Abstract
An important consideration for the development of biosensors is the adsorption of the bio recognition element to the surface of a substrate. As the first step in the immobilization process, adsorption affects most immobilization routes and much attention is given into the research of this process to maximize the overall activity of the bio sensor. The use of nanomaterials, specifically nanoparticles and nanostructured films, offers advantageous properties that can be fine-tuned for interaction with specific proteins to maximize activity, minimize structural changes, and enhance the catalytic step. In the biosensor field, protein-nanomaterial interactions are an emerging trend that span across many disciplines. This review addresses recent publications about the proteins most frequently used, their most relevant characteristics, and the conditions required to adsorb them to nanomaterials. When relevant and available, subsequent analytical figures of merits are discussed for selected biosensors. The general trend amongst the research papers allows concluding that the use of nanomaterials has already provided significant improvements in the analytical performance of many biosensors and that this research field will continue to grow.
1-INTRODUCTION
The adsorption of proteins to surfaces is a central concern for the rational design and application of materials[1]. As it will be later specifically addressed, the rate and strengths of the initial physical interactions between proteins and surfaces dictate (to a large degree) the final conformation, stability, and activity of such proteins. This issue, that plays a major role in determining the biocompatibility of materials[2, 3], can also dictate the analytical performance of almost every analytical device that uses a biorecognition element (antigen, antibody, enzyme, nucleic acids, or even whole cells)[4]. The topic has become even more relevant in the last decade because an increasing number of applications of biosensors and other protein-based analytical devices have been presented, spanning across a wide array of applications including healthcare, security, environmental, agriculture, food control, process control, and microbiology[5, 6]. Modern biosensors are inexpensive, simple to operate, fast, and provide enough selectivity to be applied in the analysis of relatively complex samples. However, and despite the body of research currently available, only a few biosensors are commercially available andcan compete with more complex techniques in terms of sensitivity and limits of detection. Aiming to address these shortcomings, a series of strategies have been recently proposed[7-10]. Among those, and reflecting on the progress made in the techniques available for their synthesis and characterization, the use of nanomaterials (defined as materials with at least one feature or component having dimensions between 1-100 nm) has emerged as one of the leading trends for the development of biosensors and other bioanalytical devices [11]. Their unique chemical, mechanical, electrical, and structural properties enable tuninginteractions at the nanoscale and catering for the most suitable conditions for protein immobilization.
In general, and looking beyond the boundaries imposed by the selected transduction method (electrochemical, electrical, optical, piezoelectric, or thermal), assessing the role of the chemistry and topography of the surface[12-14], the physical and chemical characteristics of the protein to be used[15, 16], the immobilization route, and the experimental conditions selected for the coupling are fundamental to overcome current limitations. Considering these aspects, researchers currently have a variety of immobilization methods at their disposal[17-19], including covalent attachment, entrapment, encapsulation and cross-linking. While covalent attachment can provide an avenue to form a permanent bond between the functional groups of the protein and those of the substrate, the reactions are typically slow, laborious, and the experimental conditions required for such reactions can be detrimental to both the protein and electronic properties of the substrate[20-23]. The use of bifunctional reagents can be a simple and fast method to promote covalent interactions between the substrate-protein and protein-protein interface[24-26], but the bioactivity of the layer can be compromised by the poor accessibility of active sites. Alternatively, proteins can be entrapped within a highly cross-linked polymer matrix[27, 28] or encapsulated within a membrane[29, 30]. Depending on the specific conditions, these strategies can impose a limitation to the diffusion of both analytes and products.On the other side of the spectrum, adsorption can be identified as the mildest immobilization method and therefore has the greatest potential to preserve the native structure of the biorecognition element. As it is a spontaneous process driven (mainly) by hydrophobic, electrostatic, and van der Waals interactions[31-33], adsorption provides a simple and fast way to attach proteins to surfaces. Although it dictates the first interaction with the surface and consequently affects all other immobilization routes, the main drawback of this method is that the immobilized protein is (theoretically) in equilibrium with the solution and can therefore be gradually desorbed during the operation, upon changes in the solution pH, or by the addition of competing molecules (surfactants or other proteins). To minimize this possibility, it is essential to carefully choose the experimental conditions for the immobilization, maximize the initial adsorption rate, and strike a balance between the stability of the adsorbed layer and the structure of the protein. Studies of protein adsorption to solid surfaces are certainly not new[1], however analytical applications of adsorbed proteins to nanomaterials have experienced a tremendous growth in the last 10 years (Figure 1) and are projected to continue in an upward trend. These materials allow increasing the effective area of the sensor, promoting the interaction of the biorecognition element with the substrates, enhancing the detection process, and therefore improving the overall analytical performance of the sensor.
Figure 1.
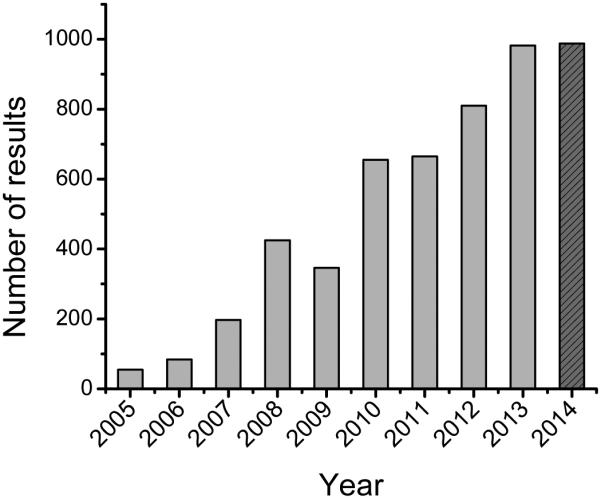
Google scholar search comprising the years of 2005 to 2014 using the keywords “nanomaterials”, “adsorption”, and “biosensor”. Search conducted on 09/25/2014.
Aiming to provide a critical overview of the advantages and limitations of the adsorption of enzymes to nanoparticles and nanostructured materials for analytical applications, this review will focus on some of the most representative reports published in the last decade. Since it is almost impossible to cover every paper published in the topic, the goal of this review is to highlight examples of the strategies leading to significant improvements of the analytical figures of merit. In order to provide context for the discussion, the review starts with a general discussion about the adsorption of proteins and the effects of the most relevant variables. Then, recent advances pertinent to the development of biosensors using adsorption of proteins to nanomaterials (nanoparticles and nanostructured films) are discussed, considering the appropriate analytical figures of merit. As a reference point, the review also includes a short description of the techniques used to investigate the adsorption of proteins as well as emerging trends with potential applications for analytical devices.
It is also important to mention that previous reviews and articles describing the adsorption of proteins to various surfaces have been presented[1, 14, 34-49] and were used as reference points to start the discussion related to the analytical aspects of the phenomenon.
2. GENERAL CONSIDERATIONS
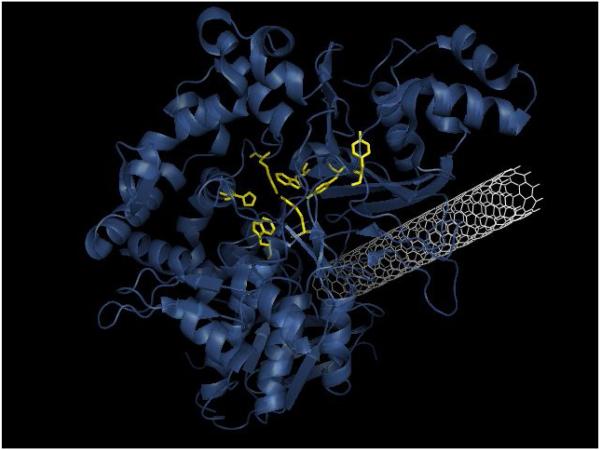
Proteins are biopolymers formed by linear sequences of amino acids (primary structure), arranged into local three-dimensional units forming (mainly) α-helices and β-sheets (secondary structure)that are folded into larger 3D structures conferring the shape of the proteins(tertiary structure). A number of these tertiary structures can be assembled originating the quaternary structure of the protein[50, 51]. As it defines the charge profile, location of hydrophobic patches, and possibility to form hydrogen bonds, the structure of the protein should be considered in the experimental design. Obtaining the structure of most proteins is quite trivial and can be done by either downloading the corresponding files from the Protein Data Bank (http://www.rcsb.org) or by the use of computational methods considering the pKa and location of each amino acid. This information has been used to gain preliminary insights about the potential adsorptive behavior of proteins[14, 52, 53] and estimate the enzymatic parameters from the structure[54]. Without requiring specific training or computational facilities, two possible approaches related to the topic of the review can be mentioned. The first case involves a docking calculation using Auto Dock Vina 4.2[55]. As recently described[20, 56], these calculations can be used to identify the most probable binding sites and the energy involved in the adsorption process (Figure 2). Dependent on the size of the system, these calculations can be completed in less than 8 h using a standard computer.
Figure 2.
Binding pose of glutamate oxidase on a CNT according to the AutoDock Vina molecular docking program. The amino acids of the enzyme’s active site are highlighted in yellow. Reprinted from [56], with permission from Wiley.
The second approach that can be used to estimate the protein concentration on a surface only requires a series of input variables (protein concentration, protein descriptors derived from primary structure, surface descriptors, pH, and ionic strength)[57]. The algorithm is freely available online(http://bad.molecularsense.com/) and has a predictive error of <5%, with respect to a validated database. In addition to those and aiming to predict the adsorptive capacity of the surface of nanomaterials, the biological surface adsorption index (BSAI) was presented by Xia et. al.[58]. This novel approach was used to characterize surface adsorption energy of nanomaterials considering five descriptors (hydrophobicity, hydrogen bonding, polarity, polarizability, and lone-pair electrons) of the nanomaterial interaction with biological components. Freely available packages to aid inthe visualization of protein structures (such as PyMol http://www.pymol.org/, Jmol http://jmol.sourceforge.net/, or Polyview 3D http://polyview.cchmc.org/polyview3d.html) can be used to highlight protein active sites and functional hot spots[59].
The protein adsorption process comprises of various steps including transport of the protein from the bulk solution into the inter facial region, attachment of the protein at the sorbent surface, relaxation of the protein on the surface, detachment from the surface, and transport of the detached protein back into the solution. These steps take place on various time-scales, ranging from a few seconds for the first protein-surface interaction to several hours to achieve steady-state conditions. Although the process is rather simple, it involves the interplay of many variables including temperature (under most conditions the adsorption is an endothermic process), charge(proteins adsorb most onto surfaces at there isoelectric point (IEP) minimizing protein-protein repulsions), size (smaller proteins diffuse quicker and arrive at the surface faster), shape (dictating the number of contact points with the substrate surface)[60], hydrophobic interactions[52, 61-63], and the ability to form π-π stacking and hydrogen bonds[44, 64]. The sum of these contributions (some favorable and some unfavorable) determines not only how, how fast, and how much proteins are adsorbed, but also under what conditions they may be removed from the substrate. It is also important to point out that adsorbed proteins are only marginally stable on the surface because the beneficial interactions that govern the native structure in solution are counterbalanced by a large entropy loss associated with going from a large ensemble of states to a more restricted set of conformations, typically affecting the original protein function[38]. Though several techniques are available to measure protein stability[65-67], differential scanning calorimetry has been mostly used to classify proteins within a range of soft and hard[68] based on their denaturation temperature (Td). Soft proteins (like bovine serum albumin, BSA, Td=57[69]) are less structurally stable, tend to experience large conformational changes (surface-induced unfolding and refolding) during the adsorption process, and can be adsorbed under a wide range of conditions. On the other side, hard proteins (such as lysozyme, Td= 73[69]− 76[70]) tend to preserve much of the native structure after the adsorption step and their adsorption requires a more careful selection of the environment. Needless to say, proteins with Td values in between that range show intermediate behavior and provide a much more challenging scenario for the prediction of their adsorptive behavior. Likewise, the properties of the substrate are just as important as those of the protein when considering adsorption. Hydrophobicity (contact angle), charge, topography, and chemical groups can have significant impacts on the adsorption process. In general, proteins tend to adsorb more readily to hydrophobic surfaces, promoting the release of water molecules and increasing the entropy[52, 57, 71-73]. Several groups have used this property to either promote or restrict the adsorption of proteins in analytical applications[74, 75]. Protein adsorption and orientation are also influenced by electrostatic interactions between the surface and the protein [14, 62, 76]. As an increased surface roughness is typically translated into an increased surface area for adsorption and an increased surface curvature, the topography and the number of contact points have also been considered[12, 77-84]. Undoubtedly, increasing the amount and preserving the catalytic activity of the immobilized proteins is essential to improve the performance of analytical devices. Therefore, using the tunable nanoscale properties associated with nanomaterials presents itself as a unique alternative to promote favorable conditions and ultimately preserve the bioactivity of the protein[12, 81, 85-90].
3. TECHNIQUES TO INVESTIGATE PROTEIN ADSORPTION
There are a number of techniques that can be used to characterize the nanomaterial/protein interface[91] and obtain valuable information about the adsorption process. Surface features and topography can be determined using atomic force microscopy (AFM) [89, 92-94], scanning electron microscopy (SEM), transmission electron microscopy (TEM) [95], and scanning tunneling microscopy (STM)[6]. AFM can render 3D images of surface topography but can be affected by a series of artifacts [96]. On the other hand, electron microscopy techniques can potentially allow the observation of the substrates at the subatomic level but are (generally) destructive and require the sample to be conductive. Recent progress in the field has allowed the inclusion of cryo-probes and the development of electron crystallography (used to determine the structure of several proteins allowing the identification, in some cases, of secondary structure motifs) [97, 98]. Other techniques [91] used to support research related to protein adsorption include capillary electrophoresis [99], mass spectrometry [100, 101] and particle mass spectrometry [102] (to identify and quantify proteins through fragmentation), various vibrational spectroscopictechniques [103, 104] (to study protein/substrate interface and elucidate the biosensing mechanism), UV-Vis spectroscopy [105], Fourier-transform infrared (FTIR) spectroscopy[106, 107] (to analyze sequential and competitive adsorption), circular dichroism (CD) spectroscopy[108, 109] (to investigate the secondary structure of adsorbed proteins), isothermal titration calorimetry [110] (to investigate thermodynamic properties of interactions), surface plasmon resonance (SPR)[37, 111-113] (for quantitation of proteins and kinetic studies but limited to gold or silver surfaces), total internal reflection fluorescence spectroscopy [114] (for determining conformational changes in adsorbed proteins), infrared spectroscopy [115] (to observe protein conformation and load but with limited use in multicomponent protein solutions because of complex spectra), and fluorescence spectroscopy[84, 116] (to observe structural change in protein and protein loading).As their interaction with proteins can lead to spectral shifts in gold nanoparticles(AuNP), various colorimetric assays have been presented to monitor either protein-protein interactions [117] or conformational changes [118]. Additionally, it was recently reported that in the low protein coverage regime, nanoparticle tracking analysis, and differential dynamic light scattering (but not differential centrifugal sedimentation)correlate with the expected plasmon frequency shift of AuNP[119].
Experiments dealing with the adsorption of proteins to nanomaterials can be performedin either static (batch experiments)[16, 108] or dynamic fashion. The latter methodology allows calculating not only the adsorbed amount but also the adsorption rate, which could be then used to calculate the probability of attachment for a protein under specific experimental conditions [120-122]. Among other techniques[123-126] that have been applied, adsorption kinetics of proteins can be investigated using quartz crystal microbalance (QCM)[92, 127-130] orellipsometry[131, 132]. QCM has the capability of quantifying extremely small amounts of proteins (mass changes) adsorbedto flat and fixed surfaces by measuring the change in frequency across a quartz crystal. QCM is highly sensitive but it is limited by the availability of appropriate substrates[133] and can also be affected by temperature gradients and pressure applied to the surface. Ellipsometry measures the amplitude ratio and phase difference upon reflectance of parallel and perpendicular components of polarized light reflecting from a surface. Since it measures the ratio of two values originated by the same signal, the measurements are highly accurate and reproducible. The measurements are complementary to the information obtained by QCM and can be used to determine the optical properties of asubstrate and the thickness of multiple layers on the surface.The technique is simple, nondestructive, has angstrom resolution, and has the capability of allowing the observation of the adsorption process inreal time[94, 134, 135]. The use of imaging ellipsometry can also provide spatial resolution of protein binding [136]. In most cases and considered a major limitation, the substrates’ optical properties should be known (at least approximately) and a comparison between an optical model and experimental data should be carefully performed to obtain meaning fulresults. Although it requires the calculation of a sensitivity factor, reflectometry allows following adsorption experiments in real time (with a higher frequency than ellipsometry) using a simpler experimental setup[137-140].
4. ADSORBEDPROTEINS
Since nanomaterials made from metals, semiconductors, and organic compounds can enhance optical, electrical, chemical, and magnetic properties that are relevant to sensing, researchers have extensively applied them for analytical purposes[141, 142]. One common problem observed in the literature is that while adsorption has been widely used to immobilize biorecognition elements to the surface of nanomaterials, only a fraction of those reports involve the corresponding adsorption study. As a result, adsorption has become (for some) a naive process in which the protein is brought into contact with the surface at conditions that may (or may not) conduce to an efficient and stable attachment. A frequent assumption is to adsorb proteins at the optimum pH for biological activity, which may not result in maximum coverage.It was also observed in some papers that the biorecognition element was adsorbed from a mixture containing BSA[143-146], which could result in competition of the two proteins for the active sites of the surface, and therefore a sensor with lower analytical figures of merit. Considering the number of reports in the area, this section of the review was organized according to the protein selected for the analytical application. A summary of the analytical figures of merit for glucose and peroxide biosensors (commonly used as model systems) is available in the Supplementary Information.
4.1. OXIDASES
Since a variety of analytical methodologies can be coupled to H2O2, oxidases have been the most popular choice for the development of biosensors. The H2O2is generated, for most cases, in stoichiometric amountswith respect to the substrate, and can be detected spectrophotometrically (e.g.using o-dianisidine in the presence of peroxidase) or electrochemically under a variety of experimental conditions.
4.1.1. Glucose Oxidase
Probably the most commonly used enzyme for analytical applications is glucose oxidase (GOx). GOxis a well-characterized enzyme that oxidizes glucose to gluconolactone and H2O2 and accounts for approximately 85% of the total biosensor market[147]. Since a variety of analytical methodologies can be coupled to H2O2, GOx has been considered a model enzyme and used for the development of a suite of electrochemical biosensors. To avoid electrostatic interactions between proteins and increase the adsorbed amount, the adsorption is usually performed at the IEP of the enzyme (4.2)[148];however, many researchers have chosen to perform the immobilization at different pH values [149, 150]. It is also important to point out that under optimized conditions, most surfaces can be saturated with GOx in less than 60 min using a solution containing ~0.1 mg·mL−1 of GOx[148, 151, 152]. Aiming at increasing the adsorbed amount, but at the risk of losing enzymatic activity or increasing cost, a number of papers reported the immersion of the substrates for 8 – 24 h[149, 153, 154] and the use of solutions containing as much as 20 mg·mL−1[150] or 40 mg·mL−1[155] of the enzyme. Due to their exquisite electronic properties and adsorptive capacity, some of the most common nanomaterials used for the immobilization of GOx are carbon-based nanomaterials[156-163]. Using these substrates, GOx can be readily adsorbed by a combination of hydrophobic and (to a much lesser degree) electrostatic interactions forming a closely packed and irreversibly bound layer of protein with a thickness comparable to the size of the protein [148]. Despite the fact that several authors have reported direct electron transfer [164-167], Wooten et al.[168] concluded that GOx adsorbed on carbon nanotubes (CNT) yields a pair of surface-confined current peaks at −0.48 V that are not compatible with the direct electron transfer between the enzymatically active GOx and CNT. In these cases, signal transduction relied on the decrease in the O2 reduction current and an increase in the H2O2 reduction current. Multiple examples of glucose biosensors built on CNT have been presented [169-179]. CNT can also be chemically modified to tweak their optical, electronic, and chemical properties [180-182] and therefore improve the performance of the resulting sensors. For example, Deng et al. [164] prepared a novel glucose biosensor using nitrogen doped carbon nanotubes (CNx-MWNTs) and adsorbed GOx. The CNx-MWNTs were prepared using a previously-reported method[183] and further modified to get a 4.6% nitrogen doping. The CNx-MWNTs were spotted onto a polished glassy carbon electrode (GCE) followed by an immersion in a solution of GOx (2 mg·mL−1, 20 h, and 4 °C) to promote adsorption and then spotted with Nafion to improve stability. Electrochemical impedance spectroscopy (EIS) showed that CNx-MWCTs-GCE exhibits a good electron-transfer rate (Ks=4.6 s−), bioactivity, reproducibility (RSD = 5%, n=10), and stability (96% of initial response after 20 days of storage). Nitrogen-doping has also been used for the modification of free-standing carbon fibers and the development of glucose sensors [184]. Instead of nitrogen doping, Zhang et al.[185] prepared a biosensor by immobilizing GOx (12h immersion) onto gold-platinum nanoparticle (AuPt NP)-MWNT-ionic liquid (i.e., 1-octyl-3-methylimidazolium hexafluorophosphate, [OMIM] PF6) composite. Using EIS, they established that the AuPt3-MWNT-[OMIM] PF6/GCE had the least electron transfer resistance when compared to GCE, MWNT/GCE, and MWNT-[OMIM] PF6/GCE. These electrodes enabled obtaining an apparent Michaelis-Menten constant of 76 μM, a sensitivity of 3.47 μA·mM−1, and a linear range of response in the 0.01 – 9.49 mM. GOx can also be adsorbed to gold surfaces. Among those, Wang et al.[186] utilized AuNP (20 – 30 nm) electrodeposited on indium tin oxide (ITO)for the development of a disposable sensor and noted that the addition ofcysteine yielded significant improvements in the performance and stability of the composite. A glucose biosensor obtained by adsorbing GOx onto nanostructured Au thin films was also reported by Qiu et al.[154]. These films were prepared electrochemically by displacing nanostructured thin films of deposited cobalt with gold on the surface of a GCE. Then, the enzyme adsorption was carried out submerging the Au/GCE in a solution containing GOx for 24 h. It is also common to observe that nanocomposites of GOx adsorbed to AuNPs are prepared with organic polymers. For example, German et al.[187] utilized AuNP(3.5, 6, and 13 nm) embedded in a polymerized pyrrole layer deposited on a carbon rod (CR) electrode. The use of the AuNP (13 nm)provided the basis to obtain a response that was twice as large as the one obtained with the bare GOx-CR electrode. Other polymers such as AgCl-polyaniline [153] have also been explored to support AuNP and adsorb GOx. Aiming to create a low mass transport barrier and improve conduction pathways to directelectron transfer of the mediator toward the electrode surface, GOx adsorbed to AuNP (11 nm) has also been embedded in a porous structure of a silica-gel network[188].
Platinum electrodes and platinum nanoparticles (PtNP) have advantageous catalytic properties towards the electrochemical oxidation of H2O2 and have also been used to develop sensors based on adsorbed GOx. Combining these catalytic properties with those of ordered mesoporous carbon (OMC)[189], CNT[189, 190], or graphene [191] can lead to significant improvements in sensitivity due to the synergy between the electrochemical properties of the nanoparticles and surface area and fast electron transfer of the substrate. PtNP (4 and 28 nm) can also be used in combination with silica nanoparticles (100nm)[192] in the fabrication of a glucose biosensor that preserved up to 90% of the initial response after 23 days. The large surface area of the nanoparticles ultimately played a crucial role in the adsorption of the enzyme to improve the catalytic activity and stability. Using a similar electrode, Li et al.[193] obtained a wide linear range for glucose (0.001 −2.6 mM).
Following the examples described with platinum, the system GOx-AuNP has also been combined with CNT[177, 194], OMC[195], and graphene oxide [196]. Kang et al.[197] synthesized wrinkled graphene sheets with carboxylic and hydroxyl groups which were used as a “nanofiller” on a chitosan-GCE. Although the layer of GOx deposited on the graphene-chitosan GCE slightly affected the electron transfer, this electrode was able to preserve the bioactivity of the enzyme, retaining 95% of its initial response after being stored for a week at 4 °C.The remarkable properties of graphene towards the adsorption of GOx have been combined with quantum dots. In this regard, it is worth highlighting a report from Zhiguo et al.[198], who combined graphene nanosheets, AuNP(10 nm) and a CdTe-CdS core shell of quantum dots to develop one of the top analytical glucose sensors in terms of response time (0.045 s), limit of detection (3 × 10−12 M), sensitivity (5,762,800 μA·mM−1·cm−2), stability (95% of its initial signal after 26 weeks) and (5.24 nM) indicating a fast reaction rate even at low concentrations. Figure 3 shows the assembly and the reaction scheme for such a biosensor.
Figure 3.
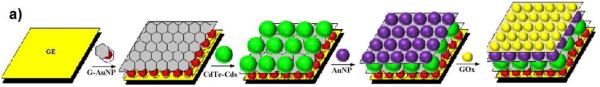
Fabrication of the biosensor (a) and oxidation of glucose at the GOx/AuNP/CdTe-CdS/G-AuNP/GE surface (b). Reprinted from [198], with permission from Elsevier.
Similarly, Wang et al. [199] utilized a previously developed mixture of GOx with graphene-CdS-Nafion nanocomposite [200] to modify the surface of a GCE. After verifying that the structure of the enzyme was not affected during the preparation of the sensor, they confirmed the existence of a synergistic effect in the modified electrode. The biosensor also showed good reproducibility by successively detecting 5.0 mM glucose 6 times with a RSD of 5.3% and acceptable stability by maintaining 93% of its initial response after being stored for a 30 days at 4 °C.
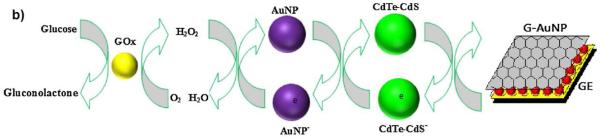
For simplicity, biosensors can be fabricated bythe adsorption of GOx to commercially available screen printed electrodes (SPE). For example, Gao et al. [201] prepared a biosensor by layer-by-layer (LbL) assembly of single-walled carbon nanotubes (SWCNTs)-GOx and synthesized osmium (bpy)2-complexed poly (1-vinylimidazole) (PVI-Os) on screen-printed carbon electrodes (SPCE) by means of electrodeposition. CV was used to determine that the electron transfer resistance increased if more than 4 bilayers were deposited. In a similar process, Deng et al. [165] prepared a LbL assembly of adsorbed GOx and poly(ethylenimine) (PEI) on a polished GCE modified with multi-walled carbon nanotubes (MWCNT). A GCE with CNTs and only GOx was prepared without LbLassembly for comparison. They determined that three bilayers of PEI/GOx (with a single outer layer of PEI for stability) showed the maximum peak currents for the electroactive GOx, showing no decreases in peak current after continuously scanning for 30 cycles and retainingmore than 95% of its initial response after storage for 20 days. Other composites have also been used by other authors [202]. CNT and graphene remain popular substrates for the immobilization of GOx, and other carbon-based substrates are also emerging for the immobilization of Gox. Using silica (SBA-15 and FDU-5) as templates, You et al. [203] developed mesoporous carbon materials and used them for the immobilization of GOx (Figure 4). They concluded that the highly branched intertwined nanostructure of the material enabled larger loading of GOx that was able to preserve >85% of the activity after being stored for 45 days.
Figure 4.
Schematic diagram of the electron transfer (ET) and the bioelectrocatalytic process of GOx/3D-CMM electrode. Reprinted from[203], with permission from Elsevier.
In addition to the previously-described CNT and graphene, optically transparent carbon electrodes (OTCE) [204] have been used as substrates to immobilize GOx. These electrodes can be fabricated by reduction at high temperature of a variety of organic precursors (including adsorbed proteins [205] or photoresist[206]) and have enabled the development of glucose sensors (OTCE immersed for 1 hin 4 mg·mL−1 under mild agitation).
Thelow-toxicity, potential biocompatibility, and electrochemical activity of other nanoparticles such as silver (AgNPs, 14 nm)[207], iron oxide [208] or zinc oxide [209-211] has sparked the development of other biosensors. The latter is particularly interesting due to its high IEP(about 9.5) which serves as a positively charged substrate for the immobilization of GOx (and other negatively-charged proteins) via electrostatic interactions. With the intention of preserving the native structure of the enzyme, Bhakta et al. recently demonstrated the potential advantages of nanoporous substrates fabricated using block co-polymers of polystyrene-block-poly(2-vinylpyridine) for the immobilization of GOx [94].
Peroxidase
Peroxidase isozymes are monomeric proteins (~40 kDa)that generally contain a polypeptide chain, a hemin group plus Ca2+, and carbohydrate side chains[212]. Their IEP is in the 5.5 −9.0range[213]. The enzymes can catalyze reactions involving the breakdown of R-OO-R’ and more commonly, catalyze reactions involving the reduction/oxidation of H2O2. The enzyme is often used as a conjugate or in tandem with other biosensing systems (e.g. immunoglobin labeling[214, 215] and oxidases[216]) because its reaction produces an observable electrochemical or even chemiluminescence [217] signal. Even though they areless common, stand-alone peroxide biosensors can play an important role and provide relevant information for the fabrication of biosensors that use conjugated systems.
There are many research papers that highlight the adsorption of peroxidase to nanocomposites containing AuNPs including nano-Au/poly2,6-pyridinediamine[218], toluidine blue-AuNP [219], nano-Au/thionine/poly (p-aminobenzene sulfonic acid)[220], AuNP (3-mercaptopropyl)-trimethoxysilane (MPS) sol-gel[221], nano-Au sol-gelderived carbon ceramic electrode[222], AuNP-thionine-AuNP-MWCNs-chitosan [223], Au colloid-cysteine-nafion-Pt [224], AuNP-chitosanhydrogel modified Au electrode[225], Au nano-seeds dotted TiO2 nanocomposite [226] and AuNP-ITO[227]. Among those it is worth highlighting the work byChen et al.[219], who used a LbL assembly of AuNPs and toluidine blue (TB) for the adsorption of horseradish peroxidase (HRP) to fabricate a peroxide biosensor. With the use of a cysteine aqueous solution and gold colloid solution, the Au electrode surface was deposited with a negatively charged precursor film. Then, 4 h of immersion time for each positively charged TB and negatively charged nano-Au solution was used to create the bilayers in the form of (TB/Au)n (n>6 to give maximum peak currents using CV). Then the electrode was submerged in a solution of HRP at pH = 6.0 for 8 h to promote the adsorption of HRP to form the final modified electrode.They determined that the linear range of response was between 150 nM to 8.6 mM and the limit of the detection was 70 nM. Their biosensor retained 85% of its original response after 30 days of storage and decreasing current signal by 8.0% after 100 consecutive scans (0.1 mM H2O2). Wang et al. [226] fabricated a peroxide biosensor by dotting and drying a mixture of Au nano-seeds, TiO2 dispersion, and HRP on a polished GCE followed by a Nafion coat for stability. It was concluded by CV measurements that the nanocomposite facilitates a fast electron-transfer reaction. After 100 consecutive scans, no obvious decrease in signal was observed and after two weeks of storage, an acceptable 90.1% of original response was retained.
Similar to Au, MWCNTs can also provide a favorable substrate for adsorption of HRP. Li et al.[223] used a MWCNT-chitosan composite with thionine and AuNP to fabricate a peroxide biosensor. A polished GCE was spotted with a AuNP-MWCNT-chitosan nanocomposite followed by electrochemical deposition of a thionine solution. The final modified electrode was produced by pipetting a mixture of HRP and AuNP on the substrate surface (Figure 5). They determined that the linear range of response was between 500 nM and 1.5 mM and the limit of the detection 37.5 nM. Overall, the biosensor showed acceptable stability by retaining 97% of its original response after 2 weeks of storage and decreasing current signal by 5.6% after 100 consecutive scans (H2O2 concentration not listed). Reproducibility was good showing an RSD less than 4% of independently prepared electrodes (n=10). Similarly, Zhang et al. [228] used a biosensor fabricated by the copolymerization of PtNP and neutral red on a polished GCE modified with MWCNTs, followed by immersion in a solution of HRP for 8 h. The biosensor presented byLi et al.[223] showed acceptable stability and reproducibility, but better limit of detection and linear range.
Figure 5.
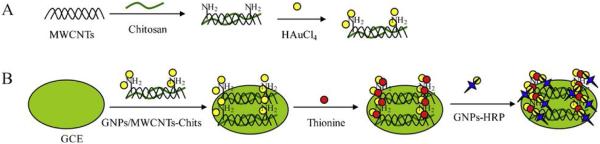
The fabricating procedures for GNPs/MWCNTs–Chits composite (A) and the biosensor (B).Reprinted from [223], with permission from Elsevier.
In addition to Au and MWCNTs, HRP can be adsorbed to ZnO nanorods [229], AgNP-cysteamine-Au electrode[230], and other nanocomposite systems [231-234]. Among those, Gu et al. [229] used an optimized method to hydrothermally grow ZnO nanostructures on gold wires. The final modified electrode was fabricated by LbL assembly of negatively charged poly(sodium 4-styrenesulfonate) (PSS) and positively charged HRP solution (5 h immersion time). Through CV, they concluded more layers (up to 5) resulted in higher HRP loading and sensitivity. The was the lowest when 3 layers were adsorbed to the substrate and while the limit of detection was comparable, stability and reproducibility tests were not shown. Desorption of the bio-recognition element is probably one of the most important disadvantages of the immobilization process carried out by physical interactions between the biomolecules and the substrate. In order to improve the stability of the adsorbed biologically active molecules, different methods based on adsorption processes were widely explored to advance the field of biosensors such as self-assembled monolayers (SAM), Langmuir-Blodgett (LB) films, and LbL films. Recently, Wang et al.[235] developed an amperometric biosensor to detect hydrogen peroxide in which HRP was adsorbed onto SWCNTs and then the resulted composite (HRP/SWCNTs) was immobilized onto a L-cysteine SAM. The biosensor was acquired following the steps shown in Figure 6. At the outset, SWCNTs were oxidized with a H2O2-HCl mixture and sonicated for6 h in order to obtain SWCNTsfunctionalized with carboxylic groups. After purification, the oxidized SWCNTs were dispersed in asolution of sodium cholate to avoid the aggregation. Next, HRP was allowed to adsorb on the SWCNTs and the sodium cholate replaced by the enzyme was removed by dialysis. Furthermore, a gold electrode was modified by a self-assembled L-cystine monolayer with the aim of attaching the obtained HRP-SWCNTs to the gold surface covalently. The electrode performance showed that the most significant aspects of the biosensor werethe high biocatalytic response (in the pH range of 7.0 – 8.0) and the low detection limit of the device.
Figure 6.
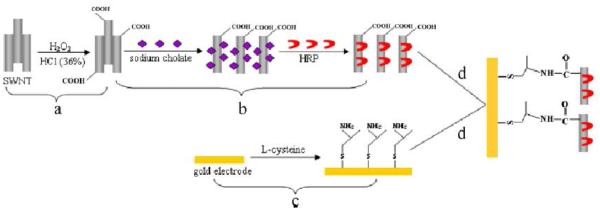
Schematic of preparation of enzyme electrodes. Reprinted from [235], with permission from Elsevier.
Another oxidoreductase enzyme linked to H2O2 iscatalase. This enzymecatalyzes the disproportionation of two molecules of H2O2 to molecular oxygen and water. Catalase is a hard enzyme with a MW of 240 kDa, an IEP of 5.4, and one of highest turnover numbers in nature[236]. Catalase has been adsorbed to a variety of substrates using solutions with pH values of 5.6 to 6.5 [237] and for a period of time that ranges from 30 min [238] to 14 h [72]. Catalase adsorbed onto Mn2O3 nanoparticles [239], nickel oxide nanoparticles [240] or CNT [241] has been used to facilitate the amperometric detection of H2O2. In the latter cases, authors discussed the possibility of even achievingdirect electrochemistry. Performing experiments at the IEP of the protein, Felhofer et al. [242] demonstrated that not only the adsorbed amount but also the initial adsorption rate were determinants of the catalytic activity of a CNT-based nanocomposite (Figure 7) and that such scaffolds can be saturated in less than 30 min, when the most favorable experimental conditions are selected. In line with other studies addressing the curvature of the nanomaterial, Zhang reported that the curvature of the CNT (single-wall, multiple wall, or oxidized) can also influence the activity of adsorbed catalase [243].
Figure 7.
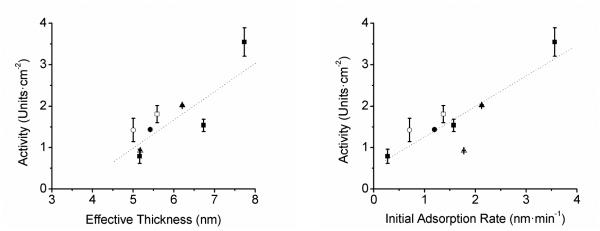
Activity as a function of effective thickness (left) and initial adsorption rate (right) for pH = 5.4 (■), pH = 4.4 (▲), pH = 6.4 (●), pH = 4.4 + 100 mmol·L−1 NaCl (△), pH = 5.4 + 100 mmol·L−1 NaCl (□), and pH 6.4 + 100 mmol·L−1 NaCl (○). Dashed lines are drawn to guide the eye. Reprinted from [242], with permission from American Chemical Society.
Lactate Oxidase
Another example of a biosensor developed based on the electrochemical detection of H2O2 produced by an enzyme involves lactate oxidase (LOx). This enzyme is able to oxidize L-lactate to pyruvate and H2O2 and shows maximum catalytic activity at pH = 6.5 and 37 °C.The enzyme has an IEPof approximately 4.6 [244] and a molecular weight of about 50 kDa (commonly found as a multimers containing 4-8 subunits). Although the most common form is obtained fromnatural sources (including Pediococcus sp.), other variants have also been engineered, characterized, and used for analytical purposes [245]. As an example, Mingli et al. [246] prepared a lactate biosensor by incorporating LOx in hydrothermally-processed porous hydrogen titanate (H2Ti3O7) nanotubes (TNTs). As observed by SEM, TEM, and X-ray diffraction (XRD), the TNTs formed a 3D porous network with high surface area for the support of LOx that provided electron transport channels to the electrode surface, promoting direct electron transfer between the FAD active site of the LOx and the gold surface. The biosensor also showed good reproducibility by successively detecting 6 mM lactic acid 6 times with a RSD of 2.47%.Platinum nanoparticles and MWCNT were also used as a composite film for the detection of lactate [247]. The RSD of the sensor was extremely low at 0.4% after five consecutive measurements. Other sensors based on LOx immobilized on ZnO nanorods [244], ZnO-CNT [248] or nitrogen-doped CNT [249] have also been presented.
Xanthine Oxidase
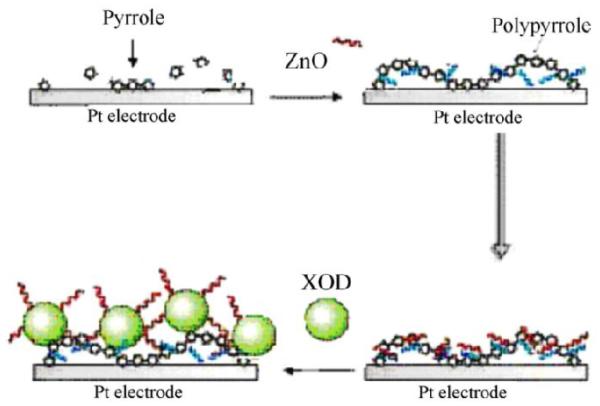
Another enzymeused in the fabrication of biosensors is xanthine oxidase (XOx). XOx catalyzes the hydroxylation of hypoxanthine to xanthine and H2O2 and then xanthine to uric acid (which can be electrochemically oxidized to allantoin) at an optimum pH of 7.5. XOx can also act on other groups such as 1-methylxanthine (a metabolite of caffeine). XOx is normally isolated as a mixture of two molybdenum-containing subunits of approximately 150kDaand has 2 flavin molecules (bound as FAD). Despite the fact that no recent papers have described the adsorption process, XOx can be adsorbed at pH in the range of 6to7 [146, 250] during a period of 2 h [251] – 8h [252]. XOx was adsorbed (along with PDDA) on an electrode containing AuNP – single – walled carbon nanohorns (5 – 8 nm) hybrids[253], resulting in outstanding sensitivity (141.1 mA·M−1·cm−2) and detection limits. Physical adsorption between XOx and ZnO nanoparticles in a polypyrrole composite film (66 mg·mL−1, 12h and 4 oC) was also used to fabricate a biosensor and later optimized for the detection of xanthine by testing different experimental conditions[254].Figure 8 displays the scheme for the reaction occurring at the biosensor surface.
Figure 8.
Schematic representation of chemical reaction at xanthine oxidase (XOD in figure)/ZnO-NPs-Ppy/Pt. Reprinted from [254], with permission from Elsevier.
Shan et al. used 18.5 nm laponite (hydrous sodium lithium magnesium silicate) nanoparticles to immobilize xanthine oxidase on a biosensor[255]. The laponite nanoparticles supplied a high surface area for protein loading, as well as high surface energy that favored the conformation of the protein and the electron transfer. The reproducibility was outstanding with a relative standard deviation of 1.2% after 12 different sensors were independently tested. Other xanthine sensors based on nano-CaCO3[256] and layered double hydroxides [257] have also been described.
Glutamate Oxidase
Due to the increased consumer awareness of levels of glutamate added to foods and its critical role in brain functions, several biosensors based on L-glutamate oxidase (GluOx) have been presented. The most common form (obtained from Streptomyces sp.) is a hexameric structure with a molecular weight of 140 kDa and IEP of 6.2 [258]. This variant provides high substrate specificity and high stability (~80 oC)[259] and though the enzyme is active between pH 4 and10, the optimum catalytic activity is obtained at a pH of7.5. These properties have been used to promote adsorption using phosphate buffer (pH=7.4). Evans et al. adsorbed GluOx, separately and along with GOx and LOx, to 15 nm silica nanoparticles entrapped in cellulose fibers for the fabrication of a microfluidic paper-based analytical device (μPAD). Their modified μPAD detected three analytes by way of colorimetric detection.
The nanoparticles were used to immobilize the protein to the paper and a 61% increase in color intensity was observed when the nanoparticles were incorporated into the device versus native paper [122]. A novel glutamate amperometric biosensor was made through the assembly of PtNP on modified ordered three-dimensional gold nanowire arrays[260]. Adsorbing the enzyme to a nanocomposite of oxygen-rich ceria and titania nanoparticles dispersed within a semi-permeable chitosan membrane allowed the detection in hypoxic environments with detection limits of 0.5 μM [261]. In a simpler fabrication process, Khan et al. [56] prepared a biosensor to detect glutamateusing SPE containing CNT modified with GluOx and a flow injection analysissystem. The system enabled obtaining limits of detection in the nM level, fast response time (≤5 s), and good operational and long-term stability. Even though adsorption was not considered, it is worth mentioning that the effect of several immobilization methods has been recently compared [262].
Tyrosinases
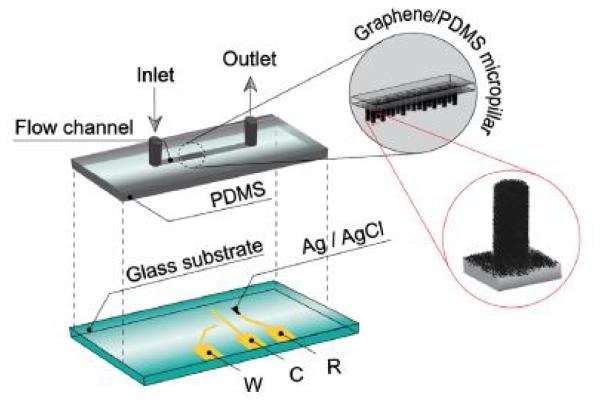
Also known as polyphenol oxidase, tyrosinasesarea copper-containing enzyme which is extensively distributed in microorganisms, animals, and plants[263]. Due to its high availability, the mushroom tyrosinaseis mostly used. With an IEP of 4.2 [264], Tyrosinasecatalyzes the oxidation ofphenols to the correspondingo-quinones. Subsequently, the quinones can be used to electrochemically follow the enzymatic reaction as a result of their electroactive behavior[265]. Baptista-Pires et al.[71] used oxidized graphene oxide (oGO) and reduced graphene oxide (rGO) to generate electroactive platforms for biosensing applications. Graphene oxide was deposited by drop casting onto graphite SPE. Then, the electrodes were used to adsorb the enzyme. The amperometric measurements were achieved applying a potential of −0.1 V. The results showed a remarkably higher response of SPE/rGO/Tyrosinase biosensor than the SPE/oGO/Tyrosinase biosensor. That behavior was attributed to the fact that the electrostatic interactions, as the driving force for enzyme binding to oGO, severely affected the activity of tyrosinase. Also, the higher conductivity of the rGO promoted high charge transfer. Additionally, Liu et al. [266] also used graphene oxide (GO) to adsorb tyrosinaseonto 3D graphene micropillar electrodes. The micropillars were patterned in a polydimethylsiloxane (PDMS) substrate and modified with 3-aminopropyltriethoxysilane (APTES). Then, GO flakes were allowed to self-assemble onto the micropillars. Because the oxidative process during the GO synthesis produced negatively-charged functional groups (carboxylic acid, hydroxyl, and epoxy), graphene was adsorbed onto the substrate by strong electrostatic interactions between the negatively charged GO flakes and the positively-charged micropillars modified with APTES. The sensing device was designed with a microchannel for the sample injection, a 3D graphene micropillar, two Au electrodes, and one Ag/AgCl electrode as shown in Figure 9.
Figure 9.
Illustration of a 3D graphene micropillar incorporated electrochemical sensor device. The microfluidic channel was patternedin the PDM Slayer for sample loading, and the graphene micropillar was fabricatedin the middle of the micro channel. The working (W), counter(C), and reference(R)electrodes were deposited on a glass substrate. Reprinted from [266], with permission from Elsevier.
The enzymatic reaction was measured following the increase in o-quinone produced during the enzymatic reaction by amperometry at −0.1 V. Also, Tyrosinase was immobilized by adsorbing onto ZnO nanorods as reported by Chen et al.[267]. Because the ZnO nanorods have a high IEP (9.5), the immobilization of the enzyme wascarried out taking advantage of electrostatic interactions. The biosensors were achieved by casting the nanorod-enzyme suspension on the GCE. Then, a Nafion solution was spotted on the biosensors and the film was allowed dry overnight. The resulting electrode was used successfully to detect phenol and catechol. The of tyrosinase adsorbed on the ZnO nanorods was found to be 0.24 mM for phenol and 1.75 mM for catechol. The value observed in the case of phenol was slightly lower than that obtained by encapsulation into TiO2 sol-gel films (0.29 mM[268]) and in copolymer grafting silica sol-gel composites (0.37 mM[269]). Although the activity of the enzyme can be determined by Vmax/KM, Chen et al. stated that the lower value obtained for tyrosinaseled to improved bioactivity.[267]. Additionally, a phenol biosensor has been described by Pavinatto et al.[270] incorporating tyrosinasein a LB film of arachidic acid and lutetium bisphthalocyanine (LuPc2). In the same way, Apetrei et al.[271] used the biosensor based on the LB films developed by Pavinatto et al. to compare the tyrosinaseactivity on the detection of one monophenol (vanillic acid), two diphenols (catechol and caffeic acid), and two triphenols (gallic acid and pyrogallol). The experimental results demonstrated a degree of difference in the sensitivity toward the phenols and showed a tendency to decrease in the order of diphenols>triphenols>monophenols.
Laccase
Laccase, also named benzenediol: oxygen oxidoreductase, is a glycoprotein and belongs to the group of multi-nuclear copper-containing proteins[272]. This enzyme has an IEP of 4.4, a molecular weight of 62 kDa[273], and can catalyze the oxidation of a wide range of substrates, which present similar chemical characteristics to p-diphenol[274]. The detection of the quinone generated by the enzyme reaction can be monitored using electrochemical techniques by the detection of the product on the electrode surface or through a mediator. Portaccio et al.[275] modified the surface of a SPE by using a nanocomposite formed by carbon black and thionine as the electrochemical mediators in order to improve the analytical performance of the biosensor. Thus, the enzymatic reaction produced an increase in the amount of oxidized thionine and consequently the increase in the cathodic current. Laccase was immobilized by adsorption onto the electrode and used to quantify bisphenol A. The obtained biosensor showed better results when compared with sensors based on carbon paste [276] or GCE [277]. On the other hand, gold electrodes were also used as an immobilization platform for laccase as reported by Casero et al.[278] In this case, hydroquinone (HQ) was oxidized enzymatically to quinone and then reduced again to HQ on the electrode surface. The cathodic peak current was recorded by cyclic voltammetry and used to follow the evolution of the enzymatic reaction.
Other Oxidases
Because highly conductive materials like metals or graphite produce an undesirable degradation of the proteinadsorbed on those substrates, metal oxide semiconductors (TiO2, WO3, SnO2, ZnO, and tin-doped ITO) have ignited significant interest in the immobilization of biomolecules such as glucose dehydrogenase (GDH) and sulfite oxidase [279] or cytochrome c [280-282] for not only their conductivity but also their transparency (enhancing their optical properties). It is important to point out that while ITO is the most widely used substrate, the relatively low abundance of indium has sparked the development of promising materials with lower indium content[283, 284]. In addition, antimony-doped tin oxide (ATO) showed an improved conductivity conferred by the Sb dopant and maintained the high transparency observed in ITO. Consequently, ATO has been employed for the immobilization of proteins such as cytochrome c and azurin[285]. Recently, Frasca et al.[286] also used thin films of ATO with two different porosities (ATO-F127 and pl-ATO) as platforms to adsorb Human sulfite oxidase (hSO) by immersion for 2 h. The response of the obtained biosensor was carried out using a flow injection/amperometric detection system while the electrode was polarized at 0 V.
Uricase is another important oxidase relevant to the degradation of purines because one of the metabolites of the pathway (uric acid) remains an important marker for disorders such as gout, hyperuricemia and the Lesch–Nyhan syndrome. For these reasons, a number of biosensors have been presented using uricase adsorbed to nanomaterials, including ZnO nanoparticles [287], LB films of fatty acids [288], and carbon felt [289]. In agreement with previous strategies, the adsorption performed at pH values in the 6 – 7 range, from a solution containing 0.02 μg·mL−1 – 1 mg·mL−1, yielded the best results. Other alternatives for the immobilization of the enzyme have also been presented [290].
Using ellipsometry, Mora et al.[291] investigated the interaction of D-amino acid oxidase (DAAO) with SWCNT. In this case, dynamic adsorption experiments, performed as a function of the protein concentration, pH, and ionic strength, allowed them to conclude that DAAO can adopt multiple orientations on the surface and that the maximum catalytic activity can be obtained by controlling the adsorption conditions.
4.2. ANTIBODIES
A very important group of biosensors has been developed by the adsorption of a variety of antibodies[111]. Their properties and specific considerations to promote not only the immobilization but also the correct orientation have been recently studied [62, 292] and therefore will only be briefly discussed. Although different immobilization routes have been presented, each one with its own advantages and limitations, physical adsorption of antibodies avoids the use reagents and decreases the chances of protein denaturation. Probably the most significant advantage is the exquisite selectivity of these sensors. Besides stability, one particular limitation for the rational development of immunosensors is the cost of these proteins, which in most cases imposes a financial burden for the corresponding adsorption studies.
Inorganic nanoparticles are of high interest to researchers because many of these materials are presentin vertebrates. Specifically, hydroxyapatite (HA) is biodegradable, biocompatible, inexpensive, as well as soluble and less toxic than silica, quantum dots, CNT, and magnetic particles.In a recent report, the potential of HA as a carrier for human immunoglobulin G (IgG) has been investigated. IgG was adsorbed to 30 nm HA nanocrystals at pH = 7.4[293]. The adsorption capacity of the HA nanocrystals reached a plateau at an adsorbed amount of 2.3 mg·m−2 IgG. When measuring the adsorbed amount they found two populations of IgG bound with different strengths, with most of IgG having a strong link with HA.A reusable sensor for the detection of IgG was fabricated after adsorption times, ion strength, and pH were optimized [294]. A 0.2 mg·mL−1 solution of anti-hIgG at pH = 7.7 was adsorbed to the magnetic hydroxyapatite (HAP)/γ-Fe2O3/Au nanocomposite for 2.5 h and studied with magnetic separation-assisted quartz crystal microbalance (QCM). By manipulating the magnetic field, the sensor can be regenerated through direct adsorption of the antibody to the transducer surface; therefore avoiding time-consuming chemical linkers and improving the detection of the analyte.
Nanomaterials have also been utilized as electrochemical signal probes on immunosensors made from disposable SPCE for the detection of an antigen associated with human lung cancer. A polyethylene glycol layer was adsorbed on the surface of the electrode, followed by polyclonal secondary anti-ENO1-antibody-tagged AuNPs to improve the sensitivity of the assay. Square wave voltammetry revealed the novel electrochemical immunosensor could detect as low as 2.38 pg·mL−1 of ENO1[295]. Another electrochemical sensor was developed through the adsorption of anti-carcinoembryonic antigen (CEA) antibodies on the surface of a staphylococcal protein A-AuNP modified gold electrode[296]. A rapid increase in current was seen (Figure 10) when the electrode was measured with 15 nm AuNPs as opposed to 40 nm AuNPs, while a current plateau was first noticed at 0.6 mg·mL−1 when adsorbing different concentrations from 0.1 mg·mL−1 to 1.0 mg·mL−1.Saturation conditions occurred at 90 min. It is also important to note that natural polymers such as chitin, chitosan, and silk fibroin have also been used by means of immobilization matricesin order to improve the stabilityof the biorecognition element. As anexample of this strategy, a LbL immunosensor prepared by immobilizing the peptide NS5A-1 derived from hepatitis C virus (HCV) NS5A protein and silk fibroin (SF)was reported by Moraes et al.[297].
Figure 10.
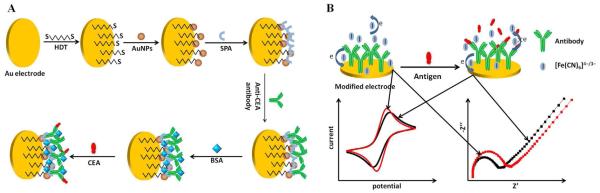
(A) The schematic description of the preparation procedures of immunosensor and (B) the detection principle of antibody-based electrochemical immunosensor. Reprinted from [296], with permission from Elsevier.
4.3. OTHER PROTEINS
Many other proteins have also been combined with nanomaterials to produce biosensors. For example, Costa Rama et al. [298] recently fabricated a biosensor for detection of amyloid-beta 1-42 (Aβ1-42, a biomarker for Alzheimer’s disease) using a SPCE with the incorporation of AuNP. TheseAuNP were generated on the surface of the SPCE by means of electrodeposition and the final modified electrode was created according to the schematic shown in Figure 11. As previously published, gold nanostructured surfaces show better sensitivity than non-nanostructured surfaces. The conditions for the experiment were optimized (non-specific binding at the electrode was avoided by adding BSA to the solution of the antibody and Aβ1-42 concentration of 300 ng·mL−1 was chosen because of reproducibility). The reproducibility was measured by six independently prepared electrodes achieving a maximum signal of 61 ± 3 μA. While these types of biosensors show the versatility and variety in the industry, a shortcoming of thesespecific electrodes is that they are only good for single use measurements. Even so and according to the authors, there has only been one other published biosensor for Aβ peptides using similar methods and its limit of detectionis about 4.5 μg·mL−1[299] compared to their LOD of 0.1 ng·mL−1.
Figure 11.
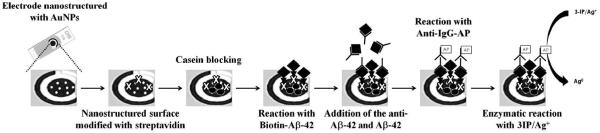
Schematic representation of the immunosensing strategy for the detection of Aβ1-42 Reprinted from[298], with permission from Elsevier.
The shape-selective properties of zeolites have been utilized as strategy to preferentially adsorb certain molecules. That property is related to the crystalline structure of zeolites that may produce linked cages, cavities, and channels which can control the entry of molecules according to their size[300]. Recently, potentiometric biosensors prepared adsorbing urease on different zeolite (silicalite, nano beta zeolite, and zeolite L) monolayers were reported by Shelyakina et al.[301]. The zeolite substrates were obtained by immobilizing the zeolite particles onto the surface of pH-sensitive field-effect transistors (ISFET). The functioning of the biosensor is based on the reaction of urea cleavage to NH +ions and consequently the consumption of protons. Thus, the enzymatic reaction was followed by changing the pH inside the selective membrane which is recorded by the ISFET. It was observed that the additionof zeolites resulted in anincreasedlinear range (mainly with silicalite and nano-zeolite L), reduced limit of detection forurea determination, improved reproducibility, and decreased time of analysis.
Biosensors based on landscape phages adsorbed on the surface of a quartz crystal microbalance have been used for the detection of β-galactosidase from Escherichia coli[302]. These sensorsprovided a detection limit in the nM range and a response time of a ~100 s over the range of 0.003–210 nM.
Even though it does not provide specific recognition properties, albumin is a very important protein for analytical devices. Albumin (and most commonly BSA) is a heart-shaped soft protein with a molecular weight of 66.5 kDa and an IEP of 4.8 [108]. Because BSA typically exhibits significant surface-induced spreading upon adsorption, a stable layer of the protein can be formed under a wide number of conditions in less than 1 h [135, 205, 303]. For these reasons (and its low cost), it has been extensively used as a model protein to study adsorption [68, 80, 110, 134, 137, 139, 304, 305], to aid in the suspension of CNT [306], and to block the remaining sites of surfaces after the immobilization of a biorecognition element [250, 260, 294, 307]. BSA has also been adsorbed to the surface of PMMA nanoparticles to enhance the subsequent attachment of GOx by electrostatic interactions, retaining most of the activity of the free enzyme[308, 309]. It is also important to mention that Benavidez et al. [310] recently reported the possibility to induce the accumulation of multiple layers of BSA by the application of an external potential, a phenomenon that is compatible with the polarization of the protein described by other authors [311].
5-EMERGING TRENDS
The biorecognition element for biosensors is not only limited to enzymes and antibodies but can also span a vast array of compounds including DNA, cells, microorganisms, organelles, and plant or animal tissues[4]. Point-of-care monitoring, short analyses times, easy sample preparation, and cost are currently of the upmost importance in diagnostic testing. Infectious diseases and agents, food intoxication, and contaminated water are still major worldwide concerns[312]. To cater to the constant need for rapid onsite monitoring in healthcare, environment, agriculture, food control, process control, and microbiology, the research industry is heading towards designing biosensors and fabricating nanomaterials tailored to the needs of the active biorecognition element[313-315]. Proteins can be considered the “nanoscale building blocks” for nanomaterials. In addition, affinity based biosensors are emerging as in-vivo monitoring of the human physiology has become more vital. More and more nanotechnology-based immunosensors, DNA sensors, aptasensors, and phage-basedbiosensors are being developed in tandem with novel electrochemical, optical, and mass sensitive detection techniques[316]. It is also important to highlight that the relatively recent development of single-domain antibodies (nanobodies) can provide unique alternatives to traditional immunosensors in terms of stability and availability [317]. The implementation of nanomaterials during the development of biosensors provides a path to improve selectivity and sensitivity, portability, and cost leading to low detection limits while keeping a wide linear range of detection.
6-CONCLUSIONS
The adsorption of proteins to solid surfaces are mostly regulated by a combination of hydrophobic and electrostatic interactions. Other factors such as solution/solvent properties, ion co-adsorption, and structural rearrangements can also play important roles in this process. The latter is driven by favorable entropic and unfavorable enthalpic changes to reduce the overall free energy of the system and may be conducive to significant losses in the structural properties of the biorecognition element. A detailed description of the adsorption process is required to achieve the full potential of the protein, however, adequate coverage of the substrate can be often obtained by immersing the surface in a 1 mg·mL−1 solution of the protein at the IEPfor approximately 1h. While researchers should be encouraged to continue looking beyond GOx, this protein remains dominantin the biosensor field. From the surface standpoint, nanoparticles and other nanomaterials have beenthe driving force behind the development of sophisticated biosensors in recent years. The use of metal nanoparticles, such as gold and platinum, is widely observed in literature because the particles can improve the rate of electron transfer at the biointer face. By altering conditions such as pH, nanomaterial size and shape, and temperature, analytical figures of merit have been improved drastically. The formation of nanofilms for the detection of various analytes is also a rapidly growing area of biosensors. The use of nanotubes and their tunable features are just a few ways researchers are improving the analytical performance of biosensors. The importance of nanomaterials is growing as the demand for quick, selective, inexpensive, stable, and reproducible analytical devices continues to surge. The fabrication of the above mentioned biosensors all show very unique methodologies to improve bioactivity of the enzyme/material nanocomposites, especially if more than one type of nanomaterial is used. The compatibility of nanomaterials with enzymes has become the focus of many research endeavors and is now recognized as a critical consideration to further the applicability of biosensors. The technology highlighted in this review is just a glimpse of the many sensors being fabricated and used in the medical, environmental, food, and biological industries. In summary, it is expected that the use of nanomaterials as sorbent surfaces for the development of biosensors will continue to grow and provide new avenues to bolster the field.
Supplementary Material
7. ACKNOWLEDGEMENTS
The authors gratefully acknowledge the financial support provided by the University of Texas at San Antonio and the National Institutes of Health through the National Institute of General Medical Sciences (2SC3GM081085), the Research Centers at Minority Institutions (G12MD007591), and the Partnership for Research and Education in Materials at the University of Texas at San Antonio (NSF-DMR-0934218).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8. REFERENCES
- [1].Nakanishi K, Sakiyama T, Imamura K. On the adsorption of proteins on solid surfaces, a common but very complicated phenomenon. J. Biosci. Bioeng. 2001;91:233–244. doi: 10.1263/jbb.91.233. [DOI] [PubMed] [Google Scholar]
- [2].Roach P, Farrar D, Perry CC. Interpretation of Protein Adsorption: Surface-Induced Conformational Changes. J. Am. Chem. Soc. 2005;127:8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- [3].Barnthip N, Parhi P, Golas A, Vogler EA. Volumetric interpretation of protein adsorption: Kinetics of protein-adsorption competition from binary solution. Biomaterials. 2009;30:6495–6513. doi: 10.1016/j.biomaterials.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Manz A, Pamme N, Iossifidis D. Bioanalytical chemistry. Imperial College Press; River Edge, NJ: 2004. [Google Scholar]
- [5].Kirsch J, Siltanen C, Zhou Q, Revzin A, Simonian A. Biosensor technology: recent advances in threat agent detection and medicine. Chem. Soc. Rev. 2013;42:8733–8768. doi: 10.1039/c3cs60141b. [DOI] [PubMed] [Google Scholar]
- [6].Qureshi A, Gurbuz Y, Niazi JH. Biosensors for cardiac biomarkers detection: A review. Sens. Actuators. 2012:62–76. B. 171–172. [Google Scholar]
- [7].Ward WK. How to design a biosensor. J. Diabetes Sci. Technol. 2007;1:201–204. doi: 10.1177/193229680700100210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Albareda-Sirvent M, Merkoçi A, Alegret S. Configurations used in the design of screen-printed enzymatic biosensors. A review, Sens. Actuators. 2000;69:153–163. B. [Google Scholar]
- [9].Turner APF. Biosensors: sense and sensibility. Chem. Soc. Rev. 2013;42:3184–3196. doi: 10.1039/c3cs35528d. [DOI] [PubMed] [Google Scholar]
- [10].Monošík R, Stred’anský M, Šturdík E. Biosensors - classification, characterization and new trends. Acta Chim Slov. 2012:109–120. [Google Scholar]
- [11].Chen A, Chatterjee S. Nanomaterials based electrochemical sensors for biomedical applications. Chem. Soc. Rev. 2013;42:5425–5438. doi: 10.1039/c3cs35518g. [DOI] [PubMed] [Google Scholar]
- [12].Lord MS, Foss M, Besenbacher F. Influence of nanoscale surface topography on protein adsorption and cellular response. Nano Today. 2010;5:66–78. [Google Scholar]
- [13].Sutherland DS, Broberg M, Nygren H, Kasemo B. Influence of Nanoscale Surface Topography and Chemistry on the Functional Behaviour of an Adsorbed Model Macromolecule. Macromol. Biosci. 2001;1:270–273. [Google Scholar]
- [14].Elter P, Lange R, Beck U. Electrostatic and Dispersion Interactions during Protein Adsorption on Topographic Nanostructures. Langmuir. 2011;27:8767–8775. doi: 10.1021/la201358c. [DOI] [PubMed] [Google Scholar]
- [15].Karajanagi SS, Vertegel AA, Kane RS, Dordick JS. Structure and function of enzymes adsorbed onto single-walled carbon nanotubes. Langmuir. 2004;20:11594–11599. doi: 10.1021/la047994h. [DOI] [PubMed] [Google Scholar]
- [16].Mathe C, Devineau S, Aude JC, Lagniel G, Chedin S, Legros V, Mathon MH, Renault JP, Pin S, Boulard Y, Labarre J. Structural determinants for protein adsorption/non-adsorption to silica surface. PloS one. 2013;8:e81346. doi: 10.1371/journal.pone.0081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jia F, Narasimhan B, Mallapragada S. Materials-based strategies for multi-enzyme immobilization and co-localization: A review. Biotechnol. Bioeng. 2014;111:209–222. doi: 10.1002/bit.25136. [DOI] [PubMed] [Google Scholar]
- [18].Jesionowski T, Zdarta J, Krajewska B. Enzyme immobilization by adsorption: a review. Adsorption. 2014;20:801–821. [Google Scholar]
- [19].Qu Z, Chen K, Gu H, Xu H. Covalent Immobilization of Proteins on 3D Poly(acrylic acid) Brushes: Mechanism Study and a More Effective and Controllable Process. Bioconjugate Chem. 2013;25:370–378. doi: 10.1021/bc400530s. [DOI] [PubMed] [Google Scholar]
- [20].Cabral MF, Barrios JD, Kataoka EM, Machado SAS, Carrilho E, Garcia CD, Ayon AA. Computational, electrochemical, and spectroscopic, studies of acetycholinesterase covalently attached to carbon nanotubes. Colloids Surf. 2013;103:624–629. doi: 10.1016/j.colsurfb.2012.08.039. B. [DOI] [PubMed] [Google Scholar]
- [21].Müller V, Rathousky J, Fattakhova-Rohlfing D. Covalent immobilization of redox protein within the mesopores of transparent conducting electrodes. Electrochim. Acta. 2014;116:1–8. [Google Scholar]
- [22].Williams R, Blanch H. Covalent immobilization of protein monolayers for biosensor applications. Biosens. Bioelectron. 1994;9:159–167. doi: 10.1016/0956-5663(94)80108-8. [DOI] [PubMed] [Google Scholar]
- [23].Zhang S, Wang N, Yu H, Niu Y, Sun C. Covalent attachment of glucose oxidase to an Au electrode modified with gold nanoparticles for use as glucose biosensor. Bioelectrochemistry. 2005;67:15–22. doi: 10.1016/j.bioelechem.2004.12.002. [DOI] [PubMed] [Google Scholar]
- [24].Wong SS. Chemistry of protein conjugation and cross-linking. CRC press; 1991. [Google Scholar]
- [25].Sheldon R, Schoevaart R, Van Langen L. Cross-linked enzyme aggregates (CLEAs): A novel and versatile method for enzyme immobilization (a review) Biocatal. Biotransform. 2005;23:141–147. [Google Scholar]
- [26].Cao L, van Rantwijk F, Sheldon RA. Cross-linked enzyme aggregates: a simple and effective method for the immobilization of penicillin acylase. Org. Lett. 2000;2:1361–1364. doi: 10.1021/ol005593x. [DOI] [PubMed] [Google Scholar]
- [27].Avnir D, Braun S, Lev O, Ottolenghi M. Enzymes and other proteins entrapped in sol-gel materials. Chem. Mater. 1994;6:1605–1614. [Google Scholar]
- [28].Gupta R, Chaudhury N. Entrapment of biomolecules in sol–gel matrix for applications in biosensors: Problems and future prospects. Biosens. Bioelectron. 2007;22:2387–2399. doi: 10.1016/j.bios.2006.12.025. [DOI] [PubMed] [Google Scholar]
- [29].Hasanzadeh M, Shadjou N, Eskandani M, Soleymani J, Jafari F, de la Guardia M. Dendrimer-encapsulated and cored metal nanoparticles for electrochemical nanobiosensing. TrAC, Trends Anal. Chem. 2014;53:137–149. [Google Scholar]
- [30].Graça JS, de Oliveira RF, de Moraes ML, Ferreira M. Amperometric glucose biosensor based on layer-by-layer films of microperoxidase-11 and liposome-encapsulated glucose oxidase. Bioelectrochemistry. 2014;96:37–42. doi: 10.1016/j.bioelechem.2014.01.001. [DOI] [PubMed] [Google Scholar]
- [31].Andrade J, Hlady V. Biopolymers/Non-Exclusion HPLC. Springer; 1986. Protein adsorption and materials biocompatibility: a tutorial review and suggested hypotheses; pp. 1–63. [Google Scholar]
- [32].Norde W. Adsorption of proteins from solution at the solid-liquid interface. Adv. Colloid Interface Sci. 1986;25:267–340. doi: 10.1016/0001-8686(86)80012-4. [DOI] [PubMed] [Google Scholar]
- [33].Haynes CA, Norde W. Globular proteins at solid/liquid interfaces. Colloids Surf. 1994;2:517–566. B. [Google Scholar]
- [34].Baszkin A, Norde W. Physical chemistry of biological interfaces. CRC Press; 1999. [Google Scholar]
- [35].Siqueira JR, Jr, Caseli L, Crespilho FN, Zucolotto V, Oliveira ON., Jr Immobilization of biomolecules on nanostructured films for biosensing. Biosens. Bioelectron. 2010;25:1254–1263. doi: 10.1016/j.bios.2009.09.043. [DOI] [PubMed] [Google Scholar]
- [36].Rabe M, Verdes D, Seeger S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011;162:87–106. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- [37].Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein-Nanoparticle Interactions: Opportunities and Challenges. Chem. Rev. 2011;111:5610–5637. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- [38].Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3:40–47. [Google Scholar]
- [39].Lyklema J. Proteins at solid—liquid interfaces A colloid-chemical review. Colloids Surf. 1984;10:33–42. [Google Scholar]
- [40].Johnson BJ, Russ Algar W, Malanoski AP, Ancona MG, Medintz IL. Understanding enzymatic acceleration at nanoparticle interfaces: Approaches and challenges. Nano Today. 2014;9:102–131. [Google Scholar]
- [41].Jesionowski T, Zdarta J, Krajewska B. Enzyme immobilization by adsorption: a review. Adsorption. 2014:1–21. [Google Scholar]
- [42].Lu Y, Rajaraman S, Ward WK, Vigersky RA, Reifman J. Predicting human subcutaneous glucose concentration in real time: a universal data-driven approach. Conference proceedings : … Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2011. pp. 7945–7948. IEEE Engineering in Medicine and Biology Society. Conference. [DOI] [PubMed] [Google Scholar]
- [43].Bhushan B, Schricker SR. A review of block copolymer-based biomaterials that control protein and cell interactions. J. Biomed. Mater. Res. Part A. 2014;102:2467–2480. doi: 10.1002/jbm.a.34887. [DOI] [PubMed] [Google Scholar]
- [44].Scida K, Stege PW, Haby G, Messina GA, Garcia CD. Recent applications of carbon-based nanomaterials in analytical chemistry: Critical review. Anal. Chim. Acta. 2011;691:6–17. doi: 10.1016/j.aca.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lazzara TD, Mey I, Steinem C, Janshoff A. Benefits and Limitations of Porous Substrates as Biosensors for Protein Adsorption. Anal. Chem. 2011;83:5624–5630. doi: 10.1021/ac200725y. [DOI] [PubMed] [Google Scholar]
- [46].Singh R, Mukherjee MD, Sumana G, Gupta RK, Sood S, Malhotra BD. Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sensors and Actuators B: Chemical. 2014;197:385–404. [Google Scholar]
- [47].Sin ML, Mach KE, Wong PK, Liao JC. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert review of molecular diagnostics. 2014;14:225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pedrero M, Campuzano S, Pingarrón JM. Electrochemical Biosensors for the Determination of Cardiovascular Markers: a Review. Electroanal. 2014;26:1132–1153. [Google Scholar]
- [49].Pundir CS, Devi R. Biosensing methods for xanthine determination: A review. Enzyme and microbial technology. 2014;57:55–62. doi: 10.1016/j.enzmictec.2013.12.006. [DOI] [PubMed] [Google Scholar]
- [50].Schulz GE, Schirmer RH. Principles of protein structure. Springer-Verlag KG.; 1979. [Google Scholar]
- [51].Norde W. My voyage of discovery to proteins in flatland …and beyond. Colloids Surf. 2008;61:1–9. doi: 10.1016/j.colsurfb.2007.09.029. B. [DOI] [PubMed] [Google Scholar]
- [52].Raffaini G, Ganazzoli F. Protein Adsorption on a Hydrophobic Surface: A Molecular Dynamics Study of Lysozyme on Graphite. Langmuir. 2009;26:5679–5689. doi: 10.1021/la903769c. [DOI] [PubMed] [Google Scholar]
- [53].Benavidez TE, Torrente D, Marucho M, Garcia CD. Adsorption and Catalytic Activity of Glucose Oxidase Accumulated on OTCE upon the Application of External Potential. J. Colloid Interface Sci. 2014 doi: 10.1016/j.jcis.2014.08.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Stein M, Gabdoulline RR, Wade RC. Calculating enzyme kinetic parameters from protein structures. Biochem. Soc. Trans. 2008;36:51–54. doi: 10.1042/BST0360051. [DOI] [PubMed] [Google Scholar]
- [55].Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Khan R, Gorski W, Garcia CD. Nanomolar Detection of Glutamate at a Biosensor Based on Screen-Printed Electrodes Modified with Carbon Nanotubes. Electroanal. 2011;23:2357–2363. doi: 10.1002/elan.201100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Vasina EN, Paszek E, Nicolau JDV, Nicolau DV. The BAD project: data mining, database and prediction of protein adsorption on surfaces. Lab Chip. 2009;9:891–900. doi: 10.1039/b813475h. [DOI] [PubMed] [Google Scholar]
- [58].Xia XR, Monteiro-Riviere NA, Mathur S, Song X, Xiao L, Oldenberg SJ, Fadeel B, Riviere JE. Mapping the Surface Adsorption Forces of Nanomaterials in Biological Systems. ACS nano. 2011;5:9074–9081. doi: 10.1021/nn203303c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Porollo A, Meller J. Versatile annotation and publication quality visualization of protein complexes using POLYVIEW-3D. BMC Bioinf. 2007;8:316. doi: 10.1186/1471-2105-8-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Puleo DA, Bizios R. Biological interactions on materials surfaces : understanding and controlling protein, cell, and tissue responses. Springer, Dordrecht; London: 2009. [Google Scholar]
- [61].Park M, Park SS, Selvaraj M, Zhao D, Ha C-S. Hydrophobic mesoporous materials for immobilization of enzymes. Microporous Mesoporous Mater. 2009;124:76–83. [Google Scholar]
- [62].Wiseman ME, Frank CW. Antibody Adsorption and Orientation on Hydrophobic Surfaces. Langmuir. 2011;28:1765–1774. doi: 10.1021/la203095p. [DOI] [PubMed] [Google Scholar]
- [63].Puddu V, Perry CC. Peptide Adsorption on Silica Nanoparticles: Evidence of Hydrophobic Interactions. ACS nano. 2012;6:6356–6363. doi: 10.1021/nn301866q. [DOI] [PubMed] [Google Scholar]
- [64].Zuo G, Zhou X, Huang Q, Fang HP, Zhou RH. Adsorption of Villin Headpiece onto Graphene, Carbon Nanotube, and C60: Effect of Contacting Surface Curvatures on Binding Affinity. J. Phys. Chem. C. 2011;115:23323–23328. [Google Scholar]
- [65].Ghaemmaghami S, Oas TG. Quantitative protein stability measurement in vivo. Nat. Struct. Mol. Biol. 2001;8:879–882. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- [66].Robertson AD, Murphy KP. Protein Structure and the Energetics of Protein Stability. Chem. Rev. 1997;97:1251–1268. doi: 10.1021/cr960383c. [DOI] [PubMed] [Google Scholar]
- [67].Geer MA, Fitzgerald MC. Energetics-Based Methods for Protein Folding and Stability Measurements. Annu. Rev. Anal. Chem. 2014;7:209–228. doi: 10.1146/annurev-anchem-071213-020024. [DOI] [PubMed] [Google Scholar]
- [68].Efimova YM, Haemers S, Wierczinski B, Norde W, Well A.A.v. Stability of globular proteins in H2O and D2O. Biopolymers. 2007;85:264–273. doi: 10.1002/bip.20645. [DOI] [PubMed] [Google Scholar]
- [69].Norde W, Giacomelli CE. Conformational changes in proteins at interfaces: From solution to the interface, and back. Macromolecular Symposia. 1999;145:125–136. [Google Scholar]
- [70].Norde W, Lyklema J. Interfacial behaviour of proteins, with special reference to immunoglobulins. A physicochemical study. Adv. Colloid Interface Sci. 2012;179–182:5–13. doi: 10.1016/j.cis.2012.06.011. [DOI] [PubMed] [Google Scholar]
- [71].Baptista-Pires L, Pérez-López B, Mayorga-Martinez CC, Morales-Narváez E, Domingo N, Esplandiu MJ, Alzina F, Torres CMS, Merkoçi A. Electrocatalytic tune of biosensing response through electrostatic or hydrophobic enzyme – graphene oxide interactions. Biosens. Bioelectron. 2014;61:655–662. doi: 10.1016/j.bios.2014.05.028. [DOI] [PubMed] [Google Scholar]
- [72].Pamuła E, Rouxhet PG. Influence of surface properties of carbon fibres on the adsorption of catalase. Carbon. 2005;43:1432–1438. [Google Scholar]
- [73].Roach P, Farrar D, Perry CC. Interpretation of protein adsorption: Surface-induced conformational changes. J. Am. Chem. Soc. 2005;127:8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- [74].Jungbauer A, Machold C, Hahn R. Hydrophobic interaction chromatography of proteins: III. Unfolding of proteins upon adsorption. J. Chromatogr. A. 2005;1079:221–228. doi: 10.1016/j.chroma.2005.04.002. [DOI] [PubMed] [Google Scholar]
- [75].Huang B, Wu H, Kim S, Zare RN. Coating of poly(dimethylsiloxane) with n-dodecyl-[small beta]-d-maltoside to minimize nonspecific protein adsorption. Lab Chip. 2005;5:1005–1007. doi: 10.1039/b509251e. [DOI] [PubMed] [Google Scholar]
- [76].Hartvig RA, van de Weert M, Østergaard J, Jorgensen L, Jensen H. Protein Adsorption at Charged Surfaces: The Role of Electrostatic Interactions and Interfacial Charge Regulation. Langmuir. 2011;27:2634–2643. doi: 10.1021/la104720n. [DOI] [PubMed] [Google Scholar]
- [77].Lundqvist M, Sethson I, Jonsson BH. Protein adsorption onto silica nanoparticles: Conformational changes depend on the particles' curvature and the protein stability. Langmuir. 2004;20:10639–10647. doi: 10.1021/la0484725. [DOI] [PubMed] [Google Scholar]
- [78].Hao D-X, Huang Y-D, Wang K, Wei Y-P, Zhou W-Q, Li J, Ma G-H, Su Z-G. Multiscale evaluation of pore curvature effects on protein structure in nanopores. J. Mater. Chem. B. 2014;2:1770–1778. doi: 10.1039/c3tb21714k. [DOI] [PubMed] [Google Scholar]
- [79].Cai K, Bossert J, Jandt KD. Does the nanometre scale topography of titanium influence protein adsorption and cell proliferation? Colloids Surf. 2006;49:136–144. doi: 10.1016/j.colsurfb.2006.02.016. B. [DOI] [PubMed] [Google Scholar]
- [80].Roach P, Farrar D, Perry CC. Surface tailoring for controlled protein adsorption: Effect of topography at the nanometer scale and chemistry. J. Am. Chem. Soc. 2006;128:3939–3945. doi: 10.1021/ja056278e. [DOI] [PubMed] [Google Scholar]
- [81].Song W, Chen H. Protein adsorption on materials surfaces with nano-topography. Chin. Sci. Bull. 2007;52:3169–3173. [Google Scholar]
- [82].Gagner JE, Qian X, Lopez MM, Dordick JS, Siegel RW. Effect of gold nanoparticle structure on the conformation and function of adsorbed proteins. Biomaterials. 2012;33:8503–8516. doi: 10.1016/j.biomaterials.2012.07.009. [DOI] [PubMed] [Google Scholar]
- [83].Faghihi F, Eslaminejad M. Baghaban. The effect of nano-scale topography on osteogenic differentiation of mesenchymal stem cells, Biomedical papers of the Medical Faculty of the University Palacky. Olomouc, Czechoslovakia. 2014;158:5–16. doi: 10.5507/bp.2013.013. [DOI] [PubMed] [Google Scholar]
- [84].Qian X, Levenstein A, Gagner JE, Dordick JS, Siegel RW. Protein Immobilization in Hollow Nanostructures and Investigation of the Adsorbed Protein Behavior. Langmuir. 2014;30:1295–1303. doi: 10.1021/la4048006. [DOI] [PubMed] [Google Scholar]
- [85].Nel AE, Mädler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- [86].Roach P, Eglin D, Rohde K, Perry CC. Modern biomaterials: a review—bulk properties and implications of surface modifications. J. Mater. Sci. - Mater. Med. 2007;18:1263–1277. doi: 10.1007/s10856-006-0064-3. [DOI] [PubMed] [Google Scholar]
- [87].Fendler JH. John Wiley & Sons; 2008. Nanoparticles and Nanostructured Films: Preparation, Characterization, and Applications. [Google Scholar]
- [88].Scopelliti PE, Borgonovo A, Indrieri M, Giorgetti L, Bongiorno G, Carbone R, Podesta A, Milani P. The effect of surface nanometre-scale morphology on protein adsorption. PloS one. 2010;5:e11862. doi: 10.1371/journal.pone.0011862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhou D, Wang X, Birch L, Rayment T, Abell C. AFM study on protein immobilization on charged surfaces at the nanoscale: toward the fabrication of three-dimensional protein nanostructures. Langmuir. 2003;19:10557–10562. [Google Scholar]
- [90].Kane RS, Stroock AD. Nanobiotechnology: Protein-Nanomaterial Interactions. Biotechnol. Prog. 2007;23:316–319. doi: 10.1021/bp060388n. [DOI] [PubMed] [Google Scholar]
- [91].Kim J-H, Yoon J-Y. Protein adsorption on polymer particles. CRC Press; 2002. [Google Scholar]
- [92].Casero E, Vazquez L, Parra-Alfambra AM, Lorenzo E. AFM, SECM and QCM as useful analytical tools in the characterization of enzyme-based bioanalytical platforms. Analyst. 2010;135:1878–1903. doi: 10.1039/c0an00120a. [DOI] [PubMed] [Google Scholar]
- [93].Schaefer J, Schulze C, Marxer EEJ, Schaefer UF, Wohlleben W, Bakowsky U, Lehr C-M. Atomic Force Microscopy and Analytical Ultracentrifugation for Probing Nanomaterial Protein Interactions. ACS Nano. 2012;6:4603–4614. doi: 10.1021/nn202657q. [DOI] [PubMed] [Google Scholar]
- [94].Bhakta SA, Benavidez TE, Garcia CD. Immobilization of glucose oxidase to nanostructured films of polystyrene-block-poly(2-vinylpyridine) J. Colloid Interface Sci. 2014;430:351–356. doi: 10.1016/j.jcis.2014.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Flegler SL, Heckman JW, Klomparens KL, Klomparens KL, Klomparens KL. Scanning and transmission electron microscopy: an introduction. WH Freeman; New York: 1993. [Google Scholar]
- [96].Gołek F, Mazur P, Ryszka Z, Zuber S. AFM image artifacts. Appl. Surf. Sci. 2014;304:11–19. [Google Scholar]
- [97].Russo CJ, Passmore LA. Controlling protein adsorption on graphene for cryo-EM using low-energy hydrogen plasmas. Nat. Meth. 2014;11:649–652. doi: 10.1038/nmeth.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Goldie K, Abeyrathne P, Kebbel F, Chami M, Ringler P, Stahlberg H. Cryo-electron Microscopy of Membrane Proteins. In: Kuo J, editor. Electron Microscopy. Humana Press; 2014. pp. 325–341. [DOI] [PubMed] [Google Scholar]
- [99].de Jong S, Krylov SN. Pressure-Based Approach for the Analysis of Protein Adsorption in Capillary Electrophoresis. Anal. Chem. 2012;84:453–458. doi: 10.1021/ac2030333. [DOI] [PubMed] [Google Scholar]
- [100].Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time evolution of the nanoparticle protein corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- [101].Casiano-Maldonado M, Lim GT, Li X, Reneker DH, Puskas JE, Wesdemiotis C. Protein adsorption on thermoplastic elastomeric surfaces: A quantitative mass spectrometry study. Int. J. Mass Spectrom. 2013;354–355:391–397. [Google Scholar]
- [102].Xiong C, Zhou X, Zhang N, Zhan L, Chen S, Wang J, Peng W-P, Chang H-C, Nie Z. Quantitative Assessment of Protein Adsorption on Microparticles with Particle Mass Spectrometry. Anal. Chem. 2014;86:3876–3881. doi: 10.1021/ac4042312. [DOI] [PubMed] [Google Scholar]
- [103].Osawa M. Handbook of Vibrational Spectroscopy. John Wiley & Sons; 2006. Surface-Enhanced Infrared Absorption Spectroscopy. [Google Scholar]
- [104].Fesenko O, Yatsenko L, Brodin M. Springer Proceedings in Physics. Springer; New York, NY: 2012. Nanomaterials Imaging Techniques, Surface Studies, and Applications. [Google Scholar]
- [105].Wang Y, Ni YN. Combination of UV-vis spectroscopy and chemometrics to understand protein-nanomaterial conjugate: A case study on human serum albumin and gold nanoparticles. Talanta. 2014;119:320–330. doi: 10.1016/j.talanta.2013.11.026. [DOI] [PubMed] [Google Scholar]
- [106].Griffiths PR, De Haseth JA. Fourier transform infrared spectrometry. John Wiley & Sons; 2007. [Google Scholar]
- [107].Perevedentseva E, Cai PJ, Chiu YC, Cheng CL. Characterizing Protein Activities on the Lysozyme and Nanodiamond Complex Prepared for Bio Applications. Langmuir. 2011;27:1085–1091. doi: 10.1021/la103155c. [DOI] [PubMed] [Google Scholar]
- [108].Giacomelli CE, Norde W. The Adsorption–Desorption Cycle. Reversibility of the BSA–Silica System. J. Colloid Interface Sci. 2001;233:234–240. doi: 10.1006/jcis.2000.7219. [DOI] [PubMed] [Google Scholar]
- [109].Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- [110].Baier G, Costa C, Zeller A, Baumann D, Sayer C, Araujo PHH, Mailänder V, Musyanovych A, Landfester K. BSA Adsorption on Differently Charged Polystyrene Nanoparticles using Isothermal Titration Calorimetry and the Influence on Cellular Uptake. Macromol. Biosci. 2011;11:628–638. doi: 10.1002/mabi.201000395. [DOI] [PubMed] [Google Scholar]
- [111].Vashist SK, Dixit CK, MacCraith BD, O'Kennedy R. Effect of antibody immobilization strategies on the analytical performance of a surface plasmon resonance-based immunoassay. Analyst. 2011;136:4431–4436. doi: 10.1039/c1an15325k. [DOI] [PubMed] [Google Scholar]
- [112].Souto DEP, Silva JV, Martins HR, Reis AB, Luz RCS, Kubota LT, Damos FS. Development of a label-free immunosensor based on surface plasmon resonance technique for the detection of anti-Leishmania infantum antibodies in canine serum. Biosens. Bioelectron. 2013;46:22–29. doi: 10.1016/j.bios.2013.01.067. [DOI] [PubMed] [Google Scholar]
- [113].Sciacca B, Monro TM. Dip Biosensor Based on Localized Surface Plasmon Resonance at the Tip of an Optical Fiber. Langmuir. 2014;30:946–954. doi: 10.1021/la403667q. [DOI] [PubMed] [Google Scholar]
- [114].Woods DA, Bain CD. Total internal reflection spectroscopy for studying soft matter. Soft Matter. 2014;10:1071–1096. doi: 10.1039/c3sm52817k. [DOI] [PubMed] [Google Scholar]
- [115].Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta, Bioenerg. 2007;1767:1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- [116].Shang L, Brandholt S, Stockmar F, Trouillet V, Bruns M, Nienhaus GU. Effect of Protein Adsorption on the Fluorescence of Ultrasmall Gold Nanoclusters. Small. 2012;8:661–665. doi: 10.1002/smll.201101353. [DOI] [PubMed] [Google Scholar]
- [117].Tsai C-S, Yu T-B, Chen C-T. Gold nanoparticle-based competitive colorimetric assay for detection of protein-protein interactions. Chem. Commun. 2005:4273–4275. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- [118].Tsai D-H, DelRio FW, Keene AM, Tyner KM, MacCuspie RI, Cho TJ, Zachariah MR, Hackley VA. Adsorption and Conformation of Serum Albumin Protein on Gold Nanoparticles Investigated Using Dimensional Measurements and in Situ Spectroscopic Methods. Langmuir. 2011;27:2464–2477. doi: 10.1021/la104124d. [DOI] [PubMed] [Google Scholar]
- [119].Bell NC, Minelli C, Shard AG. Quantitation of IgG protein adsorption to gold nanoparticles using particle size measurement. Anal. Methods. 2013;5:4591–4601. [Google Scholar]
- [120].Mora MF, Reza Nejadnik M, Baylon-Cardiel JL, Giacomelli CE, Garcia CD. Determination of a setup correction function to obtain adsorption kinetic data at stagnation point flow conditions. J. Colloid Interface Sci. 2010;346:208–215. doi: 10.1016/j.jcis.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Benavidez TE, Garcia CD. Potential-Assisted Adsorption of Bovine Serum Albumin onto Optically Transparent Carbon Electrodes. Langmuir. 2013;29:14154–14162. doi: 10.1021/la4029657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Evans E, Moreira Gabriel EF, Benavidez TE, Tomazelli Coltro WK, Garcia CD. Modification of microfluidic paper-based devices with silica nanoparticles. Analyst. 2014 doi: 10.1039/c4an01147c. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Kalashnikova IV, Ivanova ND, Evseeva TG, Menshikova AY, Vlakh EG, Tennikova TB. Study of dynamic adsorption behavior of large-size protein-bearing particles. J. Chromatogr. A. 2007;1144:40–47. doi: 10.1016/j.chroma.2006.10.060. [DOI] [PubMed] [Google Scholar]
- [124].Lionello A, Josserand J, Jensen H, Girault HH. Dynamic protein adsorption in microchannels by "stop-flow" and continuous flow. Lab Chip. 2005;5:1096–1103. doi: 10.1039/b506009e. [DOI] [PubMed] [Google Scholar]
- [125].Ueda T, Ishihara K, Nakabayashi N. Adsorption–Desorption of proteins on phospholipid polymer surfaces evaluated by dynamic contact angle measurement. J. Biomed. Mater. Res. 1995;29:381–387. doi: 10.1002/jbm.820290313. [DOI] [PubMed] [Google Scholar]
- [126].Santos O, Nylander T, Paulsson M, Trägårdh C. Whey protein adsorption onto steel surfaces—effect of temperature, flow rate, residence time and aggregation. J. Food Eng. 2006;74:468–483. [Google Scholar]
- [127].Enarsson L-E, WÃ¥gberg L. Adsorption Kinetics of Cationic Polyelectrolytes Studied with Stagnation Point Adsorption Reflectometry and Quartz Crystal Microgravimetry. Langmuir. 2008;24:7329–7337. doi: 10.1021/la800198e. [DOI] [PubMed] [Google Scholar]
- [128].Jia X, Xie Q, Zhang Y, Yao S. Simultaneous Quartz Crystal Microbalance-Electrochemical Impedance Spectroscopy Study on the Adsorption of Anti-human Immunoglobulin G and Its Immunoreaction at Nanomaterial-modified Au Electrode Surfaces. Anal. Sci. 2007;23:689–696. doi: 10.2116/analsci.23.689. [DOI] [PubMed] [Google Scholar]
- [129].Teichroeb JH, Forrest JA, Jones LW, Chan J, Dalton K. Quartz crystal microbalance study of protein adsorption kinetics on poly(2-hydroxyethyl methacrylate) J. Colloid Interface Sci. 2008;325:157–164. doi: 10.1016/j.jcis.2008.05.052. [DOI] [PubMed] [Google Scholar]
- [130].Zhang Y, Fung Y, Sun H, Zhu D, Yao S. Study of protein adsorption on polymer coatings surface by combining quartz crystal microbalance with electrochemical impedance methods. Sens. Actuators. 2005;108:933–942. B. [Google Scholar]
- [131].Fujiwara H. Principles and applications. J. Wiley & Sons; West Sussex, England: 2007. Spectroscopic ellipsometry. [Google Scholar]
- [132].Mora MF, Wehmeyer J, Synowicki R, Garcia CD. Investigating the Adsorption of Proteins Via Spectroscopic Ellipsometry. In: Bizios R, Puleo D, editors. Biological Interactions on Material Surfaces: Understanding and Controlling Protein, Cell, and Tissue Responses. Springer; New York, NY: 2009. [Google Scholar]
- [133].Valenti LE, Herrera E, Stragliotto MF, Martins VL, Torresi RM, Giacomelli CE. Optimizing the Bioaffinity Interaction between His-Tag Proteins and Ni(II) Surface Sites. Proteins at Interfaces III State of the Art, American Chemical Society. 2012:37–53. [Google Scholar]
- [134].Wehmeyer JL, Synowicki R, Bizios R, Garcia CD. Dynamic adsorption of albumin on nanostructured TiO2 thin films. Mater. Sci. Eng. C. 2010;30:277–282. doi: 10.1016/j.msec.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Chumbimuni-Torres KY, Coronado RE, Mfuh AM, Castro-Guerrero C, Silva MF, Negrete GR, Bizios R, Garcia CD. Adsorption of proteins to thin-films of PDMS and its effect on the adhesion of human endothelial cells. RSC Adv. 2011;1:706–714. doi: 10.1039/C1RA00198A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Ross AM, Zhang D, Deng X, Chang SL, Lahann J. Chemical-Vapor-Deposition-Based Polymer Substrates for Spatially Resolved Analysis of Protein Binding by Imaging Ellipsometry. Anal. Chem. 2011;83:874–880. doi: 10.1021/ac102535j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].de Vos WM, Biesheuvel PM, de Keizer A, Kleijn JM, Cohen Stuart MA. Adsorption of the Protein Bovine Serum Albumin in a Planar Poly(acrylic acid) Brush Layer As Measured by Optical Reflectometry. Langmuir. 2008;24:6575–6584. doi: 10.1021/la8006469. [DOI] [PubMed] [Google Scholar]
- [138].García G, García CD, Ortiz PI, De Pauli CP. Reflectometry applied to electrochemically generated phenoxy radical adsorption monitoring. J. Electroanal. Chem. 2002;519:53–59. [Google Scholar]
- [139].Valenti LE, Fiorito PA, Garcia CD, Giacomelli CE. The adsorption-desorption process of bovine serum albumin on carbon nanotubes. J. Colloid Interface Sci. 2007;307:349–356. doi: 10.1016/j.jcis.2006.11.046. [DOI] [PubMed] [Google Scholar]
- [140].Carot ML, Torresi RM, Garcia CD, Esplandiu MJ, Giacomelli CE. Electrostatic and Hydrophobic Interactions Involved in CNT Biofunctionalization with Short ss-DNA. J. Phys. Chem. C. 2010;114:4459–4465. doi: 10.1021/jp9085359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Chan CPY, Mak WC, Cheung KY, Sin KK, Yu CM, Rainer TH, Renneberg R. Evidence-Based Point-of-Care Diagnostics: Current Status and Emerging Technologies. Annu. Rev. Anal. Chem. 2013;6:191–211. doi: 10.1146/annurev-anchem-062012-092641. [DOI] [PubMed] [Google Scholar]
- [142].Carregal-Romero S, Caballero-Díaz E, Beqa L, Abdelmonem AM, Ochs M, Hühn D, Suau BS, Valcarcel M, Parak WJ. Multiplexed Sensing and Imaging with Colloidal Nano- and Microparticles. Annu. Rev. Anal. Chem. 2013;6:53–81. doi: 10.1146/annurev-anchem-062012-092621. [DOI] [PubMed] [Google Scholar]
- [143].Kong T, Chen Y, Ye Y, Zhang K, Wang Z, Wang X. An amperometric glucose biosensor based on the immobilization of glucose oxidase on the ZnO nanotubes. Sensors and Actuators B: Chemical. 2009;138:344–350. [Google Scholar]
- [144].Schwinté P, Ball V, Szalontai B, Haikel Y, Voegel JC, Schaaf P. Secondary Structure of Proteins Adsorbed onto or Embedded in Polyelectrolyte Multilayers. Biomacromolecules. 2002;3:1135–1143. doi: 10.1021/bm025547f. [DOI] [PubMed] [Google Scholar]
- [145].Hossain MF, Park JY. Amperometric Glucose Biosensor Based on Pt-Pd Nanoparticles Supported by Reduced Graphene Oxide and Integrated with Glucose Oxidase. Electroanal. 2014;26:940–951. [Google Scholar]
- [146].Oztürk FÖ, Erden PE, Kaçar C, Kiliç E. Amperometric biosensor for xanthine determination based on Fe3O4 nanoparticles. Acta Chim Slov. 2014;61:19–26. [PubMed] [Google Scholar]
- [147].Wang J. Electrochemical Glucose Biosensors. Chem. Rev. 2007;108:814–825. doi: 10.1021/cr068123a. [DOI] [PubMed] [Google Scholar]
- [148].Nejadnik MR, Francis L, Garcia CD. Adsorption of Glucose Oxidase to 3-D Scaffolds of Carbon Nanotubes: Analytical Applications. Electroanal. 2011;23:1462–1469. doi: 10.1002/elan.201000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Goran JM, Mantilla SM, Stevenson KJ. Influence of Surface Adsorption on the Interfacial Electron Transfer of Flavin Adenine Dinucleotide and Glucose Oxidase at Carbon Nanotube and Nitrogen-Doped Carbon Nanotube Electrodes. Analytical Chemistry. 2013;85:1571–1581. doi: 10.1021/ac3028036. [DOI] [PubMed] [Google Scholar]
- [150].Hobbs JM, Patel NN, Kim DW, Rugutt JK, Wanekaya AK. Glucose Determination in Beverages Using Carbon Nanotube Modified Biosensor: An Experiment for the Undergraduate Laboratory. J. Chem. Educ. 2013;90:1222–1226. [Google Scholar]
- [151].Hibbert DB, Gooding JJ, Erokhin P. Kinetics of Irreversible Adsorption with Diffusion: Application to Biomolecule Immobilization. Langmuir. 2002;18:1770–1776. [Google Scholar]
- [152].Calvo EJ, Etchenique R, Pietrasanta L, Wolosiuk A, Danilowicz C. Layer-by-layer self-assembly of glucose oxidase and Os(Bpy)2CIPyCH2NH-poly(allylamine) bioelectrode. Anal. Chem. 2001;73:1161–1168. doi: 10.1021/ac0011686. [DOI] [PubMed] [Google Scholar]
- [153].Yan W, Feng X, Chen X, Hou W, Zhu J-J. A super highly sensitive glucose biosensor based on Au nanoparticles–AgCl@polyaniline hybrid material. Biosens. Bioelectron. 2008;23:925–931. doi: 10.1016/j.bios.2007.09.002. [DOI] [PubMed] [Google Scholar]
- [154].Qiu C, Wang X, Liu X, Hou S, Ma H. Direct electrochemistry of glucose oxidase immobilized on nanostructured gold thin films and its application to bioelectrochemical glucose sensor. Electrochim. Acta. 2012;67:140–146. [Google Scholar]
- [155].German N, Ramanaviciene A, Voronovic J, Ramanavicius A. Glucose biosensor based on graphite electrodes modified with glucose oxidase and colloidal gold nanoparticles. Microchim. Acta. 2010;168:221–229. [Google Scholar]
- [156].Hecht DS, Hu L, Irvin G. Emerging Transparent Electrodes Based on Thin Films of Carbon Nanotubes, Graphene, and Metallic Nanostructures. Adv. Mat. 2011;23:1482–1513. doi: 10.1002/adma.201003188. [DOI] [PubMed] [Google Scholar]
- [157].Han Z, Fina A. Thermal conductivity of carbon nanotubes and their polymer nanocomposites: A review. Prog. Polym. Sci. 2011;36:914–944. [Google Scholar]
- [158].Bose S, Khare RA, Moldenaers P. Assessing the strengths and weaknesses of various types of pre-treatments of carbon nanotubes on the properties of polymer/carbon nanotubes composites: A critical review. Polymer. 2010;51:975–993. [Google Scholar]
- [159].Singh P, Campidelli S, Giordani S, Bonifazi D, Bianco A, Prato M. Organic functionalisation and characterisation of single-walled carbon nanotubes. Chem. Soc. Rev. 2009;38:2214–2230. doi: 10.1039/b518111a. [DOI] [PubMed] [Google Scholar]
- [160].Yang W, Thordarson P, Gooding JJ, Ringer SP, Braet F. Carbon nanotubes for biological and biomedical applications. Nanotechnology. 2007:412001. [Google Scholar]
- [161].Liliane B. Multiwall carbon nanotube elastomeric composites: A review. Polymer. 2007;48:4907–4920. [Google Scholar]
- [162].Belin T, Epron F. Characterization methods of carbon nanotubes: a review. Mater. Sci. Eng. B. 2005;119:105–118. [Google Scholar]
- [163].Lin Y, Taylor S, Li H, Fernando KAS, Qu L, Wang W, Gu L, Zhou B, Sun Y-P. Advances toward bioapplications of carbon nanotubes. J. Mater. Chem. 2004;14:527–541. [Google Scholar]
- [164].Deng S, Jian G, Lei J, Hu Z, Ju H. A glucose biosensor based on direct electrochemistry of glucose oxidase immobilized on nitrogen-doped carbon nanotubes. Biosens. Bioelectron. 2009;25:373–377. doi: 10.1016/j.bios.2009.07.016. [DOI] [PubMed] [Google Scholar]
- [165].Deng C, Chen J, Nie Z, Si S. A sensitive and stable biosensor based on the direct electrochemistry of glucose oxidase assembled layer-by-layer at the multiwall carbon nanotube-modified electrode. Biosens. Bioelectron. 2010;26:213–219. doi: 10.1016/j.bios.2010.06.013. [DOI] [PubMed] [Google Scholar]
- [166].Holland JT, Lau C, Brozik S, Atanassov P, Banta S. Engineering of Glucose Oxidase for Direct Electron Transfer via Site-Specific Gold Nanoparticle Conjugation. J. Am. Chem. Soc. 2011;133:19262–19265. doi: 10.1021/ja2071237. [DOI] [PubMed] [Google Scholar]
- [167].Ueda A, Kato D, Kurita R, Kamata T, Inokuchi H, Umemura S, Hirono S, Niwa O. Efficient Direct Electron Transfer with Enzyme on a Nanostructured Carbon Film Fabricated with a Maskless Top-Down UV/Ozone Process. J. Am. Chem. Soc. 2011;133:4840–4846. doi: 10.1021/ja108614d. [DOI] [PubMed] [Google Scholar]
- [168].Wooten M, Karra S, Zhang M, Gorski W. On the Direct Electron Transfer, Sensing, and Enzyme Activity in the Glucose Oxidase/Carbon Nanotubes System. Anal. Chem. 2014;86:752–757. doi: 10.1021/ac403250w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Cai C, Chen J. Direct electron transfer of glucose oxidase promoted by carbon nanotubes. Anal. Biochem. 2004;332:75–83. doi: 10.1016/j.ab.2004.05.057. [DOI] [PubMed] [Google Scholar]
- [170].Gao Q, Guo Y, Liu J, Yuan X, Qi H, Zhang C. A biosensor prepared by co-entrapment of a glucose oxidase and a carbon nanotube within an electrochemically deposited redox polymer multilayer. Bioelectrochemistry. 2011;81:109–113. doi: 10.1016/j.bioelechem.2011.04.003. [DOI] [PubMed] [Google Scholar]
- [171].Salimi A, Compton RG, Hallaj R. Glucose biosensor prepared by glucose oxidase encapsulated sol-gel and carbon-nanotube-modified basal plane pyrolytic graphite electrode. Anal. Biochem. 2004;333:49–56. doi: 10.1016/j.ab.2004.06.039. [DOI] [PubMed] [Google Scholar]
- [172].Sato N, Okuma H. Development of single-wall carbon nanotubes modified screen-printed electrode using a ferrocene-modified cationic surfactant for amperometric glucose biosensor applications. Sens. Actuators. 2008;129:188–194. B. [Google Scholar]
- [173].Sotiropoulou S, Chaniotakis NA. Carbon nanotube array-based biosensor. Anal. Bioanal. Chem. 2003;375:103–105. doi: 10.1007/s00216-002-1617-z. [DOI] [PubMed] [Google Scholar]
- [174].Vaze A, Hussain N, Tang C, Leech D, Rusling J. Biocatalytic anode for glucose oxidation utilizing carbon nanotubes for direct electron transfer with glucose oxidase. Electrochem. Com. 2009;11:2004–2007. doi: 10.1016/j.elecom.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Monosik R, Stredansky M, Luspai K, Magdolen P, Sturdik E. Amperometric glucose biosensor utilizing FAD-dependent glucose dehydrogenase immobilized on nanocomposite electrode. Enzyme Microb. Technol. 2012;50:227–232. doi: 10.1016/j.enzmictec.2012.01.004. [DOI] [PubMed] [Google Scholar]
- [176].Rahman MM, Umar A, Sawada K. Development of amperometric glucose biosensor based on glucose oxidase co-immobilized with multi-walled carbon nanotubes at low potential. Sens. Actuators. 2009;137:327–333. B. [Google Scholar]
- [177].Rakhi RB, Sethupathi K, Ramaprabhu S. A Glucose Biosensor Based on Deposition of Glucose Oxidase onto Crystalline Gold Nanoparticle Modified Carbon Nanotube Electrode. J. Phys. Chem. B. 2009;113:3190–3194. doi: 10.1021/jp810235v. [DOI] [PubMed] [Google Scholar]
- [178].Wang J. Carbon-Nanotube Based Electrochemical Biosensors: A Review. Electroanal. 2005;17:7–14. [Google Scholar]
- [179].Rivas GA, Rubianes MD, Rodríguez MC, Ferreyra NF, Luque GL, Pedano ML, Miscoria SA, Parrado C. Carbon nanotubes for electrochemical biosensing. Talanta. 2007;74:291–307. doi: 10.1016/j.talanta.2007.10.013. [DOI] [PubMed] [Google Scholar]
- [180].Zhao J, Chen X, Xie JRH. Optical properties and photonic devices of doped carbon nanotubes. Anal. Chim. Acta. 2006;568:161–170. doi: 10.1016/j.aca.2006.02.006. [DOI] [PubMed] [Google Scholar]
- [181].Deng C, Chen J, Chen X, Xiao C, Nie L, Yao S. Direct electrochemistry of glucose oxidase and biosensing for glucose based on boron-doped carbon nanotubes modified electrode. Biosens. Bioelectron. 2008;23:1272–1277. doi: 10.1016/j.bios.2007.11.009. [DOI] [PubMed] [Google Scholar]
- [182].Soetedjo H, Mora MF, Garcia CD. Optical properties of single-wall carbon nanotube films deposited on Si/SiO2 wafers. Thin Solid Films. 2010;518:3954–3959. doi: 10.1016/j.tsf.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Chen H, Yang Y, Hu Z, Huo K, Ma Y, Chen Y, Wang X, Lu Y. Synergism of C5N Six-Membered Ring and Vapor–Liquid–Solid Growth of CNx Nanotubes with Pyridine Precursor. J. Phys. Chem. B. 2006;110:16422–16427. doi: 10.1021/jp062216e. [DOI] [PubMed] [Google Scholar]
- [184].Liu D, Zhang X, You T. Electrochemical Performance of Electrospun Free-Standing Nitrogen-Doped Carbon Nanofibers and Their Application for Glucose Biosensing. ACS Appl Mater Interfaces. 2014;6:6275–6280. doi: 10.1021/am501713g. [DOI] [PubMed] [Google Scholar]
- [185].Zhang Y, Guo G, Zhao F, Mo Z, Xiao F, Zeng B. A Novel Glucose Biosensor Based on Glucose Oxidase Immobilized on AuPt Nanoparticle – Carbon Nanotube – Ionic Liquid Hybrid Coated Electrode. Electroanal. 2010;22:223–228. [Google Scholar]
- [186].Wang J, Wang L, Di J, Tu Y. Disposable biosensor based on immobilization of glucose oxidase at gold nanoparticles electrodeposited on indium tin oxide electrode. Sens. Actuators. 2008;135:283–288. B. [Google Scholar]
- [187].German N, Ramanavicius A, Voronovic J, Ramanaviciene A. Glucose biosensor based on glucose oxidase and gold nanoparticles of different sizes covered by polypyrrole layer. Colloids Surf., A. 2012;413:224–230. [Google Scholar]
- [188].Zhang S, Wang N, Niu Y, Sun C. Immobilization of glucose oxidase on gold nanoparticles modified Au electrode for the construction of biosensor. Sens. Actuators. 2005;109:367–374. B. [Google Scholar]
- [189].Jiang X, Wu Y, Mao X, Cui X, Zhu L. Amperometric glucose biosensor based on integration of glucose oxidase with platinum nanoparticles/ordered mesoporous carbon nanocomposite. Sens. Actuators. 2011;153:158–163. B. [Google Scholar]
- [190].Tang H, Chen J, Yao S, Nie L, Deng G, Kuang Y. Amperometric glucose biosensor based on adsorption of glucose oxidase at platinum nanoparticle-modified carbon nanotube electrode. Anal. Biochem. 2004;331:89–97. doi: 10.1016/j.ab.2004.05.005. [DOI] [PubMed] [Google Scholar]
- [191].Wu H, Wang J, Kang X, Wang C, Wang D, Liu J, Aksay IA, Lin Y. Glucose biosensor based on immobilization of glucose oxidase in platinum nanoparticles/graphene/chitosan nanocomposite film. Talanta. 2009;80:403–406. doi: 10.1016/j.talanta.2009.06.054. [DOI] [PubMed] [Google Scholar]
- [192].Ren X, Meng X, Tang F, Zhang L. Biosensor enhanced by glucose oxidase biomimetic membrane containing the platinum and silica nanoparticles. Mater. Sci. Eng. C. 2009;29:2234–2238. [Google Scholar]
- [193].Li H, He J, Zhao Y, Wu D, Cai Y, Wei Q, Yang M. Immobilization of glucose oxidase and platinum on mesoporous silica nanoparticles for the fabrication of glucose biosensor. Electrochim. Acta. 2011;56:2960–2965. [Google Scholar]
- [194].Zhang H, Meng Z, Wang Q, Zheng J. A novel glucose biosensor based on direct electrochemistry of glucose oxidase incorporated in biomediated gold nanoparticles–carbon nanotubes composite film. Sens. Actuators. 2011;158:23–27. B. [Google Scholar]
- [195].Wang L, Bai J, Bo X, Zhang X, Guo L. A novel glucose sensor based on ordered mesoporous carbon–Au nanoparticles nanocomposites. Talanta. 2011;83:1386–1391. doi: 10.1016/j.talanta.2010.11.022. [DOI] [PubMed] [Google Scholar]
- [196].Zhou L, Jiang Y, Gao J, Zhao X, Ma L. Graphene Oxide as a Matrix for the Immobilization of Glucose Oxidase. Appl. Biochem. Biotechnol. 2012;168:1635–1642. doi: 10.1007/s12010-012-9884-4. [DOI] [PubMed] [Google Scholar]
- [197].Kang X, Wang J, Wu H, Aksay IA, Liu J, Lin Y. Glucose Oxidase–graphene–chitosan modified electrode for direct electrochemistry and glucose sensing. Biosens. Bioelectron. 2009;25:901–905. doi: 10.1016/j.bios.2009.09.004. [DOI] [PubMed] [Google Scholar]
- [198].Zhiguo G, Shuping Y, Zaijun L, Xiulan S, Guangli W, Yinjun F, Junkang L. An ultrasensitive electrochemical biosensor for glucose using CdTe-CdS core–shell quantum dot as ultrafast electron transfer relay between graphene-gold nanocomposite and gold nanoparticle. Electrochim. Acta. 2011;56:9162–9167. [Google Scholar]
- [199].Wang K, Liu Q, Guan Q-M, Wu J, Li H-N, Yan J-J. Enhanced direct electrochemistry of glucose oxidase and biosensing for glucose via synergy effect of graphene and CdS nanocrystals. Biosens. Bioelectron. 2011;26:2252–2257. doi: 10.1016/j.bios.2010.09.043. [DOI] [PubMed] [Google Scholar]
- [200].Wang K, Liu Q, Wu X-Y, Guan Q-M, Li H-N. Graphene enhanced electrochemiluminescence of CdS nanocrystal for H2O2 sensing. Talanta. 2010;82:372–376. doi: 10.1016/j.talanta.2010.04.054. [DOI] [PubMed] [Google Scholar]
- [201].Gao Q, Guo Y, Zhang W, Qi H, Zhang C. An amperometric glucose biosensor based on layer-by-layer GOx-SWCNT conjugate/redox polymer multilayer on a screen-printed carbon electrode. Sens. Actuators. 2011;153:219–225. B. [Google Scholar]
- [202].Rogaleva N, Korpan Y, Biloivan O. Glucose Biosensor Based on Screen-Printed Electrodes and Glucose Oxidase Layer Modified by MWCNT-NH2. Sens. Lett. 2011;9:2356–2359. [Google Scholar]
- [203].You C, Xu X, Tian B, Kong J, Zhao D, Liu B. Electrochemistry and biosensing of glucose oxidase based on mesoporous carbons with different spatially ordered dimensions. Talanta. 2009;78:705–710. doi: 10.1016/j.talanta.2008.12.032. [DOI] [PubMed] [Google Scholar]
- [204].Dai Y, Swain GM, Porter MD, Zak Jerzy. New Horizons in Spectroelectrochemical Measurements: Optically Transparent Carbon Electrodes. Anal. Chem. 2008;80:14–22. doi: 10.1021/ac085996r. [DOI] [PubMed] [Google Scholar]
- [205].Alharthi SA, Benavidez TE, Garcia CD. Ultrathin Optically Transparent Carbon Electrodes Produced from Layers of Adsorbed Proteins. Langmuir. 2013;29:3320–3327. doi: 10.1021/la3049136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Benavidez TE, Garcia CD. Spectroscopic and electrochemical characterization of nanostructured optically transparent carbon electrodes. Electrophoresis. 2013;34:1998–2006. doi: 10.1002/elps.201300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Ngeontae W, Janrungroatsakul W, Maneewattanapinyo P, Ekgasit S, Aeungmaitrepirom W, Tuntulani T. Novel potentiometric approach in glucose biosensor using silver nanoparticles as redox marker. Sens. Actuators. 2009;137:320–326. B. [Google Scholar]
- [208].Kaushik A, Khan R, Solanki PR, Pandey P, Alam J, Ahmad S, Malhotra BD. Iron oxide nanoparticles–chitosan composite based glucose biosensor. Biosens. Bioelectron. 2008;24:676–683. doi: 10.1016/j.bios.2008.06.032. [DOI] [PubMed] [Google Scholar]
- [209].Zhao ZW, Chen XJ, Tay BK, Chen JS, Han ZJ, Khor KA. A novel amperometric biosensor based on ZnO:Co nanoclusters for biosensing glucose. Biosens. Bioelectron. 2007;23:135–139. doi: 10.1016/j.bios.2007.03.014. [DOI] [PubMed] [Google Scholar]
- [210].Gupta V. ZnO based third generation biosensor. Thin Solid Films. 2010;519:1141–1144. [Google Scholar]
- [211].Fang B, Zhang C, Wang G, Wang M, Ji Y. A glucose oxidase immobilization platform for glucose biosensor using ZnO hollow nanospheres. Sens. Actuators. 2011;155:304–310. B. [Google Scholar]
- [212].Veitch NC. Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry. 2004;65:249–259. doi: 10.1016/j.phytochem.2003.10.022. [DOI] [PubMed] [Google Scholar]
- [213].Lavery CB, MacInnis MC, MacDonald MJ, Williams JB, Spencer CA, Burke AA, Irwin DJG, D’Cunha GB. Purification of Peroxidase from Horseradish (Armoracia rusticana) Roots. J. Agric. Food. Chem. 2010;58:8471–8476. doi: 10.1021/jf100786h. [DOI] [PubMed] [Google Scholar]
- [214].Liu S, Yuan R, Chai Y, Su H. A label-free amperometric immunosensor based on horseradish peroxidase functionalized carbon nanotubes and bilayer gold nanoparticles. Sens. Actuators. 2011;156:388–394. B. [Google Scholar]
- [215].Lim SY, Ahn J, Lee JS, Kim M-G, Park CB. Graphene-Oxide-Based Immunosensing through Fluorescence Quenching by Peroxidase-Catalyzed Polymerization. Small. 2012;8:1994–1999. doi: 10.1002/smll.201200379. [DOI] [PubMed] [Google Scholar]
- [216].Cao X, Li Y, Zhang Z, Yu J, Qian J, Liu S. Catalytic activity and stability of glucose oxidase/horseradish peroxidase co-confined in macroporous silica foam. Analyst. 2012;137:5785–5791. doi: 10.1039/c2an36237f. [DOI] [PubMed] [Google Scholar]
- [217].Díaz AN, Sanchez FG, García J. Hydrogen peroxide assay by using enhanced chemiluminescence of the luminol-H2O2-horseradish peroxidase system: Comparative studies. Anal. Chim. Acta. 1996;327:161–165. [Google Scholar]
- [218].Cao S, Yuan R, Chai Y, Zhang L, Li X, Gao F. A mediator-free amperometric hydrogen peroxide biosensor based on HRP immobilized on a nano-Au/poly 2, 6-pyridinediamine-coated electrode. Bioprocess. Biosyst. Eng. 2007;30:71–78. doi: 10.1007/s00449-006-0100-1. [DOI] [PubMed] [Google Scholar]
- [219].Chen S, Yuan R, Chai Y, Xu L, Wang N, Li X, Zhang L. Amperometric Hydrogen Peroxide Biosensor Based on the Immobilization of Horseradish Peroxidase (HRP) on the Layer-by-Layer Assembly Films of Gold Colloidal Nanoparticles and Toluidine Blue. Electroanal. 2006;18:471–477. [Google Scholar]
- [220].Gao F, Yuan R, Chai Y, Chen S, Cao S, Tang M. Amperometric hydrogen peroxide biosensor based on the immobilization of HRP on nano-Au/Thi/poly p-aminobenzene sulfonic acid)-modified glassy carbon electrode. J. Biochem. Bioph. Methods. 2007;70:407–413. doi: 10.1016/j.jbbm.2006.09.007. [DOI] [PubMed] [Google Scholar]
- [221].Jia J, Wang B, Wu A, Cheng G, Li Z, Dong S. A method to construct a third-generation horseradish peroxidase biosensor: self-assembling gold nanoparticles to three-dimensional sol-gel network. Anal. Chem. 2002;74:2217–2223. doi: 10.1021/ac011116w. [DOI] [PubMed] [Google Scholar]
- [222].Lei C-X, Hu S-Q, Gao N, Shen G-L, Yu R-Q. An amperometric hydrogen peroxide biosensor based on immobilizing horseradish peroxidase to a nano-Au monolayer supported by sol–gel derived carbon ceramic electrode. Bioelectrochemistry. 2004;65:33–39. doi: 10.1016/j.bioelechem.2004.06.002. [DOI] [PubMed] [Google Scholar]
- [223].Li S, Zhu X, Zhang W, Xie G, Feng W. Hydrogen peroxide biosensor based on gold nanoparticles/thionine/gold nanoparticles/multi-walled carbon nanotubes–chitosans composite film-modified electrode. Appl. Surf. Sci. 2012;258:2802–2807. [Google Scholar]
- [224].Liu Y, Yuan R, Chai Y, Tang D, Dai J, Zhong X. Direct electrochemistry of horseradish peroxidase immobilized on gold colloid/cysteine/nafion-modified platinum disk electrode. Sens. Actuators. 2006;115:109–115. B. [Google Scholar]
- [225].Luo X-L, Xu J-J, Zhang Q, Yang G-J, Chen H-Y. Electrochemically deposited chitosan hydrogel for horseradish peroxidase immobilization through gold nanoparticles self-assembly. Biosens. Bioelectron. 2005;21:190–196. doi: 10.1016/j.bios.2004.07.029. [DOI] [PubMed] [Google Scholar]
- [226].Wang Y, Ma X, Wen Y, Xing Y, Zhang Z, Yang H. Direct electrochemistry and bioelectrocatalysis of horseradish peroxidase based on gold nano-seeds dotted TiO2 nanocomposite. Biosens. Bioelectron. 2010;25:2442–2446. doi: 10.1016/j.bios.2010.04.002. [DOI] [PubMed] [Google Scholar]
- [227].Wang J, Wang L, Di J, Tu Y. Electrodeposition of gold nanoparticles on indium/tin oxide electrode for fabrication of a disposable hydrogen peroxide biosensor. Talanta. 2009;77:1454–1459. doi: 10.1016/j.talanta.2008.09.034. [DOI] [PubMed] [Google Scholar]
- [228].Zhang Y, Yuan R, Chai Y, Xiang Y, Hong C, Ran X. An amperometric hydrogen peroxide biosensor based on the immobilization of HRP on multi-walled carbon nanotubes/electro-copolymerized nano-Pt-poly (neutral red) composite membrane. Biochem. Eng. J. 2010;51:102–109. [Google Scholar]
- [229].Gu B, Xu C, Zhu G, Liu S, Chen L, Wang M, Zhu J. Layer by layer immobilized horseradish peroxidase on zinc oxide nanorods for biosensing. J. Phys. Chem. B. 2009;113:6553–6557. doi: 10.1021/jp900048m. [DOI] [PubMed] [Google Scholar]
- [230].Ren C, Song Y, Li Z, Zhu G. Hydrogen peroxide sensor based on horseradish peroxidase immobilized on a silver nanoparticles/cysteamine/gold electrode. Anal. Bioanal. Chem. 2005;381:1179–1185. doi: 10.1007/s00216-004-3032-0. [DOI] [PubMed] [Google Scholar]
- [231].Liu Y, Lei J, Ju H. Amperometric sensor for hydrogen peroxide based on electric wire composed of horseradish peroxidase and toluidine blue-multiwalled carbon nanotubes nanocomposite. Talanta. 2008;74:965–970. doi: 10.1016/j.talanta.2007.08.001. [DOI] [PubMed] [Google Scholar]
- [232].Yuan P, Zhuo Y, Chai Y, Ju H. Dendritic silver/silicon dioxide nanocomposite modified electrodes for electrochemical sensing of hydrogen peroxide. Electroanal. 2008;20:1839–1844. [Google Scholar]
- [233].Furkan Y, Emre Ç, Mehmet Ş, Abdülhadi B. Development of an Amperometric Hydrogen Peroxide Biosensor based on the Immobilization of Horseradish Peroxidase onto Nickel Ferrite Nanoparticle-Chitosan Composite. Nano-Micro Lett. 2011;3:91–98. [Google Scholar]
- [234].Zhang Q, Yang S, Zhang J, Zhang L, Kang P, Li J, Xu J, Zhou H, Song X-M. Fabrication of an electrochemical platform based on the self-assembly of graphene oxide–multiwall carbon nanotube nanocomposite and horseradish peroxidase: direct electrochemistry and electrocatalysis. Nanotechnology. 2011;22:494010. doi: 10.1088/0957-4484/22/49/494010. [DOI] [PubMed] [Google Scholar]
- [235].Wang Y, Du J, Li Y, Shan D, Zhou X, Xue Z, Lu X. A amperometric biosensor for hydrogen peroxide by adsorption of horseradish peroxidase onto single-walled carbon nanotubes. Colloids Surf. 2012;90:62–67. doi: 10.1016/j.colsurfb.2011.09.045. B. [DOI] [PubMed] [Google Scholar]
- [236].Brigelius-Flohé R, Kipp A. Glutathione peroxidases in different stages of carcinogenesis. Biochim. Biophys. Acta, Gen. Subj. 2009;1790:1555–1568. doi: 10.1016/j.bbagen.2009.03.006. [DOI] [PubMed] [Google Scholar]
- [237].Bayramoglu G, Karagoz B, Yilmaz M, Bicak N, Arica MY. Immobilization of catalase via adsorption on poly(styrene-d-glycidylmethacrylate) grafted and tetraethyldiethylenetriamine ligand attached microbeads. Bioresour. Technol. 2011;102:3653–3661. doi: 10.1016/j.biortech.2010.12.029. [DOI] [PubMed] [Google Scholar]
- [238].Fooladsaz K, Negahdary M, Rahimi G, Habibi-Tamijani A, Parsania S, Akbari-dastjerdi H, Sayad A, Jamaleddini A, Salahi F, Asad A. Dopamine Determination with a Biosensor Based on Catalase and Modified Carbon Paste Electrode with Zinc Oxide Nanoparticles. Int. J. Electrochem. Sci. 2012;7:9892–9908. [Google Scholar]
- [239].Nikan EA, Negahdary M, Nabi-abdoluosefi E, Ghassamipour S, Motaghi O. A new hydrogen peroxide biosensor by using modified Carbon Paste Electrode with catalase enzyme and Mn2O3 Nanoparticles. Ann. Biol. Res. 2012;3:5242–5251. [Google Scholar]
- [240].Salimi A, Sharifi E, Noorbakhsh A, Soltanian S. Direct electrochemistry and electrocatalytic activity of catalase immobilized onto electrodeposited nano-scale islands of nickel oxide. Biophys. Chem. 2007;125:540–548. doi: 10.1016/j.bpc.2006.11.004. [DOI] [PubMed] [Google Scholar]
- [241].Wang L, Wang J, Zhou F. Direct Electrochemistry of Catalase at a Gold Electrode Modified with Single-Wall Carbon Nanotubes. Electroanal. 2004;16:627–632. [Google Scholar]
- [242].Felhofer JL, Caranto JD, Garcia CD. Adsorption Kinetics of Catalase to Thin Films of Carbon Nanotubes. Langmuir. 2010;26:17178–17183. doi: 10.1021/la103035n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Zhang C, Luo S, Chen W. Activity of catalase adsorbed to carbon nanotubes: Effects of carbon nanotube surface properties. Talanta. 2013;113:142–147. doi: 10.1016/j.talanta.2013.03.027. [DOI] [PubMed] [Google Scholar]
- [244].Ibupoto ZH, Shah SM, Khun K, Willander M. Electrochemical L-lactic acid sensor based on immobilized ZnO nanorods with lactate oxidase. Sensors (Basel, Switzerland) 2012;12:2456–2466. doi: 10.3390/s120302456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Taurino I, Reiss R, Richter M, Fairhead M, Thöny-Meyer L, De Micheli G, Carrara S. Comparative study of three lactate oxidases from Aerococcus viridans for biosensing applications. Electrochim. Acta. 2013;93:72–79. [Google Scholar]
- [246].Mingli Y, Jin W, Huaqing L, Jian-Guo Z, Nianqiang Nick W. A lactate electrochemical biosensor with a titanate nanotube as direct electron transfer promoter. Nanotechnology. 2008;19:075502. doi: 10.1088/0957-4484/19/7/075502. [DOI] [PubMed] [Google Scholar]
- [247].Huang J, Li J, Yang Y, Wang X, Wu B, Anzai J.-i., Osa T, Chen Q. Development of an amperometric l-lactate biosensor based on l-lactate oxidase immobilized through silica sol–gel film on multi-walled carbon nanotubes/platinum nanoparticle modified glassy carbon electrode. Mater. Sci. Eng. C. 2008;28:1070–1075. [Google Scholar]
- [248].Wang YT, Yu L, Wang J, Lou L, Du WJ, Zhu ZQ, Peng H, Zhu JZ. A novel l-lactate sensor based on enzyme electrode modified with ZnO nanoparticles and multiwall carbon nanotubes. J. Electroanal. Chem. 2011;661:8–12. [Google Scholar]
- [249].Goran JM, Lyon JL, Stevenson KJ. Amperometric Detection of l-Lactate Using Nitrogen-Doped Carbon Nanotubes Modified with Lactate Oxidase. Anal. Chem. 2011;83:8123–8129. doi: 10.1021/ac2016272. [DOI] [PubMed] [Google Scholar]
- [250].Agüí L, Manso J, Yáñez-Sedeño P, Pingarrón JM. Amperometric biosensor for hypoxanthine based on immobilized xanthine oxidase on nanocrystal gold–carbon paste electrodes. Sens. Actuators. 2006;113:272–280. B. [Google Scholar]
- [251].Zardeneta G, Mukai H, Marker V, Milam SB. Protein interactions with particulate teflon: Implications for the foreign body response. J. Oral. Maxillofac. Surg. 1996;54:873–878. doi: 10.1016/s0278-2391(96)90540-6. [DOI] [PubMed] [Google Scholar]
- [252].Devi R, Narang J, Yadav S, Pundir CS. Amperometric determination of xanthine in tea, coffee, and fish meat with graphite rod bound xanthine oxidase. J. Anal. Chem. 2012;67:273–277. doi: 10.1039/c1an15838d. [DOI] [PubMed] [Google Scholar]
- [253].Zhang L, Lei J, Zhang J, Ding L, Ju H. Amperometric detection of hypoxanthine and xanthine by enzymatic amplification using a gold nanoparticles-carbon nanohorn hybrid as the carrier. Analyst. 2012;137:3126–3131. doi: 10.1039/c2an35284b. [DOI] [PubMed] [Google Scholar]
- [254].Devi R, Thakur M, Pundir CS. Construction and application of an amperometric xanthine biosensor based on zinc oxide nanoparticles–polypyrrole composite film. Biosens. Bioelectron. 2011;26:3420–3426. doi: 10.1016/j.bios.2011.01.014. [DOI] [PubMed] [Google Scholar]
- [255].Shan D, Wang Y-N, Xue H-G, Cosnier S, Ding S-N. Xanthine oxidase/laponite nanoparticles immobilized on glassy carbon electrode: Direct electron transfer and multielectrocatalysis. Biosens. Bioelectron. 2009;24:3556–3561. doi: 10.1016/j.bios.2009.05.009. [DOI] [PubMed] [Google Scholar]
- [256].Shan D, Wang Y, Xue H, Cosnier S. Sensitive and selective xanthine amperometric sensors based on calcium carbonate nanoparticles. Sens. Actuators. 2009;136:510–515. B. [Google Scholar]
- [257].Shan D, Wang Y, Zhu M, Xue H, Cosnier S, Wang C. Development of a high analytical performance-xanthine biosensor based on layered double hydroxides modified-electrode and investigation of the inhibitory effect by allopurinol. Biosens. Bioelectron. 2009;24:1171–1176. doi: 10.1016/j.bios.2008.07.023. [DOI] [PubMed] [Google Scholar]
- [258].Kusakabe H, Midorikawa Y, Fujishima T, Kuninaka A, Yoshino H. Purification and Properties of a New Enzyme, L-Glutamate Oxidase, from Streptomyces sp. X-119-6 Grown on Wheat Bran. Agric. Biol. Chem. 1983;47:1323–1328. [Google Scholar]
- [259].Arima J, Sasaki C, Sakaguchi C, Mizuno H, Tamura T, Kashima A, Kusakabe H, Sugio S, Inagaki K. Structural characterization of l-glutamate oxidase from Streptomyces sp. X-119-6. FEBS J. 2009;276:3894–3903. doi: 10.1111/j.1742-4658.2009.07103.x. [DOI] [PubMed] [Google Scholar]
- [260].Jamal M, Xu J, Razeeb KM. Disposable biosensor based on immobilisation of glutamate oxidase on Pt nanoparticles modified Au nanowire array electrode. Biosens. Bioelectron. 2010;26:1420–1424. doi: 10.1016/j.bios.2010.07.071. [DOI] [PubMed] [Google Scholar]
- [261].Özel RE, Ispas C, Ganesana M, Leiter JC, Andreescu S. Glutamate oxidase biosensor based on mixed ceria and titania nanoparticles for the detection of glutamate in hypoxic environments. Biosens. Bioelectron. 2014;52:397–402. doi: 10.1016/j.bios.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Vasylieva N, Maucler C, Meiller A, Viscogliosi H, Lieutaud T, Barbier D, Marinesco S. Immobilization Method to Preserve Enzyme Specificity in Biosensors: Consequences for Brain Glutamate Detection. Anal. Chem. 2013;85:2507–2515. doi: 10.1021/ac3035794. [DOI] [PubMed] [Google Scholar]
- [263].Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis. J. Biol. Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- [264].Espín JC, Wichers HJ. Slow-Binding Inhibition of Mushroom (Agaricus bisporus) Tyrosinase Isoforms by Tropolone. J. Agric. Food. Chem. 1999;47:2638–2644. doi: 10.1021/jf981055b. [DOI] [PubMed] [Google Scholar]
- [265].Seo S-Y, Sharma VK, Sharma N. Mushroom Tyrosinase: Recent Prospects. J. Agric. Food. Chem. 2003;51:2837–2853. doi: 10.1021/jf020826f. [DOI] [PubMed] [Google Scholar]
- [266].Liu F, Piao Y, Choi JS, Seo TS. Three-dimensional graphene micropillar based electrochemical sensor for phenol detection. Biosens. Bioelectron. 2013;50:387–392. doi: 10.1016/j.bios.2013.06.055. [DOI] [PubMed] [Google Scholar]
- [267].Chen L, Gu B, Zhu G, Wu Y, Liu S, Xu C. Electron transfer properties and electrocatalytic behavior of tyrosinase on ZnO nanorod. J. Electroanal. Chem. 2008;617:7–13. [Google Scholar]
- [268].Yu J, Liu S, Ju H. Mediator-free phenol sensor based on titania sol–gel encapsulation matrix for immobilization of tyrosinase by a vapor deposition method. Biosens. Bioelectron. 2003;19:509–514. doi: 10.1016/s0956-5663(03)00227-6. [DOI] [PubMed] [Google Scholar]
- [269].Wang B, Zhang J, Dong S. Silica sol–gel composite film as an encapsulation matrix for the construction of an amperometric tyrosinase-based biosensor. Biosens. Bioelectron. 2000;15:397–402. doi: 10.1016/s0956-5663(00)00096-8. [DOI] [PubMed] [Google Scholar]
- [270].Pavinatto F, Fernandes E, Alessio P, Constantino C, De Saja J, Zucolotto V, Apetrei C, Oliveira O, Jr, Rodriguez-Mendez M. Optimized architecture for Tyrosinase-containing Langmuir–Blodgett films to detect pyrogallol. J. Mater. Chem. 2011;21:4995–5003. [Google Scholar]
- [271].Apetrei C, Alessio P, Constantino CJL, de Saja JA, Rodriguez-Mendez ML, Pavinatto FJ, Fernandes EGR, Zucolotto V, Oliveira ON., Jr Biomimetic biosensor based on lipidic layers containing tyrosinase and lutetium bisphthalocyanine for the detection of antioxidants. Biosens. Bioelectron. 2011;26:2513–2519. doi: 10.1016/j.bios.2010.10.047. [DOI] [PubMed] [Google Scholar]
- [272].Piontek K, Antorini M, Choinowski T. Crystal Structure of a Laccase from the FungusTrametes versicolor at 1.90-Å Resolution Containing a Full Complement of Coppers. J. Biol. Chem. 2002;277:37663–37669. doi: 10.1074/jbc.M204571200. [DOI] [PubMed] [Google Scholar]
- [273].Zouari-Mechichi H, Mechichi T, Dhouib A, Sayadi S, Martínez AT, Martínez MJ. Laccase purification and characterization from Trametes trogii isolated in Tunisia: decolorization of textile dyes by the purified enzyme. Enzyme Microb. Technol. 2006;39:141–148. [Google Scholar]
- [274].Claus H. Laccases: structure, reactions, distribution. Micron. 2004;35:93–96. doi: 10.1016/j.micron.2003.10.029. [DOI] [PubMed] [Google Scholar]
- [275].Portaccio M, Di Tuoro D, Arduini F, Moscone D, Cammarota M, Mita DG, Lepore M. Laccase biosensor based on screen-printed electrode modified with thionine–carbon black nanocomposite, for Bisphenol A detection. Electrochim. Acta. 2013;109:340–347. [Google Scholar]
- [276].Mita D, Attanasio A, Diano N, Grano V, Bencivenga U, Rossi S, Canciglia P, Mita L, Portaccio M, Arduini F. Bioremediation and biodetermination of bisphenol A (BPA) in aqueous solutions. In: Marino M, Mita DG, editors. The endocrine disruptors. Reserach Singpost; 2007. pp. 159–179. [Google Scholar]
- [277].Dempsey E, Diamond D, Collier A. Development of a biosensor for endocrine disrupting compounds based on tyrosinase entrapped within a poly(thionine) film. Biosens. Bioelectron. 2004;20:367–377. doi: 10.1016/j.bios.2004.02.007. [DOI] [PubMed] [Google Scholar]
- [278].Casero E, Petit-Domínguez MD, Vázquez L, Ramírez-Asperilla I, Parra-Alfambra AM, Pariente F, Lorenzo E. Laccase biosensors based on different enzyme immobilization strategies for phenolic compounds determination. Talanta. 2013;115:401–408. doi: 10.1016/j.talanta.2013.05.045. [DOI] [PubMed] [Google Scholar]
- [279].Sarauli D, Riedel M, Wettstein C, Hahn R, Stiba K, Wollenberger U, Leimkühler S, Schmuki P, Lisdat F. Semimetallic TiO2 nanotubes: new interfaces for bioelectrochemical enzymatic catalysis. J. Mater. Chem. 2012;22:4615–4618. [Google Scholar]
- [280].Deng Z, Gong Y, Luo Y, Tian Y. WO3 nanostructures facilitate electron transfer of enzyme: Application to detection of H2O2 with high selectivity. Biosens. Bioelectron. 2009;24:2465–2469. doi: 10.1016/j.bios.2008.12.037. [DOI] [PubMed] [Google Scholar]
- [281].Hulko M, Hospach I, Krasteva N, Nelles G. Cytochrome C biosensor—A model for gas sensing. Sensors. 2011;11:5968–5980. doi: 10.3390/s110605968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Frasca S, Richter C, von Graberg T, Smarsly BM, Wollenberger U. Electrochemical switchable protein-based optical device. Eng. Life Sci. 2011;11:554–558. [Google Scholar]
- [283].Aksu Y, Frasca S, Wollenberger U, Driess M, Thomas A. A molecular precursor approach to tunable porous tin-rich indium tin oxide with durable high electrical conductivity for bioelectronic devices. Chem. Mater. 2011;23:1798–1804. [Google Scholar]
- [284].Renault C, Andrieux CP, Tucker RT, Brett MJ, Balland V.r., Limoges B. Unraveling the Mechanism of Catalytic Reduction of O2 by Microperoxidase-11 Adsorbed within a Transparent 3D-Nanoporous ITO Film. J. Am. Chem. Soc. 2012;134:6834–6845. doi: 10.1021/ja301193s. [DOI] [PubMed] [Google Scholar]
- [285].Kwan P, Schmitt D, Volosin AM, McIntosh CL, Seo D-K, Jones AK. Spectroelectrochemistry of cytochrome c and azurin immobilized in nanoporous antimony-doped tin oxide. Chem. Commun. 2011;47:12367–12369. doi: 10.1039/c1cc14881h. [DOI] [PubMed] [Google Scholar]
- [286].Frasca S, Molero Milan A, Guiet A, Goebel C, Pérez-Caballero F, Stiba K, Leimkühler S, Fischer A, Wollenberger U. Bioelectrocatalysis at mesoporous antimony doped tin oxide electrodes—Electrochemical characterization and direct enzyme communication. Electrochim. Acta. 2013;110:172–180. [Google Scholar]
- [287].Akgöl S, Öztürk N, Alev Karagözler A, Aktaş Uygun D, Uygun M, Denizli A. A new metal-chelated beads for reversible use in uricase adsorption. J. Mol. Catal. B: Enzym. 2008;51:36–41. [Google Scholar]
- [288].Zanon NCM, Oliveira Jr ON, Caseli L. Immbolization of uricase enzyme in Langmuir and Langmuir-Blodgett films of fatty acids: Possible use as a uric acid sensor. J. Colloid Interface Sci. 2012;373:69–74. doi: 10.1016/j.jcis.2011.07.095. [DOI] [PubMed] [Google Scholar]
- [289].Wang Y, Hasebe Y. Uricase-adsorbed carbon-felt reactor coupled with a peroxidase-modified carbon-felt-based H2O2 detector for highly sensitive amperometric flow determination of uric acid. J. Pharm. Biomed. Anal. 2012;57:125–132. doi: 10.1016/j.jpba.2011.08.021. [DOI] [PubMed] [Google Scholar]
- [290].Lin W, Zhang H, Wu J, Wang Z, Sun H, Yuan J, Chen S. A novel zwitterionic copolymer with a short poly(methyl acrylic acid) block for improving both conjugation and separation efficiency of a protein without losing its bioactivity. J. Mater. Chem. B. 2013;1:2482–2488. doi: 10.1039/c3tb00474k. [DOI] [PubMed] [Google Scholar]
- [291].Mora MF, Giacomelli CE, Garcia CD. Interaction of D-Amino Acid Oxidase to Carbon Nanotubes: Implications in the Design of Biosensors. Anal. Chem. 2009;81:1016–1022. doi: 10.1021/ac802068n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Trilling AK, Beekwilder J, Zuilhof H. Antibody orientation on biosensor surfaces: a minireview. Analyst. 2013;138:1619–1627. doi: 10.1039/c2an36787d. [DOI] [PubMed] [Google Scholar]
- [293].Iafisco M, Varoni E, Di Foggia M, Pietronave S, Fini M, Roveri N, Rimondini L, Prat M. Conjugation of hydroxyapatite nanocrystals with human immunoglobulin G for nanomedical applications. Colloids Surf. 2012;90:1–7. doi: 10.1016/j.colsurfb.2011.09.033. B. [DOI] [PubMed] [Google Scholar]
- [294].Zhang Y, Wang H, Yan B, Zhang Y, Li J, Shen G, Yu R. A reusable piezoelectric immunosensor using antibody-adsorbed magnetic nanocomposite. J. Immunol. Meth. 2008;332:103–111. doi: 10.1016/j.jim.2007.12.019. [DOI] [PubMed] [Google Scholar]
- [295].Ho J.-a.A., Chang H-C, Shih N-Y, Wu L-C, Chang Y-F, Chen C-C, Chou C. Diagnostic Detection of Human Lung Cancer-Associated Antigen Using a Gold Nanoparticle-Based Electrochemical Immunosensor. Anal. Chem. 2010;82:5944–5950. doi: 10.1021/ac1001959. [DOI] [PubMed] [Google Scholar]
- [296].Zhou J, Du L, Zou L, Zou Y, Hu N, Wang P. An ultrasensitive electrochemical immunosensor for carcinoembryonic antigen detection based on staphylococcal protein A—Au nanoparticle modified gold electrode. Sens. Actuators. 2014;197:220–227. B. [Google Scholar]
- [297].Moraes ML, Lima LR, Silva RR, Cavicchioli M, Ribeiro SJL. Immunosensor Based on Immobilization of Antigenic Peptide NS5A-1 from HCV and Silk Fibroin in Nanostructured Films. Langmuir. 2013;29:3829–3834. doi: 10.1021/la304404v. [DOI] [PubMed] [Google Scholar]
- [298].Rama EC, González-García MB, Costa-García A. Competitive electrochemical immunosensor for amyloid-beta 1-42 detection based on gold nanostructurated Screen-Printed Carbon Electrodes. Sens. Actuators. 2014;201:567–571. B. [Google Scholar]
- [299].Chikae M, Fukuda T, Kerman K, Idegami K, Miura Y, Tamiya E. Amyloid-β detection with saccharide immobilized gold nanoparticle on carbon electrode. Bioelectrochemistry. 2008;74:118–123. doi: 10.1016/j.bioelechem.2008.06.005. [DOI] [PubMed] [Google Scholar]
- [300].Ardakani MM, Akrami Z, Kazemian H, Zare HR. Electrocatalytic characteristics of uric acid oxidation at graphite–zeolite-modified electrode doped with iron (III) J. Electroanal. Chem. 2006;586:31–38. [Google Scholar]
- [301].Shelyakina M, Soldatkin O, Arkhypova V, Kasap B, Akata B, Dzyadevych S. Study of zeolite influence on analytical characteristics of urea biosensor based on ion-selective field-effect transistors. Nanoscale Res. Lett. 2014;9:124. doi: 10.1186/1556-276X-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Nanduri V, Sorokulova IB, Samoylov AM, Simonian AL, Petrenko VA, Vodyanoy V. Phage as a molecular recognition element in biosensors immobilized by physical adsorption. Biosens. Bioelectron. 2007;22:986–992. doi: 10.1016/j.bios.2006.03.025. [DOI] [PubMed] [Google Scholar]
- [303].Nejadnik MR, Garcia CD. Staining proteins: A simple method to increase the sensitivity of ellipsometric measurements in adsorption studies. Colloids Surf. B. 2011;82:253–257. doi: 10.1016/j.colsurfb.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Bhan C, Mandlewala R, Gebregeorgis A, Raghavan D. Adsorption–Desorption Study of BSA Conjugated Silver Nanoparticles (Ag/BSA NPs) on Collagen Immobilized Substrates. Langmuir. 2012;28:17043–17052. doi: 10.1021/la303539n. [DOI] [PubMed] [Google Scholar]
- [305].Yu CH, Al-Saadi A, Shih S-J, Qiu L, Tam KY, Tsang SC. Immobilization of BSA on Silica-Coated Magnetic Iron Oxide Nanoparticle. J. Phys. Chem. C. 2008;113:537–543. [Google Scholar]
- [306].Edri E, Regev O. pH Effects On BSA-Dispersed Carbon Nanotubes Studied by Spectroscopy-Enhanced Composition Evaluation Techniques. Anal. Chem. 2008;80:4049–4054. doi: 10.1021/ac800124x. [DOI] [PubMed] [Google Scholar]
- [307].Cowsill BJ, Waigh TA, Eapen S, Davies R, Lu JR. Interfacial structure and history dependent activity of immobilised antibodies in model pregnancy tests. Soft Matter. 2012;8:9847–9854. [Google Scholar]
- [308].He C, Liu J, Xie L, Zhang Q, Li C, Gui D, Zhang G, Wu C. Activity and Thermal Stability Improvements of Glucose Oxidase upon Adsorption on Core–Shell PMMA–BSA Nanoparticles. Langmuir. 2009;25:13456–13460. doi: 10.1021/la9019124. [DOI] [PubMed] [Google Scholar]
- [309].He C, Liu J, Zhang Q, Wu C. A novel stable amperometric glucose biosensor based on the adsorption of glucose oxidase on poly(methyl methacrylate)–bovine serum albumin core–shell nanoparticles. Sens. Actuators. 2012;166–167:802–808. B. [Google Scholar]
- [310].Benavidez TE, Garcia CD. Potential-Assisted Adsorption of Bovine Serum Albumin onto Optically-Transparent Carbon Electrodes. Langmuir. 2013;29:14154–14162. doi: 10.1021/la4029657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Koutsioubas A, Lairez D, Zalczer G, Cousin F. Slow and remanent electric polarization of adsorbed BSA layer evidenced by neutron reflection. Soft Matter. 2012;8:2638–2643. [Google Scholar]
- [312].Kaittanis C, Santra S, Perez JM. Emerging nanotechnology-based strategies for the identification of microbial pathogenesis. Adv Drug Deliver Rev. 2010;62:408–423. doi: 10.1016/j.addr.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Srinivasan N, Kumar S. Ordered and disordered proteins as nanomaterial building blocks. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2012;4:204–218. doi: 10.1002/wnan.1160. [DOI] [PubMed] [Google Scholar]
- [314].Hattori T, Umetsu M, Nakanishi T, Sawai S, Kikuchi S, Asano R, Kumagai I. A High-Affinity Gold-Binding Camel Antibody: Antibody Engineering for One-Pot Functionalization of Gold Nanoparticles as Biointerface Molecules. Bioconjugate Chem. 2012;23:1934–1944. doi: 10.1021/bc300316p. [DOI] [PubMed] [Google Scholar]
- [315].Lee SJ, Scotti N, Ravasio N, Chung IS, Song H. Bovine Serum Albumin as an Effective Surface Regulating Biopolymer for Morphology Control of Gold Polyhedrons. Cryst. Growth Des. 2013;13:4131–4137. [Google Scholar]
- [316].Arugula MA, Simonian A. Novel trends in affinity biosensors: current challenges and perspectives. Measurement Science and Technology. 2014;25 032001. [Google Scholar]
- [317].Muyldermans S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.