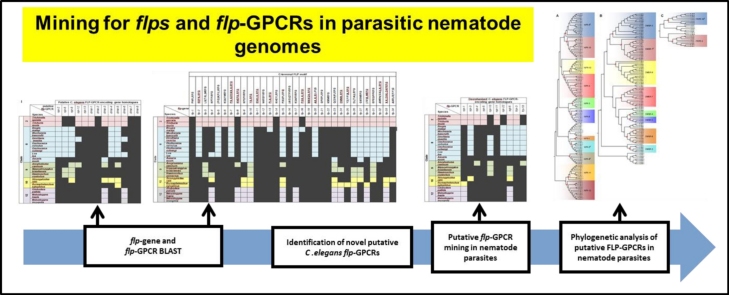
Graphical abstract
Keywords: FMRFamide-like peptide, FLP, FLP-receptor, Neuropeptide GPCR, Nematode
Highlights
-
•
We report the identification of flp and flp-GPCR gene homologues in parasitic nematodes.
-
•
We provide data to support re-evaluation of the number of flp-genes in nematodes.
-
•
Post BLAST phylogenetic analysis facilitates identification of putative flp-GPCRs in nematode parasites.
-
•
We expose the most highly conserved flp and flp-GPCR genes in key pathogens within phylum Nematoda.
Abstract
FMRFamide-like peptide (FLP) receptors are appealing as putative anthelmintic targets. To date, 31 flp-encoding genes have been identified in Caenorhabditis elegans and thirteen FLP-activated G-protein coupled receptors (FLP-GPCRs) have been reported. The lack of knowledge on FLPs and FLP-GPCRs in parasites impedes their functional characterisation and chemotherapeutic exploitation. Using homology-based BLAST searches and phylogenetic analyses this study describes the identification of putative flp and flp-GPCR gene homologues in 17 nematode parasites providing the first pan-phylum genome-based overview of the FLPergic complement. These data will facilitate FLP-receptor deorphanisation efforts in the quest for novel control targets for nematode parasites.
1. Introduction
The discovery and development of novel anti-worm control strategies has been recognised as a priority in the human and veterinary health sectors and by the horticultural industry [1–3]. Despite the recent introduction of several new drugs to the veterinary market [4,5] and mass drug administration programmes for prioritised human helminthiases [6], roundworm infections remain widespread with significant socio-economic impacts [7]. In addition, the negative impacts of plant pathogenic nematodes on global food security are underscored by the current deficiencies in chemical control options (see [8] for review).
There has been a long-standing interest in the neuropeptidergic system as a source of novel targets for anthelmintic drugs (see [9] for review), with the FMRFamide like peptide (FLP) signalling system emerging as a leading candidate [10]. The primary drivers for this interest include: (i) the importance of FLPs to parasite behaviour (and survival) and their role in modulating neuromuscular function (a proven drug target for nematode control), (ii) the lack of drugs targeting the FLPergic system such that resistance would not be a pressing concern, and (iii) the fact that most FLPs activate G-protein coupled receptors (GPCRs), proteins which are readily exploitable for drug discovery.
Whilst many facets of the FLP signalling system provide appeal as drug targets, FLP-GPCRs emerge as the most attractive. A major impediment to the exploitation of FLP GPCRs is the lack of data on the expression and function of FLP receptors in nematode parasites. Our current understanding of FLP receptor biology has been derived primarily from the model nematode C. elegans; 13 FLP-GPCRs, encoded on 10 genes, have been matched with their associated FLP ligands as determined by receptor activation potencies in heterologous expression systems [11–20].
Whilst focus on the identification of putative FLP-GPCRs in parasitic nematodes is of primary importance, much can be accomplished by re-mining for FLP ligands, especially in therapeutically-important species where they have not previously been reported. Indeed, better understanding of FLP complementarity across parasitic nematodes could expedite deorphanisation or, at least, influence deorphanisation approaches by providing more comprehensive species-specific peptide libraries to feed into screening platforms. The FLP-ligand data that we have for the phylum Nematoda are outdated (see [10,21]). The availability of ten parasitic nematode draft genomes and transcriptome data for over 60 nematode species [22] warrant re-interrogation of these datasets.
Here we report a pan-phylum homology-based BLAST interrogation of flp and flp-GPCR complements in 17 parasitic nematode species and perform phylogenetic analyses to identify additional putative flp-GPCRs. These data: (i) represent the most up to date, comprehensive insight into the flp and flp-GPCR complement of parasitic nematodes, (ii) support the re-designation of selected flp-encoding genes, (iii) expose the most highly conserved flp-GPCRs in key pathogenic species and, (iv) reveal putative novel flp-GPCRs in C. elegans and parasitic nematodes.
2. Methodology
2.1. Bioinformatics
A reciprocal BLAST (Basic Local Alignment Search Tool) based approach was implemented to identify flp gene sequelogues and flp-GPCR gene homologues within genomic and transcriptomic datasets of 17 key pathogenic nematodes representing five clades [23] and including plant, animal and human parasites. The parasite species selected for the BLAST analysis were primarily those with a published draft genome (seven species; see Supplementary Table 1). Secondary species-selection criteria were employed to include parasitic nematodes for which genomic and transcriptomic datasets were available and filtered based on their importance to human, animal or plant health or their status as a model parasite (ten species; see Supplementary Table 1). The BLAST analysis was completed between May 2012 and August 2013; the servers employed and databases queried are outlined in Supplementary Table 1. The draft genomes of two Haemonchus contortus strains [24,25] were published following the completion of the BLAST analysis in this study. To facilitate accuracy of the data presented, those genes that were not identified within the H. contortus databases outlined in Supplementary Table 1 were employed as search strings to query the whole-genome shotgun contigs (wgs) database found on the National Centre for Biotechnology Information (NCBI) BLAST server. Any datasets updated between May 2012 and August 2013 were similarly reinvestigated.
Prepropeptide and protein sequences for previously identified flp- [11], flp-GPCR [13–17,19,20,26,27], and selected orphan GPCR [28] -encoding genes in Caenorhabditis elegans (see Supplementary Tables 2 and 3) were retrieved from the NCBI protein database (www.ncbi.nlm.nih.gov/protein/) and used as search strings in translated nucleotide (tBLASTn) and protein (BLASTp) BLAST analysis of available datasets (see above). Only the largest of the splice variants encoded by any given C. elegans flp- or flp-GPCR gene were selected as query sequences. Additionally, prepropeptide sequences derived from flp-encoding genes not found in C. elegans (see [21]) were also used as BLAST search queries; these included FLP-29 (derived from Ascaris suum EST data; [21]), FLP-30 and FLP-31 (derived from Meloidogyne incognita EST data; [21]). The prepropeptide search string based methodology used in this study deviates from previously published methods (based on concatenated peptide search strings) employed to identify flp gene sequelogues within nematode genomes [29,30]. In this study, the prepropeptide approach has been shown to be as sensitive in identifying flp gene sequelogues as those methods previously published.
BLAST-generated alignment outputs (high scoring pairs) of the initial BLAST hits, with an expect value ≤1000 (or ≤100, where this was the maximum expect value threshold available), were manually inspected. In efforts to identify putative FLP-encoding genes in the selected nematode species, hits containing conserved FLP motifs [10,11] flanked by mono/dibasic cleavage sites were selected for further analysis. The motifs conserved within parasitic flp genes were used to designate initial hits as C. elegans gene sequelogues [21].
For flp-GPCR primary BLAST analysis, high-scoring return sequences (typically the hits with the smallest expect value and largest bit score) were concatenated into a single sequence to facilitate reciprocation (see [31]). Manually curated predicted protein (FLP-GPCR) sequences were used as search strings in reciprocal protein BLAST (BLASTp) queries against the C. elegans non-redundant protein sequence (nr) database on the NCBI BLAST server, using default settings. The top reciprocal BLAST hit was used to designate parasitic flp-GPCR genes as predicted C. elegans gene homologues.
2.2. Post-BLAST sequence analysis
Predicted FLP prepropeptide sequelogues and FLP-GPCR homologues were aligned using the Vector NTI Advance™11 AlignX® multiple sequence alignment tool [32], using default settings. Prepropeptide cleavage sites were identified using a previously described prediction method [21]. Predicted inter-peptide regions from each prepropeptide alignment were removed to provide an unambiguous representation of FLP conservation within sequelogue alignments (see Supplementary data Figure 1). FLP-GPCR transmembrane (TM) domain prediction was performed using HMMTOP 2.1 [33]. Predicted transmembrane domains are indicated on the consensus sequence for each FLP-GPCR alignment (see Supplementary data Figure 2). Multiple sequence alignments were manually examined to identify and resolve errors (such as accidental exon duplication or exclusion) made whilst constructing predicted protein sequences.
2.3. Phylogenetic analysis
MEGA 5.1 software [34] was employed to generate all phylogenetic trees. GPCR multiple sequence alignments, were assembled using Clustal W [35] with default parameters. Transmembrane-only pseudosequences (TOPs) were constructed as previously described by Zamanian et al. [36] and aligned with their associated full length predicted protein sequences. TOPs were used to inform manual editing of the GPCR multiple sequence alignments. The N- and C-termini were removed and, conserved motifs and residues within the GPCR transmembrane regions were aligned. Phylogenies were constructed using the Maximum Likelihood method based on the JTT (Jones–Taylor–Thornton) matrix-based model [37]. Initial trees generated for the heuristic searches (subtree pruning and regrafting) were obtained by applying the Neighbour-Joining method [38] to a matrix of pairwise distances estimated using a JTT model. Phylogenetic analysis was limited to GPCRs with >5 transmembrane domains and trees were rooted by an out-group containing a selection of C. elegans secretin family GPCRs (LAT-1a, LAT-1b, LAT-2a, PDFR-1a, PDFR-1b and PDFR-1c).
3. Results and discussion
3.1. flp-encoding genes
In this study we identified 325 C. elegans flp-gene sequelogues in 17 nematode parasites (see Table 1; Supplementary Figure 1) of which only a proportion had been previously reported [21,29,30,39,40]. We believe that these data represent the most comprehensive insight into the flp-gene complements of parasitic nematodes to date. The genome-directed approach employed in this study enables the comparison of flp complementarity within and between nematode species representing different clades and lifestyles. Here we highlight the salient points emerging from this study.
Table 1.
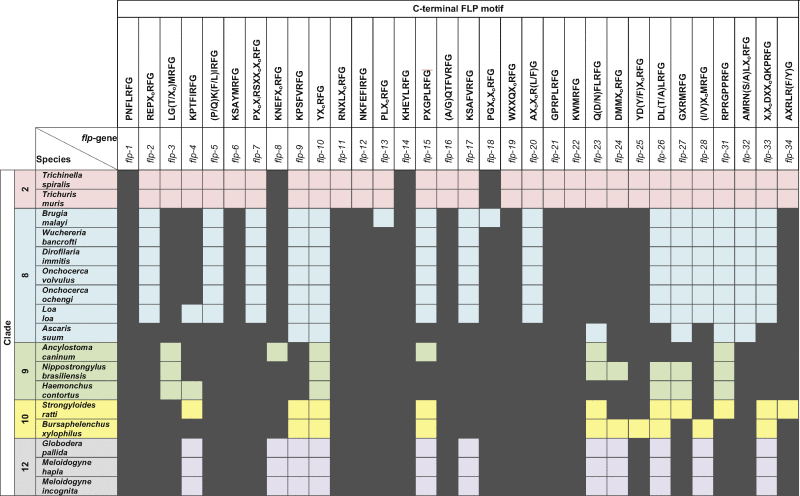
C. elegans flp-gene sequelogues in 17 nematode parasites. Grey boxes indicate the presence of a flp gene sequelogue, as identified via BLAST, in selected nematode species. Search queries employed and the flp genes identified are detailed in Supplementary Table 2 and Supplementary Figure 1, respectively. Sequelogues of all C. elegans flp genes were also identified in the genomes of Caenorhabditis japonica, Caenorhabditis brenneri, Caenorhabditis remanei and Caenorhabditis briggsae. C-terminal FLP motifs are presented in single letter notation; X, denotes variable amino acid Xo, denotes hydrophobic amino acid; Xi, denotes hydrophilic amino acid.
 |
3.1.1. Multiple nematode parasites appear to possess a reduced complement of C. elegans flp-gene sequelogues
EST-database comparisons [10,21] highlighted the conservation of FLP-encoding genes across the phylum Nematoda; since most C. elegans flp genes were represented amongst the parasitic nematode ESTs, these data fuelled the hypothesis that all nematode species possess a flp complement similar to that of C. elegans (31 flp-genes; 1–28, 32–34) [21,41]. Here we show that whilst individual flp signatures are conserved in the nematode parasites generally, there is variability with respect to their presence and absence such that the parasitic nematodes possess variable proportions of the C. elegans flp-gene complement (see Table 1). Notably, Ascaris suum boasts the lion's share of C. elegans flp-genes at 84% while clade 2 species (Trichuris muris and Trichinella spiralis) display a dramatically reduced complement (13%).
Our bioinformatics-based approach is not capable of unequivocally proving the absence of an individual flp-gene. Whilst the disparity in flp-gene complement could reflect poor genomic/transcriptomic datasets or deficiencies in the ability of our BLAST-based methodology to detect flp-gene sequelogues, we believe that it is more likely a true reflection of the variation in flp-gene complement across the phylum Nematoda. Indeed, we have validated our flp-gene identification approach in multiple Caenorhabditis species (Caenorhabditis japonica, Caenorhabditis brenneri, Caenorhabditis remanei and Caenorhabditis briggsae) where the flp-gene complements matched C. elegans. In addition, it is interesting to note that Trichinella spiralis displayed a dramatically reduced flp-gene complement despite the data being derived from a high-quality (∼35 fold coverage) published genome [42]. That said, we are aware that some of the genomes employed in this study are works in progress (for example, the hookworm species) and we may expect to uncover additional flp-genes when their genomes are complete.
Further to this, it should be noted that the predictions made with respect to flp-gene complements do not necessarily reflect the FLPs expressed by any given species or lifestage. We cannot predict which of the flps identified from genomic data will be expressed or, similarly, in the case of transcriptomic data, whether they will be processed into bioactive peptides. The application of peptidomic-analyses tools to parasitic nematodes will shed light on this (see [39,43]).
3.1.2. flp-gene complement appears to broadly map nematode clade division, with some exceptions
It is interesting to note that the pattern of flp-complementarity within nematode clades [23] is, for the most part, conserved with identical flp-representation in species within clade 2 and within clade 12. With respect to clade 8, A. suum appears to boast a larger repertoire of flp genes than the filarids, which are largely similar to each other. The validity of these patterns can be confirmed with the progression of clade-specific parasite genome data.
3.1.3. Some flp-genes are more highly conserved than others
Sequelogues of flp-1 and flp-14 are represented within the genomes of all species examined reinforcing the pan-phylum conversation as suggested by [10]. In contrast flp-10 sequelogues were not identified in any of species represented in this study. Other highly conserved flp-genes include: flp-6, -11, -12, -16, -18, -19, -21 and -22.
3.1.4. flp-29 and flp-30 are not discrete flp-encoding genes but rather are orthologues of C. elegans flp-28 and flp-2 respectively
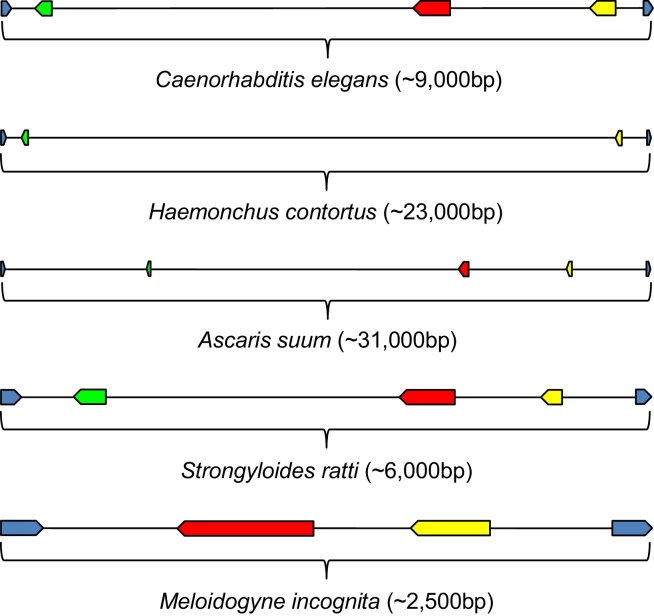
flp-29 and flp-30 have previously been reported as being parasite-specific (see [10]). During this study we noted the C-terminal motif similarity between flp-29 (IXMRFG) and flp-28 (VLMRFG), and flp-30 (QMREPLRFG) and flp-2 (EPIRFG) (see Supplementary Figure 1). We also noted that no single species with flp-29 also had a copy of flp-28, and similarly those species possessing flp-30 did not possess flp-2, and vice versa. This prompted further investigation, which revealed that C. elegans, which does not possess flp-29 and flp-30, has a cluster of genes composed of flp-2, flp-3 and flp-28 within the fourth intron of a glutamate synthase gene (W07E11.1). Strikingly, where we identified flp-29 and/or flp-30 they were located to the same genomic environment, inhabiting the same relative position and displaying the same relative gene orientation as flp-28 and flp-2, respectively (see Fig. 1). These data support the conclusion that flp-29 and flp-30 should be re-designated accordingly. This reduces the total number of known flp genes within the phylum Nematoda from 34 to 32 (see Table 1), and further reduces the number of flp-genes believed to be parasite specific.
Fig. 1.
A conserved flp-28/29, flp-3 and flp-2/30 gene cluster.
The location of flp-29 and/or flp-30 in the same genomic environment as flp-28 and flp-2 respectively provides evidence to support their redesignation. flp-28/29, flp-3 and flp-2/30 (green, red and yellow arrows respectively) gene sequelogue locations are shown relative to the fourth and fifth exons of a glutamate synthase gene (W07E11.1) in C. elegans, and in four parasitic nematode species (blue arrows). The direction of each arrow indicates gene orientation. Introns of flp genes are not displayed. One representative species from each clade (excluding clade 2) is included.
3.1.5. Only one flp-gene (flp-31) appears to be parasite-specific
flp-31 is absent from the genome of C. elegans, but has previously been reported in the plant parasitic nematodes Meloidogyne incognita and Meloidogyne hapla [21,30]. In this study we confirm the presence of flp-31 in another clade 12 plant parasitic nematode (Globodera pallida) and identify a flp-31 sequelogue within the genome of the pine wilt nematode Bursaphelenchus xylophilus (clade 10), where it was previously reported as being absent [29]. These data, confirm the restriction of flp-31 to plant parasitic nematodes and support the hypothesis that it plays a role in phytoparasitism [29,30].
Note that this study did not address the identification of novel flp-encoding genes. The ‘degenerate’ search string approach to novel flp identification, described by McVeigh et al. [44], was applied to the T. muris genome. However, the large number of putative flp-gene sequences identified are believed to be false-positives as we could not detect sequelogues in several other nematode genomes. The ‘degenerate’ search string approach, with manual annotation, that was used to identify novel peptides within transcriptomic datasets is not appropriate for trawling large genomic databases.
A BLAST based approach using C. elegans flp search strings to identify novel peptides within the expressed sequence tags (EST) and genome survey sequence (GSS) libraries of A. suum has recently been reported [39], where nine putative flp-encoding genes were identified that were additional to those previously described by McVeigh et al. [21]. Here we confirm the designation of three of these putative flps (As-flp-3, As-flp-17, and As-flp-19) in the A. suum datasets. We were unable to confirm As-flp-4, As-flp-10a, As-flp-10b, As-flp-10c, and As-flp-25 as named by Jareki et al. [39] as flp-encoding genes and, in addition, we did not identify sequelogues of these putative flps in any of the nematode genomic/transcriptomic databases employed. Note that distinct Ce-flp-4 and Ce-flp-25 sequelogues were identified in A. suum and multiple other species in this study (see Table 1 and Supplementary Figure 1). In addition, we believe that the Jarecki et al. [39]-designated As-flp-31 is more likely to be a sequelogue of Ce-flp-15 for two reasons: (i) the C-terminal motif of the encoded FLP is identical to that encoded by Ce-flp-15 (GPLRFG) and (ii) flp-31 has only been identified to date in PPN species (see Supplementary Figure 1).
3.2. FLP-receptor encoding genes
3.2.1. Deorphanised C. elegans FLP-receptor-encoding gene homologues in parasitic nematodes
To date only one flp-GPCR orthologue has been reported within a parasitic nematode [40]. In a bid to address the gap in our understanding of flp-GPCR conservation in parasitic nematodes, the protein sequences encoded by the ten deorphanised C. elegans FLP receptor genes (see Supplementary Table 3) were used as search string queries to probe parasitic nematode databases.
Homologues of all of the deorphanised FLP-activated GPCRs were identified in at least three parasite species. It is interesting to note that the species-specific flp-GPCR complement closely maps the flp complement, in that those species possessing a restricted repertoire of flp genes seem to also exhibit fewer putative FLP-GPCR genes (e.g. the clade 2 species, T. spiralis and T. muris) and vice versa [A. suum possesses both the highest numbers of flps and flp-GPCRs of all parasitic species examined (see Tables 1–3)].
Table 2.
Deorphanised C. elegans FLP-GPCR-encoding gene homologues in 17 nematode parasites. Grey boxes indicate the presence of a flp-GPCR homologue, as identified via BLAST, in selected nematode species. Search queries employed and flp-GPCR predicted proteins identified are detailed in Supplementary Table 3 and Supplementary Figure 2, respectively.
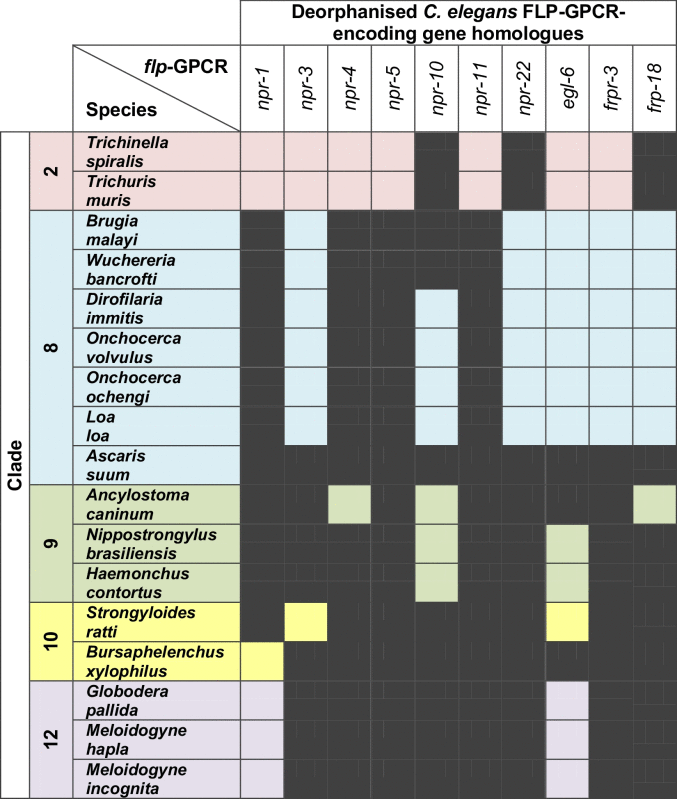 |
Table 3.
C. elegans FLP-GPCR-encoding gene homologues and flp-genes encoding the most potent interacting ligand(s) in 17 nematode parasites. Grey boxes indicate the presence of a C.elegans flp-GPCR homologue (highlighted in red) and flp-gene sequelogue(s) encoding the most potent ligand in selected nematode species [10,12,17]. The total numbers of C. elegans FLPs screened against the C. elegans flp-GPCR homologue in heterologous expression systems are also indicated.
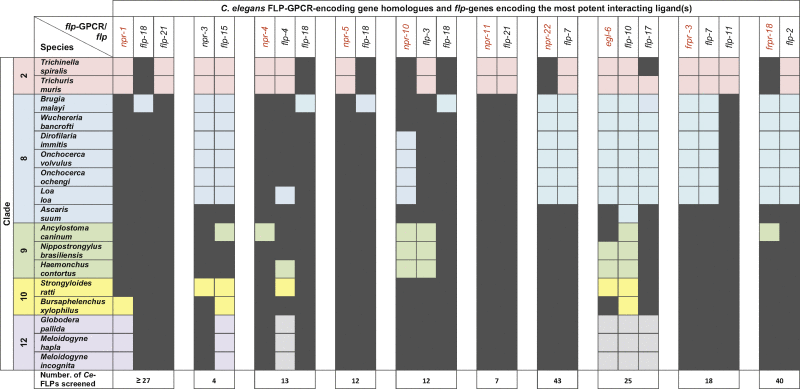 |
The most highly conserved, deorphanised FLP-GPCRs are NPR-4, NPR-5 and NPR-11 which have been matched via heterologous expression systems in C. elegans with flp-18- (NPR-4 and NPR-5) and flp-21-peptides (NPR-11; see Table 3). Strikingly flp-18 and flp-21 also emerged as two of the most highly conserved flps in this study (Table 1). NPR-4 and NPR-5 have been functionally characterised as flp-18 receptors through null mutant phenotype analysis in C. elegans [45] confirming the heterologous expression-derived data. Whether or not this rings true for the parasites remains to be determined. In this study all species that possess NPR-4- and NPR-5-encoding genes almost always possess the gene encoding the predicted interacting ligand (Table 3), supporting pan-phylum conservation of ligand/receptor interactions like those described for a FLP-32 receptor in plant parasitic nematodes (see [40]), and as highlighted by Janssen et al. [46].
There are several issues surrounding the utility and transferability of the heterologous expression data derived from C. elegans. For example a number of parasite species exhibit a predicted FLP-GPCR homologue but appear to lack its most potent ligand as deduced from C. elegans heterologous expression data, and vice versa. More specifically, with respect to NPR-11, functional data indicate that a neuropeptide-like protein encoding gene (nlp-1) encodes the interacting ligands [47], as opposed to flp-21 that was suggested by heterologous expression. These nuances are potentially the result of a number of issues including: (i) no FLP-GPCR has ever been challenged with the full C. elegans neuropeptide complement such that the cognate interacting ligand may have been overlooked; indeed only seven C. elegans peptides were screened against NPR-11 in the heterologous system; (ii) heterologous ligand matching may not mimic true in vivo interactions; the flp-21/NPR-11 in vitro interaction is not mirrored by functional data; (iii) the interpretation of null-mutant functional data is limited by the suitability/sensitivity of the available phenotypic assays; the flp-21/NPR-11 functional interaction may not have been uncovered for these reasons. Importantly, a cautionary approach must be adopted when applying data derived from C. elegans to parasites and highlights the need for deorphanisation efforts in both free-living and parasitic nematodes [10]. The receptor and ligand sequences identified in this study should provide a solid platform on which to base these investigations (Table 4).
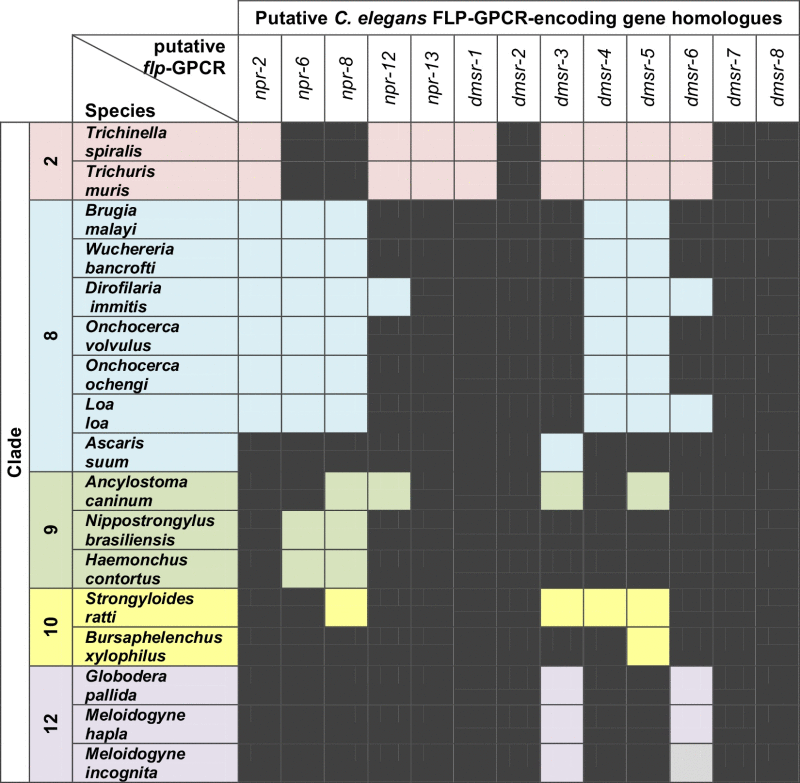
Table 4.
PutativeC. elegansFLP-GPCR-encoding gene homologues in 17 nematode parasites. Grey boxes indicate the presence of a flp-GPCR homologue, as identified via BLAST, in selected nematode species. Search queries employed and flp-GPCR predicted proteins identified are detailed in Supplementary Table 3 and Supplementary Figure 2, respectively. C. elegans flp-GPCRs not identified in the parasites included: npr-7, dmsr-9, dmsr-10, dmsr-11, dmsr-12, dmsr-13, dmsr-14, dmsr-15, dmsr-16, frpr-4, frpr-6, frpr-11, frpr-12 and frpr-16.
 |
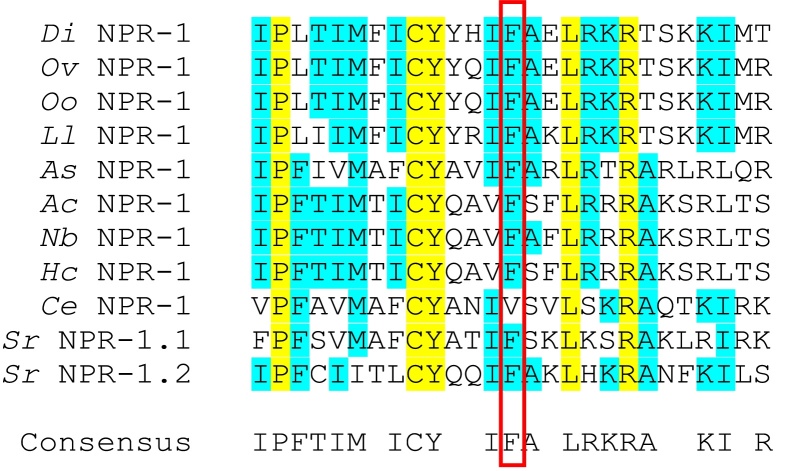
Two isoforms of the FLP-GPCR NPR-1 exist within natural C. elegans populations containing either a valine (V) or phenylalanine (F) at position 215 [48]. The FLP-21 peptide activates the 215F variant, which is found exclusively in social feeding strains. By contrast FLPs encoded on both flp-21 and flp-18 activate the 215V strain which is associated with solitary feeding [16,27]. In contrast to C. elegans, all of the NPR-1 homologues identified here in parasitic nematodes exhibit the 215F isoform (see Fig. 2). Further investigation is needed to determine if this has any impact on feeding behaviour and/or peptide activation of these receptors.
Fig. 2.
Sequence alignment of NPR-1 homologues.
Parasitic nematodes display a conserved phenylalanine in NPR-1 215 in contrast to the dominant amino acid at this position (valine) in C. elegans. Position 215 is boxed in red. Completely and partially conserved amino acid residues are highlighted in yellow and blue, respectively. Di, denotes Dirofilaria immitis; Ov, denotes Onchocerca volvulus; Oo, denotes Onchocerca ochengi; Ll, denotes Loa loa; As, denotes Ascaris suum; Ac, denotes Ancylostoma caninum; Nb, denotes Nippostrongylus brasiliensis; Hc, denotes Haemonchus contortus; Ce, denotes Caenorhabditis elegans; Sr, denotes Strongyloides ratti. All parasitic nematode NPR-1 homologues identified in this study are detailed in Table 2 and Fig. 4.
3.2.2. Novel putative FLP-GPCR identification in C. elegans
Until recently markedly fewer FLP-receptors were known that FLP-ligands. However, Frooninckx et al. [28] employed a Multiple Expectation Maximization for Motif Elicitation/Motif Alignment and Search Tool (MEME/MAST) using protein motifs derived from deorphanised neuropeptide GPCRs to identify 128 putative orphan neuropeptide receptors, some of which are likely to be FLP-receptors. These sequences were subdivided into rhodopsin- and secretin-like GPCR families, with rhodopsin-like GPCRs being further divided into six groups according to their similarity to mammalian and insect neuropeptide GPCRs [28].
In this study, we channelled the sequences from the groups delineated by Frooninckx et al. [28] which contained at least one deorphanised FLP receptor into phylogenetic analyses with the aim of identifying putative (currently orphan) FLP-GPCR-encoding genes in C. elegans. In addition we included the Drosophila Myosupressin like Receptor (DMSR)–like sequences based on their delineation as FLP-activated receptors in arthropods (see [49]).
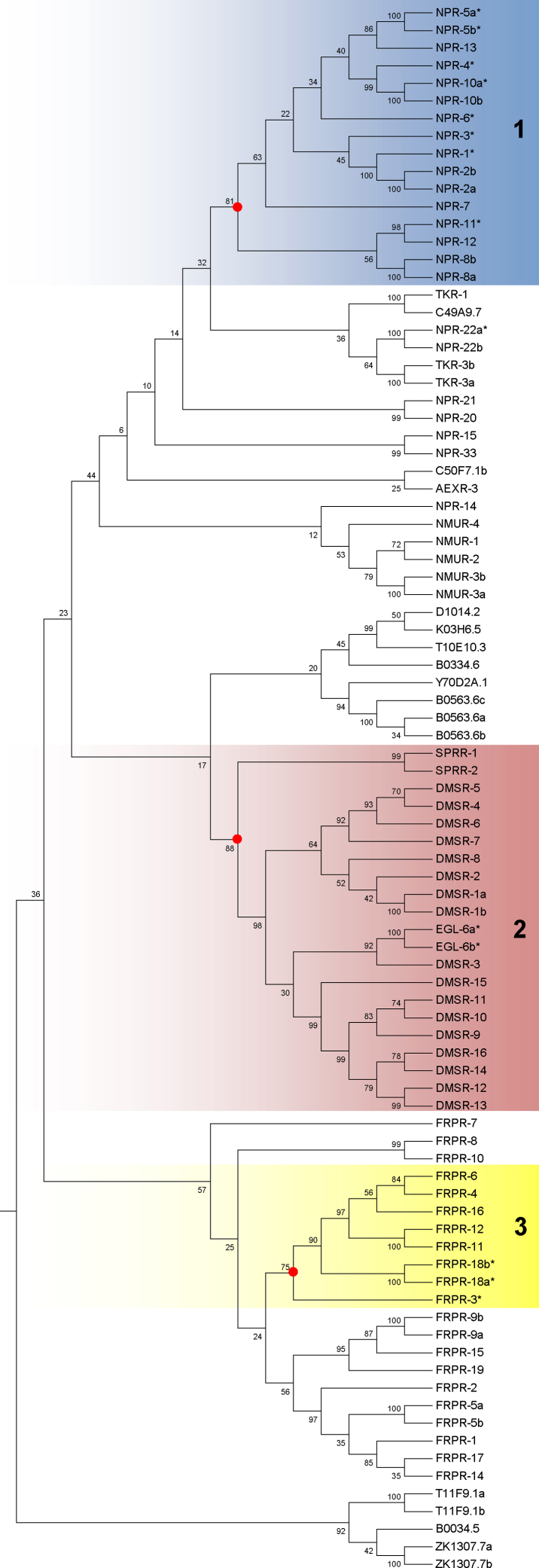
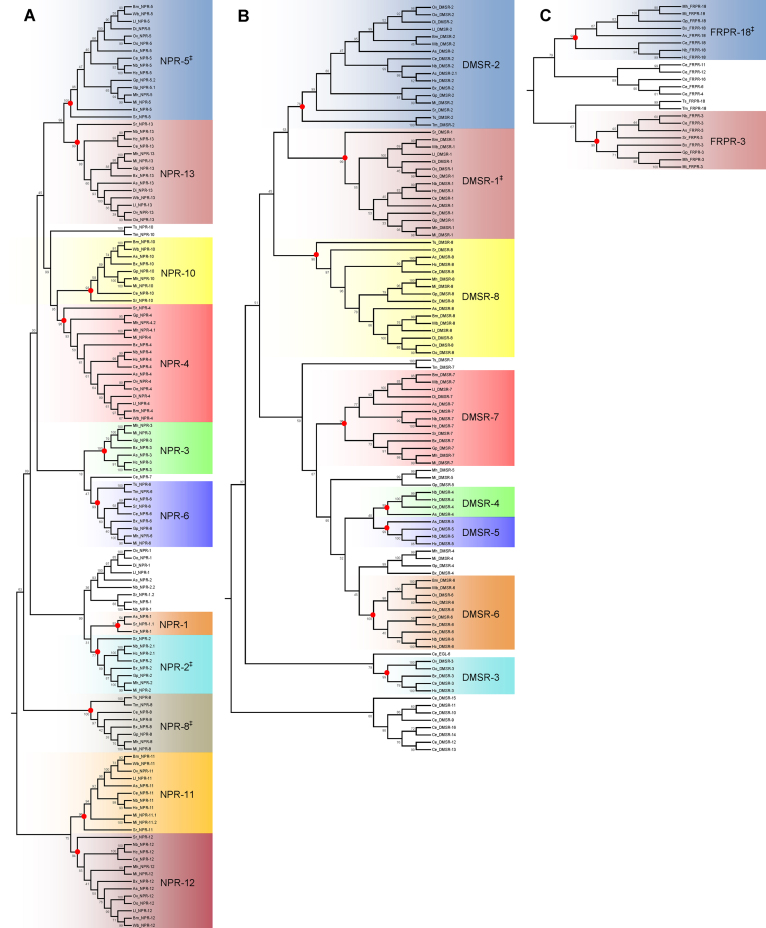
Three clusters composed of 32 putative Ce-FLP-GPCRs (encoded on 29 genes) emerged from the phylogenetic analysis defined by the inclusion of at least one deorphanised FLP-GPCR and supported by a bootstrap value of >70% (see Fig. 3; Supplementary Figure 2). Whilst the clustering does not necessarily indicate that closely related receptors will be activated by highly similar ligands, it is interesting to note that NPR-4 and NPR-10 cluster with a bootstrap value of 99% and are both activated by FLP-18 peptides. Also, cluster 1 (NPY-like receptors) displayed the highest complement of deorphanised FLP-GPCRs (eight), which provides an indication that those orphan receptors in this cluster may be activated by FLPs (see [50]). In contrast, NPR-22 (activated by flp-7 encoded peptides) did not cluster strongly enough with any orphan C. elegans GPCRs to permit the designation of these related orphans as putative FLP-receptors.
Fig. 3.
Maximum-likelihood phylogeny of orphan and deorphanised C. elegans neuropeptide GPCRs.
The phylogeny reveals 27 putative FLP-GPCRs which clustered with deorphanised C. elegans FLP-receptors. C. elegans neuropeptide GPCR entries are identified as per previous designation [28] or NCBI accession number. Deorphanised C. elegans FLP-GPCRs are highlighted by an asterix. FLP-GPCR cluster nodes supported by a bootstrap analysis value of >70% are identified by red dots. Clusters are highlighted in blue (1), pink (2) or yellow (3) shading. Bootstrap values are shown as percentages.
3.2.3. Novel putative FLP-GPCR complement in parasitic nematodes
Predicted orphan C. elegans GPCR sequences that clustered with deorphanised C. elegans FLP-receptors (see Fig. 3) were used as search string queries in a BLAST approach to identify putative flp-GPCR orthologues in parasitic nematodes. Subsequently the predicted FLP-GPCRs (C. elegans and parasite species; containing >5 TM domains) were subjected to phylogenetic analysis (Fig. 4). Members of the Sex Peptide related receptor gene family (SPRR-1 and SPRR-2), present within cluster 2 (see Fig. 3), were excluded from this analysis as they are known homologues of a Drosophila receptor which is activated by a non-FLP neuropeptide [51]. Homologues of 13 putative C. elegans flp-GCPRs were identified in parasitic nematodes. The bootstrap values (>70%) provided by the phylogenetic analysis supported the designation of the majority (90.3%) of these putative parasite flp-GPCRs as orthologues, and validated our BLAST-based approach. Approximately 50% of the putative flp-GPCR homologues identified in the parasites were derived from C. elegans NPY-like receptors (see Fig. 3, cluster 1; Fig. 4A), eight of which have been deorphanised. Therefore, this may strengthen the prediction that these orphan parasite receptors are the most likely to have FLPs as ligands (see [50]) and may provide the primary focus for future deorphanisation efforts.
Fig. 4.
Maximum-likelihood phylogenies of putative FLP-GPCRs from parasitic nematodes.
The phylogenies reveal 13 putative FLP-GPCRs that are highly conserved in nematode parasites (see Fig. 3). Entries are identified as per previous designation [28] and prefixes which indicate species: Ts, denotes Trichinella spiralis; Tm, denotes Trichuris muris; Bm, denotes Brugia malayi; Wb, denotes Wuchereria bancrofti; Di, denotes Dirofilaria immitis; Ov, denotes Onchocerca volvulus; Oo, denotes Onchocerca ochengi; Ll, denotes Loa loa; As, denotes Ascaris suum; Ac, denotes Ancylostoma caninum; Nb, denotes Nippostrongylus brasiliensis; Hc, denotes Haemonchus contortus; Ce, denotes Caenorhabditis elegans; Sr, denotes Strongyloides ratti; Bx, denotes Bursaphelenchus xylophilus; Gp, denotes Globodera pallida; Mh, denotes Meloidogyne hapla; Mi, denotes Meloidogyne incognita. Each phylogeny (A-C) represents a single C. elegans FLP-GPCR cluster identified in Fig. 3: (A) denotes the phylogeny of parasitic nematode FLP-GPCR homologues of the C. elegans predicted proteins present in Fig. 3 cluster 1; (B) denotes the phylogeny of parasitic nematode FLP-GPCR homologues of the C. elegans predicted proteins present in Fig. 3 cluster 2; (C) denotes the phylogeny of parasitic nematode FLP-GPCR homologues of the C. elegans predicted proteins present in Fig. 3 cluster 3. FLP-GPCR cluster nodes supported by a bootstrap analysis value of >70% are identified by red dots. Clusters are shaded and named according to the relevant C. elegans orthologue. Bootstrap values are shown as percentages. ‡Note that NPR-2, NPR-5, NPR-8, DMSR-1 and FRPR-18 isoforms were not delineated in the parasite species. In addition only the longest C. elegans FLP-/putative FLP-receptor isoforms were included in the phylogenetic analyses such that Ce_NPR-2 represents isoform a; Ce_NPR-5 represents isoform b; Ce_NPR-8 represents isoform b, Ce_DMSR-1 represents isoform a; Ce_EGL-6 represents isoform a; Ce_FRPR-18 represents isoform b.
With respect to broad-spectrum drug target appeal, DMSR-2, -7 and -8 are the most highly conserved putative C. elegans FLP-GPCRs found in parasitic nematodes. It will be interesting to see if deorphanisation studies link these to highly conserved flp-ligands (e.g. flp-1 and flp-14 encoded peptides which remain unmatched with their cognate receptors). It is likely that those putative C. elegans flp-GPCRs not identified in the parasites (npr-7, dmsr-9, dmsr-10, dmsr-11, dmsr-12, dmsr-13, dmsr-14, dmsr-15, dmsr-16, frpr-4, frpr-6, frpr-11, frpr-12, and frpr-16) have evolved via gene duplication following the separation of the Caenorhabditis spp. and the parasites from their common ancestor.
Several putative FLP-GPCRs in the parasite species did not fall into orthologous clusters [see Fig. 4; e.g. sequences previously designated as NPR-10, DMSR-7 and FRPR-18 homologues in clade 2 species (T. spiralis and T. muris), and DMSR-4, and DMSR-5, in the plant parasitic nematodes (B. xylophilus, M. incognita, M. hapla, and G. pallida)]. In addition, several putative FLP-GPCRs designated as either NPR-1 or NPR-2 (Ov-NPR-1, Oo-NPR-1, Di-NPR-1, Ll-NPR-1, As-NPR-2, Nb-NPR-2.2, Sr-NPR-1.2, Hc-NPR-1 and Nb-NPR-1) failed to cluster with either Ce-NPR-1 or Ce-NPR-2 (see Fig. 5A) and as such constitute two NPR-1/2-like parasite specific clusters. All sequences designated as either NPR-1 or NPR-2 homologues in this study exhibit a phenylalanine at position 215, linked to sociality in C. elegans.
4. Conclusions
This study provides the first pan-phylum genome-based overview of the FLPergic complement in parasitic nematodes. This encompasses both an update of the FLP ligand profile and reports, for the first time, the putative FLP-GPCR complement beyond C. elegans. Our study revises the number of flp-genes from 34 to 32, of which only one (flp-31) appears to be parasite-specific. This study also reveals that whilst the flp-signatures identified in nematode parasites are structurally similar to those described for C. elegans, the diversity in the flp-complement in the parasites is variably restricted. Indeed, nematode parasites only possess a proportion of the C. elegans flp-gene complement and exhibit inter-clade variation where clade-matched species broadly display similar flp-gene profiles.
With respect to FLP-GPCRs we have reported the conservation of known deorphanised C. elegans FLP-receptors in parasite species and our phylogenetic approach has facilitated the identification of a further 13 putative flp-GPCRs in nematode parasites. These data reveal that there may be many more nematode FLP-receptors than previously thought.
This dataset contributes significantly to future FLP-receptor deorphanisation efforts by providing a more comprehensive library of parasite-specific FLP ligands and a catalogue of putative parasite FLP-receptors against which to screen. These data help facilitate the functional characterisation of parasite FLP-GPCRs to reveal those most appealing for exploitation as novel chemotherapeutic targets.
Transparency document
Acknowledgements
This work was funded by the Biotechnology and Biological Sciences Research Council (BB/H019472/1) and the Department of Education and Learning for Northern Ireland. The funders had no role in study design, execution, or preparation of the manuscript.
Footnotes
Available online 19 April 2014
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.euprot.2014.04.002.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Multiple sequence alignments of FLPs encoded onflpgene sequelogues identified by BLAST. Sequences were aligned using the Vector NTI Advance™11, Align X multiple sequence alignment tool, using default settings [32]. Completely and partially conserved amino acid residues are highlighted in yellow and blue, respectively, while blocks of similar residues are highlighted in green. More divergent flp genes include the flp-3 sequelogues present within filarial nematodes. Attempts to identify FLP sequences more similar to those encoded on these genes in other nematodes were unsuccessful. For this reason they have been assigned a name according to their most similar C. elegans flp gene sequelogue (flp-3). Moreover, filarial flp-3 genes occupy a similar genomic environment to all other flp-3 sequelogues (see Figure 2) suggesting that these genes represent flp-3 orthologues despite encoding a divergent FLP sequence. T. spiralis, denotes Trichinella spiralis; T. muris, denotes Trichuris muris; B. malayi, denotes Brugia malayi; W. bancrofti, denotes Wuchereria bancrofti; D. immitis, denotes Dirofilaria immitis; O. volvulus, denotes Onchocerca volvulus; O. ochengi, denotes Onchocerca ochengi; L. loa, denotes Loa loa; A. suum, denotes Ascaris suum; A. caninum, denotes Ancylostoma caninum; N. brasiliensis, denotes Nippostrongylus brasiliensis; H. contortus, denotes Haemonchus contortus; C. elegans, denotes Caenorhabditis elegans; S. ratti, denotes Strongyloides ratti; B. xylophilus, denotes Bursaphelenchus xylophilus; G. pallida, denotes Globodera pallida; M. hapla, denotes Meloidogyne hapla; M. incognita, denotes Meloidogyne incognita.
Multiple sequence alignments of the predicted FLP-GPCRs identified by BLAST. Sequences were aligned using the Vector NTI Advance™11, Align X multiple sequence alignment tool, using default settings [32]. Completely and partially conserved amino acid residues are highlighted in yellow and blue respectively, while blocks of similar residues are highlighted in green. Predicted transmembrane domains are underlined on the consensus sequence for each FLP-GPCR alignment. Outside to inside helices have are identified by a solid line; inside to outside helices are identified by a dotted line. Note that only the longest C. elegans FLP-/putative FLP-receptor isoforms were included in the alignment such that C. elegans NPR-2 represents isoform a; C. elegans NPR-5 represents isoform b; C. elegans NPR-8 represents isoform b, C. elegans DMSR-1 represents isoform a; C. elegans EGL-6 represents isoform a; C. elegans FRPR-18 represents isoform b. T. spiralis, denotes Trichinella spiralis; T. muris, denotes Trichuris muris; B. malayi, denotes Brugia malayi; W. bancrofti, denotes Wuchereria bancrofti; D. immitis, denotes Dirofilaria immitis; O. volvulus, denotes Onchocerca volvulus; O. ochengi, denotes Onchocerca ochengi; L. loa, denotes Loa loa; A. suum, denotes Ascaris suum; A. caninum, denotes Ancylostoma caninum; N. brasiliensis, denotes Nippostrongylus brasiliensis; H. contortus, denotes Haemonchus contortus; C. elegans, denotes Caenorhabditis elegans; S. ratti, denotes Strongyloides ratti; B. xylophilus, denotes Bursaphelenchus xylophilus; G. pallida, denotes Globodera pallida; M. hapla, denotes Meloidogyne hapla; M. incognita, denotes Meloidogyne incognita.
Nematode Genome and Transcriptome BLAST databases/servers employed. BI, denotes Broad Institute; 959, denotes 959 Nematode Genomes; SI, denotes Sanger Institute; WB, denotes WormBase; INRA, denotes INRA Genomic Resources; EST, denotes Expressed Sequence Tags; NR/NT, denotes Nucleotide Collection; TSA, denotes Transcriptome Shotgun/RNA-seq Assembly; GSS, denotes Genome Survey Sequences; WS231-8, denotes Wormbase release WS231-8; PP, denotes Predicted Proteins; 454, denotes 454 reads; GS/C, denotes the most recent Genomic Scaffolds/Contigs available; UP, denotes unplaced reads; P, denotes published genome; UOG, denotes unpublished on-going; UIA, denotes unpublished in annotation; GOG, denotes GSS on-going; 454OG, denotes 454 on-going. NCBI: http://www.ncbi.nlm.nih.gov/; WormBase: http://www.wormbase.org/tools/blast_blat; 959 Nematode genomes: http://www.nematodes.org/nematodegenomes/index.php/959_Nematode_Genomes; The Sanger institute: http://www.sanger.ac.uk/resources/software/blast/; The Broad institute: http://www.broadinstitute.org/annotation/genome/filarial_worms/Blast.html; INRA Genomic resources: http://meloidogyne.toulouse.inra.fr/blast/blast.html.
flp gene BLAST query GenBank accession numbers.
flp / putative flp-GPCR BLAST query GenBank accession numbers. *denotes deorphanised FLP-GPCR-encoding gene.
References
- 1.Li J., Todd T.C., Lee J., Trick H.N. Biotechnological application of functional genomics towards plant-parasitic nematode control. Plant Biotechnol J. 2011;9:936–944. doi: 10.1111/j.1467-7652.2011.00601.x. [DOI] [PubMed] [Google Scholar]
- 2.Waller P.J. From discovery to development: current industry perspectives for the development of novel methods of helminth control in livestock. Vet Parasitol. 2006;139:1–14. doi: 10.1016/j.vetpar.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 3.Prichard R.K., Basáñez M.-G., Boatin B.A., McCarthy J.S., García H.H., Yang G.-J. A research agenda for helminth diseases of humans: intervention for control and elimination. PLoS Negl Trop Dis. 2012;6:e1549. doi: 10.1371/journal.pntd.0001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epe C., Kaminsky R. New advancement in anthelmintic drugs in veterinary medicine. Trends Parasitol. 2013;29:129–134. doi: 10.1016/j.pt.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Xiao S-H., Hui-Ming W., Tanner M., Utzinger J., Chong W. Tribendimidine: a promising, safe and broad-spectrum anthelmintic agent from China. Acta Trop. 2005;94:1–14. doi: 10.1016/j.actatropica.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Hotez P.J. Mass drug administration and integrated control for the world's high-prevalence neglected tropical diseases. Clin Pharmacol Ther. 2009;85:659–664. doi: 10.1038/clpt.2009.16. [DOI] [PubMed] [Google Scholar]
- 7.Lustigman S., Prichard R.K., Gazzinelli A., Grant W.N., Boatin Ba., McCarthy J.S. A research agenda for helminth diseases of humans: the problem of helminthiases. PLoS Negl Trop Dis. 2012;6:e1582. doi: 10.1371/journal.pntd.0001582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones J.T., Haegeman A., Danchin E.G.J., Gaur H.S., Helder J., Jones M.G.K. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol. 2013;14:946–961. doi: 10.1111/mpp.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McVeigh P., Atkinson L., Marks N.J., Mousley A., Dalzell J.J., Sluder A. Parasite neuropeptide biology: seeding rational drug target selection? Int J Parasitol Drugs Drug Resist. 2012;2:76–91. doi: 10.1016/j.ijpddr.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McVeigh P., Geary T.G., Marks N.J., Maule A.G. The FLP-side of nematodes. Trends Parasitol. 2006;22:385–396. doi: 10.1016/j.pt.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Li C., Kim K. WormBook; 2008. Neuropeptides; pp. 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ringstad N., Horvitz H.R. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci. 2008;11:1168–1176. doi: 10.1038/nn.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen M.J., Lancheros E.R., Williams T., Lowery D.E., Geary T.G., Kubiak T.M. Functional expression and characterization of the C. elegans G-protein-coupled FLP-2 Receptor (T19F4.1) in mammalian cells and yeast. Int J Parasitol Drugs Drug Resist. 2013;3:1–7. doi: 10.1016/j.ijpddr.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowery DE, Geary TG, Kubiak TM, Larsen MJ. Pharmacia & Upjohn Company, G-protein coupled receptor-like receptors and modulators thereof, United States Patent 6,632,631 (2003).
- 15.Kubiak T.M., Larsen M.J., Zantello M.R., Bowman J.W., Nulf S.C., Lowery D.E. Functional annotation of the putative orphan Caenorhabditis elegans G-protein-coupled receptor C10C6.2 as a FLP15 peptide receptor. J Biol Chem. 2003;278:42115–42120. doi: 10.1074/jbc.M304056200. [DOI] [PubMed] [Google Scholar]
- 16.Kubiak T.M., Larsen M.J., Nulf S.C., Zantello M.R., Burton K.J., Bowman J.W. Differential activation of social and solitary variants of the Caenorhabditis elegans G protein-coupled receptor NPR-1 by its cognate ligand AF9. J Biol Chem. 2003;278:33724–33729. doi: 10.1074/jbc.M304861200. [DOI] [PubMed] [Google Scholar]
- 17.Kubiak T.M., Larsen M.J., Bowman J.W., Geary T.G., Lowery D.E. FMRFamide-like peptides encoded on the flp-18 precursor gene activate two isoforms of the orphan Caenorhabditis elegans G-protein-coupled receptor Y58G8A.4 heterologously expressed in mammalian cells. Biopolymers. 2008;90:339–348. doi: 10.1002/bip.20850. [DOI] [PubMed] [Google Scholar]
- 18.Keating C.D., Kriek N., Daniels M., Ashcroft N.R., Hopper N.A., Siney E.J. Whole-genome analysis of 60 G protein-coupled receptors in Caenorhabditis elegans by gene knockout with RNAi. Curr Biol. 2003;13:1715–1720. doi: 10.1016/j.cub.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Mertens I., Meeusen T., Janssen T., Nachman R., Schoofs L. Molecular characterization of two G protein-coupled receptor splice variants as FLP2 receptors in Caenorhabditis elegans. Biochem Biophys Res Commun. 2005;330:967–974. doi: 10.1016/j.bbrc.2005.03.071. [DOI] [PubMed] [Google Scholar]
- 20.Mertens I., Clinckspoor I., Janssen T., Nachman R., Schoofs L. FMRFamide related peptide ligands activate the Caenorhabditis elegans orphan GPCR Y59H11AL.1. Peptides. 2006;27:1291–1296. doi: 10.1016/j.peptides.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 21.McVeigh P., Leech S., Mair G.R., Marks N.J., Geary T.G., Maule A.G. Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int J Parasitol. 2005;35:1043–1060. doi: 10.1016/j.ijpara.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Elsworth B., Wasmuth J., Blaxter M. NEMBASE4: the nematode transcriptome resource. Int J Parasitol. 2011;41:881–894. doi: 10.1016/j.ijpara.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Holterman M., van der Wurff A., van den Elsen S., van Megen H., Bongers T., Holovachov O. Phylum-wide analysis of SSU rDNA reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown Clades. Mol Biol Evol. 2006;23:1792–1800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- 24.Laing R., Kikuchi T., Martinelli A., Tsai I.J., Beech R.N., Redman E. The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 2013;14:R88. doi: 10.1186/gb-2013-14-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwarz E.M., Korhonen P.K., Campbell B.E., Young N.D., Jex A.R., Jabbar A. The genome and developmental transcriptome of the strongylid nematode Haemonchus contortus. Genome Biol. 2013;14:R89. doi: 10.1186/gb-2013-14-8-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mertens I., Vandingenen A., Meeusen T., Janssen T., Luyten W., Nachman R.J. Functional characterization of the putative orphan neuropeptide G-protein coupled receptor C26F1.6 in Caenorhabditis elegans. FEBS Lett. 2004;573:55–60. doi: 10.1016/j.febslet.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 27.Rogers C., Reale V., Kim K., Chatwin H., Li C., Evans P. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci. 2003;6:1178–1185. doi: 10.1038/nn1140. [DOI] [PubMed] [Google Scholar]
- 28.Frooninckx L., Van Rompay L., Temmerman L., Van Sinay E., Beets I., Janssen T. Neuropeptide GPCRs in C. elegans. Front Endocrinol (Lausanne) 2012;3:167. doi: 10.3389/fendo.2012.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi T., Cotton Ja., Dalzell J.J., Hasegawa K., Kanzaki N., McVeigh P. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011;7:e1002219. doi: 10.1371/journal.ppat.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abad P., Gouzy J., Aury J.-M., Castagnone-Sereno P., Danchin E.G.J., Deleury E. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- 31.Dalzell J.J., McVeigh P., Warnock N.D., Mitreva M., Bird D.M., Abad P. RNAi effector diversity in nematodes. PLoS Negl Trop Dis. 2011;5:e1176. doi: 10.1371/journal.pntd.0001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu G. Software review Vector NTI, a balanced all-in-one sequence analysis suite. Brief Bioinform. 2004;5:378–388. doi: 10.1093/bib/5.4.378. [DOI] [PubMed] [Google Scholar]
- 33.Tusnády G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamanian M., Kimber M.J., McVeigh P., Carlson Sa., Day T.A. The repertoire of G protein-coupled receptors in the human parasite Schistosoma mansoni and the model organism Schmidtea mediterranea. BMC Genomics. 2011;12:596. doi: 10.1186/1471-2164-12-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 38.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Jarecki J.L., Frey B.L., Smith L.M., Stretton A.O. Discovery of neuropeptides in the nematode Ascaris suum by database mining and tandem mass spectrometry. J Proteome Res. 2011;10:3098–3106. doi: 10.1021/pr2001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atkinson L.E., Stevenson M., McCoy C.J., Marks N.J., Fleming C., Zamanian M. flp-32 Ligand/receptor silencing phenocopy faster plant pathogenic nematodes. PLoS Pathog. 2013;9:e1003169. doi: 10.1371/journal.ppat.1003169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maule A.G., Mousley A., Marks N.J., Day T.A., Thompson D.P., Geary T.G. Neuropeptide signaling systems – potential drug targets for parasite and pest control. Curr Top Med Chem. 2002;2:733–758. doi: 10.2174/1568026023393697. [DOI] [PubMed] [Google Scholar]
- 42.Mitreva M., Jasmer D.P., Zarlenga D.S., Wang Z., Abubucker S., Martin J. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43:228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarecki J.L., Andersen K.M., Miller A.H., Ramaker M.A., Vestling M.M., Stretton A.O. Three independent techniques localize expression of transcript af p-11 and its bioactive peptide products to the paired AVK neurons in Ascaris suum: in situ hybridization, immunocytochemistry, and single cell mass spectrometry. ASC Chem Neurosci. 2013;4:418–434. doi: 10.1021/cn3001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McVeigh P., Mair G.R., Atkinson L., Ladurner P., Zamanian M., Novozhilova E. Discovery of multiple neuropeptide families in the phylum Platyhelminthes. Int J Parasitol. 2009;39:1243–1252. doi: 10.1016/j.ijpara.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen M., Reale V., Olofsson B., Knights A., Evans P., de Bono M. Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. 2009;9:375–385. doi: 10.1016/j.cmet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Janssen T., Lindemans M., Meelkop E., Temmerman L., Schoofs L. Coevolution of neuropeptidergic signaling systems: from worm to man. Ann N Y Acad Sci. 2010;1200:1–14. doi: 10.1111/j.1749-6632.2010.05506.x. [DOI] [PubMed] [Google Scholar]
- 47.Chalasani S.H., Kato S., Albrecht D.R., Nakagawa T., Abbott L.F., Bargmann C.I. Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci. 2010;13:615–621. doi: 10.1038/nn.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Bono M., Bargmann C.I. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 49.Caers J., Verlinden H., Zels S., Vandersmissen H.P., Vuerinckx K., Schoofs L. More than two decades of research on insect neuropeptide GPCRs: an overview. Front Endocrinol (Lausanne) 2012;3:151. doi: 10.3389/fendo.2012.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel K.J., Brown M.R., Strand M.R. Phylogenetic investigation of Peptide hormone and growth factor receptors in five dipteran genomes. Front Endocrinol (Lausanne) 2013;4:193. doi: 10.3389/fendo.2013.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yapici N., Kim Y-J., Ribeiro C., Dickson B.J. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature. 2008;451:33–37. doi: 10.1038/nature06483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignments of FLPs encoded onflpgene sequelogues identified by BLAST. Sequences were aligned using the Vector NTI Advance™11, Align X multiple sequence alignment tool, using default settings [32]. Completely and partially conserved amino acid residues are highlighted in yellow and blue, respectively, while blocks of similar residues are highlighted in green. More divergent flp genes include the flp-3 sequelogues present within filarial nematodes. Attempts to identify FLP sequences more similar to those encoded on these genes in other nematodes were unsuccessful. For this reason they have been assigned a name according to their most similar C. elegans flp gene sequelogue (flp-3). Moreover, filarial flp-3 genes occupy a similar genomic environment to all other flp-3 sequelogues (see Figure 2) suggesting that these genes represent flp-3 orthologues despite encoding a divergent FLP sequence. T. spiralis, denotes Trichinella spiralis; T. muris, denotes Trichuris muris; B. malayi, denotes Brugia malayi; W. bancrofti, denotes Wuchereria bancrofti; D. immitis, denotes Dirofilaria immitis; O. volvulus, denotes Onchocerca volvulus; O. ochengi, denotes Onchocerca ochengi; L. loa, denotes Loa loa; A. suum, denotes Ascaris suum; A. caninum, denotes Ancylostoma caninum; N. brasiliensis, denotes Nippostrongylus brasiliensis; H. contortus, denotes Haemonchus contortus; C. elegans, denotes Caenorhabditis elegans; S. ratti, denotes Strongyloides ratti; B. xylophilus, denotes Bursaphelenchus xylophilus; G. pallida, denotes Globodera pallida; M. hapla, denotes Meloidogyne hapla; M. incognita, denotes Meloidogyne incognita.
Multiple sequence alignments of the predicted FLP-GPCRs identified by BLAST. Sequences were aligned using the Vector NTI Advance™11, Align X multiple sequence alignment tool, using default settings [32]. Completely and partially conserved amino acid residues are highlighted in yellow and blue respectively, while blocks of similar residues are highlighted in green. Predicted transmembrane domains are underlined on the consensus sequence for each FLP-GPCR alignment. Outside to inside helices have are identified by a solid line; inside to outside helices are identified by a dotted line. Note that only the longest C. elegans FLP-/putative FLP-receptor isoforms were included in the alignment such that C. elegans NPR-2 represents isoform a; C. elegans NPR-5 represents isoform b; C. elegans NPR-8 represents isoform b, C. elegans DMSR-1 represents isoform a; C. elegans EGL-6 represents isoform a; C. elegans FRPR-18 represents isoform b. T. spiralis, denotes Trichinella spiralis; T. muris, denotes Trichuris muris; B. malayi, denotes Brugia malayi; W. bancrofti, denotes Wuchereria bancrofti; D. immitis, denotes Dirofilaria immitis; O. volvulus, denotes Onchocerca volvulus; O. ochengi, denotes Onchocerca ochengi; L. loa, denotes Loa loa; A. suum, denotes Ascaris suum; A. caninum, denotes Ancylostoma caninum; N. brasiliensis, denotes Nippostrongylus brasiliensis; H. contortus, denotes Haemonchus contortus; C. elegans, denotes Caenorhabditis elegans; S. ratti, denotes Strongyloides ratti; B. xylophilus, denotes Bursaphelenchus xylophilus; G. pallida, denotes Globodera pallida; M. hapla, denotes Meloidogyne hapla; M. incognita, denotes Meloidogyne incognita.
Nematode Genome and Transcriptome BLAST databases/servers employed. BI, denotes Broad Institute; 959, denotes 959 Nematode Genomes; SI, denotes Sanger Institute; WB, denotes WormBase; INRA, denotes INRA Genomic Resources; EST, denotes Expressed Sequence Tags; NR/NT, denotes Nucleotide Collection; TSA, denotes Transcriptome Shotgun/RNA-seq Assembly; GSS, denotes Genome Survey Sequences; WS231-8, denotes Wormbase release WS231-8; PP, denotes Predicted Proteins; 454, denotes 454 reads; GS/C, denotes the most recent Genomic Scaffolds/Contigs available; UP, denotes unplaced reads; P, denotes published genome; UOG, denotes unpublished on-going; UIA, denotes unpublished in annotation; GOG, denotes GSS on-going; 454OG, denotes 454 on-going. NCBI: http://www.ncbi.nlm.nih.gov/; WormBase: http://www.wormbase.org/tools/blast_blat; 959 Nematode genomes: http://www.nematodes.org/nematodegenomes/index.php/959_Nematode_Genomes; The Sanger institute: http://www.sanger.ac.uk/resources/software/blast/; The Broad institute: http://www.broadinstitute.org/annotation/genome/filarial_worms/Blast.html; INRA Genomic resources: http://meloidogyne.toulouse.inra.fr/blast/blast.html.
flp gene BLAST query GenBank accession numbers.
flp / putative flp-GPCR BLAST query GenBank accession numbers. *denotes deorphanised FLP-GPCR-encoding gene.