Abstract
In S phase, the replication and transcription of genomic DNA need to accommodate each other, otherwise their machineries collide, with chromosomal instability as a possible consequence. Here, we characterized the human replication fork barrier (RFB) that is present downstream from the 47S pre-rRNA gene (ribosomal DNA [rDNA]). We found that the most proximal transcription terminator, Sal box T1, acts as a polar RFB, while the other, Sal box T4/T5, arrests replication forks bidirectionally. The fork-arresting activity at these sites depends on polymerase I (Pol I) transcription termination factor 1 (TTF-1) and a replisome component, TIMELESS (TIM). We also found that the RFB activity was linked to rDNA copies with hypomethylated CpG and coincided with the time that actively transcribed rRNA genes are replicated. Failed fork arrest at RFB sites led to a slowdown of fork progression moving in the opposite direction to rRNA transcription. Chemical inhibition of transcription counteracted this deceleration of forks, indicating that rRNA transcription impedes replication in the absence of RFB activity. Thus, our results reveal a role of RFB for coordinating the progression of replication and transcription activity in highly transcribed rRNA genes.
INTRODUCTION
The rRNA gene, ribosomal DNA (rDNA), encodes RNA components of ribosomes. In the human genome, there are ∼400 copies of rDNA encoding the 47S pre-rRNA. These copies are distributed over five clusters of tandemly repeated rDNA on the short arms of acrocentric chromosomes 13, 14, 15, 21, and 22. To meet the vast demand for cellular ribosomes in proliferating human cells, rDNA is heavily transcribed by RNA polymerase I (Pol I). The transcription activity of Pol I fluctuates during the cell cycle; in S phase, the activity is especially vigorous (1). Since both replication and transcription can occur in the same region on the genomic DNA, cells require mechanisms that coordinate these processes.
The replication fork barrier (RFB) site near the 3′ end of the pre-rRNA coding region has been identified in many organisms, including yeasts, plants, frogs, and mammals (2). In these organisms, with the exception of humans, the RFB predominantly inhibits progression of the replication fork in the opposite direction to pre-rRNA transcription (head-on direction), whereas replication in the same direction (codirection) is not obstructed. Therefore, it is assumed that the RFB arrests the replication fork before it enters the coding region from downstream and thereby prevents the replication fork from colliding with pre-rRNA transcription. In contrast, RFBs in humans are reported to be bidirectional (3).
The RFB is formed by a tight complex between certain DNA sequences and proteins that bind to these elements. Fob1 in budding yeast (Saccharomyces cerevisiae) is the best characterized RFB binding protein (4, 5). Deletion of FOB1 allows the replication fork to enter the 35S pre-rRNA coding region from the downstream direction (6, 7). However, when normal numbers of rDNA copies are present, collision of replication with transcription does not occur in the absence of Fob1 because not all repeats are actively transcribed. In fact, the fob1 mutation stabilizes the rDNA cluster because fork arrests or DNA double-strand breaks at the RFB no longer occur (7–11). In contrast, in a fob1 mutant with a low rDNA copy number, transcription of most rDNA copies is activated and the movement of the replication fork is slowed down within the coding region due to collision with the Pol I transcription machinery (12). This collision results in the production of extrachromosomal rDNA circles (ERCs) and a change in rDNA copy number, thus indicating an important role of the RFB for the suppression of rDNA instability in budding yeast. In other organisms, the relevant role of RFBs has not yet been unraveled, although they are assumed to have a similar role by the polarity of fork arrests.
In mice, downstream from the 47S pre-rRNA-coding region, there are 10 repeated transcription terminator elements, called Sal boxes T1 to T10 (13). The Pol I-specific transcription termination factor 1 (TTF-1), the ortholog of fission yeast (Schizosaccharomyces pombe) Reb1, binds to the Sal boxes and terminates pre-rRNA transcription (14–16). An in vitro cell-free replication assay using simian virus 40 (SV40) T antigen revealed that, of the 10 Sal boxes, Sal box T2 causes polar arrest of the SV40 replication fork (17). This polar arrest requires TTF-1 binding to T2, while a unique stretch of GC sequence preceding T2, not present at the other Sal box elements, is also essential for this element to act as an RFB. The KU complex, which binds to this GC stretch, is also implicated in the arrest of the SV40 replication fork (18). In contrast to these in vitro observations, in several mouse cell lines, fork arrests were detected at multiple sites near the repeating Sal boxes (19). This inconsistency calls for a more precise in vivo analysis to assess the RFB model established by the SV40 replication studies.
In human cells, multiple Sal boxes are located downstream from the 47S pre-rRNA-coding region (20). The number of Sal boxes differs depending on the number of R repeat segments in each rDNA copy (21, 22). As in mice, replication fork arrest in human cells occurs within these repeated regions, but unlike other organisms, replication is blocked in both directions (3, 23).
Apart from proteins that specifically bind to the RFB, two replication factors also contribute to replication fork arrest. These form a complex, Tof1-Csm3 in budding yeast and Swi1-Swi3 in fission yeast (24–27). In budding yeast, the Tof1-Csm3 complex counteracts the removal of nonhistone proteins that are bound to DNA by the Rrm3 helicase, thereby facilitating replication fork arrest at RFBs associated with Fob1 (27). Both orthologous protein complexes are also implicated in the stabilization of replication forks and the intra-S-phase-checkpoint pathway, which is activated by DNA damage or nucleotide depletion (28–30). Their mammalian counterpart, the TIMELESS (TIM)-TIPIN complex, has been shown to play similar roles during S phase (31–33). However, it was unclear whether the TIM-TIPIN complex is required for replication fork-blocking activity at RFB sites.
In this study, we performed detailed characterization of RFBs in mammalian rDNA using two-dimensional gel electrophoresis. We found that the Pol I transcription terminator complex, Sal box/TTF-1, acts as an RFB whose activity is linked to the epigenetic status of the rDNA. Our results revealed that RFBs play a role in coordinating replication progress and pre-rRNA transcription activity in human cells.
MATERIALS AND METHODS
Cell lines.
HeLa and 293E cells were grown in Dulbecco's modified Eagle's medium with high glucose (DMEM-HG) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Pen-Strep). Mouse embryonic stem (ES) cells were cultured as previously described (34).
2-D gel analysis of replication intermediates.
To preserve the structure of replication intermediates, agarose plugs were used for DNA purification and restriction enzyme digestions. Cultured cells were embedded in 0.6% low-melting-point agarose and then treated with 1 mg/ml proteinase K, 1% sarcosyl, 0.5 M EDTA (pH 9.5) for 24 to 48 h at 50°C. Before restriction enzyme digestion, the agarose plugs were treated with 10 μM phenylmethylsulfonyl fluoride for 1 h and washed with Tris-EDTA (TE) buffer. DNA was then digested with the appropriate restriction enzyme for 16 to 20 h at 37°C. Neutral/neutral two-dimensional (2-D) gel electrophoresis was carried out as described previously (35, 36). In neutral/alkaline 2-D gel electrophoresis (37), the second dimension was run under alkaline conditions (40 mM NaOH, 1 mM EDTA) over 1% agarose at 0.5 V/cm for 36 h. The separated DNA was transferred to a nylon membrane by Southern blotting and hybridized to PCR-amplified fragments labeled with [32P]dCTP using the Random Primer DNA labeling kit (TaKaRa Bio). The probes used annealed to 28S (amplified with primer set YA650/YA651 [YA650/-1]) (see Table S1 in the supplemental material), Sal boxes (YA847/-51), oriP-1 (YA780/-1), oriP-2 (YA872/-3), oriP-3 (YA914/-5), 3′ external transcribed spacer (3′ETS) (YA592/-3), or mouse 28S (YA646/-7). Radioactive signals were detected with a Typhoon FLA9000 (GE) phosphorimager, and the images obtained were analyzed with ImageJ software (NIH). The hybridization signals of replication intermediates indicated in the figures were normalized to a common not-saturated linear signal. Upon TIM depletion, the signals were further normalized with respect to the area around the descending Y arc (signal of Y forks) without the RFB signals. The ratios between the results for targeted and corresponding control knockdowns are shown.
Plasmids.
The HindIII fragment containing the SV40 promoter and origin was removed from the pSOP plasmid, which was kindly provided by Kenji Moriyama and Hisao Masai (Rinshoken, Tokyo, Japan) (38), to obtain the OriP vector used in this study. Fragments of human class I rDNA were PCR amplified from genomic DNA using primer set YA794/-5 or YA879/-51 (see Table S1 in the supplemental material), sequenced to confirm the absence of PCR-induced mutations, and cloned into the OriP vector as indicated below. Mutagenesis of AG to GA in the Sal box consensus sequence was done with the QuikChange multisite-directed mutagenesis kit (Agilent Technologies). Individual Sal boxes, formed by annealing complementary oligonucleotides encompassing the sequences given in Fig. 2C, were cloned in the BstZ17I site of the OriP vector. The mouse terminator DNA fragment (mR1+mR2 [see Fig. 4C]), isolated as a NarI-DpnI fragment from pMr3′Eco (a kind gift from Ingrid Grummt, DKFZ-ZMBH Alliance, Germany), was digested with AflIII to obtain mR1 and mR2 fragments. These fragments were inserted into the HindIII site of the OriP vector. The orientation of all DNA fragments in the OriP vector was determined by sequencing analysis. To express small hairpin RNAs (shRNAs) in mouse ES cells, with the target sequences being shTTF1-1 (5′-GGATATGGAAACTGGGATCAT), shTTF1-2 (5′-GCCCTGGAAGCTCGTGTACTA), and shCTRL (5′-CGTACGCGGAATACTTCGA) (39), the pSuper vector (Oligoengine) was used according to the manufacturer's instructions.
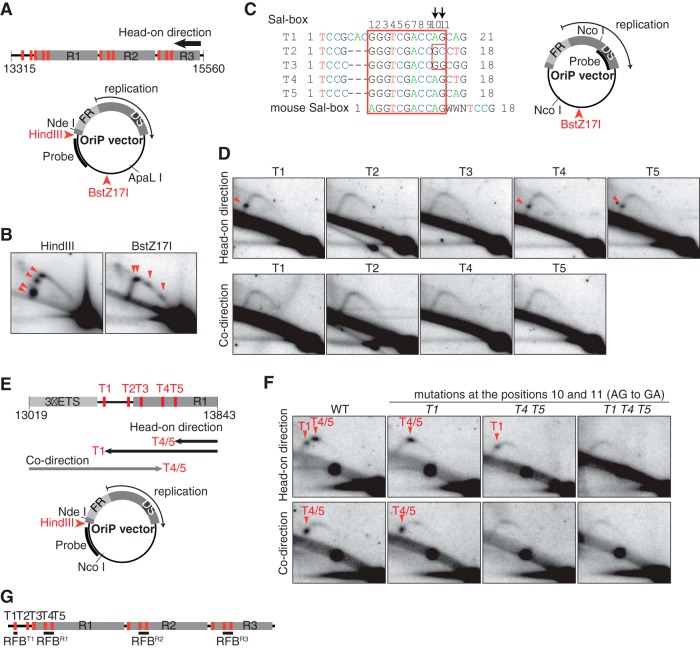
FIG 2.
The canonical Sal box functions as an RFB. (A) The intergenic spacer sequence corresponding to nucleotide positions 13315 to 15560 in the sequence with GenBank accession number U13369 was inserted into the OriP vector either at the HindIII or the BstZ17I site. Red bars represent the locations of Sal boxes. In the OriP vector, FR and DS regions in the OriP element are indicated. (B) 2-D gel analyses of ApaLI-NdeI fragments derived from the OriP vector bearing the class I sequence in the head-on direction at the HindIII or BstZ17I site. The oriP-1 probe, indicated in panel A, was used for hybridization. Arrowheads indicate accumulation of Y forks due to RFB activity. (C) Alignment of Sal box sequences. The conserved 11-bp sequence is within the red box. Each of these Sal box sequences was inserted into the OriP vector at the BstZ17I site in either head-on or codirectional orientation. (D) 2-D gel analyses of NcoI fragments from OriP vectors with Sal boxes shown in panel C. Replication in the head-on direction, as well as the codirection, was examined, with accumulated Y forks (arrowheads) visualized by hybridization to the oriP-2 probe. (E and F) The Sal box T1-to-T5 region (nt 13019 to 13843 in the sequence with accession number U13369), cloned in the HindIII site of the OriP vector, was mutagenized as indicated (E), and NcoI-NdeI fragments were analyzed by 2-D gel analysis (F). Y fork accumulations due to RFB activity of T1 or T4/T5 (arrowheads), visualized by hybridization to the oriP-3 probe, are indicated (F) and illustrated (E). (G) Summary of RFB locations of in human class I rDNA.
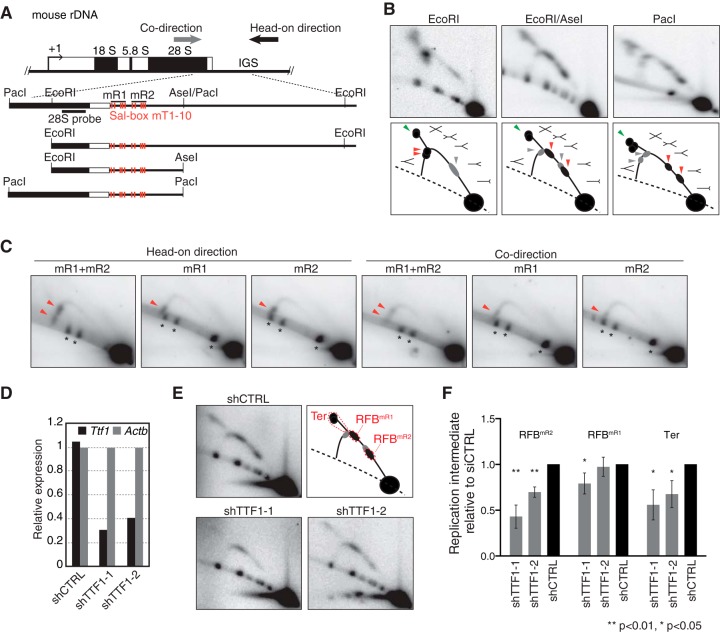
FIG 4.
The mR1 and mR2 repeats in mouse rDNA each have RFB activity in a TTF-1-dependent manner. (A) Structure of mouse rDNA with the restriction sites used and mouse 28S probe indicated. (B) 2-D gel analyses of replication intermediates of Sal box-containing regions in mouse ES cells (top) and corresponding explanatory diagrams (bottom). Arrowheads point to the accumulation of Y forks during head-on directional replication (red) or codirectional replication (gray) or to converging forks (green) associated with replication termination. (C) The mR1 and mR2 sequences arrest replication forks on episomally replicating DNA in 293E cells. The indicated sequences were cloned into the OriP vector at the HindIII site in either orientation. Replication intermediates of ApaLI-NdeI fragments from these plasmids were visualized by Southern blotting with the oriP-1 probe (Fig. 2A). Arrowheads indicate accumulated Y forks representing RFB activity. Asterisks mark cross-hybridization with nonreplicating linear DNA molecules. (D) Knockdown of Ttf-1 mRNA levels as verified by RT-PCR in mouse ES cells transfected with shRNA-expressing plasmids. Values are normalized with respect to the level of Actb expression. (E) 2-D gel analyses of EcoRI-AseI fragments in TTF-1 knockdown cells. Top right, diagram showing the typical pattern of replication intermediates, with RFB positions indicated. RFBmR1 and RFBmR2 represent Y fork accumulation at the respective RFB sites during head-on directional replication. (F) Quantification of replication intermediates detected as shown in panel E was as described for Fig. 3. The mean results and SD from four independent experiments are shown.
Nucleotide sequence accession number.
The sequence of class I rDNA determined in this study was deposited in the DDBJ under accession number AB972464.
RESULTS
Identification of RFB sites in human rDNA.
In human rDNA, the intergenic spacer (IGS) region around the transcription terminator shows heterogeneity in length due to variation in the number of R repeats (21, 22). In HeLa cells, we detected two restriction fragments encompassing this region after digestion of genomic DNA with AflII. These two restriction length variations, which we call class I and class II, contained three and two R repeats, respectively (Fig. 1A and B).
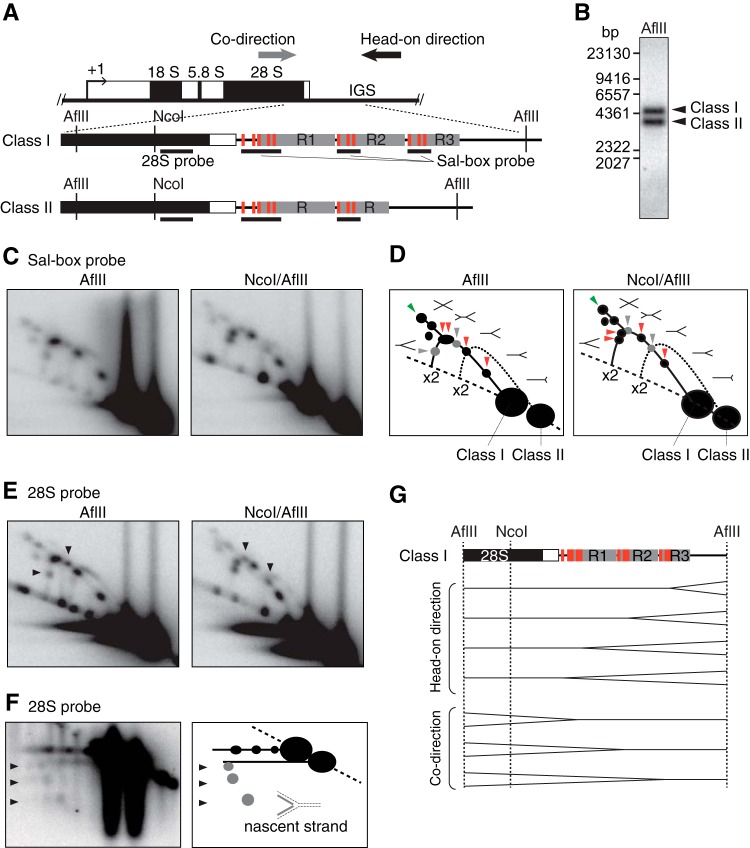
FIG 1.
Human cells contain multiple RFB sites near Sal boxes downstream from the 47S pre-rRNA-coding region. (A) Structure of human rDNA. AflII fragments containing the transcription terminator region with its upstream coding region are magnified, showing Sal boxes (red bars), R repeats (gray rectangles), the positions of 28S or Sal box probes (black lines), and the directions of replication (arrows). Class I and class II rDNAsw differ in the number of R repeats. (B) Digestion of genomic DNA from HeLa cells with AflII reveals two restriction length variations, class I and class II, that, after Southern blotting, were visualized by hybridization with the 28S probe. (C) 2-D gel analysis of replication intermediates on AflII and AflII/NcoI fragments visualized with the Sal box probe. Note that multiple Y forks accumulate on the Y arc of class I but not on that of class II. (D) Schematic diagram of results shown in panel C. Shapes of replication intermediates and Y-, double-Y-, and X-shaped molecules are represented. Nonreplicating molecules appear on the dashed line. Red arrowheads indicate RFB signals of accumulated Y forks formed by head-on directional replication, whereas gray arrowheads indicate Y fork accumulation during codirectional replication. Green arrowheads indicate termination of replication by the convergence of forks. The solid line indicates replication intermediate signals from class I rDNA. The Y arc from class II rDNA is depicted with a dotted line. The positions of duplicated DNA are indicated by ×2. (E) 2-D gel analysis of AflII and AflII/NcoI fragments hybridized to the 28S probe. Arrowheads point to enhanced signals of accumulating Y forks moving in the codirection. (F) Left, neutral/alkaline 2-D gel analysis of AflII fragments, visualized with the 28S probe; right, the results are diagrammed. Arrowheads indicate the accumulation of nascent strands from Y forks moving in the codirection. (G) Inferred positions of replication fork arrests in class I rDNA.
To investigate RFB activity in human rDNA, replication intermediates of class I and class II fragments were analyzed by neutral/neutral two-dimensional (2-D) gel electrophoresis followed by Southern blotting (Fig. 1C). Among the typical patterns of replication molecules observed (Fig. 1D) in class I rDNA but not in class II rDNA, several spots corresponding to accumulated Y-shaped replication fork molecules (Y forks) were detected. Thus, class I rDNA contains a number of specific sites where the replication fork is arrested that represent RFB activity. We also detected signals of double-Y- and X-shaped molecules (Fig. 1D, green arrowheads), suggesting that fork arrest at the RFB sites frequently leads to replication termination by the convergence of replication forks. Comparison between the patterns obtained for AflII- and NcoI/AflII-digested fragments revealed the direction of fork arrest at the RFB sites (see Fig. S1 in the supplemental material for the strategy). The accumulation of Y forks in the AflII digests shifted in a counterclockwise direction on the Y arc after the length of the fragment was reduced by double digestion with NcoI (Fig. 1D, red arrowheads). Thus, in class I rDNA, four RFBs arrest the replication fork, moving in the head-on direction, i.e., opposite to pre-rRNA transcription. The locations of RFB sites can be estimated from the position of arrested intermediates in the 2-D gel analysis (Fig. 1G) and were mapped near the clustered Sal boxes, which therefore might be linked to the RFB activities.
Consistent with previous findings (3), arrest of replication forks moving in the codirection, i.e., the same direction as pre-rRNA transcription, was also detected for class I rDNA, although the related signals were weak (Fig. 1C and D, gray arrowheads). In this case, the spots with accumulated Y forks in the AflII digests shifted in the clockwise direction on the Y arc after double digestion with NcoI. Hybridization to a probe recognizing the 28S rRNA-coding region (28S probe) enhanced the intensity of these signals, as this probe will anneal to the already-duplicated branches of the Y forks (Fig. 1E, arrowheads). Nascent strands from codirectional replication forks were resolved by neutral/alkaline 2-D gel analysis and visualized by hybridization with the 28S probe. In the second dimension, consisting of alkaline conditions, single-stranded nascent DNA migrates faster than its template strands. Three spots corresponding to the accumulation of nascent strands were detected (Fig. 1F, arrowheads), indicating that codirectional replication halted at three specific sites in class I rDNA. Intermediates related to an arrest at one of these sites could only be detected using neutral/alkaline separation. Due to insufficient resolution in neutral/neutral 2-D gel analysis, the corresponding spot was overshadowed by intense signals of nearby migrating forks arrested during head-on replication. Codirectional replication was arrested at positions that were mapped around the clustered Sal boxes (Fig. 1G). Thus, the Sal boxes might be involved in the arrest of both codirectional and head-on replication, yet, as described below, not in the same manner.
In summary, around the Sal boxes in class I rDNA, we identified four sites with pronounced RFB activity toward head-on directional replication and three sites that affected codirectional replication. While the bidirectional fork arrest occurs in class I rDNA, arrest in the head-on direction is predominant.
Sal box T1 acts as a polar RFB, while Sal box T4/T5 arrests replication forks moving in both directions.
To determine more precisely the roles of the Sal boxes in the arrest of replication forks, we set up an in vivo system in which class I rDNA sequences could be analyzed for their effect on replication. For this, the replication system from the Epstein-Barr virus (40) was employed to create a vector (OriP) that was able to replicate extrachromosomally and in a unidirectional manner in human 293E cells. These cells stably expressed the viral EBNA-1 protein that initiates replication from OriP by recruiting cellular replication factors. After transfection of 293E cells with recombinant OriP plasmids, the RFB activity of particular DNA segments could be analyzed using plasmid-specific probes (see Fig. S2A to D in the supplemental material).
When the class I fragment was cloned into the OriP vector in the head-on direction, accumulation of arrested replication forks was observed near the Sal boxes (Fig. 2A and B), showing that their RFB activities were recapitulated on the plasmid. This result also indicated that replication fork arrests at the RFB sites occur independently of pre-rRNA transcription, as suggested previously (17). To localize more precisely the elements responsible for the fork arrests, we dissected the class I rDNA sequence by testing the RFB activity of distinct fragments in the OriP context (see Fig. S2E and F in the supplemental material). We observed fork arrest only in head-on replication with Sal box-containing fragments and found that two independent RFB activities in the transcription terminator region of the pre-rRNA were linked to the Sal boxes of the first termination sequences (T1 to T2) and the R repeat (T3 to T5).
To directly determine which of the Sal boxes T1 to T5 was required for the arrest of replication forks, 18 to 21 bp of each sequence was cloned into the OriP vector and tested for RFB activity in head-on replication (Fig. 2C and D). The presence of T1, T4, and T5 but not T2 and T3 led to the accumulation of Y forks. T2 and T3 contain natural point mutations in the consensus sequence at positions 10 and 11 (AG to GC in T2 and A to G in T3) (Fig. 2C) that likely account for their lack of RFB activity (Fig. 2D). Previously, it was demonstrated that T2 and T3 had no affinity for their binding protein and failed to terminate Pol I transcription (41). When we replaced the AG duplets in the active Sal boxes T1, T4, and T5 with GA in a fragment encompassing the Sal box T1-to-T5 region, arrested replication forks corresponding to RFB activity at these elements were no longer observed (Fig. 2E and F). Similarly to T2 and T3, mutation of the consensus sequence inactivated these Sal boxes. Taken together, these results indicate that, in addition to their role in transcription termination, canonical Sal boxes, together with their binding factor TTF-1, act as RFB sites in head-on replication. Importantly, when individual Sal boxes were replicated in the codirection, no RFB activity was detected (Fig. 2D). This result revealed an intrinsic polarity of Sal boxes in arresting a replication fork; head-on but not codirectional replication progress is inhibited by a Sal box.
Although a Sal box is ineffective by itself in blocking codirectional replication, in the natural context of the class I fragment, such activity was observed on chromosomal DNA (3) (Fig. 1) and episomal DNA (see Fig. S2G in the supplemental material). Dissection demonstrated that the RFB activity resided in a region shared by all three repeats, which contained T3 to T5 (see Fig. S2G). To determine their contribution to codirectional RFB activity, the mutant forms of T4 and T5 were tested in the context of the Sal box T1-to-T5 region. As shown by the data in Fig. 2E and F, the wild-type fragment accumulated one arrested replication fork, which, as expected, was not affected upon mutation of T1. However, mutations in T4 and T5 eliminated codirectional replication fork arrest.
Taken together, our results demonstrate that Sal boxes T4 and T5 are required for the arrest of both head-on and codirectional replication, whereas T1 only acts in the head-on direction (Fig. 2E). Since individual Sal boxes do not arrest codirectional replication (Fig. 2D) and T4 and T5 are only 39 bp apart in class I rDNA, we speculate that they act cooperatively. These two elements are present in all three R repeats (R1 to R3) in which, because of their sequence similarity, replication fork arrests can be expected to occur via the same mechanism. Here, we refer to the RFB sites at T4 and T5 in the three R repeats, at which replication forks are arrested bidirectionally, as RFBR1, RFBR2, and RFBR3 (Fig. 2G). The unique RFB site at T1 is the polar barrier, only blocking replication in one direction, and is called RFBT1.
Knockdown of TTF-1 attenuated RFB activity in human rDNA.
The Sal box binding protein TTF1 associates only with canonical Sal boxes (41) that mediate RFB activity, which strongly suggests that binding of TTF-1 is needed to arrest replication forks at RFB sites. To determine a role of TTF-1 for RFB activity, we treated cells with two independent small interfering RNAs (siRNAs) against TTF-1 (siTTF1-1 and siTTF1-2). Cellular depletion of TTF-1 by these siRNAs was successful, as the protein could no longer be detected by Western blotting (Fig. 3A), although ∼20 to 30% of mRNA expression remained (Fig. 3B). 2-D gel analysis revealed a significant (P < 0.05) decrease in the accumulation of Y forks at all RFB sites except RFBT1/R1 in siTTF1-treated cells (Fig. 3C and D), indicating that TTF-1 is required for replication fork arrest at these sites. The signals representing converging replication forks also decreased upon knockdown of TTF-1, suggesting that replication termination is associated with RFB activity. In contrast, the accumulations of Y forks at RFBT1/R1 in control cells and those treated with siTTF1 were similar. We consider that this result is, on one hand, due to the difficulty of completely depleting TTF-1, while on the other hand, we might be witnessing a counterbalancing effect. Although all RFB sites could be affected equally by depletion of TTF-1, head-on directional replication forks more frequently reached the distal RFBT1/R1 due to reduced fork arrests at RFBR2 or RFBR3. This, however, was not observed as an increased accumulation of Y forks since the decreased level of TTF-1 allowed more replication forks to pass through RFBT1/R1. This explanation is consistent with the fact that in siTTF-1-treated cells, the arrest of replication forks was relatively less affected at RFBR2 than at RFBR3. Furthermore, it cannot be excluded that TTF-1 binds RFBT1/R1 with a higher affinity than the other sites for as-yet-unknown reasons. Alternatively, a mechanism not dependent on the binding of TTF-1 might cause a specific arrest of the replication fork at RFBT1/R1.
FIG 3.
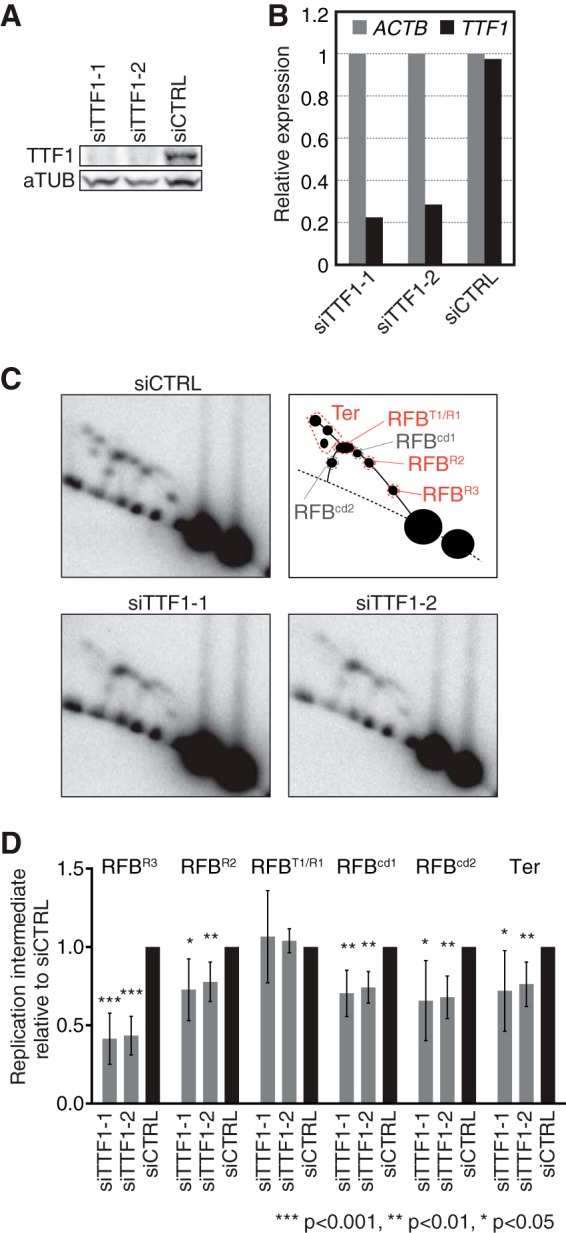
TTF-1 functions in replication fork arrest at RFBs in human rDNA. (A) Levels of cellular TTF-1 as examined by Western blotting 96 h after transfection of HeLa cells with siTTF1-1, siTTF1-2, and siCTRL. (B) TTF-1 mRNA expression as examined by reverse transcription (RT)-PCR 72 h after siRNA transfection. Values were normalized to the expression level of ACTB. (C) Replication intermediates in TTF-1-depleted cells were analyzed by 2-D gel analyses 96 h after siRNA transfection. Southern blots of AflII-digested DNA were hybridized to the 28S probe. Top right, diagram showing the typical pattern of replication intermediates generated at various RFB positions (RFBT1/R1, RFBR2, and RFBR3) during head-on directional replication. RFBcd1 and RFBcd2 represent Y fork accumulation during codirectional replication at RFBR1 and RFBR3, respectively. (D) Quantification of Y forks arrested at indicated RFBs and of replication termination (Ter) detected as shown in panel C relative to the results for corresponding replication intermediates from siCTRL cells. The mean results and standard deviations (SD) from ≥6 independent experiments are shown. The P values were calculated using a one-sample t test with a hypothetical mean of 1.
The mouse mR1 and mR2 repeats act as RFB sites.
To establish whether the arrest of replication forks by TTF-1 and Sal boxes is governed by a conserved mechanism, we did similar experiments to analyze the RFB activity in mouse cells. By sequence comparison, we found that in mouse cells, the Sal box-containing IGS region contains two repeats, one consisting of mouse Sal box T1 (mT1) to mT5 and the other of mT6 to mT10 (see Fig. S3A and B in the supplemental material), which are herein called mR1 and mR2, respectively.
To investigate in detail the mouse RFB sites in vivo, we examined replication intermediates of this IGS region by 2-D gel analysis followed by Southern blotting (Fig. 4A and B). The typical patterns of replication intermediates on the descending part of the Y arc as observed after EcoRI digestion (Fig. 4B, bottom) were as previously reported (19). Upon further digestion with AseI, we found that the signals of accumulated Y forks separated into two spots (Fig. 4B, red arrowheads) and shifted clockwise on the Y arc, showing that replication forks moving in the head-on direction had been arrested at two RFB sites, which were mapped near the mR1 and mR2 repeats. Since the hybridization signals at both RFB sites had an elongated shape, it appears that replication was arrested at multiple sites within the mR1 and mR2 repeats, consistent with the clustering of Sal boxes in each repeat (see Fig. S3B in the supplemental material). Double-Y- and X-shaped molecules that represent converging replication forks were also observed in the 2-D gel analysis (Fig. 4B, green arrowheads), indicating that replication fork arrest at these RFBs frequently resulted in replication termination.
In contrast to the previous in vivo and in vitro studies (17, 19), we detected Y forks that had been arrested during codirectional replication, although the hybridization signals were fairly weak (Fig. 4B, gray arrowheads). In the EcoRI digest, these intermediates formed an elongated signal on the ascending Y arc, which was separated into two spots that had shifted in the counterclockwise direction after the fragment was shortened by digestion with EcoRI/AseI or PacI. Again, the positions where the replication forks arrested were mapped near mR1 and mR2. These results suggest that the progress of codirectional replication is also impeded by the mR1 and mR2 repeats.
To determine whether the mR1 and mR2 repeats were each able to block replication, we cloned these sequences into the OriP vector and let them replicate in 293E cells. Accumulation of Y forks was detected in either direction of replication in the case of each repeat, although the signals were much fainter for codirectional pauses than for forks moving in the head-on direction (Fig. 4C). Thus, natural events that occur during mouse DNA replication could be reproduced in human cells and on the plasmids, strongly suggesting that comparable mechanisms involving Sal boxes regulate RFB activity.
To confirm that mouse TTF-1 is required for blocking replication forks, TTF-1 was knocked down in ES cells by using two small hairpin RNAs, shTTF1-1 and shTTF1-2. Ttf-1 mRNA expression levels were reduced by 60 to 70%, with the former shRNA being slightly more effective than the latter (Fig. 4D). 2-D gel analysis of DNA isolated from shTTF1-1 and shTTF1-2 cells showed that the accumulations of Y forks at mR2 and of converging replication forks were significantly reduced (Fig. 4E and F). TTF-1 depletion by shTTF1-1 also reduced Y fork accumulation at mR1, although to a lesser extent than at mR2. The finding that RFB activity in the head-on direction was stronger nearer the 3′ end of the pre-rRNA coding region was also seen in human cells after TTF-1 depletion (Fig. 3C and D). These results suggest that TTF-1, as in human cells, is required for arresting replication forks at RFB sites in mouse cells.
The KU complex has been found to be important for RFB activity at mT2 during SV40 replication in vitro (18). However, when we examined the levels and positions of Y forks in the absence of KU70, we did not see a significant difference between Ku70 knockout and wild-type cells (see Fig. S3C in the supplemental material), showing that the KU complex is dispensable for arresting replication forks at mR1 and mR2 in vivo.
In human cells, RFB activity relates to epigenetic status.
The timing of DNA replication is linked to the epigenetic status of the genomic locus, and we wondered how RFB activity near the 3′ end of the pre-rRNA coding region was regulated during S phase. We therefore analyzed replication intermediates of class I and II fragments in synchronized human cells (Fig. 5A). In the case of class I rDNA, the accumulation of Y forks at the RFB sites and of double-Y- and X-shaped molecules was highest early in S phase. These signals gradually decreased as S phase progressed, and only very weak RFB activity was seen in the mid- to late S phase, though the continuous Y arc signal indicated ongoing replication of class I rDNA. In contrast, Y forks of class II rDNA, as indicated by a faint spot, only accumulated early in S phase and not later on (Fig. 5A, early-S, arrowhead). Thus, the early-replicating rDNA copies, which mainly consisted of class I rDNA, were proficient for arresting replication forks at specific RFB sites. In late-replicating rDNA, however, replication forks were not blocked. For class II rDNA, the Y arc signal became continuous in the mid- and late S phase. Notably, late in S phase, the Y arc signal of class II rDNA was more intense than that of class I rDNA (Fig. 5A, late-S, arrowhead). These results suggest that a significant fraction of class II rDNA replicates mainly in the mid- to late S phase and is devoid of RFB activity. By sequencing analysis, we confirmed that the canonical Sal box sequence is present at T1, T4, and T5 in class II rDNA (Y. Akamatsu and T. Kobayashi, unpublished results), which implies that the RFB sites are epigenetically inactivated.
FIG 5.
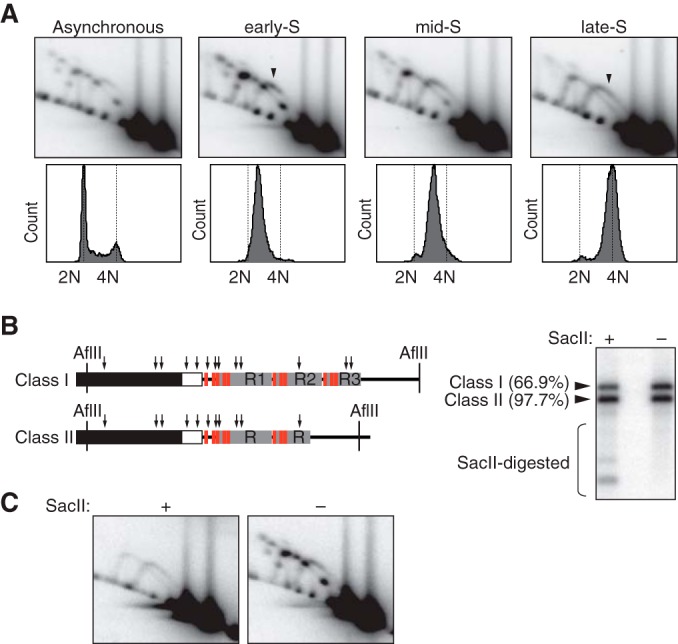
RFB activity is epigenetically regulated in human rDNA. (A) HeLa cells released from a double thymidine block were collected at early, mid-, and late S phase and analyzed by 2-D gel electrophoresis. Replication intermediates of AflII fragments were hybridized to the 28S probe. The progression of S phase is shown below the gels. Cells were stained with propidium iodide and analyzed by flow cytometry. Arrowheads indicate replication intermediates of class II rDNA. (B) Significant fractions of class I and class II rDNA are resistant to cleavage by SacII. Left, AflII rDNA fragments contain multiple SacII sites (arrows); right, HeLa genomic DNA, digested by AflII with or without SacII, was hybridized to the 28S probe. Percentages given for SacII-resistant fractions are the average results for two independent samples. (C) RFB activity is linked to SacII-cleavable rDNA. 2-D gel analyses of samples digested as described for panel B are shown.
In mammalian rDNA, two kinds of epigenetically distinct rDNA copies can be discerned, of which one set is actively transcribed (active rDNA) and the other is transcriptionally silent (silent rDNA) (42). It has been shown that these also differ in their timing of replication during S phase; active rDNA replicates early in S phase, whereas silent rDNA replicates late in S phase (43). Therefore, our results suggest a close link between active rDNA and the blocking of replication forks at RFB sites, which mostly occurs in class I rDNA, whereas silent rDNA is devoid of RFB activity. To address this hypothesis, the CpG methylation status of class I and II rDNA was examined by restriction digestion with SacII. Cleavage by this enzyme is inhibited not only by methylation but also by hemimethylation of the CpG dinucleotide in the CCGC/GG recognition sequence (44). As shown by the results in Fig. 5B, 66.9% of class I and 97.6% of class II rDNA fragments generated by AflII digestion remained intact after cleavage with SacII despite the presence of multiple SacII sites within the AflII fragments. Thus, most of the class II and a fraction of class I rDNAs were methylated. 2-D gel analysis of AflII fragments (Fig. 5C) revealed that after SacII digestion, none of the replication forks arrested at the RFB sites could be detected and only a continuous Y arc indicative of uninterrupted replication was observed. This result showed that RFB sites are only active in nonmethylated rDNA copies, which, apart from HeLa cells, was also found when the relation between methylation status and replication was analyzed for HEK293 and VA13/WI38 cells (see Fig. S4 in the supplemental material). These results are consistent with our hypothesis that replication forks are arrested at the RFB sites in active rDNA but not in silent rDNA.
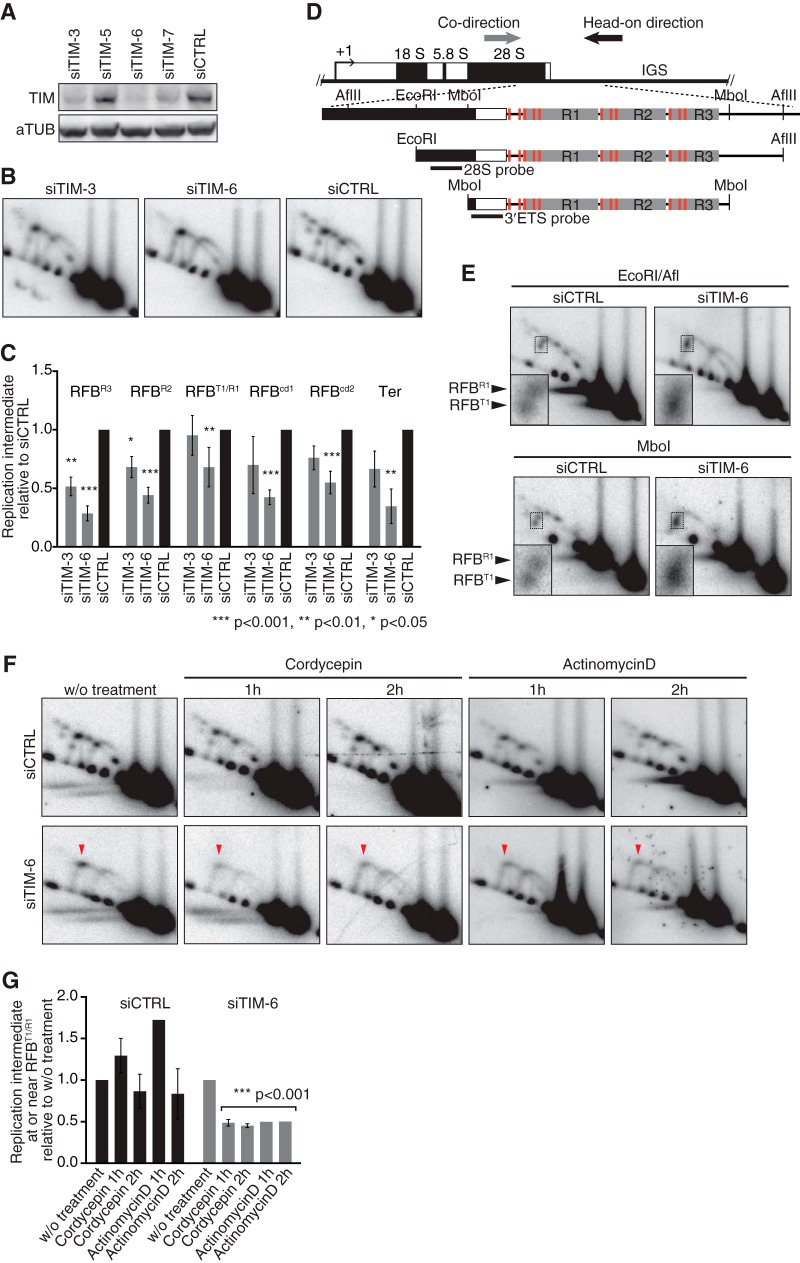
In TIM-depleted cells, replication collides with transcription due to failed fork arrests.
It has been shown in yeast that stable arrest of the replication fork at the RFB site depends on Tof1/Swi1, an accessory factor of the replication machinery (24–27). We investigated whether the human homologue TIM is required for replication fork arrests at RFB sites. We analyzed replication intermediates by 2-D gel electrophoresis in cells in which TIM had been significantly knocked down with siRNAs siTIM-3 and siTIM-6, with the latter being the most effective (Fig. 6A). In siTIM-3-treated cells, a significant reduction in fork arrests was seen at RFBR2 and RFBR3, whereas siTIM-6 treatment decreased fork arrests at all RFB sites (Fig. 6B and C). Thus, the role of TIM in arresting replication forks at RFB sites is evolutionarily conserved from yeasts to human. It seemed that, after depletion of TIM, fork arrest at RFBT1/R1 was less affected than that at RFBR2 or RFBR3 (Fig. 6B and C), as was observed in the case of TTF-1 depletion (Fig. 3). To examine this in detail, we reduced the fragment length by digestion with EcoRI/AflII or MboI, which gave sufficient resolution to separate the Y forks accumulating at RFBT1 and RFBR1 in the case of the control cells (Fig. 6D and E). In siTIM-6-treated cells, however, these Y forks were merged, as revealed by an unprecedented broad signal (Fig. 6E, insets), indicating that the progress of the replication fork was slowed down around these sites. This pattern was not seen after the knockdown of TTF-1 (see Fig. S5 in the supplemental material). Since the forks in these novel signals were moved anticlockwise on the Y arc, consistent with arrest in the head-on direction, and Sal box T1 is only 19 bp downstream from the 3′ end of the 47S pre-rRNA-coding region, we hypothesized that pre-rRNA transcription becomes an obstacle for replication fork movement when the fork fails to arrest at the RFB sites. To test this, we treated cells with transcription inhibitors cordycepin and actinomycin D and analyzed the fork arrests near RFBT1/R1 (Fig. 6F and G). Consistent with the observation that RFB sites are functional on plasmids, the accumulation of Y forks at the RFB sites was not affected by inhibition of pre-rRNA transcription in the control cells. In contrast, after depletion of TIM, the signal of Y forks arrested near RFBT1/R1 was diminished upon treatment with cordycepin or actinomycin D, indicating that the replication was proceeding into the 47S pre-rRNA-coding region when rRNA synthesis was shut off. These results support our hypothesis that active pre-rRNA transcription impairs replication progress, thus revealing the importance of RFBs for the coordination of DNA replication with pre-rRNA transcription.
FIG 6.
TIM is required for replication fork arrest at RFBs to coordinate the progression of replication with transcription activity. (A) Cellular depletion of TIM in HeLa cells was verified by Western blotting. (B) 2-D gel analysis of AflII fragments from TIM-depleted cells 96 h after transfection. (C) Quantification of replication intermediates detected as shown in panel B was as described in the legend to Fig. 3. The mean results and SD from independent experiments (n = 3 for siTIM-3 and n = 7 for siTIM-6 and siCTRL) are presented. (D) Diagram of class I rDNA fragments with restriction sites for AflII, MboI, and EcoRI and the positions of probes indicated. (E) 2-D gel analysis of EcoRI-AflII and MboI fragments, which were hybridized to the 28S probe and the 3′ETS probe, respectively. Insets show magnifications of the Y forks accumulating around RFBR1 and RFBT1. (F) Effect of transcription inhibition on replication. 2-D gel analyses of AflII fragments from cells that were treated with cordycepin (50 μM) or actinomycin D (50 ng/ml) for the indicated periods. Replication intermediates were visualized with the 28S probe. Arrowheads indicate transcription-dependent Y fork accumulation. (G) Quantification of replication intermediates at or near RFBR1/T1 detected as shown in panel F. Values relative to that of the signal in cells without (w/o) treatment are shown. For siTIM-6-treated cells, all values from inhibitor-treated cells were combined to calculate the P values using a one-sample t test with hypothetical mean of 1.
DISCUSSION
Our analyses were able to determine the exact positions and directions of RFBs, both in human cells and in mouse cells. The finding that canonical Sal boxes and their binding factor TTF-1 are required for replication fork arrests strongly suggests that a Sal box/TTF-1 complex acts as an RFB in mammalian rDNA. We also demonstrated that Sal box T1 causes a polar replication fork arrest, while the Sal box T4/T5 region arrests replication forks bidirectionally. The polarity of T1 fits the putative biological role of this RFB site in preventing collision of the replication fork with pre-rRNA transcription. Indeed, when RFB activity is reduced by depletion of TIM, pre-rRNA transcription impedes the progress of replication. Moreover, the RFB sites are only seen on transcriptionally active rDNA and not on rDNA that is silenced, indicating that RFBs are active on actively transcribed rDNA repeats. Thus, our results show the importance of RFB sites in coordinating the progression of replication with pre-rRNA transcription by preventing a replication fork from entering a highly transcribed region.
Some of our results seem contradictory to those of a previous report describing the identification of a mouse RFB site using an in vitro SV40 replication system (17). We demonstrated that two repeats of Sal boxes, mR1 and mR2, inhibited replication fork progress in vivo, which was not dependent on KU70 (or the continuous GC stretch it binds to) as reported for inhibition of SV40 replication (18). Moreover, in several mouse cell lines, fork arrests were detected at multiple sites near the repeating Sal boxes (19). These contradictions are likely due to differences between the SV40 and the cellular replication systems. In SV40 replication, T antigen acts as the replicative helicase, whereas in normal cellular replication, the replicative helicase consists of CMG (a complex formed by Cdc45, MCM2-7, and GINS) and this is restrained by the TIM/TIPIN fork protection complex (45, 46). We show that TIM contributes to fork arrest at RFBs similarly to Tof1/Mrc1 and Swi1/3 in yeasts. This role may be independent of the role in fork protection or it may contribute to the fork arrest at the rDNA RFBs.
In humans and mice, multiple Sal boxes are clustered together within larger, repeated (m)R segments. Recently, it was reported that clustering of Sal boxes in chromatin-coated DNA increased their binding affinity to TTF-1 protein and that they cooperatively terminated Pol I transcription (47). Their RFB activity might operate in a similar manner. Indeed, we found that two closely located functional Sal boxes, T4 and T5, become an obstacle for the codirectional replication fork that is able to pass through a single Sal box. Thus, the closely located Sal boxes within (m)R-repeats might form one functional unit that serves as an RFB site. Currently, we do not know whether the blocking of codirectional replication has a physiological function.
Our observations also suggest that RFB activity is closely related to the epigenetic status of the rDNA; the fork efficiently arrests at RFB on hypomethylated rDNA copies but not on methylated silent copies. We registered the highest RFB activity in early S phase, when actively transcribed rDNA is replicated (43). Thus, RFB sites are activated predominantly on transcriptionally active rDNA copies. This agrees with the idea that RFB sites are important for the coordination of replication fork progress with pre-rRNA transcription. Since the RFB activity is reconstituted on the plasmids and chemical inhibition of transcription did not affect the fork arrests on RFB sites, transcription activity itself is dispensable for RFB function (17; this study). It is known that, at the transcription terminator region, TTF-1 localizes selectively to nonmethylated DNA and not to methylated DNA (48). Therefore, recognition of the Sal boxes by TTF-1 might be affected by epigenetic marking of the terminator region and RFB activity on silent rDNA copies is missing.
Failure of replication fork arrest at RFB sites upon depletion of TIM leads to unprecedented accumulation of replication intermediates near the 3′ end of the 47S rRNA-coding region. A similar observation was reported for fission yeast (24). Fork arrest at RFP4 in fission yeast rDNA does not depend on the TIM homologue Swi1. As RFP4 does not block replication on a plasmid, pre-rRNA transcription has been speculated to be responsible for the fork arrest at this site. In this study, we demonstrated that, in human TIM-depleted cells, the unprecedented fork accumulation was dependent on active transcription. Thus, in the absence of TIM, pre-rRNA transcription impedes replication fork movement by head-on collision. Although the molecular mechanism of transcription-dependent replication fork arrest remains to be elucidated, it has been suggested that topological stress generated ahead of transcription and replication, DNA-RNA hybrids at the 3′ terminus of the coding region, or Pol I transcription machinery still bound to the rDNA might be responsible. These events also lead to genome instability (49).
The rDNA clusters in cancer cells are quite unstable (50, 51). Cancer cell survival is essentially supported through hyperactivated rRNA transcription, which might cause the collision of replication forks with rRNA transcription and, thereby, rDNA instability. We consider that RFB activity, by coordinating the progress of replication with elevated transcription, is important for faithful replication of rDNA clusters containing actively transcribed units and, possibly, for the structural integrity of rDNA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ingrid Grummt, Kenji Moriyama, and Hisao Masai for kindly providing plasmids.
This work is supported by Grant-in-Aid for Scientific Research on Innovative Areas from JSPS grants 23114001 and 20114002 for T.K. and Grant-in-Aid for Young Scientists (B) from JSPS grants 23770009 and 26840114 for Y.A.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01521-14.
REFERENCES
- 1.Klein J, Grummt I. 1999. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc Natl Acad Sci U S A 96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalgaard JZ, Godfrey EL, MacFarlane RJ. 2011. Eukaryotic replication barriers: how, why and where forks stall, chap 13 In Seligmann H. (ed), DNA replication: current advances. InTech, Rijeka, Croatia. doi: 10.5772/20383. [DOI] [Google Scholar]
- 3.Little RD, Platt TH, Schildkraut CL. 1993. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol 13:6600–6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi T. 2003. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol 23:9178–9188. doi: 10.1128/MCB.23.24.9178-9188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi T. 2011. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol Life Sci 68:1395–1403. doi: 10.1007/s00018-010-0613-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi T, Horiuchi T. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell 3:447–455. doi: 10.1016/S1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 9.Kaeberlein M, McVey M, Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weitao T, Budd M, Hoopes LL, Campbell JL. 2003. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J Biol Chem 278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- 11.Burkhalter MD, Sogo JM. 2004. rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell 15:409–421. doi: 10.1016/j.molcel.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi Y, Horiuchi T, Kobayashi T. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grummt I, Maier U, Ohrlein A, Hassouna N, Bachellerie JP. 1985. Transcription of mouse rDNA terminates downstream of the 3′ end of 28S RNA and involves interaction of factors with repeated sequences in the 3′ spacer. Cell 43:801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- 14.Grummt I, Rosenbauer H, Niedermeyer I, Maier U, Ohrlein A. 1986. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell 45:837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- 15.Bartsch I, Schoneberg C, Grummt I. 1988. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol 8:3891–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evers R, Smid A, Rudloff U, Lottspeich F, Grummt I. 1995. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J 14:1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber JK, Gogel E, Berger C, Wallisch M, Muller F, Grummt I, Grummt F. 1997. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 90:559–567. doi: 10.1016/S0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 18.Wallisch M, Kunkel E, Hoehn K, Grummt F. 2002. Ku antigen supports termination of mammalian rDNA replication by transcription termination factor TTF-I. Biol Chem 383:765–771. doi: 10.1515/BC.2002.080. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-estrano C, Schvartzman JB, Krimer DB, Hernandez P. 1998. Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J Mol Biol 277:249–256. doi: 10.1006/jmbi.1997.1607. [DOI] [PubMed] [Google Scholar]
- 20.Bartsch I, Schoneberg C, Grummt I. 1987. Evolutionary changes of sequences and factors that direct transcription termination of human and mouse ribosomal genes. Mol Cell Biol 7:2521–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Volpe A, Simeone A, D'Esposito M, Scotto L, Fidanza V, de Falco A, Boncinelli E. 1985. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol 183:213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- 22.Erickson JM, Schmickel RD. 1985. A molecular basis for discrete size variation in human ribosomal DNA. Am J Hum Genet 37:311–325. [PMC free article] [PubMed] [Google Scholar]
- 23.Lebofsky R, Bensimon A. 2005. DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol Cell Biol 25:6789–6797. doi: 10.1128/MCB.25.15.6789-6797.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krings G, Bastia D. 2004. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A 101:14085–14090. doi: 10.1073/pnas.0406037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev 19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. 2005. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 27.Mohanty BK, Bairwa NK, Bastia D. 2006. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foss EJ. 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157:567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 30.Noguchi E, Noguchi C, McDonald WH, Yates JR III, Russell P. 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol 24:8342–8355. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. 2007. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol 27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. 2005. Coupling of human circadian and cell cycles by the Timeless protein. Mol Cell Biol 25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou DM, Elledge SJ. 2006. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci U S A 103:18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akamatsu Y, Jasin M. 2010. Role for the mammalian Swi5-Sfr1 complex in DNA strand break repair through homologous recombination. PLoS Genet 6:e1001160. doi: 10.1371/journal.pgen.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman KL, Brewer BJ. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol 262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 36.Ide S, Kobayashi T. 2010. Analysis of DNA replication in Saccharomyces cerevisiae by two-dimensional and pulsed-field gel electrophoresis. Curr Protoc Cell Biol Chapter 22:Unit 22.14. doi: 10.1002/0471143030.cb2214s49. [DOI] [PubMed] [Google Scholar]
- 37.Nawotka KA, Huberman JA. 1988. Two-dimensional gel electrophoretic method for mapping DNA replicons. Mol Cell Biol 8:1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriyama K, Yoshizawa-Sugata N, Obuse C, Tsurimoto T, Masai H. 2012. Epstein-Barr nuclear antigen 1 (EBNA1)-dependent recruitment of origin recognition complex (Orc) on oriP of Epstein-Barr virus with purified proteins: stimulation by Cdc6 through its direct interaction with EBNA1. J Biol Chem 287:23977–23994. doi: 10.1074/jbc.M112.368456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Jasin M. 2011. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol 18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frappier L. 2012. EBNA1 and host factors in Epstein-Barr virus latent DNA replication. Curr Opin Virol 2:733–739. doi: 10.1016/j.coviro.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Pfleiderer C, Smid A, Bartsch I, Grummt I. 1990. An undecamer DNA sequence directs termination of human ribosomal gene transcription. Nucleic Acids Res 18:4727–4736. doi: 10.1093/nar/18.16.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McStay B, Grummt I. 2008. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Santoro R, Koberna K, Grummt I. 2005. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 24:120–127. doi: 10.1038/sj.emboj.7600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson PS, Papas TS, Schweinfest CW. 1993. Restriction endonuclease cleavage of 5-methyl-deoxycytosine hemimethylated DNA at high enzyme-to-substrate ratios. Nucleic Acids Res 21:681–686. doi: 10.1093/nar/21.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An P, Saenz Robles MT, Pipas JM. 2012. Large T antigens of polyomaviruses: amazing molecular machines. Annu Rev Microbiol 66:213–236. doi: 10.1146/annurev-micro-092611-150154. [DOI] [PubMed] [Google Scholar]
- 46.Cho WH, Kang YH, An YY, Tappin I, Hurwitz J, Lee JK. 2013. Human Tim-Tipin complex affects the biochemical properties of the replicative DNA helicase and DNA polymerases. Proc Natl Acad Sci U S A 110:2523–2527. doi: 10.1073/pnas.1222494110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diermeier SD, Nemeth A, Rehli M, Grummt I, Langst G. 2013. Chromatin-specific regulation of mammalian rDNA transcription by clustered TTF-I binding sites. PLoS Genet 9:e1003786. doi: 10.1371/journal.pgen.1003786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemeth A, Guibert S, Tiwari VK, Ohlsson R, Langst G. 2008. Epigenetic regulation of TTF-I-mediated promoter-terminator interactions of rRNA genes. EMBO J 27:1255–1265. doi: 10.1038/emboj.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helmrich A, Ballarino M, Nudler E, Tora L. 2013. Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol 20:412–418. doi: 10.1038/nsmb.2543. [DOI] [PubMed] [Google Scholar]
- 50.Stults DM, Killen MW, Shelton BJ, Pierce AJ. 2011. Recombination phenotypes of the NCI-60 collection of human cancer cells. BMC Mol Biol 12:23. doi: 10.1186/1471-2199-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, Arnold SM, Moscow JA, Pierce AJ. 2009. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res 69:9096–9104. doi: 10.1158/0008-5472.CAN-09-2680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.