Ehleringer JR. Potential for selection on plants for water-use efficiency as estimated by carbon isotope discrimination. Am J Bot. 1994;81:927–935. [Google Scholar]
- Endler JA. Geographic Variation, Speciation and Clines. Princeton Univ. Press; Princeton, NJ: 1977. [PubMed] [Google Scholar]
- Endler JA. Natural Selection in the Wild. Princeton Univ. Press; Princeton, NJ: 1986. [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlötterer C, et al. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 2012;21:4748–4769. doi: 10.1111/j.1365-294X.2012.05731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4. Longman; Harlow, Essex, U. K: 1996. [Google Scholar]
- Flatt T. Survival costs of reproduction in Drosophila. Exp Gerontol. 2011;46:369–375. doi: 10.1016/j.exger.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Flatt T, Heyland A, editors. The Genetics and Physiology of Life History Traits and Trade-Offs. Oxford Univ. Press; Oxford, U. K: 2011. Mechanisms of Life History Evolution. [Google Scholar]
- Flatt T, Amdam GV, Kirkwood TBL, Omholt SW. Life-History Evolution and the Polyphenic Regulation of Somatic Maintenance and Survival. Quart Rev Biol. 2013;88:185–218. doi: 10.1086/671484. [DOI] [PubMed] [Google Scholar]
- Folguera G, Ceballos S, Spezzi L, Fanara JJ, Hasson E. Clinal variation in developmental time and viability, and the response to thermal treatments in two species of Drosophila. Biol J Linn Soc. 2008;95:233–245. [Google Scholar]
- French V, Feast M, Partridge L. Body size and cell size in Drosophila: the developmental response to temperature. J Insect Physiol. 1998;44:1081–1089. doi: 10.1016/s0022-1910(98)00061-4. [DOI] [PubMed] [Google Scholar]
- Ghalambor CK, Huey RB, Martin PR, Tewksbury JJ, Wang G. Are mountain passes higher in the tropics? Janzen’s hypothesis revisited. Integr Comp Biol. 2006;46:5–17. doi: 10.1093/icb/icj003. [DOI] [PubMed] [Google Scholar]
- Geber MA. The cost of meristem limitation in Polygonum arenastrum: negative genetic correlations between fecundity and growth. Evolution. 1990;44:799–819. doi: 10.1111/j.1558-5646.1990.tb03806.x. [DOI] [PubMed] [Google Scholar]
- Gibert P, Moreteau B, Moreteau JC, David JR. Genetic variability of quantitative traits in Drosophila melanogaster (fruit fly) natural populations: analysis of wild-living flies and of several laboratory generations. Heredity. 1998;80:326–335. [Google Scholar]
- Gomez F, Hirbo J, Tishkoff SA. Genetic Variation and Adaptation in Africa: Implications for Human Evolution and Disease. Cold Spring Harb Perspect Biol. 2014;2014:6, a008524. doi: 10.1101/cshperspect.a008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille SM, Cooper CB. Elevational trends in life histories: revising the pace-of-life framework. Biol Rev. 2014 doi: 10.1111/brv.12106. In Press. [DOI] [PubMed] [Google Scholar]
- Hodkinson ID. Terrestrial insects along elevation gradients: species and community responses to altitude. Biol Rev. 2005;80:489–513. doi: 10.1017/s1464793105006767. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Weeks AR. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica. 2007;129:133–147. doi: 10.1007/s10709-006-9010-z. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Anderson A, Hallas R. Opposing clines for high and low temperature resistance in Drosophila melanogaster. Ecol Lett. 2002;5:614–618. [Google Scholar]
- Hoffmann AA, Scott M, Partridge L, Hallas R. Overwintering in Drosophila melanogaster: outdoor field cage experiments on clinal and laboratory selected populations help to elucidate traits under selection. J Evol Biol. 2003;16:614–623. doi: 10.1046/j.1420-9101.2003.00561.x. [DOI] [PubMed] [Google Scholar]
- Huxley J. Clines: an auxiliary taxonomic principle. Nature. 1938;142:219–220. [Google Scholar]
- James AC, Partridge L. Thermal evolution of rate of larval development in Drosophila melanogaster in laboratory and field populations. J Evol Biol. 1995;8:315–330. [Google Scholar]
- James AC, Azevedo RB, Partridge L. Genetic and environmental responses to temperature of Drosophila melanogaster from a latitudinal cline. Genetics. 1997;146:881–890. doi: 10.1093/genetics/146.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen DH. Why mountain passes are higher in the tropics. Am Nat. 1967;101:233–249. [Google Scholar]
- Kapun M, van Schalkwyk H, McAllister B, Flatt T, Schlötterer C. Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol Ecol. 2014;23:1813–1827. doi: 10.1111/mec.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller I, Alexander JM, Holderegger R, Edwards PJ. Widespread phenotypic and genetic divergence along altitudinal gradients in animals. J Evol Biol. 2013;26:2527–2543. doi: 10.1111/jeb.12255. [DOI] [PubMed] [Google Scholar]
- King R. Ovarian development in Drosophila melanogaster. Academic Press; New York: 1970. [Google Scholar]
- Klepsatel P, Galikova M, De Maio N, Huber CD, Schlötterer C, Flatt T. Variation in thermal performance and reaction norms among populations of Drosophila melanogaster. Evolution. 2013;67:3573–3587. doi: 10.1111/evo.12221. [DOI] [PubMed] [Google Scholar]
- Klepsatel P, Gálikova M, Huber CD, Flatt T. Similarities and differences in altitudinal versus latitudinal variation for morphological traits in Drosophila melanogaster. Evolution. 2014;68:1385–1398. doi: 10.1111/evo.12351. [DOI] [PubMed] [Google Scholar]
- Knibb WR. Chromosome inversion polymorphisms in Drosophila melanogaster II. Geographic clines and climatic associations in Australasia, North America and Asia. Genetica. 1982;58:213–221. [Google Scholar]
- Kohn MH, Shapiro J, Wu CI. Decoupled differentiation of gene expression and coding sequence among Drosophila populations. Genes Genet Syst. 2008;83:265–273. doi: 10.1266/ggs.83.265. [DOI] [PubMed] [Google Scholar]
- Kooyers NJ, Gage LR, Al-Lozi A, Olsen KM. Aridity shapes cyanogenesis cline evolution in white clover (Trifolium repens L.) Mol Ecol. 2014;23:1053–1070. doi: 10.1111/mec.12666. [DOI] [PubMed] [Google Scholar]
- Körner C. Why are there global gradients in species richness? Mountains might hold the answer. Trends Ecol Evol. 2000;15:513–514. [Google Scholar]
- Körner C. The use of ‘altitude’ in ecological research. Trends Ecol Evol. 2007;22:569–574. doi: 10.1016/j.tree.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Lack JB, Cardeno CM, Crepeau MW, Taylor W, Corbett-Detig RB, Stevens KA, et al. The Drosophila Genome Nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics. 2015 doi: 10.1534/genetics.115.174664. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Neutral Theory of Quantitative Genetic Variance in an Island Model with Local Extinction and Colonization. Evolution. 1992;46:381–389. doi: 10.1111/j.1558-5646.1992.tb02046.x. [DOI] [PubMed] [Google Scholar]
- Leinonen T, McCairns RJS, O’Hara RB, Merilä J. QST-FST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nature Rev Genet. 2013;14:179–190. doi: 10.1038/nrg3395. [DOI] [PubMed] [Google Scholar]
- Lencioni V. Survival strategies of freshwater insects in cold environments. J Limnol. 2004;63(Suppl):45–55. [Google Scholar]
- Louis J, David JR, Rouault J, Capy P. Altitudinal variations of Afro-tropical D. melanogaster populations. Droso Inf Serv. 1982;58:100–101. [Google Scholar]
- Merilä J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. J Evol Biol. 2001;14:892–903. [Google Scholar]
- Miller JR, Wood BP, Hamilton MB. FST and QST Under Neutrality. Genetics. 2008;180:1023–1037. doi: 10.1534/genetics.108.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovski P, Hoffmann AA. Postponed reproduction as an adaptation to winter conditions in Drosophila melanogaster: evidence for clinal variation under semi-natural conditions. Proc Roy Soc London B. 2001;268:2163–2168. doi: 10.1098/rspb.2001.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munjal AK, Karan D, Gibert P, Moreteau B, Parkash R, David JR. Thoracic trident pigmentation in Drosophila melanogaster: latitudinal and altitudinal clines in Indian populations. Genet Sel Evol. 1997;29:601–610. [Google Scholar]
- Norry FM, Bubliy OA, Loeschcke V. Developmental time, body size and wing loading in Drosophila buzzatii from lowland and highland populations in Argentina. Hereditas. 2001;135:35–40. doi: 10.1111/j.1601-5223.2001.t01-1-00035.x. [DOI] [PubMed] [Google Scholar]
- Norry FM, Sambucetti P, Scannapieco AC, Loeschcke V. Altitudinal patterns for longevity, fecundity and senescence in Drosophila buzzatii. Genetica. 2006;128:81–93. doi: 10.1007/s10709-005-5537-7. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Schmidt PS. Dissecting the genetics of longevity in Drosophila melanogaster. Fly (Austin) 2009;3:29–38. doi: 10.4161/fly.3.1.7771. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Blacket MJ, Hoffmann AA, Schmidt PS. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol Ecol. 2010;19:760–774. doi: 10.1111/j.1365-294X.2009.04508.x. [DOI] [PubMed] [Google Scholar]
- Parkash R, Rajpurohit S, Ramniwas S. Changes in body melanisation and desiccation resistance in highland vs. lowland populations of D melanogaster. J Insect Physiol. 2008;54:1050–1056. doi: 10.1016/j.jinsphys.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Partridge L, Coyne JA. Bergmann’s rule in ectotherms: is it adaptive? Evolution. 1997;51:632–635. doi: 10.1111/j.1558-5646.1997.tb02454.x. [DOI] [PubMed] [Google Scholar]
- Partridge L, Barrie B, Fowler K, French V. Evolution and development of body size and cell size in Drosophila melanogaster in response to temperature. Evolution. 1994;48:1269–1276. doi: 10.1111/j.1558-5646.1994.tb05311.x. [DOI] [PubMed] [Google Scholar]
- Pitchers W, Pool JE, Dworkin I. Altitudinal clinal variation in wing size and shape in African Drosophila melanogaster: one cline or many? Evolution. 2012;67:438–452. doi: 10.1111/j.1558-5646.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Aquadro CF. History and structure of sub-Saharan populations of Drosophila melanogaster. Genetics. 2006;174:915–929. doi: 10.1534/genetics.106.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Aquadro CF. The genetic basis of adaptive pigmentation variation in Drosophila melanogaster. Mol Ecol. 2007;16:2844–2851. doi: 10.1111/j.1365-294X.2007.03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, et al. Population Genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 2012;8:e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow DEL, Harvey PH. Living fast and dying young: A comparative analysis of life-history variation among mammals. J Zool. 1990;220:417–437. [Google Scholar]
- Rajpurohit S, Nedved O. Clinal variation in fitness related traits in tropical drosophilids of the Indian subcontinent. J Thermal Biol. 2013;38:345–354. [Google Scholar]
- Rajpurohit S, Parkash R, Ramniwas S, Singh S. Variations in body melanisation, ovariole number and fecundity in highland and lowland populations of Drosophila melanogaster from the Indian subcontinent. Insect Sci. 2008;15:553–561. [Google Scholar]
- Ramachandran S, Rosenberg NA. A Test of the Influence of Continental Axes of Orientation on Patterns of Human Gene Flow. Am J Phys Anthropol. 2011;146:515–529. doi: 10.1002/ajpa.21533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt JA, Kolaczkowski B, Jones CD, Begun DJ, Kern AD. Parallel Geographic Variation in Drosophila melanogaster. Genetics. 2014;197:361–373. doi: 10.1534/genetics.114.161463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff D. Comparing G matrices: a MANOVA approach. Evolution. 2002;56:1286–1291. [PubMed] [Google Scholar]
- Roff DA, Prokkola JM, Krams I, Rantala MJ. There is more than one way to skin a G matrix. J Evol Biol. 2012;25:1113–1126. doi: 10.1111/j.1420-9101.2012.02500.x. [DOI] [PubMed] [Google Scholar]
- Samis KE, Heath KD, Stinchcombe JR. Discordant Longitudinal Clines in Flowering Time and Phytochrome C in Arabidopsis thaliana. Evolution. 2008;62:2971–2983. doi: 10.1111/j.1558-5646.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- Samis KE, Murren CJ, Bossdorf O, Donohue K, Fenster CB, Malmberg RL, et al. Longitudinal trends in climate drive flowering time clines in North American Arabidopsis thaliana. Ecol Evol. 2012;2:1162–1180. doi: 10.1002/ece3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M, Brites D, Laayouni H. Thermal evolution of pre-adult life history traits, geometric size and shape, and developmental stability in Drosophila subobscura. J Evol Biol. 2006;19:2006–2021. doi: 10.1111/j.1420-9101.2006.01139.x. [DOI] [PubMed] [Google Scholar]
- Sarup P, Loeschcke V. Developmental acclimation affects clinal variation in stress resistance traits in Drosophila buzzatii. J Evol Biol. 2010;23:957–965. doi: 10.1111/j.1420-9101.2010.01965.x. [DOI] [PubMed] [Google Scholar]
- Saunders DS, Gilbert LI. Regulation of ovarian diapause in Drosophila melanogaster by photoperiod and moderately low temperature. J Insect Physiol. 1990;36:195–200. [Google Scholar]
- Saunders DS, Henrich V, Gilbert LI. Induction of diapause in Drosophila melanogaster: photoperiodic regulation and impact of arrhythmic clock mutations on time measurement. Proc Natl Acad Sci USA. 1989;86:3748–3752. doi: 10.1073/pnas.86.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS. Evolution and mechanisms of insect reproductive diapause: a plastic and pleiotropic life history syndrome. In: Flatt T, Heyland A, editors. Mechanisms of Life History Evolution. The Genetics and Physiology of Life History Traits and Trade-Offs. Oxford Univ. Press; Oxford, U.K: 2011. pp. 230–242. [Google Scholar]
- Schmidt PS, Paaby AB. Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution. 2008;62:1204–1215. doi: 10.1111/j.1558-5646.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Matzkin L, Ippolito M, Eanes WF. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution. 2005a;59:1721–1732. [PubMed] [Google Scholar]
- Schmidt PS, Paaby AB, Heschel MS. Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution. 2005b;59:2616–2625. [PubMed] [Google Scholar]
- Schmidt PS, Zhu CT, Das J, Batavia M, Yang L, Eanes WF. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc Natl Acad Sci USA. 2008;105:16207–16211. doi: 10.1073/pnas.0805485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgrò CM, van Heerwaarden B, Kellermann V, Wee CW, Hoffmann AA, Lee SF. Complexity of the genetic basis of ageing in nature revealed by a clinal study of lifespan and methuselah, a gene for ageing, in Drosophila from eastern Australia. Mol Ecol. 2013;22:3539–3551. doi: 10.1111/mec.12353. [DOI] [PubMed] [Google Scholar]
- Shelomi M. Where Are We Now? Bergmann’s Rule Sensu Lato in Insects. Am Nat. 2012;180:511–519. doi: 10.1086/667595. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and selection in a cline. Genetics. 1973;75:733–756. doi: 10.1093/genetics/75.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Langley CH, Johnson FM. Variance component analysis of allozyme frequency data from eastern populations of Drosophila melanogaster. Genetics. 1978;88:121–137. doi: 10.1093/genetics/88.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen JG, Norry FM, Scannapieco AC, Loeschcke V. Altitudinal variation for stress resistance traits and thermal adaptation in adult Drosophila buzzatii from the New World. J Evol Biol. 2005;18:829–837. doi: 10.1111/j.1420-9101.2004.00876.x. [DOI] [PubMed] [Google Scholar]
- Spitze K. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics. 1993;135:367–374. doi: 10.1093/genetics/135.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC. How much of the phenotype is necessary to understand evolution at the level of the gene? In: Wöhrmann K, Loeschcke V, editors. Population Biology and Evolution. Springer-Verlag; Berlin and Heidelberg, Germany: 1984. pp. 31–45. [Google Scholar]
- Stearns SC. Comparative and experimental approaches to the evolutionary ecology of development. Geobios (Lyon) 1989;12:349–355. [Google Scholar]
- Stearns SC. The Evolution of Life Histories. Oxford Univ. Press; Oxford, U. K: 1992. [Google Scholar]
- Stearns SC, Partridge L. The genetics of aging in Drosophila. In: Masor E, Austad S, editors. Handbook of the Biology of Aging. Academic Press; San Diego: 2001. pp. 353–368. [Google Scholar]
- Tatar M, Gray DW, Carey JR. Altitudinal variation for senescence in Melanoplus grasshoppers. Oecologia. 1997;111:357–364. doi: 10.1007/s004420050246. [DOI] [PubMed] [Google Scholar]
- Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–1898. doi: 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- Tewksbury JJ, Huey RB, Deutsch CA. Putting the Heat on Tropical Animals. Science. 2008;320:1296–1297. doi: 10.1126/science.1159328. [DOI] [PubMed] [Google Scholar]
- Therneau T. [Accessed December 18, 2014];coxme: Mixed Effects Cox Models Version 2.2–3. 2012 Available at http://cran.r-project.org/web/packages/coxme/index.html.
- Ujiié Y, Asami T, de Garidel-Thoron T, Liu H, Ishitani Y, de Vargas C. Longitudinal differentiation among pelagic populations in a planktic foraminifer. Ecol Evol. 2012;2:1725–1737. doi: 10.1002/ece3.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umina PA, Weeks AR, Kearney MR, McKechnie SW, Hoffmann AA. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science. 2005;308:691–693. doi: 10.1126/science.1109523. [DOI] [PubMed] [Google Scholar]
- Van’t Land J, van Putten P, Zwaan B, Kamping A, van Delden W. Latitudinal variation in wild populations of Drosophila melanogaster: heritabilities and reaction norms. J Evol Biol. 1999;12:222–232. [Google Scholar]
- Via S. The quantitative genetics of polyphagy in an insect herbivore. II Genetic correlations in larval performance within and among host plants. Evolution. 1984;38:896–905. doi: 10.1111/j.1558-5646.1984.tb00360.x. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Smith-Winberry G, Arnold LL, Thompson EM, Cooley AM, Yuan DC, et al. Local adaptation for body color in Drosophila americana. Heredity. 2011;106:592–602. doi: 10.1038/hdy.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaan BJ, Azevedo RBR, James AC, Van’t Land J, Partridge L. Cellular basis of wing size variation in Drosophila melanogaster: a comparison of latitudinal clines on two continents. Heredity. 2000;84:338–347. doi: 10.1046/j.1365-2540.2000.00677.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.