Abstract
Bone morphogenetic proteins (BMPs) play vital roles in regulating stem cell maintenance and differentiation. BMPs can induce osteogenesis and inhibit myogenesis of mesenchymal stem cells. Canonical BMP signaling is stringently controlled through reversible phosphorylation and nucleocytoplasmic shuttling of Smad1, Smad5, and Smad8 (Smad1/5/8). However, how the nuclear export of Smad1/5/8 is regulated remains unclear. Here we report that the Ran-binding protein RanBP3L acts as a nuclear export factor for Smad1/5/8. RanBP3L directly recognizes dephosphorylated Smad1/5/8 and mediates their nuclear export in a Ran-dependent manner. Increased expression of RanBP3L blocks BMP-induced osteogenesis of mouse bone marrow-derived mesenchymal stem cells and promotes myogenic induction of C2C12 mouse myoblasts, whereas depletion of RanBP3L expression enhances BMP-dependent stem cell differentiation activity and transcriptional responses. In conclusion, our results demonstrate that RanBP3L, as a nuclear exporter for BMP-specific Smads, plays a critical role in terminating BMP signaling and regulating mesenchymal stem cell differentiation.
INTRODUCTION
Bone morphogenetic proteins (BMPs), first identified by their ability to induce bone formation in bone matrix (1), are signal molecules belonging to the transforming growth factor beta (TGF-β) superfamily (2, 3). BMPs have critical roles in skeletal development by regulating osteoblast and chondrocyte differentiation (4), cartilage and bone formation, and limb development (5, 6). BMPs can determine the fate of mesenchymal stem cells by stimulating their differentiation into the chondroosteoblastic lineage and meanwhile blocking their differentiation into the myoblastic lineage (7). In response to BMP signals, critical osteogenic transcription factors such as Runx2 and Osterix are induced and drive efficient bone development (8). On the contrary, BMPs can inhibit myogenic differentiation by suppressing the expression of myogenic basic helix-loop-helix (bHLH) transcriptional factors such as MyoD, myogenin, and Myf5 (9) and/or inducing the expression of Id (inhibitory of differentiation or inhibitor of DNA binding) proteins that block the DNA-binding ability of bHLH transcription factors.
BMP ligands such as BMP2 or BMP4 can bind to type I and type II receptors on the cell surface. The type II receptors phosphorylate and activate the type I receptors, which in turn phosphorylate downstream receptor-regulated Smads (R-Smads), i.e., Smad1, Smad5, and Smad8 (Smad1/5/8) (10, 11). The activated phospho-R-Smads form complexes with Smad4 and translocate into the nucleus. The Smad complex acts as a transcriptional activator or repressor to regulate target gene expression (11–13).
BMP signaling is precisely controlled during development. The level of R-Smads in the nucleus determines the duration and strength of TGF-β superfamily signaling. R-Smads undergo nucleocytoplasmic shuttling, regulated by nuclear transport and retention proteins (14, 15). Ligand-induced phosphorylation of R-Smads facilitates dissociation from cytoplasmic retention, followed by nuclear import and nuclear retention, and conversely, the dephosphorylation and nuclear export of R-Smads shut off TGF-β signaling (16, 17). We recently provided evidence that the nuclear phosphatase PPM1A and the nuclear export factor RanBP3 cooperatively terminate the activities of Smad2/3 (18–20). Although PPM1A can dephosphorylate R-Smads in both TGF-β and BMP signaling pathways, RanBP3 is specifically responsible for the nuclear export of TGF-β-specific Smad2/3 (19). To date, how BMP-specific Smad1/5/8 are transported out of the nucleus remains unclear.
In this study, we report the identification and characterization of a RanBP3-like protein called RanBP3L that mediates the nuclear export of BMP-specific R-Smads. Biochemical and genetic evidence suggests that RanBP3L directly interacts with dephosphorylated Smad1/5/8 in the nucleus and facilitates the nuclear export of dephosphorylated Smad1/5/8. Consequently, the overexpression or knockdown of RanBP3L significantly alters BMP transcriptional responses and mesenchymal stem cell differentiation. These findings elucidate a novel mechanism underlying the termination of BMP-Smad signaling.
MATERIALS AND METHODS
Expression plasmids.
The following mammalian expression plasmids were previously described: hemagglutinin (HA)-, FLAG-, and glutathione S-transferase (GST)-tagged Smad plasmids (21); FLAG-tagged PPM1A plasmid (18); pcDNA-FLAG-Ran (22); and FLAG-tagged RanBP3 (19) plasmid. HA-, FLAG-, or Myc-tagged RanBP3L, RanBP3L-N (amino acids [aa] 1 to 301), and RanBP3L-R (aa 261 to 430) coding sequences were generated by PCR and subcloned into pRK3HA (C-terminal HA tag), pXF6F (N-terminal FLAG tag), and pXF3HM (N-terminal Myc tag) by using EcoRI and SalI sites. All mammalian vectors except pcDNA3 are derived from pRK5 (Genetech). All constructs containing PCR fragments were confirmed by DNA sequencing.
Antibodies and reagents.
Antibodies used in this study include anti-FLAG M2 (catalog number F3165; Sigma), anti-β-actin (catalog number A5441; Sigma), anti-HA (catalog number 3724; Cell Signaling), anti-FLAG M2 magnetic beads (catalog number M8823; Sigma), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (catalog number G8795; Sigma), anti-Smad1 (catalog number 6944; Cell Signaling), anti-Smad2/3 (catalog number 8685; Cell Signaling), anti-phospho-Smad1 (catalog number 9511; Cell Signaling), anti-Runx2 (catalog number D130-3; MBL), anti-Id1 (catalog number 5559-1; Epitomics), anti-Smad6 (catalog number sc-13048; Santa Cruz), anti-MyoD (catalog number sc-304; Santa Cruz), anti-myosin heavy chain (anti-MyHC) (catalog number MAB4470; R&D), anti-Myc (catalog number sc-40; Santa Cruz), anti-lamin A/C (catalog number sc-20681; Santa Cruz), anti-Osterix (catalog number ab22552; Abcam), anti-RanBP3L (catalog number ab116349; Abcam), anti-green fluorescent protein (anti-GFP) sc-9996; Santa Cruz), anti-c-Myc (catalog number 5605; Cell Signaling), anti-PAI-1 (sc-5297; Santa Cruz), antifibronectin (sc-8422; Santa Cruz), anti-RanBP3 (catalog number A301; Bethyl), and anti-collagen II (catalog number Co155; Array BioTech).
Cell lines and transfection.
HEK293T, C3H10T1/2, C2C12, and ATDC5 cells were grown in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Corning) with 10% fetal bovine serum (FBS) (Gibco). HaCaT cells were maintained in minimal essential medium (MEM) (Corning) with 10% FBS. C2C12 cells were cultured in 2% horse serum (Gibco) to induce myogenic induction. Cells were transfected with Lipofectamine 2000 (Invitrogen) and the X-treme Gene HP DNA transfection reagent (Roche). Cells stably expressing RanBP3L were selected with puromycin (2 ng/ml).
Luciferase reporter assays.
Reporter plasmids for Id1-luciferase (Id1-luc), SBE-OC-luc, and SBE-luc were previously described (19). Id1-luc and SBE-OC-luc were used to measure BMP-induced transcription, and SBE-luc was used to measure TGF-β-induced transcription. Cells were transfected with reporter plasmids together with a Renilla luciferase plasmid to normalize the transfection efficiency. Briefly, 24 h after transfection, cells were treated with BMP2 (20 ng/ml) or TGF-β (2 ng/ml) for 12 h. Cells were then harvested, and luciferase activity was measured by using a Dual-Luciferase reporter assay system (Promega). All assays were carried out in triplicates and normalized against Renilla luciferase activity.
Immunoprecipitation and Western blot analysis.
Cells were transfected with the indicated plasmids and harvested 24 h after transfection. Coimmunoprecipitation (co-IP) was carried out by using the appropriate tag antibody and protein A-Sepharose (GE Healthcare). After several washes, precipitated proteins were eluted in SDS loading buffer, separated by SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore), and detected by Western blotting with appropriate antibodies.
Immunofluorescence.
Cells grown on coverslips were fixed with 4% formaldehyde for 20 min and then incubated with 0.3% Triton X-100 and 5% bovine serum albumin (BSA) for 1 h. Cells were subsequently probed with primary antibodies and Alexa Fluor 546- or Alexa Fluor 488-conjugated secondary antibodies (Invitrogen). Florescence images were acquired by the use of a Zeiss LSM710 confocal microscope (Carl Zeiss).
RNA interference and real-time PCR.
Small interfering RNA (siRNA) was transfected into cells by using Lipofectamine RNAiMAX reagent (Invitrogen). siRNAs targeting mouse RanBP3L (siRanBP3L-1 [nucleotides {nt} 950 to 968] [GAGAAGAAACTGAACATAA] and siRanBP3L-2 [nt 552 to 570] [GCACGAGATTTCTGAAGTT]), human RanBP3L (si-hRanBP3L [nt 306 to 324] [GCAAAGTAGTGTTGATATA]), and human RanBP3 (sihRanBP3 [nt 494 to 512] [AGAGCCCCAGAAAAATGAG]) were synthesized by Riobio Co. Total RNAs were isolated by using TRIzol reagent (Life Technologies). Primers for real-time reverse transcription-PCR (RT-PCR) are listed in Table 1.
TABLE 1.
Real-time RT-PCR primers used in this study
| Primer | Sequence |
|---|---|
| m-β-actin-For | TGAGCGCAAGTACTCTGTGTGGAT |
| m-β-actin-Rev | ACTCATCGTACTCCTGCTTGCTGA |
| m-RanBP3L-For | ATACAGCCCTCCCTCAGCTCAAT |
| m-RanBP3L-Rev | ATTGACTGTCTGTGGCTCCACCTT |
| m-ALP-For | AGAAGTTCGCTATCTGCCTTGCCT |
| m-ALP-Rev | TGGCCAAAGGGCAATAACTAGGGA |
| m-Id1-For | AGAACCGCAAAGTGAGCAAGGT |
| m-Id1-Rev | GGTGGTCCCGACTTCAGACT |
| m-Id2-For | ATCCCACTATCGTCAGCCTGCAT |
| m-Id2-Rev | ATTCAGATGCCTGCAAGGACAGGA |
| m-RunX2-For | ATGGCTTGGGTTTCAGGTTAGGGA |
| m-RunX2-Rev | TGGAGTGAAGGATGAGGGCAAACT |
| m-Myf5-For | AGCATTGTGGATCGGATCACGTCT |
| m-Myf5-Rev | TGAGTGTCCTTGAGGATGCCTGT |
| m-MyHC-For | ACGCCATCAGGCTCAAGAAGAAGA |
| m-MyHC-Rev | TGAGTGTCCTTGAGGATGCCTTGT |
| m-Osterix-For | TCCCTTCTCAAGCACCAATGGACT |
| m-Osterix-Rev | AAATGAGTGAGGGAAGGGTGGGTA |
| m-Osteocalcin-For | TAGCAGACACCATGAGGACCATCT |
| m-Osteocalcin-Rev | CCTGCTTGGACATGAAGGCTTTGT |
In vitro protein binding assay.
GST fusions of Smads and deletion proteins were prepared from Escherichia coli strain DE3. In vitro translation of RanBP3L was carried out by using the Quick Coupled transcription/translation system (Promega). In vitro binding was carried out by using GST-tagged Smads incubated with in vitro-translated RanBP3L and its deletion mutants for 2 h in binding buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris-HCl, 5 mM EDTA) and monitored by Western blotting.
Nucleocytoplasmic fractionation and in vitro export assays.
Nucleocytoplasmic fractionation and in vitro export assays were carried out essentially as described previously (16, 19, 20). HeLa-GFP-Smad1 stable cells were first treated with BMP2 to induce nuclear Smad1 accumulation. Cells were permeabilized on ice for 5 min with a 50-μg/ml digitonin solution and then washed three times with transport buffer (20 mM HEPES [pH 7.3], 110 mM potassium acetate, 2 mM magnesium acetate, 2 mM dithiothreitol [DTT], and proteinase inhibitors). Cells were incubated in transport buffer along with the ATP regeneration system (0.5 mM ATP, 0.5 mM GTP, 5 mM creatine phosphate, and 20 U/ml creatine phosphokinase) and BSA or Ran for different time points at 30°C or on ice, respectively. Ran proteins were prepared from anti-FLAG immunoprecipitation of cell lysates from HEK293T cells expressing FLAG-Ran and eluted with FLAG peptide (Sigma). Cells were then rinsed three times with cold transport buffer and lysed with SDS loading buffer. The level of GFP-Smad1 was then analyzed by Western blotting.
Mouse BMSC isolation and in vitro differentiation.
Mouse bone marrow-derived mesenchymal stem cells (BMSCs) were isolated from 10-week-old male C57 mice (23). Cells were flushed out from the femur, filtered, and cultured with 15% FBS in alpha-MEM (Gibco). After several changes of the medium, adhesive cells were considered to be BMSCs, ready for in vitro differentiation (24).
To induce osteogenic differentiation, BMSCs were cultured at 100% confluence in alpha-MEM supplemented with 15% FBS, 50 ng/ml BMP2 (R&D), 50 μg/ml ascorbic acid (Sigma), 10 mM β-glycerophosphate (Sigma), and 10 nM dexamethasone (Sigma). Alkaline phosphatase (ALP) (Sigma) staining and alizarin red S (Sigma) staining were performed at various time points during differentiation.
Myogenic differentiation.
C2C12 cells were cultured in DMEM supplemented with 10% FBS. After reaching 100% confluence, cells were cultured in DMEM with the addition of 2% horse serum (Gibco) to induce myogenesis. Myogenic differentiation was measured by myogenic marker expression and myotube formation.
RESULTS
RanBP3L attenuates Smad1/5/8-mediated transcriptional responses.
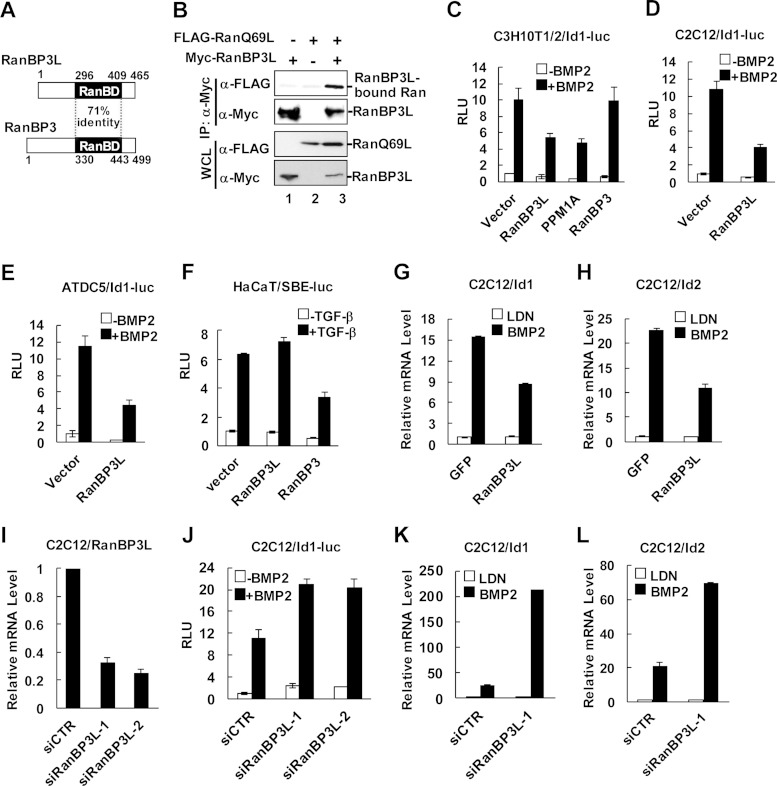
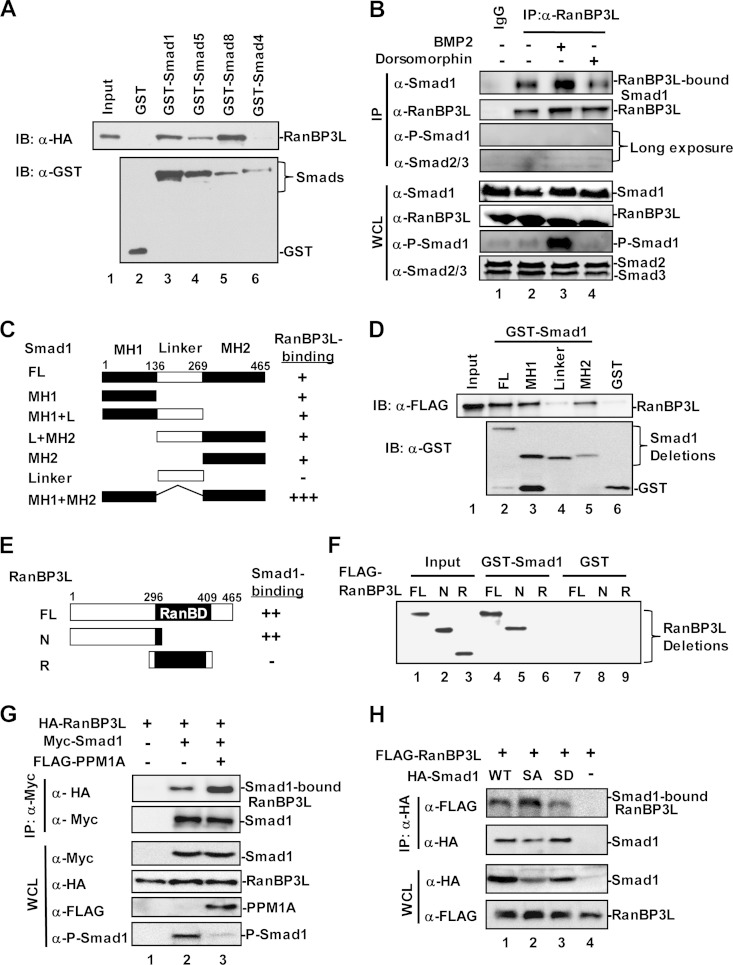
To elucidate the mechanisms underlying Smad1/5/8-dependent signaling termination, we sought to identify a nuclear export factor targeting Smad1/5/8. We reasoned that a RanBP3-like molecule should be a strong candidate for mediating the nuclear export of Smad1/5/8. In the human genome, there is indeed a RanBP3-like molecule, RanBP3L, sharing a high level of homology (71% identity) in the Ran-binding domain (RanBD) with RanBP3 (Fig. 1A). Since Ran is a central regulator of nuclear and cytoplasmic transportation (25, 26) and the Ran GTP-binding form RanGTP facilitates efficient cargo nuclear export (27), we determined whether RanBP3L binds RanGTP. Coimmunoprecipitation experiments showed that RanBP3L bound to RanQ69L (Fig. 1B), a mimetic form of RanGTP (28, 29).
FIG 1.
RanBP3L inhibits BMP-induced transcriptional responses. (A) Schematic diagram of RanBP3L and RanBP3. RanBP3L contains a Ran-binding domain (RanBD) with 71% identity to that of RanBP3. (B) RanBP3L binds with RanGTP. HEK293T cells were cotransfected with Myc-RanBP3L and FLAG-RanQ69L. RanBP3L was immunoprecipitated (IP) with anti-Myc antibody and then subjected to SDS-PAGE and Western blotting to detect RanBP3L-bound RanQ69L. WCL, whole-cell lysate. (C to E) RanBP3L inhibits BMP2-induced Id1-luc reporter activity. C3H10T1/2 (C), C2C12 (D), and ATDC5 cells (E) were transfected with RanBP3L, PPM1A, or RanBP3 together with Id1-luc reporter plasmids. BMP2 treatment and luciferase assays were done as described in Materials and Methods. RLU, relative light units. (F) RanBP3L does not affect TGF-β-induced SBE-luc reporter activity in HaCaT cells. HaCaT cells were transfected with RanBP3 or RanBP3L together with the SBE-luc reporter plasmid and then treated with 2 ng/ml TGF-β for 12 h. The experiment was carried out as described above for panels C to E. (G and H) Stable expression of hRanBP3L in C2C12 cells decreases mRNA levels of Id1 (G) and Id2 (H). Cells were treated with LDN193189 (BMP type I receptor inhibitor) (LDN) or BMP2 (50 ng/ml) for 6 h, and total RNA was extracted for qRT-PCR analysis. (I) RanBP3L knockdown efficacy was measured in C2C12 cells. C2C12 cells were transfected with RanBP3L siRNAs or control siRNA (siCTR), and total RNA was extracted for qRT-PCR analysis. (J) Knockdown of RanBP3L enhances BMP2-induced Id1-luc reporter activity. C2C12 cells were transfected with RanBP3L siRNAs or control siRNA, together with the Id1-luc reporter plasmid. (K and L) Knockdown of RanBP3L increases Id1 mRNA expression (K) and Id2 mRNA expression (L). C2C12 cells were transfected with the indicated siRNAs and treated with BMP2 (50 ng/ml) for 6 h or not treated.
We investigated the effects of RanBP3L on BMP-induced transcriptional responses using reporter assays. The C3H10T1/2 cell line is a mouse multipotent mesenchymal stem cell line that is responsive to BMP. The overexpression of RanBP3L in C3H10T1/2 cells significantly inhibited the BMP-induced transcriptional activation of both the natural Id1 promoter (Fig. 1C) and the synthetic SBE-OC promoter (containing tandem repeats of Smad-binding elements and the minimal osteocalcin promoter) (see Fig. S1A in the supplemental material). PPM1A, a pan-Smad phosphatase that negatively regulates BMP signaling (30), was included as a positive control. We further extended the study to examine the role of RanBP3L in two additional cell lines, myoblastic C2C12 cells and chondrogenic ATDC5 cells. Similar results were obtained, as RanBP3L inhibited BMP-induced Id1-luc reporter expression in both cell types (Fig. 1D and E). In sharp contrast, RanBP3L had no effect on TGF-β-induced transcriptional activation (Fig. 1F). Consistent with our previous report (19), RanBP3 inhibited TGF-β-dependent signaling but did not influence BMP signaling (Fig. 1C and F; see also Fig. S1A in the supplemental material).
To further examine the effects of RanBP3L on BMP-induced gene expression under physiological conditions, we established a C2C12 cell line stably expressing FLAG-tagged RanBP3L by lentiviral infection. Stable expression of RanBP3L in C2C12 cells clearly decreased Id1-luc expression (see Fig. S1B in the supplemental material). We then measured the endogenous mRNA levels of BMP target genes such as Id1, Id2, and alkaline phosphatase (ALP). The mRNAs of Id1 and Id2 were strongly induced by BMP in control cells (expressing GFP), but the induction of these mRNAs was reduced in RanBP3L-expressing C2C12 cells (Fig. 1G and H).
We next investigated whether the knockdown of RanBP3L in C2C12 cells potentiates BMP responses. By using two different siRNAs against RanBP3L, a 70 to 80% efficiency of RanBP3L knockdown was achieved (Fig. 1I). In the accompanying RanBP3L knockdown, BMP-induced Id1-luc reporter expression was increased (Fig. 1J). Notably, the knockdown of RanBP3L dramatically increased the expression levels of BMP downstream genes such as Id1 and Id2 in C2C12 cells (Fig. 1K and L). In contrast, the knockdown of RanBP3L in HaCaT cells had no apparent influence on TGF-β downstream gene expression (see Fig. S1C in the supplemental material). These data together strongly suggested that RanBP3L is a negative regulator of BMP signaling.
RanBP3L inhibits BMP osteogenic responses in C2C12 cells.
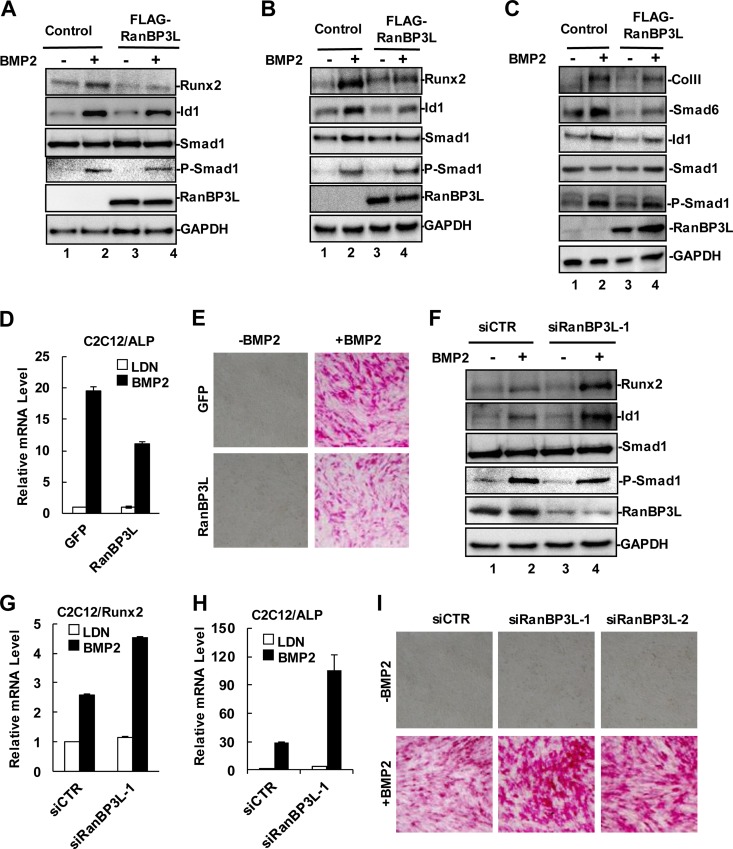
BMPs potently induce the differentiation of mesenchymal cells into osteoblast and chondrocyte lineages in vitro (6). BMP-induced osteogenic differentiation could be reproduced in fibroblasts, myoblasts, and other mesenchymal progenitor cells (31). After BMP2 treatment, C2C12 mesenchymal cells exhibit a phenotype with many osteoblast-like features. We examined the effect of RanBP3L on the expression of osteoblast marker genes. As shown in Fig. 2A, the stable expression of RanBP3L diminished the BMP-mediated induction of Id1 and Runx2 proteins. Similar results were obtained in mesenchymal C3H10T1/2 and chondrogenic ATDC5 cells stably expressing FLAG-RanBP3L (Fig. 2B and C). We further examined BMP-induced ALP levels in C2C12-RanBP3L stable cells and found that RanBP3L greatly reduced BMP-induced ALP mRNA expression (Fig. 2D) and its activity (Fig. 2E).
FIG 2.
RanBP3L inhibits BMP-induced osteoblast-like differentiation of C2C12. (A) RanBP3L decreases BMP-induced osteoblast-like differentiation in C2C12 cells. RanBP3L stable cells and GFP control cells were treated with BMP2 (50 ng/ml) for 30 h or not treated. Cell lysates were measured by Western blotting with the indicated antibodies. GAPDH served as a loading control. (B) RanBP3L decreases BMP-induced osteoblast-like differentiation in C3H10T1/2 cells. BMP2 treatment and Western blotting were done as described above for panel A. (C) RanBP3L decreases BMP-induced osteoblast-like differentiation in ATDC5 cells. BMP2 treatment and Western blotting were done as described above for panel A. (D) RanBP3L inhibits BMP2-induced ALP mRNA levels in C2C12 cells. RanBP3L-OE stable cells and GFP control cells were treated with LDN193189 (LDN) or BMP2 (50 ng/ml) for 6 h. qRT-PCR analysis was carried out as described in the legend to Fig. 1G. (E) RanBP3L decreases BMP2-induced ALP activity in C2C12 cells. The experiment was carried out as described above for panel A. (F) Depletion of RanBP3L enhances BMP-induced osteoblast-like differentiation. C2C12 cells were transfected with the indicated siRNAs and treated with BMP2 (50 ng/ml) for 30 h or not treated. (G) Knockdown of RanBP3L increases Runx2 mRNA levels in C2C12 cells. Transfection and qRT-PCR analyses were carried out as described in the legend to Fig. 1G. (H) Knockdown of RanBP3L increases ALP mRNA levels in C2C12 cells. Transfection and qRT-PCR analyses were carried out as described in the legend to Fig. 1G. (I) Knockdown of RanBP3L enhances BMP-induced ALP activity. C2C12 cells were transfected with the indicated siRNAs and treated with BMP2 (50 ng/ml) for 30 h or not treated.
Since RanBP3L inhibits BMP-induced osteoblast marker expression, we reasoned that RanBP3L depletion could enable cells to be more sensitive to undergoing BMP-induced osteoblast-like responses. As shown in Fig. 2F and G, the BMP2-mediated induction of Runx2 protein and mRNA was profoundly increased in siRanBP3L-transfected cells. In addition, RanBP3L-depleted C2C12 cells exhibit higher ALP mRNA expression levels (Fig. 2H) and activity (Fig. 2I) than did control cells after BMP treatment. These observations further support the negative role of RanBP3L in regulating osteogenic differentiation.
RanBP3L blocks osteoblast differentiation from mouse bone marrow-derived mesenchymal stem cells.
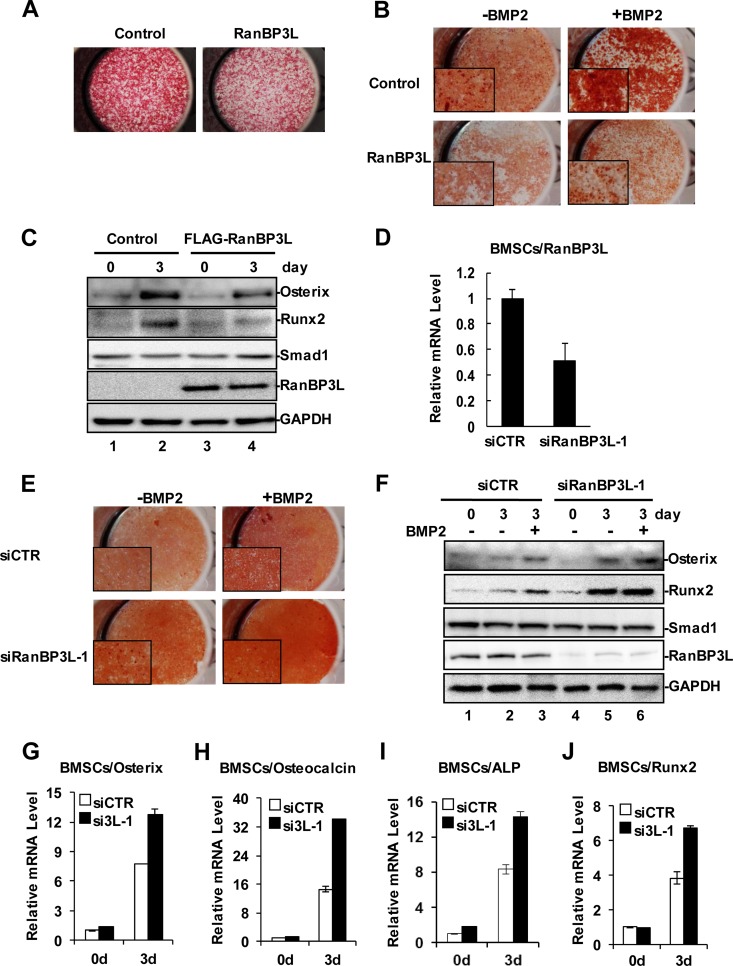
Having established the role of RanBP3L in inhibiting BMP-induced osteogenesis in cell lines, we further examined such effects in primary mouse bone marrow-derived mesenchymal stem cells (BMSCs). Ectopic expression of RanBP3L in primary BMSCs was achieved by lentiviral infection with at least a 90% infection efficiency (see Fig. S2A in the supplemental material). BMSCs can differentiate into mature osteoblasts in osteogenic induction medium, as assessed by mineral staining with alizarin red (see Fig. S2B in the supplemental material). BMP treatment significantly accelerated the osteogenesis of BMSCs. Notably, the overexpression of RanBP3L in BMSCs resulted in weakened ALP activity and alizarin red staining (Fig. 3A and B). Consistently, RanBP3L overexpression also compromised the BMP-induced expression of the preosteoblast marker Runx2 and the osteoblast marker Osterix (Fig. 3C).
FIG 3.
RanBP3L blocks BMP-induced osteogenesis in BMSCs. (A) Overexpression of RanBP3L reduces ALP production in mouse BMSCs. Pools of BMSCs with stably expressing RanBP3L were achieved by lentivirus infection. (B) Overexpression of RanBP3L reduces bone formation in mouse BMSCs. RanBP3L-OE stable cells and control cells were treated with BMP2 (50 ng/ml) for 14 days or not treated. Alizarin red staining was measured as described in Materials and Methods. (C) Overexpression of RanBP3L decreases BMP-promoted osteogenic differentiation in mouse BMSCs. RanBP3L-OE stable cells and control cells were treated with BMP2 (50 ng/ml) for 3 days or not treated. Cells were harvested, and Western blotting was conducted with the indicated antibodies. (D) Knockdown of RanBP3L enhances BMP2-induced ALP activity in BMSCs. BMSCs were transfected with the indicated siRNAs and cultured in osteogenic differentiation medium with or without BMP2 (50 ng/ml). ALP activity was detected as described in the legend to Fig. 2A. (E) Knockdown of RanBP3L enhances BMP2-induced bone formation in mouse BMSCs. BMSCs were transfected with the indicated siRNAs and treated with BMP2 for 9 days to induce osteogenic differentiation or not treated. Alizarin red staining was done as described above for panel B. (F) Depletion of RanBP3L increases BMP-induced osteoblast maturation in mouse BMSCs. BMSCs were transfected with the indicated siRNAs and then treated with BMP2 (50 ng/ml) for 3 days. The osteoblast markers were detected by Western blotting with the indicated antibodies. (G to J) Knockdown of RanBP3L enhances Runx2 (G), Osterix (H), osteocalcin (I), and ALP (J) expression. BMSCs were transfected with the indicated siRNAs, and isolated RNAs were subjected to qRT-PCR analysis as described in the legend to Fig. 1G.
We further determined the effects of RanBP3L knockdown on the osteogenesis of BMSCs. As shown in Fig. S2C in the supplemental material, we achieved an ∼50% knockdown efficiency 5 days after siRNA transfection in BMSCs. Results from ALP staining and alizarin red staining showed that the knockdown of RanBP3L accelerated the osteogenic differentiation of BMSCs (Fig. 3D and E). We also evaluated the expressions of several osteogenic markers in RanBP3L-depleted BMSCs. As assessed by Western blotting (Fig. 3F), Runx2 and Osterix proteins were markedly induced by BMP2 and were further enhanced by the knockdown of RanBP3L. In accordance, quantitative RT-PCR (qRT-PCR) analysis demonstrated that the knockdown of RanBP3L enabled BMSCs to be sensitized to BMP, as BMP2 induced higher levels of mRNAs of osteogenic markers, including Runx2, Osterix, osteocalcin, and ALP (Fig. 3G to J). Taken together, the data support the notion that RanBP3L blocks BMP-induced osteogenic differentiation of primary BMSCs.
RanBP3L promotes myogenic differentiation.
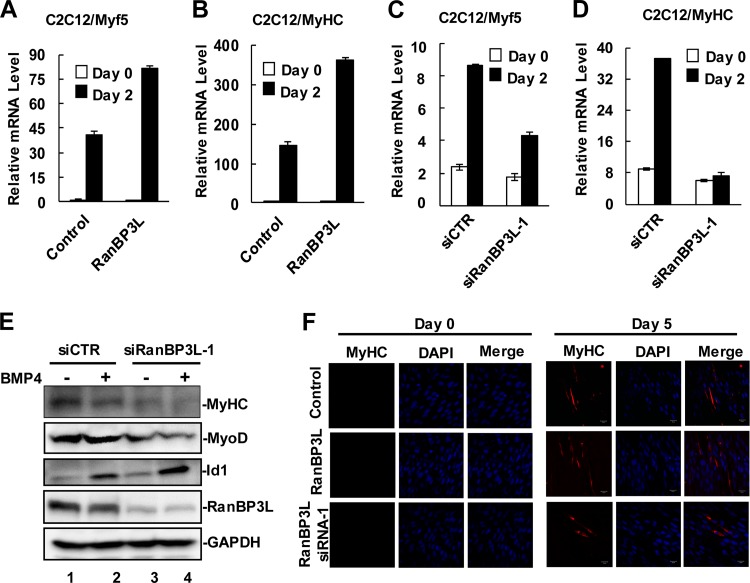
BMPs are potential inhibitors of myogenesis through the BMP-mediated suppression of myogenic bHLH transcriptional factors and the induction of Id1/Id2 expression. We thus examined whether RanBP3L, as an inhibitor of BMP signaling, promotes the myogenic differentiation of C2C12 cells. We measured the levels of myosin heavy chain (MyHC) and Myf5 as well as myotube formation, which are typical indicators of mature myogenesis, in C2C12 cells stably expressing RanBP3L. In myogenesis induction medium, C2C12 cells typically exhibited induced expression of Myf5 and MyHC (Fig. 4A and B). Notably, the mRNA levels of Myf5 and MyHC were markedly increased in C2C12 cells expressing RanBP3L in comparison to control cells (Fig. 4A and B). Accordingly, more myotube formation was observed in RanBP3L cells (Fig. 4F).
FIG 4.
RanBP3L promotes myogenic differentiation. (A and B) RanBP3L enhances mRNA levels of the myogenic markers Myf5 (A) and MyHC (B) during myogenic induction of C2C12 cells. RanBP3L-OE stable cells and control cells were cultured in DMEM with the addition of 2% horse serum (Gibco) to induce myogenesis. On day 2, total RNAs were isolated and analyzed by qRT-PCR. (C and D) Depletion of RanBP3L decreases mRNA levels of Myf5 (C) and MyHC (D) during myogenic differentiation. C2C12 cells were transfected with the indicated RanBP3L siRNAs or control siRNA. Experiments were performed as described above for panel A. (E) Knockdown of RanBP3L reduces myogenic marker gene expression. C2C12 cells were transfected with the indicated siRNAs and harvested 3 days after myogenic induction. Cell lysates were measured by Western blotting with the indicated antibodies. (F) RanBP3L promotes myotube formation. RanBP3L-OE, RanBP3L-KD, and control C2C12 cells were cultured in myogenic differentiation medium for 5 days, fixed, immunostained with anti-MyHC antibody, and imaged by confocal microscopy. DAPI, 4′,6-diamidino-2-phenylindole.
Next, we examined the effects of RanBP3L knockdown on myoblast differentiation. As shown in Fig. 4C and D, the mRNA levels of Myf5 and MyHC were reduced in RanBP3L-depleted cells. In addition, in C2C12 cells transfected with RanBP3L siRNA, we noticed that whereas Id1 was induced, MyoD and MyHC protein levels were reduced (Fig. 4E). Indeed, fewer myotubes were found in siRanBP3L-transfected cells (Fig. 4F). These data demonstrated that RanBP3L promotes myogenic differentiation through the inhibition of BMP signaling.
RanBP3L physically interacts with Smad1/5/8.
With its high level of similarity to RanBP3, which interacts with Smad2/3 and mediates Smad2/3 export (19), RanBP3L may play a role in Smad nucleocytoplasmic shuttling. We thus tested whether RanBP3L might interact with BMP-specific Smad1/5/8. We carried out an in vitro binding assay using purified recombinant GST-Smad fusion proteins and in vitro-translated RanBP3L. As shown in Fig. 5A, RanBP3L could bind to GST-Smad1/5/8 but not GST-Smad4 or GST protein (lanes 2, 3, and 6). We also carried out co-IP experiments in HEK293T cells to test their interaction in vivo. RanBP3L strongly interacted with BMP-specific Smads (Smad1/5/8) (see Fig. S3A in the supplemental material) but weakly or barely bound Smad2/3 and Smad4 (see Fig. S3A, lanes 4 to 6). In order to validate their interactions, co-IP experiments were performed to examine the endogenous RanBP3L-Smad interaction in C2C12 cells. We found that Smad1, but not Smad2/3, could be detected in the RanBP3L immunocomplex, suggesting that the RanBP3L-Smad1 interaction is specific (Fig. 5B).
FIG 5.
RanBP3L physically interacts with Smad1/5/8. (A) RanBP3L directly binds to Smad1, -5, and -8 but not Smad4 in vitro. In vitro binding was carried out with purified GST-tagged Smads and in vitro-translated RanBP3L. IB, immunoblot. (B) RanBP3L interacts with Smad1 in vivo. C2C12 cells were treated with BMP2 or dorsomorphin for 2 h. Cell lysates were immunoprecipitated with anti-RanBP3L antibody or control IgG antibody. The immunocomplexes and input were analyzed by Western blotting with the indicated antibodies. (C and D) RanBP3L binds to both the MH1 and MH2 domains of Smad1. In vitro binding assays were carried out as described above for panel A. FL, full length. (E and F) Smad1 binds to the N terminus of RanBP3L. Experiments were performed as described above for panel A. (G) PPM1A enhances the interaction between RanBP3L and Smad1. HEK293T cells were transfected with the indicated plasmids. Levels of these proteins in IP products and whole-cell lysates were analyzed by Western blotting. (H) RanBP3L has a higher binding affinity for the dephosphorylated form of Smad1. HEK293T cells were cotransfected with HA-RanBP3L, together with FLAG-Smad1 or its mutations (Smad1-2SA [unphosphorylated form] or Smad1-2SD [phosphorylation-mimicking form]). Levels of these proteins in IP products and whole-cell lysates were analyzed by Western blotting.
Smads are structurally conserved proteins consisting of an MH1 domain in the N terminus and an MH2 domain in the C terminus, linked with a relatively less conserved linker region (Fig. 5C). Our in vitro binding assay indicates that either the MH1 or MH2 domain alone, but not the linker region, can bind to RanBP3L (Fig. 5D, lanes 3 to 5). Further co-IP assays revealed that Smad1 deletion mutants with MH1 deleted (L+MH2) or MH2 deleted (MH1+L) retained the ability to bind with RanBP3L (see Fig. S3B in the supplemental material). Notably, deletion of the linker (MH1+MH2) significantly enhanced Smad1 binding to RanBP3L. These results suggest that RanBP3L binds to both the MH1 and MH2 domains.
RanBP3L contains a conserved RanBD (Fig. 5E), which is characteristic of all Ran-binding proteins. Using GST-Smad1 to pull down various mutants of RanBP3L, we found that GST-Smad1 interacted with full-length RanBP3L and the N-terminal region of RanBP3L (Fig. 5F), indicating that the N terminus of RanBP3L contains a potential Smad-binding domain to mediate its interaction with Smad1. In our previous study, we found that RanBP3 interacted with Smad2/3 mainly through its structurally conserved RanBD (19). Considering the structural similarity of RanBD on RanBP3L and RanBP3, it is understandable that RanBP3L weakly interacts Smad2/3. However, this weak interaction took place only under overexpression conditions (see Fig. S3A in the supplemental material); we could not detect their endogenous interaction in C2C12 cells (Fig. 5B).
RanBP3L prefers to bind dephosphorylated Smad1 in the nucleus.
It is generally believed that R-Smads need to be dephosphorylated before their nuclear export. Does RanBP3L bind to the dephosphorylated form of Smad1/5/8? To address this question, we analyzed the RanBP3L-Smad1 interaction in the presence of PPM1A, which is a phosphatase for Smad1 dephosphorylation (30). The results showed that PPM1A enhanced the association between RanBP3L and Smad1 (Fig. 5G, lane 3). Similarly, the overexpression of PPM1A increased the interaction between RanBP3L and Smad5 (see Fig. S3C, lane 3, in the supplemental material) or Smad8 (see Fig. S3D, lanes 3 and 4). We further tested whether phosphorylated Smad1/5/8 (P-Smad1/5/8) were in the RanBP3L immunocomplex. As shown in Fig. 5B, no P-Smad1/5/8 signal was detected even with an extremely long exposure time. Since PPM1A acts on the SXS motif of Smad1, we generated mutants with Ser-to-Ala and Ser-to-Asp substitutions, i.e., phosphorylation-defective Smad1-2SA and phosphorylation-mimicking Smad1-2SD. We found that RanBP3L preferably interacts with Smad1-2SA (Fig. 5H, lane 2). This further illustrated that dephosphorylated Smad1 is a favored cargo of RanBP3L.
RanBP3L mediates Smad1 nuclear export in a Ran-dependent manner.
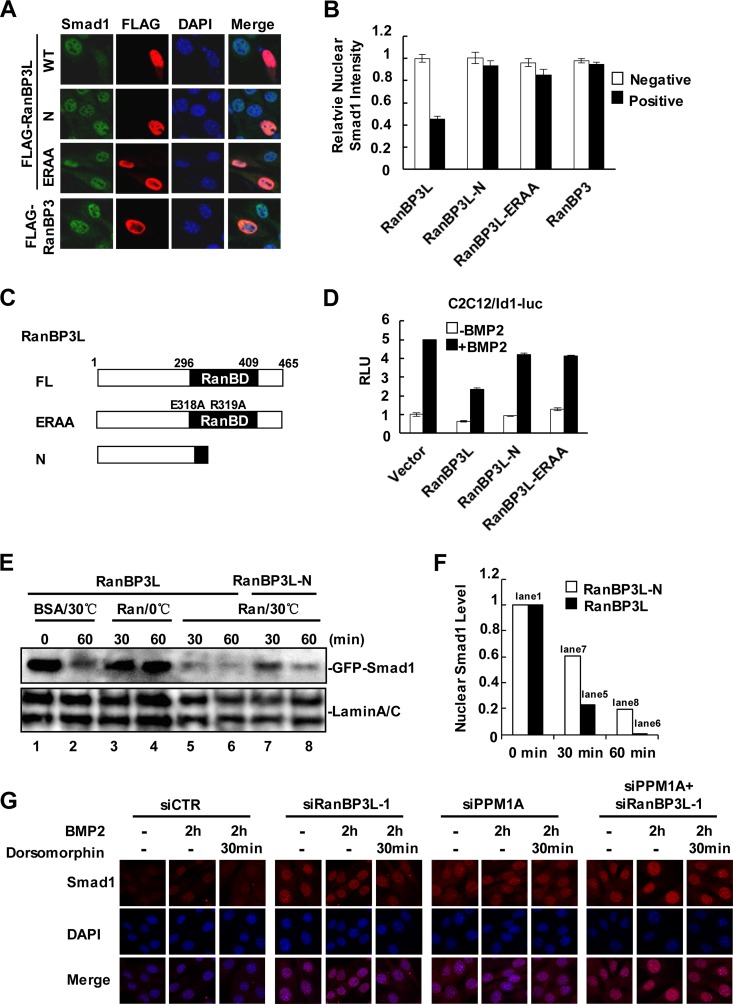
RanBPs bind to the small GTPase Ran and participate in protein transport across the nuclear membrane (32, 33). To test the role of RanBP3L in regulating Smad1 nuclear transport, we examined the subcellular distribution of endogenous Smad1 in C2C12 cells with increased or depleted expression of RanBP3L. As shown in Fig. 6A, whereas BMP induced the nuclear accumulation of endogenous Smad1, the level of nuclear Smad1 was markedly reduced in cells expressing RanBP3L. Quantitative analysis revealed that the nuclear Smad1 intensity decreased >50% in cells expressing RanBP3L (Fig. 6B). This result was also observed for C2H10T1/2 cells (see Fig. S4A in the supplemental material). In addition, we generated RanBP3L-ERAA by mutating two conserved amino acids in the RanBD of RanBP3L to disrupt its interaction with Ran-GTP. This mutant and RanBP3L-N lacking the RanBD exhibited no effect on the Smad1 nuclear distribution (Fig. 6A to C). It is noteworthy that the Ran-binding-defective mutants of RanBP3L also failed to repress BMP-induced Id1-luc expression (Fig. 6D).
FIG 6.
RanBP3L enhances the nuclear export of Smad1. (A) RanBP3L promotes Smad1 nuclear export in C2C12 cells. C2C12 cells were transfected with the indicated plasmids. After BMP2 treatment (50 ng/ml for 4 h), cells were fixed and immunostained with anti-Smad1 and anti-FLAG antibodies. DNA was stained with DAPI (4′,6-diamidino-2-phenylindole). WT, wild type. (B) Quantification of nuclear Smad1. Cells were transfected with RanBP3L, RanBP3L-N, RanBP3L-ERAA, or RanBP3 and subjected to immunostaining of Smad1. (C) Schematic diagram of RanBP3L mutants. RanBP3L-ERAA contains substitutions at E318A and R319A. (D) RanBP3L-N and RanBP3L-ERAA fail to inhibit BMP2-induced Id1-luc reporter activity. Experiments were carried out as described in the legend to Fig. 1C. (E) In vitro export assay of Smad1. HeLa cells stably expressing GFP-Smad1 were transfected with RanBP3L or the RanBP3L-N mutant, permeabilized with digitonin (50 ng/ml) for 5 min, and incubated in a reaction buffer with BSA or Ran at 30°C or on ice. Nuclear GFP-Smad1 was measured by Western blotting. (F) Relative GFP-Smad1 levels in panel E (lanes 1 and 5 to 8) were quantified by using ImageJ software. (G) C2C12 cells were transfected with the indicated siRNAs and treated with BMP2 for 2 h or not treated. The ligands were then washed away with phosphate-buffered saline, and dorsomorphin was added for 30 min before fixation. Smad1 was stained with anti-Smad1 antibody. DNA was stained with DAPI.
The requirement of Ran binding for Smad1 nuclear export was further confirmed by using an in vitro export assay (34, 35). We generated HeLa cells stably expressing GFP-Smad1 protein. GFP-Smad1 stable cells were transfected with RanBP3L and the RanBP3L-N mutant and treated with the appropriate concentration of digitonin (50 μg/ml) to permeabilize the cell membrane and yet keep the nucleus intact (see Fig. S4B in the supplemental material). After washing, the presence of GFP-Smad1 in the nucleus could be analyzed. By adding affinity-purified RanQ69L or bovine serum albumin (BSA), we evaluated the remaining nuclear GFP-Smad1. In the presence of transfected RanBP3L, nuclear GFP-Smad1 proteins were partially exported out of the nucleus by endogenous Ran-GTP (Fig. 6E, lanes 1 and 2), suggesting that this system works well. The addition of RanQ69L significantly enhanced the RanBP3L-mediated export of GFP-Smad1 (Fig. 6E, lanes 5 and 6). Regardless of the presence of RanQ69L, RanBP3L-N showed a weakened ability to transport GFP-Smad1 out of the nucleus (Fig. 6E, lanes 7 and 8, and F). These data strongly supported that RanBP3L regulates Smad1 nuclear export in a Ran-dependent manner.
In agreement with the specific role of RanBP3 in Smad2/3 nuclear export, ectopic expression of RanBP3 exhibited no effect on the Smad1 nuclear intensity (Fig. 6A and B). Conversely, the overexpression of RanBP3L did not affect the Smad2/3 nuclear localization, even in the absence of RanBP3 interference, i.e., under RanBP3 knockdown conditions (see Fig. S4C in the supplemental material). To further demonstrate the distinct functions of RanBP3L and RanBP3, we knocked down the expression of RanBP3L or RanBP3 in HaCaT cells and compared the nuclear export of Smad1/5/8 and Smad2/3. As shown in Fig. S4D in the supplemental material, the knockdown of RanBP3L but not of RanBP3 clearly enhanced the nuclear retention of Smad1 in response to BMP2. After the administration of dorsomorphin (i.e., BMP-type receptor ALK3/6 inhibitor), Smad1 rapidly diffused to the cytoplasm. The knockdown of RanBP3L but not of RanBP3 retained Smad1 in the nucleus, whereas Smad2/3 nuclear retention was enhanced only by RanBP3 knockdown (see Fig. S4E in the supplemental material).
Our previously reported data suggested that the dephosphorylation of Smads in the nucleus is a prerequisite step for Smad nuclear export (30). As shown in Fig. 6G, the knockdown of RanBP3L induced the nuclear accumulation of Smad1. Similarly, the knockdown of PPM1A also induced the nuclear accumulation of Smad1. The double knockdown of both RanBP3L and PPM1A dramatically retained Smad1 in the nucleus. These data demonstrate that both RanBP3L and PPM1A are involved in the nuclear export of BMP-specific Smads.
DISCUSSION
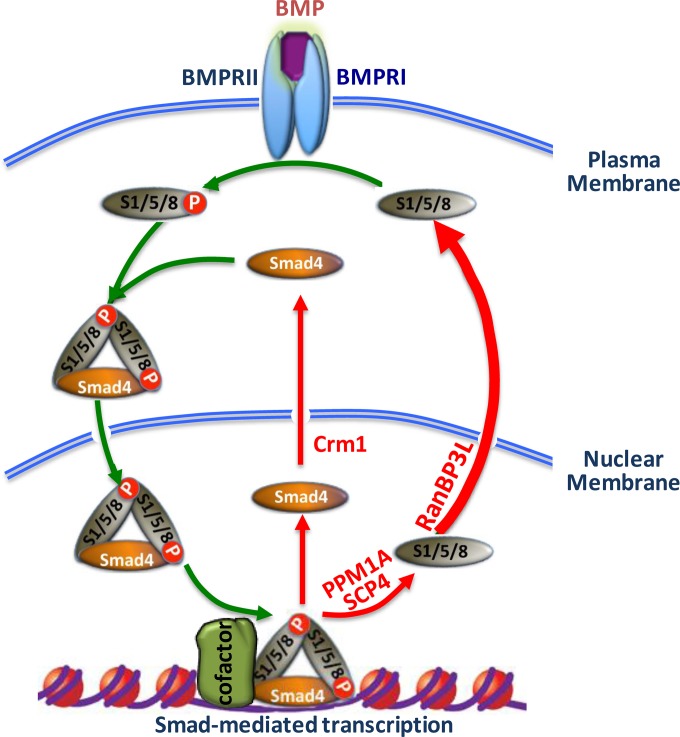
Precise control of activation and termination of TGF-β/BMP signaling is of paramount importance in maintaining proper cellular functions and tissue homeostasis. TGF-β/BMP signal termination can be regulated through several mechanisms, including reversible phosphorylation and degradation of receptors or Smad proteins and nucleocytoplasmic shuttling of Smads. We previously reported the critical role of RanBP3 in turning off TGF-β signaling (19). RanBP3 selectively targets Smad2/3 in the TGF-β pathway but not Smad1/5/8 in the BMP pathway. The exact export mechanism for Smad1/5/8 remains to be elucidated. In this study, we have identified RanBP3L as a key factor that mediates Smad1/5/8 nuclear export (Fig. 7) and regulates BMP signaling and mesenchymal stem cell differentiation.
FIG 7.
Recycling of Smad1/5/8 through RanBP3L-mediated nuclear export. Upon BMP stimulation (the BMP-bound BMPRI-BMPRII complex is shown), Smad1/5/8 become phosphorylated and form a trimeric complex with Smad4. The complex is then translocated into the nucleus, where it binds to chromatin. During signal termination (red arrows), Smad1/5/8 are dephosphorylated by PPM1A and SCP4. Whereas Smad4 is exported by a Crm1-dependent mechanism, the present study clearly demonstrates that RanBP3L drives the nuclear export of dephosphorylated Smad1/5/8. Signal transduction may be reinitiated, dependent on BMP receptor activity.
RanBP3L is an uncharacterized putative protein encoded by the human genome. Our study provides the first evidence for its molecular functions. The open reading frame contains a highly conserved RanBD, which is closely related to that of RanBP3, with an identity of 71% (Fig. 1A). Like RanBP3, RanBP3L can associate with RanGTP (Fig. 1B). RanBP3L directly binds to dephosphorylated Smad1 and functions in a Ran-dependent manner. Whereas its overexpression renders less nuclear accumulation of Smad1, its depletion by siRNA retains more Smad1 in the nucleus. Although RanBP3 (and perhaps RanBP3L) is considered a CRM1 cofactor (36), RanBP3L can mediate Smad1 nuclear export independent of CRM1. In a previous study, CRM1 was reported to mediate the nuclear export of Smad1, which can be inhibited by leptomycin B (37). However, we found that leptomycin B exhibited little effect on the nuclear accumulation of Smad1 in C3H10T1/2 cells (data not shown). In addition, the combination of RanBP3L depletion together with CRM1 inhibition did not cause more nuclear retention of Smad1 than did RanBP3L depletion alone, which implies that RanBP3L may function in a CRM1-independent manner. By using mass spectrometry analysis (data not shown), RanBP3L was found to interact with many nucleoporins and transport proteins, suggesting that RanBP3L may directly interact with the nuclear pore complex to achieve its transport function.
Given the fact that RanBP3 and RanBP3L specifically terminate TGF-β and BMP signaling, respectively, what is the underlying basis for determining the specificity? The combined MH1 and MH2 domains of Smads bind to the closely related RanBP3 or RanBP3L. However, the clear difference between RanBP3 and RanBP3L is their Smad-binding domain. The RanBD of RanBP3 binds directly to Smad2/3. How the highly conserved RanBD recognizes Smad2/3 but not Smad1/5/8 is not clear. In sharp contrast, RanBP3L binds to Smad1/5/8 via its N-terminal region, which may serve as a regulatory domain, even though its N terminus is shorter than that of RanBP3. Although the longer N terminus of RanBP3 is not used for Smad binding, it may have regulatory activities. Serine 58 in this region of RanBP3 has been reported to directly influence its transport activity (20, 38). In addition to its Smad-binding activity, other possible regulatory activities of the N-terminal domain of RanBP3L await further elucidation.
BMPs regulate mesenchymal stem cell differentiation through promoting osteogenic and chondrogenic differentiation and meanwhile blocking myogenic and adipogenic differentiation. This process is tightly controlled by a range of extracellular, intracellular, and nuclear modulators. As a nuclear export factor for Smad1/5/8, RanBP3L is anticipated to facilitate the termination of BMP-induced cellular responses during mesenchymal differentiation. By both gain-of-expression and loss-of-expression approaches, we demonstrated that RanBP3L decreases the osteogenic potential of BMSCs and C2C12 cells. In addition, we noticed that RanBP3L also inhibits chondrogenic differentiation by downregulating Sox9 and collagen II mRNA transcription in response to BMP (data not shown). These results together suggest that RanBP3L plays a critical role in balancing BMP-regulated lineage-specific differentiation of mesenchymal stem cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Di Chen for SBE-OC-luc and ATDC5 cells, Peter ten Dijke for Id1-luc, and Bert Vogelstein for SBE-luc. We are grateful to members of the Feng laboratory for helpful discussion.
This research was partly supported by grants from MOST (2012CB966600), NSFC (31090360), the NIH (R01AR053591, R01GM63773, R01CA108454, and R01DK073932), the 111 Project (B13026), Project 985, and Fundamental Research Funds for the Central Universities.
F.C. performed most of the experiments and participated in experimental design, data analysis and manuscript writing. X.L. and M.X. participated in experimental design, data analysis, and manuscript writing. Z.Z. (under the supervision of X.L.), Y.C., and C.W. (under the supervision of M.X.) provided essential research materials. P.X., J.H., and B.Z. provided research materials and participated in manuscript writing. X.-H.F. conceived of, designed, and directed the project, analyzed the data, and wrote the paper.
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00121-15.
REFERENCES
- 1.Urist MR. 1965. Bone: formation by autoinduction. Science 150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 2.Guo X, Wang XF. 2009. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res 19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyazono K, Maeda S, Imamura T. 2005. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev 16:251–263. doi: 10.1016/j.cytogfr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Yoon BS, Lyons KM. 2004. Multiple functions of BMPs in chondrogenesis. J Cell Biochem 93:93–103. doi: 10.1002/jcb.20211. [DOI] [PubMed] [Google Scholar]
- 5.Karsenty G, Wagner EF. 2002. Reaching a genetic and molecular understanding of skeletal development. Dev Cell 2:389–406. doi: 10.1016/S1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 6.Wan M, Cao X. 2005. BMP signaling in skeletal development. Biochem Biophys Res Commun 328:651–657. doi: 10.1016/j.bbrc.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Shi W, Cao X. 2007. Multiplicity of BMP signaling in skeletal development. Ann N Y Acad Sci 1116:29–49. doi: 10.1196/annals.1402.053. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. 2000. A RUNX2/PEBP2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci U S A 97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katagiri T, Akiyama S, Namiki M, Komaki M, Yamaguchi A, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. 1997. Bone morphogenetic protein-2 inhibits terminal differentiation of myogenic cells by suppressing the transcriptional activity of MyoD and myogenin. Exp Cell Res 230:342–351. doi: 10.1006/excr.1996.3432. [DOI] [PubMed] [Google Scholar]
- 10.Feng XH, Derynck R. 2005. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol 21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 11.ten Dijke P, Hill CS. 2004. New insights into TGF-beta-Smad signalling. Trends Biochem Sci 29:265–273. doi: 10.1016/j.tibs.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Wu MY, Hill CS. 2009. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell 16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Feng XH, Lin X, Derynck R. 2000. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J 19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicolas FJ, De Bosscher K, Schmierer B, Hill CS. 2004. Analysis of Smad nucleocytoplasmic shuttling in living cells. J Cell Sci 117:4113–4125. doi: 10.1242/jcs.01289. [DOI] [PubMed] [Google Scholar]
- 15.Schmierer B, Tournier AL, Bates PA, Hill CS. 2008. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci U S A 105:6608–6613. doi: 10.1073/pnas.0710134105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai F, Duan X, Liang YY, Lin X, Feng XH. 2010. Coupling of dephosphorylation and nuclear export of Smads in TGF-beta signaling. Methods Mol Biol 647:125–137. doi: 10.1007/978-1-60761-738-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Xu L. 2011. Mechanism and regulation of nucleocytoplasmic trafficking of Smad. Cell Biosci 1:40. doi: 10.1186/2045-3701-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen YG, Meng A, Feng XH. 2006. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell 125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai F, Lin X, Chang C, Feng XH. 2009. Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-beta signaling. Dev Cell 16:345–357. doi: 10.1016/j.devcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai F, Shen T, Li Z, Lin X, Feng XH. 2011. PPM1A dephosphorylates RanBP3 to enable efficient nuclear export of Smad2 and Smad3. EMBO Rep 12:1175–1181. doi: 10.1038/embor.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng XH, Liang YY, Liang M, Zhai W, Lin X. 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-β-mediated induction of the CDK inhibitor p15(Ink4B). Mol Cell 9:133–143. doi: 10.1016/S1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- 22.Giri DK, Ali-Seyed M, Li LY, Lee DF, Ling P, Bartholomeusz G, Wang SC, Hung MC. 2005. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol Cell Biol 25:11005–11018. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soleimani M, Nadri S. 2009. A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nat Protoc 4:102–106. doi: 10.1038/nprot.2008.221. [DOI] [PubMed] [Google Scholar]
- 24.Huang J, Zhao L, Xing L, Chen D. 2010. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avis JM, Clarke PR. 1996. Ran, a GTPase involved in nuclear processes: its regulators and effectors. J Cell Sci 109(Part 10):2423–2427. [DOI] [PubMed] [Google Scholar]
- 26.Moore MS. 1996. Generation of GTP-Ran for nuclear protein import. Science 272:47. doi: 10.1126/science.272.5258.47. [DOI] [PubMed] [Google Scholar]
- 27.Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. 1994. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci U S A 91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D. 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart M, Kent HM, McCoy AJ. 1998. The structure of the Q69L mutant of GDP-Ran shows a major conformational change in the switch II loop that accounts for its failure to bind nuclear transport factor 2 (NTF2). J Mol Biol 284:1517–1527. doi: 10.1006/jmbi.1998.2204. [DOI] [PubMed] [Google Scholar]
- 30.Duan X, Liang YY, Feng XH, Lin X. 2006. Protein serine/threonine phosphatase PPM1A dephosphorylates Smad1 in the bone morphogenetic protein signaling pathway. J Biol Chem 281:36526–36532. doi: 10.1074/jbc.M605169200. [DOI] [PubMed] [Google Scholar]
- 31.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. 1994. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol 127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dingwall C, Kandels-Lewis S, Seraphin B. 1995. A family of Ran binding proteins that includes nucleoporins. Proc Natl Acad Sci U S A 92:7525–7529. doi: 10.1073/pnas.92.16.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mueller L, Cordes VC, Bischoff FR, Ponstingl H. 1998. Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS Lett 427:330–336. doi: 10.1016/S0014-5793(98)00459-1. [DOI] [PubMed] [Google Scholar]
- 34.Love DC, Sweitzer TD, Hanover JA. 1998. Reconstitution of HIV-1 rev nuclear export: independent requirements for nuclear import and export. Proc Natl Acad Sci U S A 95:10608–10613. doi: 10.1073/pnas.95.18.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsay YG, Lin NY, Voss PG, Patterson RJ, Wang JL. 1999. Export of galectin-3 from nuclei of digitonin-permeabilized mouse 3T3 fibroblasts. Exp Cell Res 252:250–261. doi: 10.1006/excr.1999.4643. [DOI] [PubMed] [Google Scholar]
- 36.Lindsay ME, Holaska JM, Welch K, Paschal BM, Macara IG. 2001. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol 153:1391–1402. doi: 10.1083/jcb.153.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao Z, Watson N, Rodriguez C, Lodish HF. 2001. Nucleocytoplasmic shuttling of Smad1 conferred by its nuclear localization and nuclear export signals. J Biol Chem 276:39404–39410. doi: 10.1074/jbc.M103117200. [DOI] [PubMed] [Google Scholar]
- 38.Yoon SO, Shin S, Liu Y, Ballif BA, Woo MS, Gygi SP, Blenis J. 2008. Ran-binding protein 3 phosphorylation links the Ras and PI3-kinase pathways to nucleocytoplasmic transport. Mol Cell 29:362–375. doi: 10.1016/j.molcel.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.