Abstract
Recent human clinical trials results demonstrated successful treatment for certain genetic forms of cystic fibrosis (CF). To extend treatment opportunities to those afflicted with other genetic forms of CF disease, structural and biophysical characterization of CF transmembrane conductance regulator (CFTR) is urgently needed. In this study, CFTR was modified with various tags, including a His10 purification tag, the SUMOstar (SUMO*) domain, an extracellular FLAG epitope, or an enhanced green fluorescent protein (EGFP), each alone or in various combinations. Expressed in HEK293 cells, recombinant CFTR proteins underwent complex glycosylation, compartmentalized with the plasma membrane, and exhibited regulated chloride-channel activity with only modest alterations in channel conductance and gating kinetics. Surface CFTR expression level was enhanced by the presence of SUMO* on the N-terminus. Quantitative mass-spectrometric analysis indicated approximately 10% of the total recombinant CFTR (SUMO*-CFTRFLAG-EGFP) localized to the plasma membrane. Trial purification using dodecylmaltoside for membrane protein extraction reproducibly recovered 178 ± 56 μg SUMO*-CFTRFLAG-EGFP per billion cells at 80% purity. Fluorescence size-exclusion chromatography indicated purified CFTR was monodisperse. These findings demonstrate a stable mammalian cell expression system capable of producing human CFTR of sufficient quality and quantity to augment futrure CF drug discovery efforts, including biophysical and structural studies.
Keywords: CFTR, mammalian cell, overexpression, HEK293, biophysical analysis
INTRODUCTION
Cystic fibrosis (CF) is the most common fatal genetic disease in the western world, with an incidence of approximately 1 in 2500 births (1). CF is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene that encodes a chloride channel belonging to the ATP-binding cassette (ABC) transporter superfamily (2, 3). Defects of CFTR channel function compromise epithelial transcellular fluid regulation in the lungs, pancreas and other organs, and lead to thickening of mucus in the lungs, and eventually tissue sequelae and death. Other complications of CF include malnutrition due to pancreatic insufficiency, electrolyte imbalances, diabetes and male infertility. The most frequent genetic mutation associated with clinical CF disease is deletion of phenylalanine at position 508 (ΔF508) in CFTR. The ΔF508 mutation results in aberrant folding of the CFTR protein, retention of CFTR in the endoplasmic reticulum and premature CFTR protein degradation (4). Interestingly, wild-type CFTR (wtCFTR) appears to fold, mature, and reach the plasma membrane less efficiently compared to other ABC transporters (5-7). Cellular quality control appears to be quite stringent, and even mature CFTR at the cell-surface is endocytosed at a rate of ~10% per min in normal cells (8). N-glycosylation is intimately linked with CFTR folding in the endoplasmic reticulum, and maturation in the Golgi apparatus of glycan chains from the core-glycosylated form (band B) of CFTR to the final complex-glycosylated form (band C) often is used experimentally as a surrogate for proper CFTR folding and trafficking to the cell-surface (6, 9).
The architecture of CFTR is similar to that of other ABC transporters, consisting of transmembrane domains harboring the chloride pore, connected by cytoplasmic loops to two nucleotide binding domains (NBDs) that hydrolyze ATP (10, 11). CFTR features a unique regulatory (R) region that, when phosphorylated, regulates ATP hydrolysis-mediated channel gating (7, 12). Limited insights toward understanding this complex structure, interactions between subdomains, and preliminary functional models have been provided by low resolution structures of full-length CFTR, crosslinking experiments, nuclear magnetic resonance studies of partial molecules, and molecular modeling based on crystal structures of other ABC transporters (13-16).
Drug discovery efforts based on a variety of CFTR ‘correctors’ that improve trafficking of misfolded protein to the plasma membrane or ‘potentiators’ that improve channel function (17) were initially disappointing due to poor potency of the compounds (18-20). However, the recent FDA approval and clinical success using ivacaftor to treat CF patients with CFTR gating mutations (http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm290088.htm) firmly validates rationale for the discovery of small-molecule CF drugs. Since patients with gating mutations and other mutations that can be treated with Ivacaftor represent fewer than 15% of the over 1900 CF-causing mutations identified to date (http://www.cftr2.org/), it seems likely that numerous CF drugs will be needed to extend treatment opportunities to greater numbers of individuals afflicted with CF (21). To augment drug discovery, direct structural and biophysical characterization of wild-type and mutated forms of CFTR is urgently needed, but has been impeded by both difficulties producing significant quantities of CFTR and the limited stability of purified protein. The goal of the present study was to characterize a molecular expression strategy that was conceived to facilitate the identification and derivation of stable cell lines for high-level production of full-length CFTR. Our results demonstrated robust expression in human embryonic kidney (HEK) epithelial cells of exogenous full-length human CFTR comprising various domains and tags that facilitate the derivation of stable high-producer cell lines, assessment of CFTR biogenesis, and characterization of recombinant CFTR protein. These findings are significant as they demonstrate a mammalian cell expression system capable of producing human CFTR of sufficient quality and quantity to support biophysical and structural studies.
MATERIALS AND METHODS
CFTR expression vectors
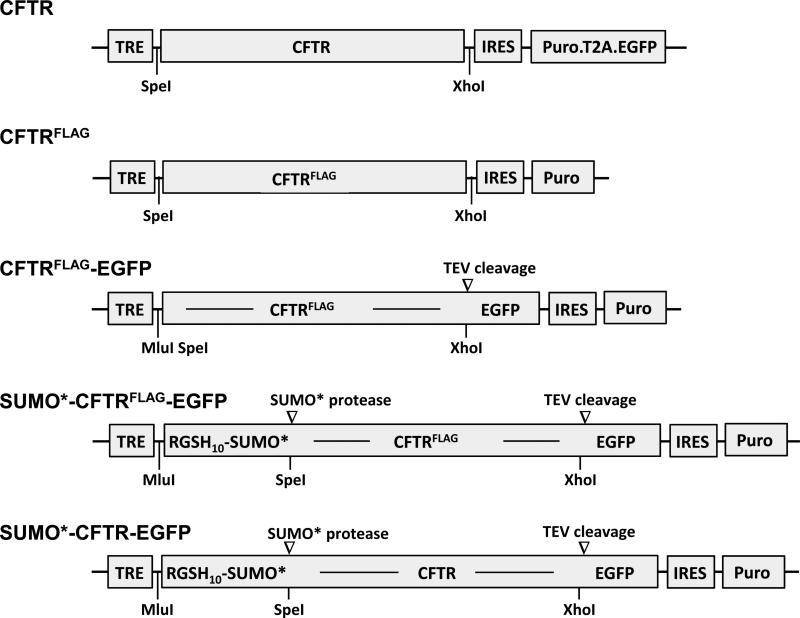
Schematics of the HIV-1-based lentiviral vectors used in these studies are shown in Fig. 1. The basic molecular genetic structure of these vectors included LTR-ψ-RRE-CTS/PPT-TRE-MCS-IRES-Puro-WPRE-ΔU3.LTR (LTR, long-terminal repeat; ψ, psi/RNA genome packaging signal; RRE, Rev response element; CTS, central termination sequence; PPT, polypurine tract; TRE, tetracycline response element; MCS, multiple cloning site; IRES, internal ribosome entry site; Puro, puromycin N-acetyl-transferase gene; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; and ΔU3.LTR, deletion in the U3 sequence of the LTR). Most of these genetic elements have been described previously (22-28). The arrangement of the TRE and IRES allows genes inserted into the MCS and those downstream of the IRES to be expressed from a single mRNA transcript.
Figure 1. Vectors used to generate stably transduced HEK293 cell lines expressing recombinant CFTR.
Illustration of expression cassettes comprising wild-type CFTR (CFTR), CFTR containing the FLAG epitope tag in the fourth extracellular loop (CFTRFLAG), CFTRFLAG fused in-frame with EGFP (CFTRFLAG-EGFP), RGSHis10-SUMO*-tagged CFTRFLAG-EGFP (SUMO*-CFTRFLAG-EGFP), and RGSHis10-SUMO*-tagged CFTR-EGFP (SUMO*-CFTR-EGFP). EGFP encodes the A206K mutation to minimize self-dimerization (25). Each of the illustrated expression cassettes represents a genetic sequence inserted into a lentiviral vector for delivery and stable integration into HEK293.M2 cells. These vectors contain a TRE promoter followed by CFTR sequence, IRES, and either the puromycin resistance gene or a puromycin-T2A-EGFP open reading frame. T2A, a member of the 2A peptide family, comprises a 19 amino acid sequence that causes a co-translational separation in the synthesis of the puromycin and EGFP polypeptides (30-32).
The TRE-CFTR-IRES-Puro.T2A.EGFP vector (K2933) was derived from a wtCFTR expression vector described previously (29) by inserting a Puro element followed by a Thosea asigna 2A-like peptide (T2A) coding sequence upstream and in-frame with enhanced green fluorescent protein (EGFP) (30-32). The CFTR FLAG-containing expression vector, TRE-CFTRFLAG-IRES-Puro (K3103) was created by polymerase chain reaction (PCR) amplification of a CFTR sequence containing the FLAG octapeptide epitope (DYKDDDDK) after residue N901 (33, 34), and its ligation into the 5’ NheI and 3’ XhoI sites of the lentiviral vector. Published studies indicate that inclusion of a FLAG tag in the 4th extracellular loop (proximal to residue 901) enables cell surface localization of CFTR without altering its expression (33, 34). The expression vector, TRE-CFTRFLAG-EGFP-IRES-Puro (K3290), was generated by ligating an A206K mutated EGFP (25) sequence in-frame and downstream of CFTRFLAG. The translational stop codon of CFTR was eliminated and a tobacco etch virus (TEV) protease cleavage site (underlined) (35) and a glycine-serine hinge were introduced between the CFTRFLAG and EGFP genes (CFTRFLAG-ENLYFQGGGGSGGSS-EGFP). The TRE-SUMO*-CFTRFLAG-EGFP-IRES-Puro expression vector (K3235) was generated by inserting a DNA segment coding for MERGSH10-LVPRGSAS-SUMOstar (synthesized by GeneArt/Life Sciences) in-frame at the 5’ end of CFTRFLAG-EGFP. The N-terminal RGSHis10 tag enables affinity purification and immunodetection of the recombinant protein. The His-tag is cleavable by the presence of a Thrombin protease cleavage site (underlined). Small ubiquitin-like modifier (SUMO, Saccharomyces cerevisiae Smt3) and SUMOstar (SUMO*) domains have been shown to enhance folding and solubility of fused recombinant proteins (36, 37), including isolated CFTR NBDs (38). SUMO* is modified at two interfacial amino acids, R64T and R71E, rendering resistance to cleavage by intrinsic eukaryotic proteases (39). The SUMO* polypeptide can be removed from its fusion partner with specific proteases (37, 40). The integrity of each of the recombinant expression vectors was confirmed by nucleotide sequence analysis. The entire ORF sequence of SUMO*-CFTRFLAG-EGFP was deposited in GenBank (accession KP202880).
Cell lines and growth conditions
HEK293 (293F; Invitrogen), HEK293.M2 (D017) (41), and cell lines derived from HEK293.M2 cells by lentiviral vector transduction were maintained as adherent cultures in DMEM/F12 medium supplemented to contain 10% fetal bovine serum (FBS) (HyClone), 100 U/mL penicillin and 0.1 mg/mL streptomycin (Life Technologies). The HEK293.M2 cell line (41) constitutively expresses a modified form of the reverse tetracycline transactivator (rtTA-M2) for specific and sensitive doxycycline (dox)-inducible gene expression under control of the tetracycline response element (42). All HEK293-derived cell lines that were adapted to serum-free suspension-culture, were maintained in CDM4HEK293 medium (HyClone) supplemented to contain 100 U/mL penicillin, 0.1 mg/mL streptomycin, 2 mM L-glutamine, 2 mM L-alanyl-L-glutamine dipeptide, 0.25 μg/mL amphotericin B, and 1:1000 (v:v) anti-clumping agent (Life Technologies). Suspension culture-adapted cells were propagated in either 1050 cm2 smooth surface roller bottles (Thermo Scientific) or a 14L autoclavable bioreactor supported by a New Brunswick BioFlo 310 benchtop fermentor system (Eppendorf) http://newbrunswick.eppendorf.com/en/products/fermentors/.
Generation of recombinant CFTR cell lines
The 293T/17 cell line (ATCC®) used for packaging of all lentiviral vector stocks was maintained in DMEM supplemented to contain 10% FBS, 100 U/mL penicillin and 0.1 mg/mL streptomycin. Lentiviral vector genomes containing the different CFTR genetic recombinants were packaged by cotransfecting 293T/17 cells with pCMVΔR8.2 packaging plasmid DNA and vesicular stomatitis virus envelope glycoprotein plasmid DNA (24, 43). Culture supernatants were collected after 60 hrs, clarified by low-speed centrifugation (250 × g, 10 min), filtered through 0.45 μm sterile filters, and concentrated by ultracentrifugation at 125,000 × g at 4°C for 2 hrs. For transduction, 105 HEK293.M2 (D017) cells in 200 μl of DMEM/F12/1% FBS were incubated with concentrated packaged vector at 37°C for 18 hrs. After 2-3 days, 1 μg/ml dox was added to the culture medium, and 24 and 48 hrs later, transgene expression was assessed by fluorescence microscopy. Cultures exhibiting a positive response in less than 40% of the cell population were discarded. Those with favorable expression were cultured for 7-9 days in the presence of 10 μg/ml puromycin. All selected cell cultures were confirmed to be >95% expression-positive for the respective transgene by monitoring either EGFP or CFTR as described below. Puromycin-selected cell populations were expanded in the absence of dox and puromycin and cryopreserved in liquid nitrogen. Subsequent studies of the different CFTR cell lines were performed with monolayer cultures, except where suspension-culture-adapted cells have been specified.
Western blot analysis
Western blot analysis of cells expressing recombinant CFTR was performed using methods reported earlier (44, 45). Briefly, cells were solubilized in sample buffer (46), sonicated, and heated at 37°C for 5 min prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 7% gels. Proteins were transferred to nitrocellulose by electroblotting, and detected using either anti-RGSHis4 (Qiagen) or R1104 anti-CFTR mouse monoclonal antibody recognizing an epitope comprising amino acids 722-734 in the CFTR regulatory (R) region (47). Horseradish peroxidase-conjugated goat anti-mouse IgG (Southern Biotech) was used as secondary antibody with Immobilon Western Chemiluminescent substrate (Millipore). The chemiluminescent signal was captured and analyzed on an ImageQuant LAS 4000 Mini luminescent image analyzer (GE Healthcare Life Sciences).
Flow cytometry
Cellular expression and plasma membrane compartmentalization of CFTR were analyzed using flow cytometry to detect EGFP expression and FLAG immunostaining. Live cells were equilibrated in room temperature (RT) staining buffer for 15 min, incubated with 5 μg/ml of SureLight APC anti-FLAG M2 mAb for 30 min at RT, washed three times with ice-cold staining buffer, detached by incubation on ice with phosphate-buffered saline containing 0.5 mM EDTA, washed once with staining buffer and resuspended in 200 μl Cytofix (BD Biosciences). Both EGFP fluorescence and FLAG/CFTR staining were measured using a BD FACSCalibur or LSRII flow cytometer (BD Biosciences). Negative/positive fluorescence boundaries were determined using HEK293.M2 cells. Flow cytometric data were analyzed using FlowJo software.
Derivation of clonal cell cultures
All recombinant CFTR cell lines analyzed in this study comprised mass populations of transduced cells unless otherwise noted. To generate clonal cell lines, cultures treated for 18-24 hrs with 1 μg/ml of dox were live-stained using SureLight allophycocyanin (APC)-conjugated anti-FLAG M2 (PerkinElmer). The most highly fluorescent cells (top 10%) were sorted individually into wells of 96-well plates using a FACS Aria (BD Biosciences). Initial screening of clonal cell cultures included analyses of cellular morphology, growth kinetics, and levels of basal and dox-induced EGFP and/or FLAG/CFTR expression using fluorescence microscopy. Clonal cultures exhibiting favorable characteristics were expanded for further analyses and cryopreservation of master stocks. Clonal cell lines adapted to serum-free suspension-culture were reanalyzed to ensure growth kinetics. Relative CFTR expression levels in suspension culture adapted cells were not predictable based on the analysis of the corresponding monolayer cells, and therefore, CFTR expression properties were re-determined in each instance. To confirm the integrity of the recombinant CFTR sequence in the D165 clonal line, PCR amplicons comprising the SUMO*-CFTRFLAG-EGFP open reading frame were generated by PCR, sequenced without cloning, and found to be identical to the transducing vector (data not shown).
Single-channel recording
Microsomal membrane fractions were prepared from the HEK293-derived cell lines overexpressing CFTR recombinant proteins or from BHK cells constitutively expressing wtCFTR, as described previously confirm citation (48). Membrane vesicles were phosphorylated by incubation with 50 nM PKA catalytic subunit (Promega) and 2 mM Na2ATP (Sigma-Aldrich) in 10 mM Hepes, pH 7.2, 0.5 mM EGTA, 2 mM MgCl2, and 250 mM sucrose at RT for 15 min. Aliquots were stored at −80°C until used. Microsomal vesicles were fused to planar lipid bilayers and single-channel currents were recorded at 30°C in symmetrical buffer (300 mM Tris HCl pH 7.2, 3 mM MgCl2, 1 mM EGTA) at −75 mV under voltage-clamp conditions as described previously (49). To maintain uniform channel orientation and optimal functional state, 2 mM Na2ATP, 50 nM PKA and 10 μl membrane vesicles were added to the one side of the bilayer only. Output signal was filtered with a cut-off frequency of 50 Hz.
Fluorescence microscopy
Cells in 8-chamber glass slides (Falcon CultureSlide) coated with bovine type I collagen (BD Biosciences) were treated with dox for 24 hrs. Live cells were immunostained without fixation by incubation on ice with 5 μg/ml anti-FLAG M2 mAb (Sigma-Aldrich) followed by cold washing and incubation with Alexa Flur® 594-conjugated goat anti-mouse IgG (H+L) antibody (Life Technologies) on ice for 30 min. Cells were washed with cold staining buffer (BD Biosciences), and then fixed for 15 min with Cytofix (BD Biosciences). Cells were washed, cured with ProLong® Gold antifade reagent containing 4’,6’-diamidino-2-phenylindole (DAPI) (Life Technologies) to stain cell nuclei, and mounted with a coverslip. Confocal images were acquired using a Zeiss LSM 710 confocal microscope with a 63x PlanApo oil immersion objective at a resolution of 1024x1024 pixels. A Helium Neon 561 nm excitation laser was used to excite Alexa Flur® 594 and image spectra at 590-700 nm. Optical sections in the z-axis (z-stack) were acquired at 0.45 micron intervals. Images of DAPI-stained cell nuclei (blue) were acquired using a 405 nm laser. For protein colocalization studies, fluorescence was analyzed using a Nikon Eclipse TE2000-S inverted microscope equipped with an Xcite 120 Fluorescent Illumination system. Images were captured and analyzed using a SPOT RT3 25.4 Color Slider camera (Diagnostics Imaging, Inc., SPOT™ Imaging Solutions division) supported with Qcapture Pro software (QImaging, Inc.).
Mass spectrometry quantitation of cell-surface CFTR
Heavy isotope-labeled CFTR was prepared from baby hamster kidney (BHK) cells expressing wtCFTR as described previously (50) using 99% atom-enriched 1,2-[13C]2-L-leucine as the isotope-labeled amino acid. A purified CFTR external standard was used to determine concentrations of labeled CFTR present in Triton X-100 cellular extracts prepared from the BHK cells (50). To specifically quantify CFTR compartmentalized in the plasma membrane, intact cells were surface biotinylated and biotinylated protein was isolated using the EZ-Link Sulfo-NHS-SS-Biotin kit (Thermo Scientific). Protein fractions (biotinylated protein or Triton X-100 cell extracts) were spiked with the L-leucine isotope-labeled cell lysate, and then subjected to SDS-PAGE, in-gel protein digestion and peptide cleanup as previously described previously (50). Three human CFTR signature peptides designated CFTR01, CFTR02 and CFTR04 of the sequences shown in Table 1 were used to quantify the isolated CFTR protein fractions using liquid chromatography multiple reaction monitoring-mass spectrometry (50, 51). The instruments used were an Eksigent NanoLC-ultra 2D Plus liquid chromatograph and 4000 QTrap mass spectrometer (ABSCIEX).
Table 1.
Mass spectrometry transitions
| Peptide | Sequence | Type of transition | Transition |
|---|---|---|---|
| Native CFTR01 | NSILTETLHR | [M+2H]2+→y7 | 592.3→869.5 |
| SILAC CFTR01 | NSI-[1,2-13C-L]-TET-[1,2-13C-L]-HR | [M+2H]2+→y7 | 594.8→873.5 |
| Native CFTR02 | LSLVDSEQGEAILPR | [M+2H]2+→y12 | 862.5→1311.7 |
| SILAC CFTR02 | [1,2-13C-L]S[1,2-13C-L]VDSEQGEAI[1,2-13C-L]PR | [M+2H]2+→y12 | 865.5→1313.7 |
| Native CFTR04 | NSILNPINSIR | [M+2H]2+→y8 | 620.8→926.5 |
| SILAC CFTR04 | NSI[1,2-13C-L]NPINSIR | [M+2H]2+→y8 | 621.9→928.5 |
SILAC: stable isotope labeling in cell culture
CFTR purification
SUMO*-CFTRFLAG-EGFP was solubilized from microsomal membranes with n-dodecyl-β-D-maltoside (DDM) and affinity purified using NiNTA resin (Qiagen) according to manufacturer-recommended procedures. In-gel fluorescence with external standards was used to quantify SUMO*-CFTRFLAG-EGFP in crude and purified fractions (52). Samples were resolved on 8% SDS-PAGE gels together with known amounts of SUMO-EGFP fusion protein (39 kD, LifeSensors). Fluorescence of EGFP-tagged proteins was imaged using a Typhoon scanner (GE Healthcare), and densitometry of fluorescent bands was performed in ImageJ (http://imagej.nih.gov/ij/).
Fluorescence size exclusion chromatography (FSEC)
FSEC of purified SUMO*-CFTRFLAG-EGFP was performed on a 3.2 mm × 30 cm Superose 6 column (GE Healthcare) in 50 mM Tris Cl pH 7.5, 0.15 M NaCl, 10% glycerol, 2.5 mM MgCl2, 0.05% DDM, 0.2 mM tris(2-carboxyethyl)phosphine, flowing at 0.04 ml/min. EGFP fluorescence was monitored at 488 nm/509 nm using a Jasco FP2020 Plus fluorimetric detector, with gain set at 100 and attenuation at 64.
RESULTS
Analysis of recombinant CFTR protein expression
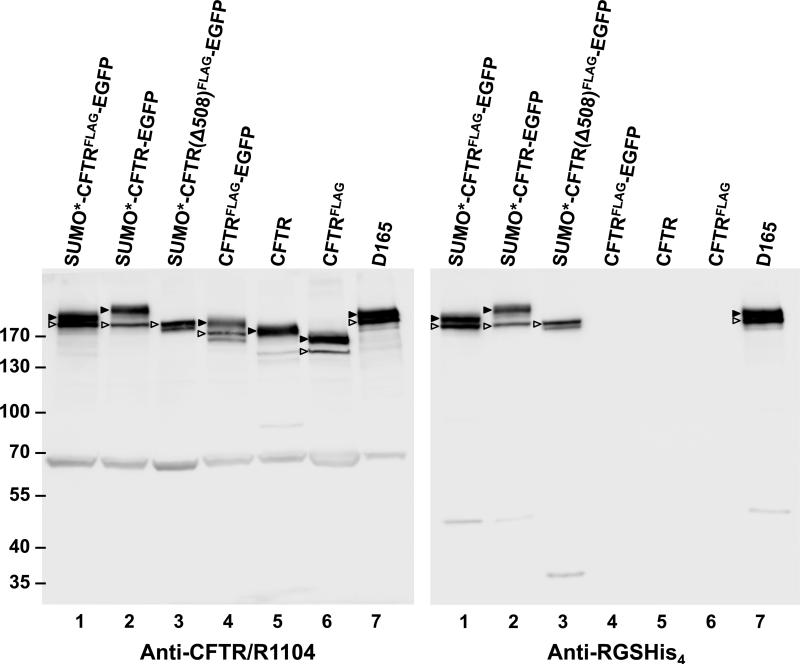
To explore the utility and performance of various in-frame domain fusions of recombinant CFTR, the HEK293.M2 cell line (41) was transduced with either the SUMO*-CFTRFLAG-EGFP, SUMO*-CFTR-EGFP, CFTRFLAG-EGFP, CFTRFLAG, or wtCFTR expression vector (Fig. 1). The resulting cultures were induced with dox for 24 hrs and CFTR expression was analyzed by Western blot (Fig. 2). Replicate blots were probed with either anti-CFTR mAb (R1104) or N-terminal-specific anti-RGSHis4 mAb. SUMO*-CFTRFLAG-EGFP, CFTRFLAG and CFTR proteins were detected at similar levels, and slightly higher than CFTRFLAG-EGFP (lane 4). SUMO*-CFTRFLAG-EGFP (lane 1) resolved into a double band, indicating expression of both core-glycosylated (band B) and complex glycosylated (band C) CFTR. Greater electrophoretic mobility of CFTRFLAG compared to CFTR was evident on SDS-gels (lane 6 vs 5, also lane 1 vs 2), and is likely due to disruption of one of the two glycosylation sites caused by insertion of the FLAG epitope (33, 34, 53, 54). The CFTRFLAG-EGFP protein exhibited both bands B and C (lane 4), while wtCFTR and CFTRFLAG (lanes 5 and 6, respectively) were expressed at high levels with a predominance of band C. Cells expressing the ΔF508 form of SUMO*-CFTRFLAG-EGFP (lane 3) exhibited reduced expression of band C, consistent with expectations for this CFTR mutant to be retained in the endoplasmic reticulum (ER) in its core-glycosylated form (4, 55). D165 (lane 7) is a clonal derivative of the SUMO*-CFTRFLAG-EGFP cell population (lane 1), which exhibited a similar immunoblot phenotype. These results provided qualitative evidence that the cell expression system utilized can produce recombinant CFTR containing various tags, including the multiply tagged SUMO*-CFTRFLAG-EGFP recombinant protein.
Figure 2. Western blot analysis of recombinant CFTR expression.
Puromycin resistant and dox-induced monolayer cultures of transduced HEK293.M2 cells were lysed in Laemmli sample buffer, adjusted to 5 × 104 cell equivalents per 10 μl, and analyzed by immunoblotting with either anti-CFTR mAb R1104 or anti-RGSHis4 mAb. Arrowheads mark complex-glycosylated (band C ▶) and core-glycosylated (band B ) forms of CFTR. In FLAG-tagged proteins, one of the two sites for N-glycosylation was disrupted. By densitometry, about 68% and 33 ± 10% (n=10) of the SUMO*-CFTRFLAG-EGFP protein was found in bands C and B, respectively. Molecular-weight markers are shown on the left in kDa. Calculated masses of the SUMO*-CFTRFLAG-EGFP, CFTRFLAG-EGFP and CFTR polypeptides are 212, 197, and 168 kDa, respectively. The 65 kDa band recognized by CFTR-specific antibody R1104 was not observed with RGS-His antibody or by in-gel EGFP fluorescence, and could represent a CFTR fragment or nonspecific cross-reactive protein. The results shown are representative of two independent analyses.
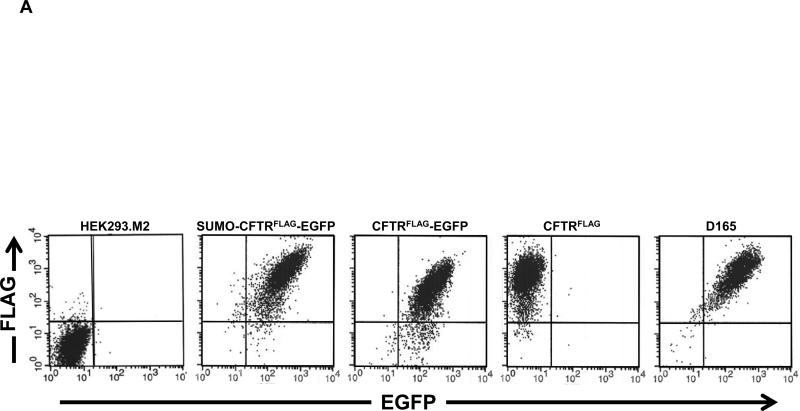
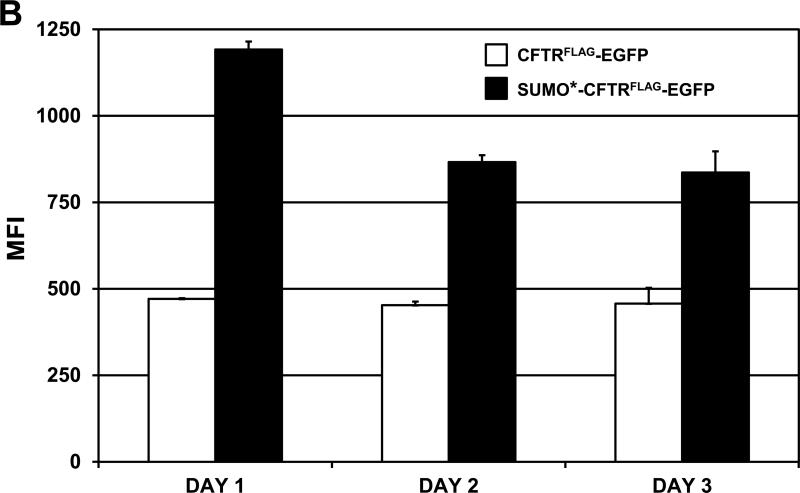
Cells expressing CFTRFLAG, CFTRFLAG-EGFP, SUMO*-CFTRFLAG-EGFP (D158) and its clonal derivative (D165), were analyzed using flow cytometric methods to compare CFTR expression levels and surface compartmentalization. Cells transduced with CFTRFLAG stained positive for the FLAG epitope, and each of the CFTRFLAG–EGFP-containing cell lines was decidedly positive for both EGFP and FLAG staining (Fig. 3A). Importantly, since immunostaining was performed on live/intact cells, positive FLAG staining indicated compartmentalization of the recombinant CFTR protein in the plasma membrane (and see Fig. 5). The overall magnitude of EGFP and FLAG-stain fluorescence was reproducibly greater for SUMO*-CFTRFLAG-EGFP (both D158 and D165 cell lines) compared to CFTRFLAG-EGFP, possibly due to enhanced stability. Similarly, cell-surface FLAG staining of CFTRFLAG appeared to be greater than that of CFTRFLAG-EGFP. The flow cytometric distribution of the clonal (D165) SUMO*-CFTRFLAG-EGFP cell population was similar to the nonclonal (D158) population, but with a greater proportion of dual-positive cells. To quantitatively compare surface expression of the SUMO*-CFTRFLAG-EGFP and CFTRFLAG-EGFP recombinant proteins, the respective cell lines were induced with dox for 24, 48 and 72 hrs, live-stained for FLAG, and analyzed by flow cytometry (Fig. 3B). Mean fluorescence intensity (MFI) values for SUMO*-CFTRFLAG-EGFP were greater at each time point compared to CFTRFLAG-EGFP, with the greatest difference observed 24 hrs after induction. These differences in surface CFTR expression level were consistent with the degree of complex glycosylation observed by Western blot analysis (Fig. 2). These results suggest that introducing the N-terminally fused SUMO* domain to CFTRFLAGEGFP increased its cell-surface expression of the protein.
Figure 3. Analysis of recombinant CFTR expression by flow cytometry.
(A) Cells were treated with 1 μg/ml of dox for 24 hrs and then live-stained for surface FLAG/CFTR expression using SureLight anti-FLAG mAb. The distribution of fluorescence intensity of EGFP and FLAG staining is shown for each of the cell populations analyzed: HEK293.M2 (control), SUMO*-CFTRFLAG-EGFP, CFTRFLAG-EGFP, CFTRFLAG and D165. D165 is a clonal derivative of the D158 cell line expressing SUMO*-CFTRFLAG-EGFP. HEK293.M2 cells that do not express CFTR were included as a negative control. (B) The indicated cultures were immunostained for cell-surface FLAG epitope 24, 48 or 72 hrs after dox induction and analyzed by flow cytometry as in (A). LinearFlow® Fluorescently labeled polystyrene beads (Molecular Probes) were analyzed in parallel to control for possible inter-assay variation of the flow cytometer. MFI values for each cell population were calculated from two independent experiments.
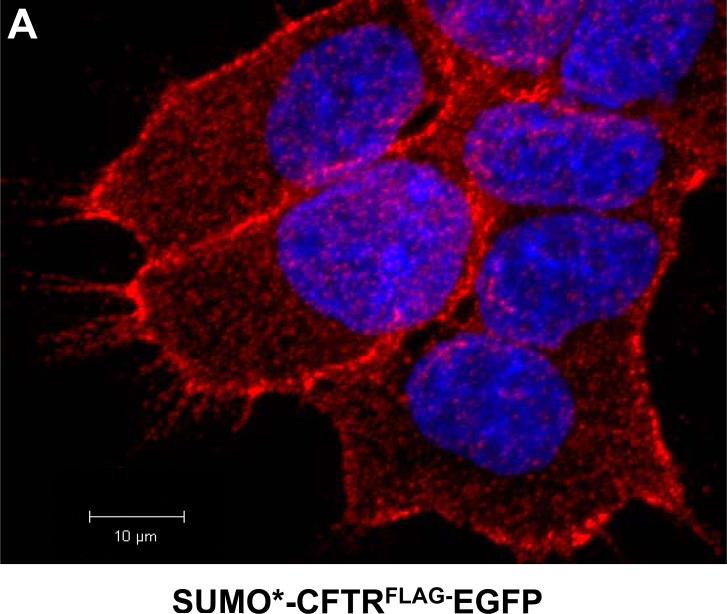
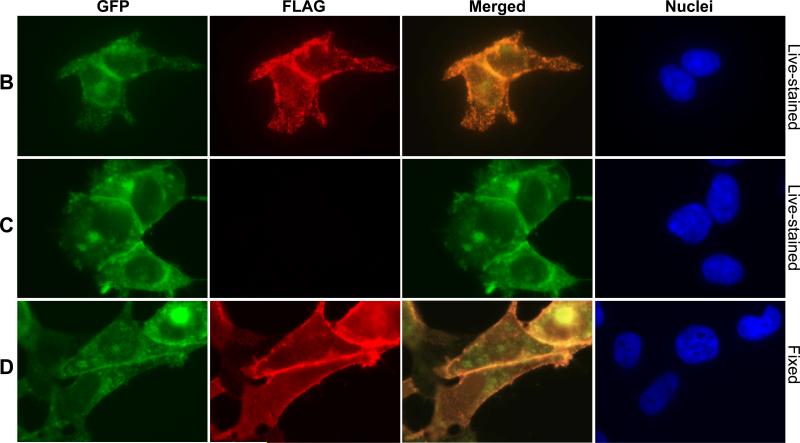
Figure 5. Immunofluorescence microscopy analysis of SUMO*-CFTRFLAG-EGFP cell-surface expression.
(A) SUMO*-CFTRFLAG-EGFP cells were treated with dox (1 μg/ml) for 24 hrs, and then live-stained for extracellular FLAG. FLAG immunostaining was analyzed by confocal laser-scanning microscopy. A z-stack of 0.45 micron optical sections was captured, and the composite image is illustrated in red with DAPI-stained cell (blue) nuclei. (B-D) For analysis of protein colocalization, expression of SUMO*-CFTRFLAG-EGFP (B, D) and SUMO*-CFTR-EGFP (C) was dox-induced (1 μg/ml) for 24 hrs and examined by wide-field fluorescence microscopy using a Nikon Eclipse TE2000-s inverted microscope. Either live/intact (B and C) or fixed and permeabilized (D) cells were immunostained to detect the FLAG epitope. Images of EGFP (green) and FLAG-stain fluorescence (red) are depicted. A composite of the two fluorescent images (merged) is shown in column 3. DAPI-stained cell nuclei are shown in blue.
Channel activity of CFTR in cell membranes
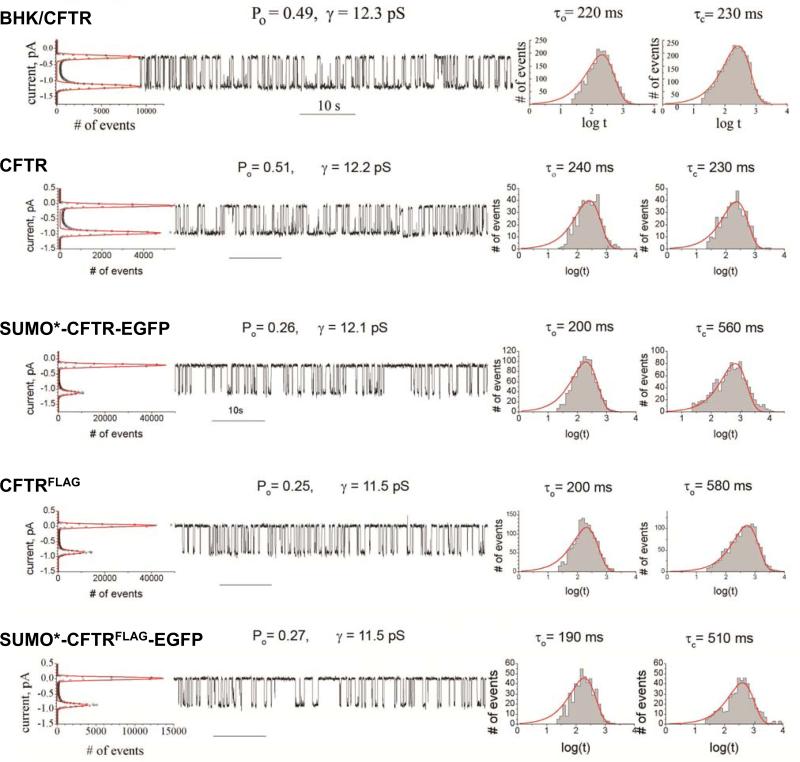
Single channel recording provides a means to validate function of recombinant CFTR function (56, 57). Expressed wtCFTR in BHK and HEK cell lines (Fig. 4) exhibited single-channel conductance (γ) of 12.3 ± 0.1 pS (n=6) and 12.2 ± 0.1 pS (n=7), respectively. Slight differences in conductance and gating kinetics were not significant, indicating the channel properties of untagged wtCFTR were independent of the cell line in which it was expressed. Modification of CFTR with both SUMO* and EGFP tags (Fig. 4) did not affect single channel conductance, but approximately doubled the mean closed time (τc) with no significant change in the mean open time (τo). As a result, the open probability (Po) decreased approximately two-fold, to 0.26. CFTRFLAG exhibited a single-channel conductance of 11.5 ± 0.1 pS (n=7), which was a statistically significant reduction compared to wt CFTR (P<0.05), and could reflect the influence of additional negative charge (due to FLAG sequence, DYKDDDDK) near the outer vestibule. Po and gating kinetics of CFTRFLAG were essentially the same as those of the SUMO*-CFTR-EGFP construct. The CFTR construct containing all three tags, SUMO*-CFTRFLAG-EGFP, had the same conductance as CFTRFLAG and gating kinetics typical of the individually SUMO*- or FLAG-tagged constructs. The influence of the latter two modifications on gating kinetics was not additive.
Figure 4. CFTR channel analysis.
Single-channel currents detected in microsomal membranes isolated from BHK cells constitutively expressing wild-type CFTR (top panel) and HEK293.M2 cells expressing either wild-type (second panel), or recombinant forms of CFTR (lower three panels), are shown in the middle section of each panel. All-points histograms used to define single-channel current as the distance between peaks on the graph and calculate single-channel conductance (γ) are shown on the left of each panel. Values for probability of the open state (Po) were calculated as the ratio of the area under the peak for the open state to the total area under both peaks on the all-points histogram. Dwell-time histograms for the mean open (τo) and closed (τc) times are shown on the right of each panel. The validity of the reduced two states kinetic model C ⇌ O is evident from the single exponential fit shown in each dwell time histogram.
Microscopic visualization of CFTR surface localization
Expression of recombinant SUMO*-CFTRFLAG-EGFP protein was initially analyzed by immunofluorescence confocal microscopy. Dox-induced cells, live-stained to specifically detect surface FLAG, exhibited a strong fluorescence pattern particularly evident along the cell periphery. A z-stack of acquired images illustrates a strong punctate pattern of FLAG staining (red) that is broadly distributed across the cells (Fig. 5A). The control non-FLAG-stained cells exhibited no Alexa Flur® fluorescence (data not shown). In fluorescence microscopy colocalization experiments, FLAG immunostaining (red) was detected with live-stained cells expressing SUMO*-CFTRFLAG-EGFP (Fig. 5B), but not (FLAG-less) SUMO*-CFTR-EGFP cells (Fig. 5C). In each cell type, EGFP was detected throughout the cell (green), with the brightest areas of fluorescence appearing along portions of the cell periphery and at cell-cell interfaces. Merging the FLAG and EGFP fluorescent images demonstrated extensive colocalization of the two domains at the plasma membrane. FLAG immunostaining of fixed, permeabilized cells expressing SUMO*-CFTRFLAG-EGFP (Fig. 5D) demonstrated a subcellular localization pattern similar to that of EGFP, further establishing colocalization of EGFP with CFTR protein. These results indicate compartmentalization at the plasma membrane of the recombinant SUMO*-CFTRFLAG-EGFP protein.
Quantitative analysis of CFTR expression by mass spectrometry
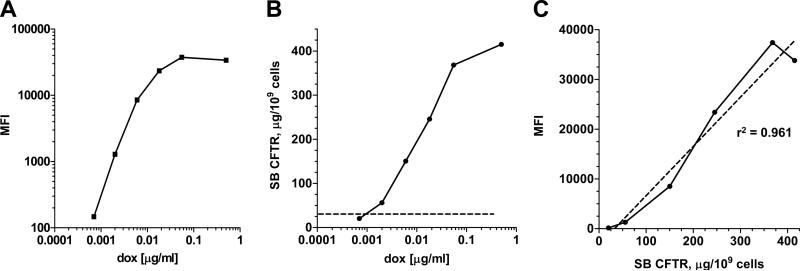
Since the rtTA-M2 enables incremental control of transcription, SUMO*-CFTRFLAG-EGFP expression was induced by treating cells with different concentrations of dox, ranging from 0.0007 to 0.5 μg/ml. Cells were collected after 24 hrs and analyzed for surface CFTR. Flow cytometric analysis demonstrated a dose-response effect of dox concentration on surface FLAG/CFTR expression level (Fig. 6A). Samples from the same cultures were further analyzed by surface biotinylation and mass spectrometry to quantify surface CFTR. In this experiment, cell-surface CFTR levels plateaued at ~400 μg per 109 cells, or 1.1 × 106 molecules CFTR per cell (Fig. 6B). In three independent experiments, the average maximum induced level of surface CFTR was 363 ± 31 μg per 109 cells. The average level of total cellular CFTR was 3581 ± μg per 109 cells (Table 2). Thus, when overexpressed at these high levels, ~1/10th of the CFTR synthesized was present on the cell-surface. Over the range of dox concentrations analyzed there was a very strong linear correlation (r2 = 0.961) between mass spectrometric and flow cytometric measurements of cell-surface SUMO*-CFTRFLAG-EGFP (Fig. 6C). These results indicated that HEK293.M2 cells biosynthesize and traffic appreciable quantities of recombinant CFTR to the plasma membrane, and flow cytometry of cells live-stained for the FLAG epitope can be used to assess CFTR surface expression.
Figure 6. Quantitative mass spectrometry analysis of cell-surface SUMO*-CFTRFLAG-EGFP.
Replicate SUMO*-CFTRFLAG-EGFP cultures (D165 cells) comprising approximately 107 cells were treated for 24 hrs with dox concentrations ranging from 0.0007 to 0.5 μg/ml. (A) Relative surface expression levels of SUMO*-CFTRFLAG-EGFP were determined using flow cytometry to analyze fluorescence from intact/live cells immunostained with SureLight anti-FLAG M2 mAb. The mean fluorescence intensities (MFI) of stained cell populations were plotted for each dox-treatment concentration. (B) In parallel replicate cultures, mass measurements of surface biotinylated (SB) SUMO*-CFTRFLAG-EGFP were determined by quantitative mass spectrometry. The background value, determined by analyzing HEK293 cells, is demarked on the plot by the horizontal dotted line. Results are expressed as μg of SB CFTR per 109 cells. (C) Correlation between mass spectrometry (SB CFTR, μg/109 cells) and flow cytometry (MFI) measurements of surface CFTR. The linear regression coefficient, r2, = 0.961.
Table 2.
Quantitative analyses of SUMO*-CFTRFLAG-EGFP expression
| μg CFTR per 109 cells | Quantitation method | |
|---|---|---|
| Total cellular CFTR | 3581 ± 674 (n=3) | mass spectrometry |
| CFTR at the cell-surface | 363 ± 31 (n=3) | mass spectrometry |
| Microsomes | 1365 ± 485 (n=9) | EGFP fluorescence |
| DDM extract1 | 905 ± 220 (n=8) | EGFP fluorescence |
| Affinity purified CFTR1 | 178 ± 56 (n=10) | EGFP fluorescence |
CFTR was extracted and purified from microsomal fractions as in Figure 7. Values are means standard deviation for n replicate experiments.
Demonstration of CFTR purification using a mild detergent
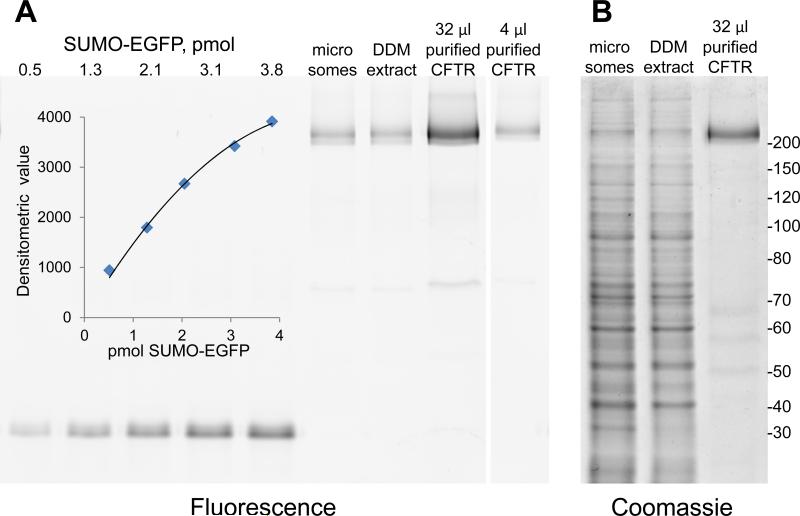
Inclusion of EGFP in the recombinant SUMO*-CFTRFLAG-EGFP protein makes it possible to quantify CFTR even in crude cell lysates using in-gel fluorescence (52). We applied this method to quantify SUMO*-CFTR-EGFP in microsomal membranes, and monitor its recovery through the trial protein purification shown in Fig. 7. The starting material, a microsomal fraction of 0.15x109 cells harvested from a 1 liter serum-free suspension-culture, contained 235 μg of SUMO*-CFTRFLAG-EGFP (1600 μg per 109 cells). DDM is a preferred mild detergent for structural biology, but it has been successfully applied for CFTR purification only with low-expressing mammalian cell lines (57, 58). Extraction of microsomes with DDM solubilized 221 μg of SUMO*-CFTRFLAG-EGFP (1470 μg per 109 cells) (Fig. 7A). With this abundant starting material, and taking advantage of the engineered His10-tag, NiNTA affinity chromatography was employed to isolate 42 μg SUMO*-CFTRCFTR-EGFP (280 μg per 109 cells) with an overall recovery of 18% (Fig. 7A). Coomassie blue staining of the same gel indicated that ~80% pure CFTR was achieved in a single chromatography step (Fig. 7B).
Figure 7. Facile detergent solubilization and affinity purification of SUMO*-CFTRFLAG-EGFP.
Microsomes isolated from D165 suspension-culture cells were solubilized with DDM and the extract was bound to NiNTA. After washing, SUMO*-CFTRFLAG-EGFP was eluted with imidazole. (A) Samples of microsomes (8 μg), DDM extract (8 μg), and NiNTA purified (~80%) CFTR (32 μl, 4 μl) were resolved on an 8% SDS-PAGE gel; a fluorescent image of the gel is shown. Known amounts of SUMO-EGFP fusion protein were loaded on the gel to generate a standard curve (inset), allowing quantitation of fluorescent CFTR bands. (B) A section of the same gel shown in (A) stained with Coomassie blue. Masses of molecular weight markers, in kDa, are shown on the right.
These same procedures were used to quantify SUMO*-CFTRFLAG-EGFP recoveries in replicate purification trials. We found a mean value of 1365 μg of SUMO*-CFTRFLAG-EGFP per 109 cells in microsomal preparations from multiple batches of D165 cultures (Table 2). Of this, DDM solubilization and NiNTA affinity chromatography yielded 905 μg and 178 μg, respectively, per 109 cells (Table 2), representing average overall CFTR recoveries of 66% and 13%, respectively. Purity of ~80% was consistently observed (data not shown).
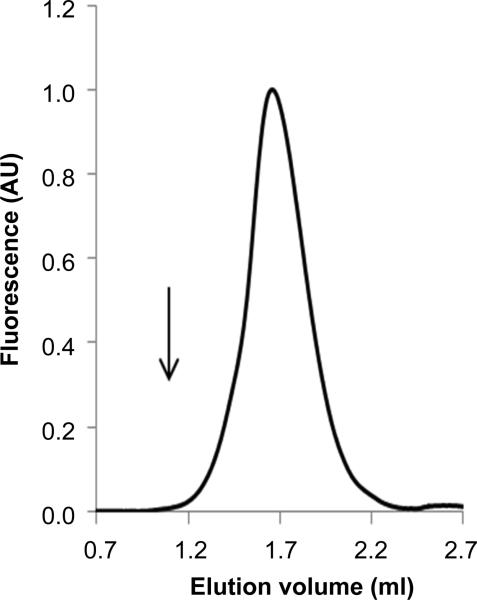
Potential aggregation of the purified SUMO*-CFTRFLAG-EGFP was assessed by FSEC. The SUMO*-CFTRFLAG-EGFP protein was eluted as a single, nearly symmetric peak without evidence of aggregated protein at the void volume (Fig. 8). The observed position of the peak would be consistent with either monomer or dimer. In a recent study, SUMO*-CFTRFLAG-EGFP purified using similar procedures was active and appeared monomeric by cryo-electron microscopy (59). These results indicated that functional SUMO*-CFTRFLAG-EGFP can be readily isolated from D165 cells in relative abundance using a mild detergent and without overt aggregation, suggesting the HEK293 mammalian cell culture system described in this study can provide a rich source of quality human CFTR protein for future biophysical investigation.
Figure 8. Monodispersity of affinity-purified SUMO*-CFTRFLAG-EGFP.
Protein eluted from NiNTA (25 μl containing 1.7 μg CFTR) was analyzed by FSEC on a 2.4 ml column. The peak of SUMO*-CFTRFLAG-EGFP (212 kDa) was eluted at 1.62 ml, near that of the monomeric P-glycoprotein-EGFP fusion protein (172 kDa), at 1.73 ml that is routinely used as a standard with FSEC assays. The arrow indicates the column void volume at 1.05 ml.
DISCUSSION
The challenge of obtaining human CFTR in sufficient quantities for biophysical studies is multifactorial, and persists as a fundamental obstacle to structure-based CF drug discovery. Mammalian cell expression systems offer the advantage of a physiologically relevant context for CFTR biogenesis so as to preserve native protein folding and post-translational modification (29). CFTR expressed constitutively in BHK cells retains chloride channel function (57), however, the cells produce only modest amounts of CFTR. In this study, we approached surmounting this obstacle by describing a linearly integrated strategy for modification of the protein and for evaluating its function and biogenesis in mammalian cells, as well as enabling production and isolation of mature, biologically active human CFTR in preparative amounts.
The TRE-SUMO*-CFTRFLAG-EGFP-IRES-Puro expression strategy (Fig. 1A) comprises several important features that facilitate consistent production of high-quality recombinant human CFTR protein in mammalian cells. Expression of CFTR is tightly regulated using the Tet-On system (60). The HEK293.M2 (D017) cells used in this study express an enhanced rtTA that is stable in eukaryotic cells, exerts minimal basal expression from the TRE promoter, and is capable of inducing high-level transactivation of gene expression upon dox binding (61). We have found the inducible expression approach circumvents transcriptional silencing of CFTR observed in HEK293 cell cultures when expression was driven constitutively by the CMV promoter (unpublished results). With dox-controlled expression, CFTR-transduced HEK293 cells grow more rapidly and reach higher densities prior to induction compared to constitutively expressing HEK293 cells. The inducible expression strategy also facilitates the generation and procurement of characterized master-stock recombinant cell lines with stable genetic and biological properties. Thus, the strategy of controlled CFTR expression is vital for optimal maintenance and expansion of “quiescent” cells, as well as for maximizing CFTR production on a per-cell basis in response to dox. Indeed, in the case of SUMO*-CFTRFLAG-EGFP, mass-spectrometric analysis indicated CFTR expression levels in the mg per 109 cell range, approximately 1/10th of which was cell-surface localized even when overexpressed to these high levels. This expression system will, therefore, enable large-scale suspension cell cultures to be grown and induced under carefully optimized and standardized conditions to produce sufficient quantities of CFTR for biophysical studies.
Recording single-channel activity provides an informative measure of CFTR function, and is traditionally used to evaluate function of recombinant CFTR proteins expressed in various cell lines (56, 57). In this study, we identified combinations of heterologous tags/domains that add utility without seriously altering CFTR gating parameters or single-channel conductance (Fig. 4). The C-terminal EGFP and N-terminal SUMO* fusions were both functionally silent, while the extracellular loop FLAG insertion caused a slight decrease in single channel conductance, whether alone or in the CFTR construct containing all three tags, SUMO*-CFTRFLAG-EGFP. Nevertheless, the differences in the gating kinetics and single-channel conductance found between the unmodified and tagged CFTR variants were not dramatic, suggesting they have the same folded state.
Recombinant SUMO fusion proteins exhibit enhanced expression, decreased susceptibility to proteolytic degradation, and improved folding in E. coli and yeast (36, 37). For CFTR fusion, we utilized the SUMO* variant containing R64T and R71E mutations to prevent its cleavage by eukaryotic proteases (39). Analysis of the N-terminus of the fusion proteins by immunoblotting with anti-RGSHis4 mAb suggested that SUMO*-CFTR fusions were intact in HEK cells (Fig. 2). The presence of this domain proved advantageous for CFTR biogenesis in our system, since the comparative expression analysis indicated a 1.5- to 2-fold increase in surface levels of SUMO*-CFTRFLAG-EGFP compared to CFTRFLAG-EGFP (Fig. 3B). If necessary for downstream applications, the SUMO* tag is removable with the highly specific SUMO* protease (36).
Cell-surface localization of CFTR is important due to multiple quality control checkpoints preclude improperly folded CFTR from reaching this compartment (6). However, since extracellular loops of CFTR are relatively small and poorly exposed, high-avidity antibody probes to extracellular regions of the protein are not available for use in cell-surface compartmentalization analyses (33). Positioning a FLAG epitope tag in the fourth extracellular loop of CFTR enabled sensitive and specific analysis of CFTR cell-surface compartmentalization and augmented the derivation of high-producer cell lines. Insertion of FLAG at this site disrupts one of two N-glycosylation sites located in this loop, and band patterns in the immunoblot analysis (Fig. 2) were consistent with hemiglycosylation of FLAG-tagged CFTR variants. Previous studies indicated that this tag placement did not compromise plasma membrane compartmentalization of CFTR or channel function (33, 34). We confirmed that FLAG-tagged variants, including those with a SUMO* N-terminal fusion, traffic successfully to the cell-surface and channel function was essentially intact with only a small decrease in conductance. Cell-surface FLAG/CFTR was quantifiable by flow cytometry, and this signal correlated strongly with cell-surface CFTR measurements made by mass spectrometric analysis of surface-biotinylated CFTR (Fig. 6C). We also used FACS to specifically isolate the highest surface FLAG/CFTR-expressing cells from the transduced population. We found this to be a specific and efficient means to derive clonal cultures (such as D165) with favorable expression characteristics from among a diverse population, and we have recently extended the approach to facilitate studies of cell lines expressing CFTR orthologs (unpublished).
The C-terminal fusion of EGFP is especially useful for both real-time monitoring of CFTR expression in live cultures and rapid evaluation of subcellular distribution by fluorescence microscopy. Flow cytometry of GFP fluorescence may be used for quantitation and facilitates cell sorting to isolate highly expressing subpopulations and derivation of clonal cell lines. Downstream, EGFP proved invaluable for CFTR quantitation both in crude cell lysates and successive protein purification steps. Using quantitative in-gel fluorescence methods, our results show that microsomal membranes contained 1.4 mg of SUMO*-CFTRFLAG-EGFP per 109 D165 suspension cells (Table 2). From this starting material, using standard methods of detergent solubilization and affinity purification, we recovered 180 μg CFTR per 109 cells at ~80% purity. Moreover, FSEC demonstrated that purified SUMO*-CFTRFLAG-EGFP was monodisperse (Fig. 8). These and recently published results (59) indicate that SUMO*-CFTRFLAG-EGFP can be readily isolated and purified in relative abundance, suggesting the mammalian cell culture system described here can provide a rich source of quality CFTR for biophysical investigations.
Our findings describe the development and validation of a robust mammalian cell expression system that facilitates reproducible and standardized large scale production of exogenous, biologically active cell-surface human CFTR protein. Importantly, the molecular expression strategy we used is vertically integrated to enhance efficiency and augment identification and derivation of stable high-producer cells, improve reproducibility and standardization of lot production, and facilitate analytical assessment at multiple steps from surface expression and cell isolation to monitoring of SUMO*-CFTRFLAG-EGFP solubilization and purification. With this expression platform in place, we anticipate that future studies with stabilizing mutations and CFTR orthologs will inform research efforts to stabilize the CFTR protein, as a prerequisite for the solution of its structure.
Acknowledgements
This study was supported by the Cystic Fibrosis Foundation (DELUCA03G0, URBATS13XX0 and YAO07XX0) and the National Institutes of Health, including R01GM095639, the Virology, Genetic Sequencing and Flow Cytometry Cores of the UAB Center for AIDS Research (P30 AI27767), the UAB Rheumatic Diseases Core Center's High Resolution Imaging Facility (5P30 AR048311) and the UAB CF Research & Translational Core Center (5P30 DK072482). Support was also provided by the UAB Gregory Fleming James Cystic Fibrosis Research Center (464-CR07). The UAB Comprehensive Cancer Center Hybridoma Core supported the preparation of R1104 mAb ascites. P-glycoprotein-EGFP was a gift of D. Swartz, TTUHSC. The authors are grateful to J. Denise Wetzel, CCHMC Medical Writer, for editing of the manuscript.
REFERENCES
- 1.Welsh MJ, Ramsey BW, Accurso F, Cutting GR. Cystic Fibrosis. In: Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, Gibson KM, Mitchell G, editors. The Online Metabolic and Molecular Bases of Inherited Disease. The McGraw-Hill Companies, Inc.; New York, NY: 2014. [Google Scholar]
- 2.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 3.Sheppard DN, Welsh MJ. Structure and function of the CFTR chloride channel. Physiol Rev. 1999;79:S23–45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 4.Lukacs GL, Mohamed A, Kartner N, Chang XB, Riordan JR, Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (delta F508) occurs in the endoplasmic reticulum and requires ATP. EMBO J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong PA, Kota P, Dokholyan NV, Forman-Kay JD. Dynamics intrinsic to cystic fibrosis transmembrane conductance regulator function and stability. Cold Spring Harb Perspect Med. 2013;3:a009522. doi: 10.1101/cshperspect.a009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pranke IM, Sermet-Gaudelus I. Biosynthesis of cystic fibrosis transmembrane conductance regulator. Int J Biochem Cell Biol. 2014;52C:26–38. doi: 10.1016/j.biocel.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Riordan JR. CFTR function and prospects for therapy. Annu Rev Biochem. 2008;77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- 8.Prince LS, Workman RB, Jr., Marchase RB. Rapid endocytosis of the cystic fibrosis transmembrane conductance regulator chloride channel. Proc Natl Acad Sci U S A. 1994;91:5192–5196. doi: 10.1073/pnas.91.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farinha CM, Matos P, Amaral MD. Control of cystic fibrosis transmembrane conductance regulator membrane trafficking: not just from the endoplasmic reticulum to the Golgi. FEBS J. 2013;280:4396–4406. doi: 10.1111/febs.12392. [DOI] [PubMed] [Google Scholar]
- 10.Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran O. On the structural organization of the intracellular domains of CFTR. Int J Biochem Cell Biol. 2014;52C:7–14. doi: 10.1016/j.biocel.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 12.Hegedus T, Aleksandrov A, Mengos A, Cui L, Jensen TJ, Riordan JR. Role of individual R domain phosphorylation sites in CFTR regulation by protein kinase A. Biochim Biophys Acta. 2009;1788:1341–1349. doi: 10.1016/j.bbamem.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Cant N, Pollock N, Ford RC. CFTR structure and cystic fibrosis. Int J Biochem Cell Biol. 2014 doi: 10.1016/j.biocel.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Dawson JE, Farber PJ, Forman-Kay JD. Allosteric Coupling between the Intracellular Coupling Helix 4 and Regulatory Sites of the First Nucleotide-binding Domain of CFTR. PLoS One. 2013;8:e74347. doi: 10.1371/journal.pone.0074347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt JF, Wang C, Ford RC. Cystic fibrosis transmembrane conductance regulator (ABCC7) structure. Cold Spring Harb Perspect Med. 2013;3:a009514. doi: 10.1101/cshperspect.a009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, Riordan JR. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci U S A. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowe SM, Verkman AS. Cystic fibrosis transmembrane regulator correctors and potentiators. Cold Spring Harb Perspect Med. 2013:3. doi: 10.1101/cshperspect.a009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galietta LJ. Managing the underlying cause of cystic fibrosis: a future role for potentiators and correctors. Paediatr Drugs. 2013;15:393–402. doi: 10.1007/s40272-013-0035-3. [DOI] [PubMed] [Google Scholar]
- 19.Hanrahan JW, Sampson HM, Thomas DY. Novel pharmacological strategies to treat cystic fibrosis. Trends Pharmacol Sci. 2013;34:119–125. doi: 10.1016/j.tips.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Lukacs GL, Verkman AS. CFTR: folding, misfolding and correcting the DeltaF508 conformational defect. Trends Mol Med. 2012;18:81–91. doi: 10.1016/j.molmed.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikpa PT, Bijvelds MJ, de Jonge HR. Cystic fibrosis: toward personalized therapies. Int J Biochem Cell Biol. 2014;52:192–200. doi: 10.1016/j.biocel.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Wu X, Levasseur DN, Liu H, Lai L, Kappes JC, Townes TM. Lentiviral vector transduction of hematopoietic stem cells that mediate long-term reconstitution of lethally irradiated mice. Stem Cells. 2000;18:352–359. doi: 10.1634/stemcells.18-5-352. [DOI] [PubMed] [Google Scholar]
- 23.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 26.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 29.McClure M, DeLucas LJ, Wilson L, Ray M, Rowe SM, Wu X, Dai Q, Hong JS, Sorscher EJ, Kappes JC, Barnes S. Purification of CFTR for mass spectrometry analysis: identification of palmitoylation and other post-translational modifications. Protein Eng Des Sel. 2012;25:7–14. doi: 10.1093/protein/gzr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Donnelly ML, Hughes LE, Luke G, Mendoza H, ten Dam E, Gani D, Ryan MD. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol. 2001;82:1027–1041. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- 32.Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 33.Howard M, DuVall MD, Devor DC, Dong JY, Henze K, Frizzell RA. Epitope tagging permits cell surface detection of functional CFTR. Am J Physiol. 1995;269:C1565–1576. doi: 10.1152/ajpcell.1995.269.6.C1565. [DOI] [PubMed] [Google Scholar]
- 34.Schultz BD, Takahashi A, Liu C, Frizzell RA, Howard M. FLAG epitope positioned in an external loop preserves normal biophysical properties of CFTR. Am J Physiol. 1997;273:C2080–2089. doi: 10.1152/ajpcell.1997.273.6.C2080. [DOI] [PubMed] [Google Scholar]
- 35.Mohanty AK, Simmons CR, Wiener MC. Inhibition of tobacco etch virus protease activity by detergents. Protein Expression and Purification. 2003;27:109–114. doi: 10.1016/s1046-5928(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Spurrier J, Butt TR, Strickler JE. Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions. Protein Expression and Purification. 2008;62:21–28. doi: 10.1016/j.pep.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panavas T, Sanders C, Butt TR. SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2009;497:303–317. doi: 10.1007/978-1-59745-566-4_20. [DOI] [PubMed] [Google Scholar]
- 38.Atwell S, Brouillette CG, Conners K, Emtage S, Gheyi T, Guggino WB, Hendle J, Hunt JF, Lewis HA, Lu F, Protasevich, Rodgers LA, Romero R, Wasserman SR, Weber PC, Wetmore D, Zhang FF, Zhao X. Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng Des Sel. 2010;23:375–384. doi: 10.1093/protein/gzq004. [DOI] [PubMed] [Google Scholar]
- 39.Peroutka RJ, Elshourbagy N, Piech T, Butt TR. Enhanced protein expression in mammalian cells using engineered SUMO fusions: secreted phospholipase A2. Protein Sci. 2008;17:1586–1595. doi: 10.1110/ps.035576.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, Blount AC, McNicholas CM, Skinner DF, Chestnut M, Kappes JC, Sorscher EJ, Woodworth B. Resveratrol enhances airway surface liquid depth in sinonasal epithelium by increasing CFTR channel open probability. PLoS ONE. 2013 doi: 10.1371/journal.pone.0081589. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci U S A. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu X, Wakefield JK, Liu H, Xiao H, Kralovics R, Prchal JT, Kappes JC. Development of a novel trans-lentiviral vector that affords predictable safety. Mol Ther. 2000;2:47–55. doi: 10.1006/mthe.2000.0095. [DOI] [PubMed] [Google Scholar]
- 44.Mulky A, Sarafianos SG, Arnold E, Wu X, Kappes JC. Subunit-specific analysis of the human immunodeficiency virus type 1 reverse transcriptase in vivo. J Virol. 2004;78:7089–7096. doi: 10.1128/JVI.78.13.7089-7096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren HY, Grove DE, De La Rosa O, Houck SA, Sopha P, Van Goor F, Hoffman BJ, Cyr DM. VX-809 corrects folding defects in cystic fibrosis transmembrane conductance regulator protein through action on membrane-spanning domain 1. Mol Biol Cell. 2013;24:3016–3024. doi: 10.1091/mbc.E13-05-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 47.Naren AP. Methods for the study of intermolecular and intramolecular interactions regulating CFTR function. Methods Mol Med. 2002;70:175–186. doi: 10.1385/1-59259-187-6:175. [DOI] [PubMed] [Google Scholar]
- 48.Aleksandrov AA, Cui L, Riordan JR. Relationship between nucleotide binding and ion channel gating in cystic fibrosis transmembrane conductance regulator. The Journal of physiology. 2009;587:2875–2886. doi: 10.1113/jphysiol.2009.170258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui L, Aleksandrov L, Hou YX, Gentzsch M, Chen JH, Riordan JR, Aleksandrov AA. The role of cystic fibrosis transmembrane conductance regulator phenylalanine 508 side chain in ion channel gating. The Journal of physiology. 2006;572:347–358. doi: 10.1113/jphysiol.2005.099457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bajrami B, Ramos A, Diego P, Coutermarsh BA, Stanton BA, Joseloff E, Yao X. High-Throughput and Accurate Quantitation of Cell Surface CFTR Using Stable Isotopes and Mass Spectrometry: A Potential Tool to Assist with CF Drug Discovery. Pediatric Pulmonology. 2011:221–221. [Google Scholar]
- 51.Jiang H, Ramos AA, Yao X. Targeted quantitation of overexpressed and endogenous cystic fibrosis transmembrane conductance regulator using multiple reaction monitoring tandem mass spectrometry and oxygen stable isotope dilution. Anal Chem. 2010;82:336–342. doi: 10.1021/ac902028f. [DOI] [PubMed] [Google Scholar]
- 52.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. Optimization of membrane protein overexpression and purification using GFP fusions. Nat Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
- 53.Chang XB, Mengos A, Hou YX, Cui L, Jensen TJ, Aleksandrov A, Riordan JR, Gentzsch M. Role of N-linked oligosaccharides in the biosynthetic processing of the cystic fibrosis membrane conductance regulator. J Cell Sci. 2008;121:2814–2823. doi: 10.1242/jcs.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howard M, Jiang X, Stolz DB, Hill WG, Johnson JA, Watkins SC, Frizzell RA, Bruton CM, Robbins PD, Weisz OA. Forskolin-induced apical membrane insertion of virally expressed, epitope-tagged CFTR in polarized MDCK cells. American journal of physiology. Cell physiology. 2000;279:C375–382. doi: 10.1152/ajpcell.2000.279.2.C375. [DOI] [PubMed] [Google Scholar]
- 55.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 56.Aleksandrov AA, Riordan JR. Regulation of CFTR ion channel gating by MgATP. FEBS Lett. 1998;431:97–101. doi: 10.1016/s0014-5793(98)00713-3. [DOI] [PubMed] [Google Scholar]
- 57.Rosenberg MF, Kamis AB, Aleksandrov LA, Ford RC, Riordan JR. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR). J Biol Chem. 2004;279:39051–39057. doi: 10.1074/jbc.M407434200. [DOI] [PubMed] [Google Scholar]
- 58.Zhang L, Aleksandrov LA, Zhao Z, Birtley JR, Riordan JR, Ford RC. Architecture of the cystic fibrosis transmembrane conductance regulator protein and structural changes associated with phosphorylation and nucleotide binding. J Struct Biol. 2009;167:242–251. doi: 10.1016/j.jsb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Hildebrandt E, Zhang Q, Cant N, Ding H, Dai Q, Peng L, Fu Y, DeLucas LJ, Kappes JC, Urbatsch IL. A Survey of Detergents for the Purification of Stable, Active Human Cystic Fibrosis Transmembrane Conductance Regulator (CFTR). Biochimica et Biophysica Acta (BBA) - Biomembranes. 2014 doi: 10.1016/j.bbamem.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 61.Pluta K, Luce MJ, Bao L, Agha-Mohammadi S, Reiser J. Tight control of transgene expression by lentivirus vectors containing second-generation tetracycline-responsive promoters. J Gene Med. 2005;7:803–817. doi: 10.1002/jgm.712. [DOI] [PubMed] [Google Scholar]