Abstract
Spinocerebellar ataxia type 7 (SCA7) is a debilitating neurodegenerative disease caused by expansion of a polyglutamine [poly(Q)] tract in ATXN7, a subunit of the deubiquitinase (DUB) module (DUBm) in the SAGA complex. The effects of ATXN7-poly(Q) on DUB activity are not known. To address this important question, we reconstituted the DUBm in vitro with either wild-type ATXN7 or a pathogenic form, ATXN7-92Q NT, with 92 Q residues at the N terminus (NT). We found that both forms of ATXN7 greatly enhance DUB activity but that ATXN7-92Q NT is largely insoluble unless it is incorporated into the DUBm. Cooverexpression of DUBm components in human astrocytes also promoted the solubility of ATXN7-92Q, inhibiting its aggregation into nuclear inclusions that sequester DUBm components, leading to global increases in ubiquitinated H2B (H2Bub) levels. Global H2Bub levels were also increased in the cerebellums of mice in a SCA7 mouse model. Our findings indicate that although ATXN7 poly(Q) expansions do not change the enzymatic activity of the DUBm, they likely contribute to SCA7 by initiating aggregates that sequester the DUBm away from its substrates.
INTRODUCTION
Spinocerebellar ataxia type 7 (SCA7) is one of nine polyglutamine [poly(Q)] expansion diseases associated with progressive neurodegeneration (1–3). SCA7 is caused by poly(Q) expansions within the N-terminal (NT) region of ATXN7. In unaffected individuals, ATXN7 contains 4 to 35 glutamine (Q) residues, whereas in SCA7 patients, ATXN7 contains from more than 36 to up to a few hundred poly(Q) repeats (2). The length of the poly(Q) expansion correlates inversely with the age of onset and the severity of the disease (4). Although it is generally agreed that aggregation of the expanded glutamine tract plays a critical role in neurotoxicity, the exact molecular mechanism of toxicity remains unclear (2).
ATXN7 is a subunit of the deubiquitinase (DUB) module (DUBm) in the highly conserved SAGA complex, which regulates gene expression by modulating histone acetylation and ubiquitination (5–7). Poly(Q) expansions in ATXN7 could affect either of these activities. Previous studies provided conflicting evidence regarding the effects of ATXN7-poly(Q) on the activity of Gcn5, the catalytic subunit of the histone acetyltransferase (HAT) module (8–11). The loss of Gcn5 accelerates cerebellar Purkinje cell and retinal degeneration in a SCA7 mouse model, indicating that Gcn5 functions are pertinent to disease progression (12). However, deletion of Gcn5 in Purkinje cells is not sufficient to cause severe ataxia, indicating that the loss of other SAGA functions contributes to SCA7 development (13). As ATXN7 is a component of the DUB module, poly(Q) expansions might affect the deubiquitination activity of SAGA.
The SAGA DUB module in Saccharomyces cerevisiae yeast is composed of Ubp8, Sgf11, Sus1, and Sgf73 (homologs of USP22, ATXNL3, ENY2, and ATXN7, respectively, in humans; Fig. 1A) (7, 14–17). Sgf73/ATXN7 serves both to tether the DUBm to SAGA through a central domain and to form an integral part of the DUBm through an N-terminal domain (18). Although Ubp8 possesses an ubiquitin (Ub)-specific hydrolase (Usp) domain, this enzyme is inactive in the absence of Sgf11, Sus1, and Sgf73 DUBm proteins (18). The crystal structure of the yeast DUBm provides a molecular model for understanding how these interactions activate Ubp8 (19, 20). Allosteric regulation of the mammalian catalytic subunit USP22 also occurs through multiple interactions with specific domains of human SAGA DUBm components. The ATXN7 zinc finger (ZnF) domain is required for association with the DUBm, and the ZnF domain in ATXN7L3 (Sgf11 in yeast) further stimulates USP22 activity (21). However, how ATXN7 poly(Q) expansions affect DUBm integrity or activity has not been addressed directly.
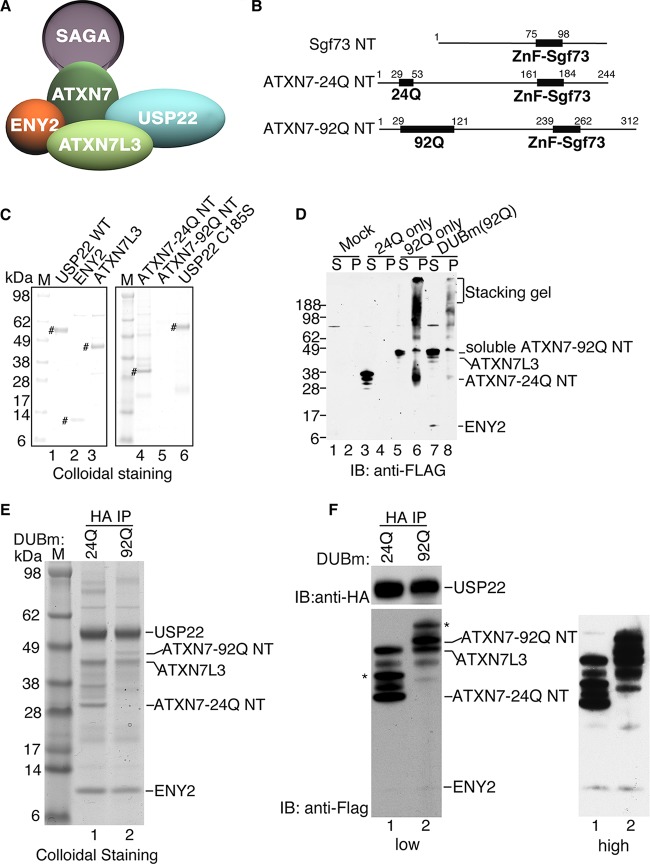
FIG 1.
Reconstitution of mammalian DUBm. (A) Schematic of the mammalian SAGA DUBm. (B) Schematic representation of ATXN7 N-terminal fragments with 24 Q residues and 92 Q residues, where Q is glutamine. (C) Colloidal staining of DUBm subunits after elution with HA or Flag peptides from immunoaffinity agarose beads. The expression levels are similar for all the subunits analyzed, with the exception of the expression level for ATXN7-92Q NT. #, purified DUBm subunits in each lane. (D) Expression of exogenous Flag-ATXN7 NT in WCEs from Sf21 insect cells after cellular fractionation at 3 days postinfection in order to assess the solubility of ATXN7-24Q NT and ATXN7-92Q NT. Lanes S, supernatant (soluble fraction); lanes P, pellet (insoluble fraction). Numbers on the left are molecular masses (in kilodaltons). (E) Colloidal staining of the reconstituted DUBm with ATXN7-24Q NT or ATXN7-92Q NT after anti-HA (USP22) immunoprecipitation. (F) Immunoblot analysis of the reconstituted DUBm for which the results are presented in panel E. ATXN7-24Q NT or ATXN7-92Q NT, ATXN7L3, and ENY2 are all Flag tagged and are detected with anti-Flag antibody; HA-USP22 is detected with anti-HA antibody. *, modified forms of ATXN7-24Q NT or ATXN7-92Q NT due to ubiquitination (data not shown). Lanes M (C and E), molecular mass markers; IB, immunoblot analysis; IP, immunoprecipitation analysis.
The best-characterized substrate for Ubp8 and USP22 is histone H2B, although other substrates have been identified in both yeast and mammalian cells (21–24). In mammalian cells, genome-wide analyses indicate that ubiquitinated H2B (H2Bub) is enriched at highly expressed genes (25), but this modification is associated with both gene activation and repression (26). Interestingly, expression of reelin, a factor important for the development and maintenance of Purkinje cells, is significantly downregulated in SCA7 astrocytes, and this downregulation is accompanied by increased levels of H2Bub at the reelin gene promoter (27). These results suggest that USP22 DUB activity may be defective in SCA7 tissues, leading to misregulation of key target genes.
In this study, we confirm that ATXN7 strongly stimulates DUB activity. Importantly, we demonstrate that ATXN7 with 92 Q residues at the N terminus (ATXN7-92Q NT) does not directly impair DUBm activity in vitro but that ATXN7-92Q NT is highly insoluble when not assembled into the DUBm. Cooverexpression of other DUBm subunits inhibits ATXN7-92Q aggregation in vitro and in human astrocytes. In addition, aggregates initiated by overexpression of ATXN7-92Q sequester ATXN7L3 and lead to increased global levels of H2Bub. Cooverexpression of ATXN7L3 and ENY2 with ATXN7-92Q ameliorates this effect. Consistent with these findings, H2Bub levels were elevated in the cerebellums of mice in a SCA7 mouse model, indicating that the impairment of DUBm activity might contribute to SCA7 disease. In conclusion, our findings suggest that ATXN7-poly(Q) causes SCA7 not by reducing USP22 activity per se but by forming aggregates that sequester DUBm components away from their substrates.
MATERIALS AND METHODS
Plasmids.
USP22 cDNA was acquired from Open Biosystems. ATXN7L3 and ENY2 cDNAs were a gift from Didier Devys. USP22, ATXN7L3, and ENY2 were subcloned into the 2.1-TOPO vector by PCR, sequenced, and then cloned into the NruI and NotI sites of the baculovirus expression vector pBacPAK8 (Clontech). A hemagglutinin (HA) tag was introduced into pBacPAK8 in BamHI and NruI, and a Flag tag was introduced into pBacPAK8 in KpnI and NruI. Mutation of USP22 cysteine 185 to serine was introduced using a site-directed mutagenesis kit (Agilent Technologies). Full-length Flag–ATXN7-24Q and Flag–ATXN7-92Q NT in the pSDM vector were provided by Patrick A. Grant. The cDNAs of human ATX7L3 and ENY2 were gifts from Didier Devys. The N termini of Flag–ATXNT7-24Q NT and Flag–ATXN7-92Q NT were subcloned into NheI and PstI of pBacPAK8.3. Production of recombinant viruses and Sf21 cell infection were performed by following the manufacturer's protocols (BD Biosciences). Packaging vectors (psPAX2 and pMD2G) and transfer vectors containing full-length Flag–ATXN7-24Q or Flag–ATXN7-92Q, which were used for production of recombinant lentivirus, were provided by Patrick A. Grant. The full-length Flag or V5-tagged ATXN7L3 and ENY2 expression constructs were generated in pSDM101 with MluI and NdeI by using a PCR cloning strategy with pBacPAK8-ATXN7L3 and pBACPAK8-ENY2 as the templates, respectively. Primer sequences are given in Table S1 in the supplemental material. Recombinant lentiviruses were produced and used according to published protocols (27).
Antibodies.
Flag antibody (M2) was purchased from Sigma. HA antibody (12CA5) was purchased from Roche. V5 antibody (R960-25) was purchased from Life Technologies. Ubiquitinated H2A (H2Aub) antibody (05-678), H2A antibody (07-146), H2B antibody (17-10054), H2Bub antibody (17–650), and acetylated H3K9 and H3K14 (H3K9/14ac) antibody (06-599) were purchased from Millipore. β-Actin antibody (I-19) was purchased from Santa Cruz. Anti-ATXN7L3 was obtained from Laszlo Tora (21). Alexa Fluor 488-conjugated anti-rabbit (A-11034) and Alexa Fluor 594-conjugated anti-mouse (A-11005) secondary antibodies for immunofluorescence were purchased from Life Technologies.
DUB module proteins and complex purifications.
To purify the DUB module proteins, recombinant baculoviruses were used to infect Sf21 cells. For DUBm complex purification, different baculoviruses were used to coinfect Sf21 cells. After 3 to 5 days of infection, cells were harvested and resuspended in buffer C (50 mM HEPES [pH 7.9], 5 mM MgCl2, 20% glycerol, 0.2% Triton X-100, 300 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) with a protease inhibitor cocktail (Roche). Then, the cells were homogenized by bead beating 3 times for 30 s each time on ice. The supernatant was recovered by centrifuging at 15,000 rpm for 10 min, and then the supernatant was further cleared by being centrifuged at 40,000 rpm for 1 h at 4°C. The supernatant was then incubated with the M2 anti-Flag–agarose (Sigma) or anti-HA matrix (Roche) equilibrated with washing buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.1% Triton X-100, 300 mM NaCl, 1 mM DTT, 1 mM PMSF) with a protease inhibitor cocktail (Roche) overnight at 4°C. After being washed with washing buffer 3 times for 10 min each time, bound proteins were eluted with the Flag peptide (0.2 mg/ml) or HA peptide (0.4 mg/ml), respectively, for 2 to 4 h at 4°C or room temperature (RT) in elution base buffer (10 mM HEPES [pH 7.9], 100 mM NaCl, 1.5 mM MgCl2, 0.05% Triton X-100, 0.2 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM PMSF) (28). The eluted DUBm complexes were further purified through a gel filtration Superdex 200 column (GE Healthcare).
Gel filtration.
The eluted DUBm complexes were loaded onto a Superdex 200 column equilibrated in elution base buffer. The column was eluted with 1.5 column volumes of buffer, and 40 fractions (each faction was 0.5 ml) were collected. Each fraction was concentrated to 30 μl by Amicon Ultra centrifugal filters (Millipore) and analyzed using Western blotting with anti-HA and anti-Flag antibodies. All these processes were performed at 4°C (29).
In vitro binding assay.
Recombinant HA-USP22 was incubated with recombinant Flag–ATXN7-24Q NT, Flag-ATXN7L3, or Flag-ENY2 at 16°C for 1 h. Then, the mixture was incubated with anti-Flag–agarose overnight at 4°C (28). After being washed 3 times with washing buffer, bound proteins were eluted with Flag peptide (0.2 mg/ml). The input and eluate were analyzed by immunoblotting with anti-Flag and anti-HA antibodies.
In vitro deubiquitination assay.
Total histones were prepared from HEK293T cells using a histone purification minikit (Active Motif). Free histones or nucleosomes (Epicypher) were incubated with purified recombinant USP22 or DUBm complex in 100 mM Tris-HCl, pH 8.0, 5% glycerol, 1 mM EDTA, 3 mM DTT for 2 h or the times specified below at 37°C as described previously (21). SDS loading buffer was added to stop the reaction, and then Western blotting was performed to analyze the products of the reaction using specific antibodies. For in vitro assays using ubiquitin vinyl sulfone (Ub-VS; Boston Biochem), the purified complexes were incubated with 1 μM or 5 μM Ub-VS in a reaction buffer (100 mM Tris-HCl, pH 8.0, 5% glycerol, 100 mM KCl, 3 mM DTT) for the times specified below at RT. Laemmli blue was added to stop the reactions, and then the reaction products were analyzed by immunoblotting. For reaction with ubiquitin–7-amido-4-methylcoumarin (Ub-AMC; Boston Biochem), the purified complexes were incubated in DUB buffer (50 mM HEPES, pH 7.9, 0.5 mM EDTA, 0.1 M NaCl, 1 mM DTT, 0.1 mg/ml bovine serum albumin) on ice for 30 min and then at RT for 5 min. Reactions were initiated by adding 0.5 μM ubiquitin-AMC in DUB buffer. The reactions were monitored continuously for 15 min by determination of the fluorescence at 25°C (excitation λ, 360 nm; emission λ, 528 nm; Biotek Synergy 2 apparatus) as described previously (30).
Preparation of cell lysates and Western blotting.
Whole-cell extracts (WCEs) were prepared for Western blotting using radioimmunoprecipitation assay buffer. The supernatants and pellets of WCEs from Sf21 cells or human astrocytes were separated after centrifuging at 15,000 rpm for 20 min at 4°C. Laemmli buffer was added to dissolve these pellets at 95°C for 5 to 10 min. Both the supernatant and pellet fractions of whole-cell lysates were analyzed by Western blotting with the indicated antibodies.
Cell culture.
Sf21 insect cells were grown in sf-900 II serum-free medium (SFM) (Life Technologies) supplemented with 1% penicillin-streptomycin (Thermo Scientific). Primary human astrocytes (N7805-100; Life Technologies) were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 1% N-2 supplement and 10% fetal bovine serum (FBS) (Life Technologies). Artificially immortalized human astrocytes were provided by Patrick A. Grant and grown in alpha minimal essential medium (Life Technologies) supplemented with 10% FBS (Life Technologies). HEK293T cells were acquired from the American Type Culture Collection and grown in high-glucose DMEM (Thermo Scientific) supplemented with 10% FBS and 1% penicillin-streptomycin (Thermo Scientific). Cells were passaged according to standard techniques.
Histone purification from ATXN7 wild-type and mutant adult mouse cerebellums.
Atxn7100Q alleles carried naturally occurring contracted mutations of the coding sequence for the 266-Q tract in Atxn7 obtained when maintaining the Atxn7266Q/5Q mouse line (34). Cerebellums from mice with all three different genotypes were collected and snap-frozen. Total histones from the cerebellums were prepared using a histone purification minikit (Active Motif). Laemmli buffer was added, and the lysates were analyzed by Western blotting. All procedures that involved animal handling were performed in accordance with the approved IACUC protocols at the M. D. Anderson Cancer Center.
Immunofluorescence microscopy.
Astrocytes were cultured on Geltrex lactate dehydrogenase-elevating virus-free, reduced growth factor basement membrane matrix (Life Technologies)-coated glass coverslips in 12-well plates for 24 to 48 h and then fixed with 3% formaldehyde, washed in phosphate-buffered saline (PBS), blocked with 10% fetal calf serum in PBS, and stained using primary antibodies (Sigma) and then secondary antibodies (Life Technologies). Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) in Vectashield mounting medium (Vector). Glass microslides with coverslips were allowed to set overnight to dry before coverslips were sealed with fingernail polish. Images were obtained using a laser scanning spectral confocal microscope (Leica STP6000).
RESULTS
Incorporation of ATXN7-poly(Q) into the DUBm promotes its solubility.
The ATXN7 N-terminal zinc finger domain is necessary and sufficient for the assembly and activation of the DUBm (Fig. 1A) (18, 31), The N-terminal part of the yeast ATXN7 homolog has been used previously for biochemical and structural analysis of the DUBm, but the yeast protein lacks the region where poly(Q) expansions occur in SCA7 disease (Fig. 1B). To define the effects of poly(Q) expansion on ATXN7 functions, we tested the assembly and enzymatic activity of an in vitro-reconstituted recombinant mammalian DUBm. We purified epitope-tagged forms of full-length USP22, ATXN7L3, and ENY2, along with two forms of the ATXN7 N-terminal domain containing either 24 Q residues (ATXN7-24Q NT; wild type [WT]) or 92 Q residues (ATXN7-92Q NT; mutant) (Fig. 1C). All of the DUBm components except for ATXN7-92Q NT were efficiently expressed as individual recombinant proteins and were readily purified using anti-Flag or anti-HA immunoprecipitation.
Since ATXN7-poly(Q) proteins form aggregates in mammalian cells (27), we reexamined our baculovirus supernatant and pellet fractions for expression of the ATXN7-24Q NT and ATXN7-92Q NT proteins. We found that ATXN7-24Q NT was soluble and readily detected in the supernatant (Fig. 1D, lanes 3 and 4), whereas the majority of ATXN7-92Q NT was insoluble and was detected mainly in the pellet (Fig. 1D, lanes 5 and 6). Importantly, the solubility of ATXN7-92Q NT was greatly increased upon coexpression with other DUBm components, as indicated by decreased amounts of the insoluble protein and a shift toward the soluble form (Fig. 1D, lanes 7 and 8). We successfully reconstituted DUBm complexes containing ATXN7-24Q NT or ATXN7-92Q NT with single affinity purification using HA-agarose beads, and we verified the proper assembly of all four subunits both by colloidal staining (Fig. 1E, lanes 1 and 2) and by immunoblotting with anti-HA and anti-Flag antibodies against the tagged subunits (Fig. 1F, lanes 1 and 2). These results indicate that the forms of ATXN7 expanded with poly(Q) do not prevent interactions with other DUB components and that the solubility and proper folding of ATXN7-92Q NT are enhanced by the incorporation into the DUBm.
ATXN7 enhances DUB activity in vitro.
The enzymatic activity of the SAGA DUB module requires the ATXN7 homolog in yeast (18), but ATXN7 was recently shown to be dispensable in the drosophila DUB module (31). A study with the mammalian components found that USP22 is partially activated in vitro upon interaction with ATXN7L3 and ENY2 and that full activity required additional interactions with ATXN7 (21), but those studies did not address the effects of poly(Q) expansions, as the construct used lacked the region that contains the poly(Q) repeats. To further address these questions, we first reconstituted subcomplexes by coinfecting various combinations of the three recombinant baculoviruses into Sf21 cells. We then isolated subcomplexes by immunoprecipitation using the HA epitope on USP22, including the catalytically inactive USP22 C185S mutant as a negative control (see Fig. S1A in the supplemental material). We found that USP22 forms tripartite subcomplexes with ATXN7L3 and ENY2 (see Fig. S1B, lane 1, and C, lane 2, in the supplemental material), with ATXN7-24Q NT and ENY2 (see Fig. S1D, lane 1), or with ATXN7-24Q NT and ATXN7L3 (see Fig. S1D, lane 2). We also tested the stability of pairwise interactions, but we found that the USP22–ATXN7-24Q NT subcomplex was the only stable two-component complex formed (see Fig. S1D, lane 4, in the supplemental material). Although USP22 interacts with either ATXN7-24Q NT or ATXN7L3, the formation of stable subcomplexes requires the presence of more than two subunits of the DUBm, showing that these interactions impact the enzymatic activity of the DUBm and, potentially, the stability of the subunits.
We next characterized the DUB activity of these subcomplexes using core histones isolated from human HEK293T cells as the substrates. No DUB activity was detected with USP22 alone (Fig. 2A, compare lanes 1 and 2) or in the presence of the USP22 C185S mutant (Fig. 2A, lanes 3 and 5), as expected. As previously reported, USP22 efficiently deubiquitinated free histone H2B when in complex with ATXN7L3 and ENY2 within 2 h (Fig. 2A, lane 4, and 2B, lane 3), whereas DUBm subcomplexes that lacked ATXN7L3 or ENY2 had no activity (Fig. 2B, lanes 2 and 4 to 6). Importantly, a time course experiment revealed that ATXN7-24Q NT further stimulated activity, as deubiquitination of both H2B and H2A (Fig. 2C and D, compare lanes 5 to 8 to lanes 9 to 12) was significantly more efficient in the presence of ATXN7. Deubiquitination of H2Aub and H2Bub in mononucleosomes was also more efficient in the presence of ATXN7-24Q NT (see Fig. S2A in the supplemental material; also data not shown).
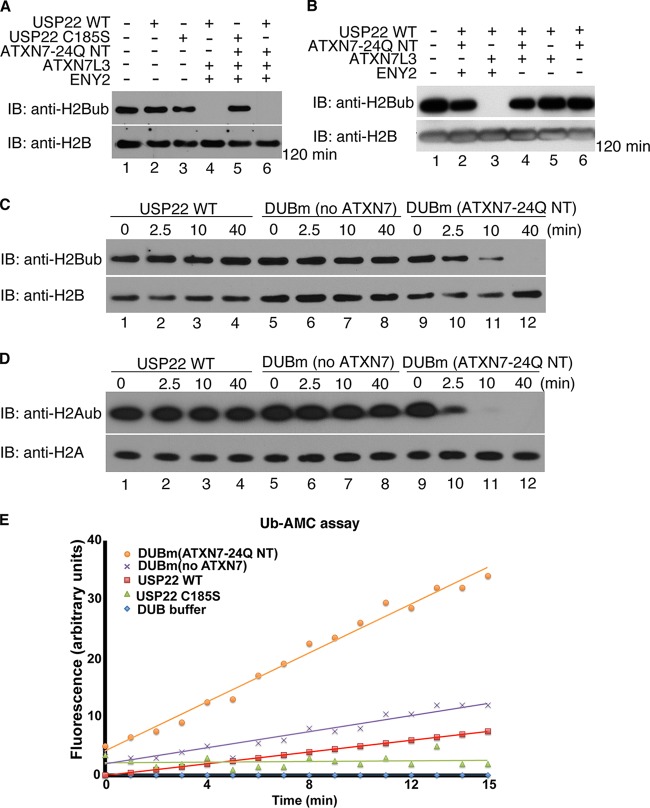
FIG 2.
ATXN7-24Q NT promotes robust DUB activity of the DUBm in vitro. (A) In vitro deubiquitination assays using core histones as the substrates were performed with USP22 alone (lanes 2 and 3), trimeric DUBm (no ATXN7-24Q NT; lane 4), and tetrameric DUBm (with ATXN7-24Q NT, lanes 5 and 6). The USP22 C185S catalytically inactive mutant served as a negative control (lanes 3 and 5). The trimeric and tetrameric complexes had similar activities on ubiquitinated H2B after 2 h. (B) In vitro DUB assays using core histones were performed for different combinations of DUBm subunits coinfected in Sf21 insect cells, in order to test potential enzymatic activity. Similar amounts of USP22 in these complexes were incubated with core histones for 2 h. No activity was observed in any dimeric complex (lanes 5 and 6), and the only trimeric complex with obvious enzymatic activity contained USP22, ATXN7L3, and ENY2 (lane 3). (C and D) In vitro DUB assays using core histones were performed with USP22 alone (lanes 1 to 4), DUBm (no ATXN7-24Q NT; lanes 5 to 8), and DUBm (with ATXN7-24Q NT; lanes 9 to 12), and samples were collected for analysis at the indicated time points. H2A (D) and H2B (C) ubiquitination was monitored with antibodies specific for these modifications. In all assays (A to D), core histones were purified from HEK293T cells. (E) Hydrolysis of ubiquitin-AMC (0.5 μM) alone or with similar amounts of recombinant USP22, DUBm (no ATXN7-24Q NT), and DUBm (with ATXN7-24Q NT). The release of AMC fluorescence was monitored by a spectrophotometer at 528 nm. The tetrameric DUBm with ATXN7-24Q NT showed the highest enzymatic activity. The USP22 WT or catalytic mutant was tagged with HA; ATXN7-24Q NT, ATXN7L3, and ENY2 were all tagged with Flag.
In order to further assess the stimulation of DUB activity by ATXN7-24Q NT, we used a fluorescent substrate, ubiquitin labeled with 7-amido-4-methylcoumarin at the C terminus (Ub-AMC), which provides a more quantitative assay for ubiquitin C-terminal hydrolase activity (32). We confirmed the requirement of ATXN7-24Q NT for robust Ub-AMC-hydrolyzing activity, as the reconstituted DUBm without ATXN7-24Q NT showed only minimal activity (Fig. 2E). Similarly, the DUBm without ATXN7L3 or ENY2 had no detectable activity on Ub-AMC (see Fig. S2B in the supplemental material). To extend these observations, we assayed the enzymatic activity of these different subcomplexes of the DUBm using ubiquitin vinyl sulfone (Ub-VS). Ub-VS is a DUB inhibitor that covalently attaches to the active site of the catalytic DUB subunit, causing a shifted migration upon gel electrophoresis (33). Although some shift of USP22 alone was observed when using higher concentrations of Ub-VS (1 μM versus 5 μM; see Fig. S2C and D in the supplemental material), both ATXN7L3 and ENY2 strongly promoted the reactivity of USP22 with Ub-VS (see Fig. S2D and E in the supplemental material), and this reactivity was further stimulated by ATXN7-24Q NT (see Fig. S2C and F). Altogether, these results demonstrate that ATXN7L3 and ENY2 allow minimal activation of USP22 but that full activation requires ATXN7.
Poly(Q)-ATXN7 does not impair the assembly or activity of the DUBm in vitro.
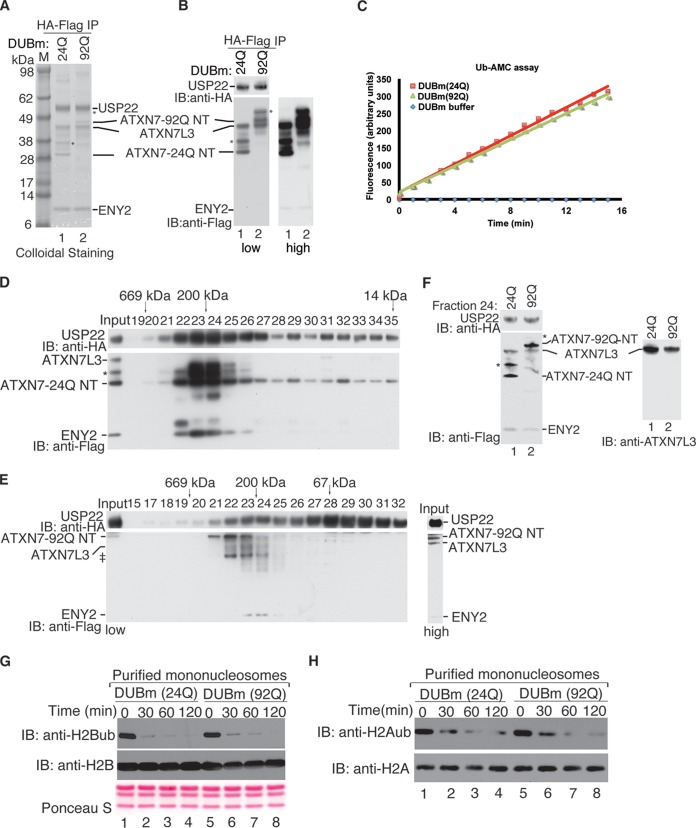
To determine the effects of ATXN7-92Q NT on DUBm activity, we performed a double-affinity purification using HA and Flag epitopes present on different DUBm subunits. A stable DUBm complex was detected both by colloidal staining (Fig. 3A, lane 2) and by immunostaining with anti-HA and anti-Flag antibodies (Fig. 3B, lane 2). Comparison of the amounts of DUBm subunits present in the ATXN7-24Q NT or ATXN7-92 NT complexes showed very similar, stoichiometric ratios (Fig. 3B, lanes 1 and 2). Importantly, Ub-AMC assays revealed no differences in the activities of these complexes (Fig. 3C). To verify these results, we combined HA immunoprecipitation of the DUBm with gel filtration. Immunoblotting of column fractions indicated that the fully assembled DUB complex eluted with an apparent molecular mass of ∼200 kDa (Fig. 3D and E). We normalized the amount of ATXN7-24Q NT with the amount of ATXN7-92Q NT collected from fractions 23 and 24, as confirmed by immunoblotting (Fig. 3F), and then carried out DUB assays using core histones (data not shown) or mononucleosomes as the substrates. We again found that ATXN7-92Q NT stimulated DUB activity to the same degree as ATXN7-24Q NT (Fig. 3G and H). Our results demonstrate that a poly(Q) expansion in ATXN7 equivalent to that found in SCA7 patients does not affect the enzymatic activity of the SAGA DUBm in vitro.
FIG 3.
ATXN7-92Q NT is not defective in promoting DUBm activity. (A) Purification of the recombinant DUBm using two sequential affinity precipitations with anti-HA– and anti-Flag–agarose beads, respectively. Comparison of the two purified DUBm complexes containing ATXN7-24Q NT or ATXN7-92Q NT with colloidal staining shows very similar ratios of the DUBm subunits. Lane M, molecular mass markers. (B) Immunoblot analysis (low and high exposures are shown) of the DUBm after double-affinity purification (A) verifies the identities and the presence of similar amounts of DUBm components. The different subunits were detected with anti-HA and anti-Flag antibodies, as indicated. *, modified forms of ATXN7-24Q NT or ATXN7-92Q NT due to ubiquitination (data not shown). (C) Comparative Ub-AMC hydrolysis assays using the reconstituted DUBm complexes with either ATXN7-24Q NT or ATXN7-92Q NT after the double-affinity purification confirm the very similar deubiquitinating activities between the two complexes. (D and E) Purification of reconstituted DUBm containing ATXN7-24Q NT (C) or ATXN7-92Q NT (D) by size exclusion chromatography using Superdex 200 gel filtration. Column fractions (numbers above the blots) were analyzed by immunoblotting with anti-HA (USP22) and anti-Flag (ATXN7, ATXN7L3, and ENY2) antibodies, and band positions for these components are as indicated. The elution profile of protein markers is indicated at the top of the blots. A complex containing all four subunits, including either ATXN7-24Q NT or ATXN7-92Q NT, is present in fractions 23 and 24 of both purifications and corresponds to the expected molecular mass for the DUBm. A darker exposure of the input lane for the ATXN7-92Q NT purification (labeled high) is provided to confirm the presence of all components in the starting material. ‡, degradation fragments. (F) Comparison of the integrity and stoichiometry of fraction 24 of the gel filtration from panels D and E was performed by immunoblotting with anti-HA and anti-Flag antibodies. Note that the amounts of the different subunits between the two reconstituted DUBm complexes are very similar. *, modified form of ATXN7-24Q NT or ATXN7-92Q NT due to ubiquitination (data not shown). (G and H) In vitro DUB assays using purified mononucleosomes as the substrate were performed with both DUB modules, and H2A and H2B ubiquitination levels were monitored with antibodies specific for these modifications. Both reconstituted complexes showed very similar enzymatic activities toward both ubiquitinated histone H2A and H2B incorporated into nucleosomes at the time points analyzed. H2A or H2B immunoblotting or Ponceau S staining served as a loading control.
ATXN7-92Q aggregates sequester DUBm components in vivo.
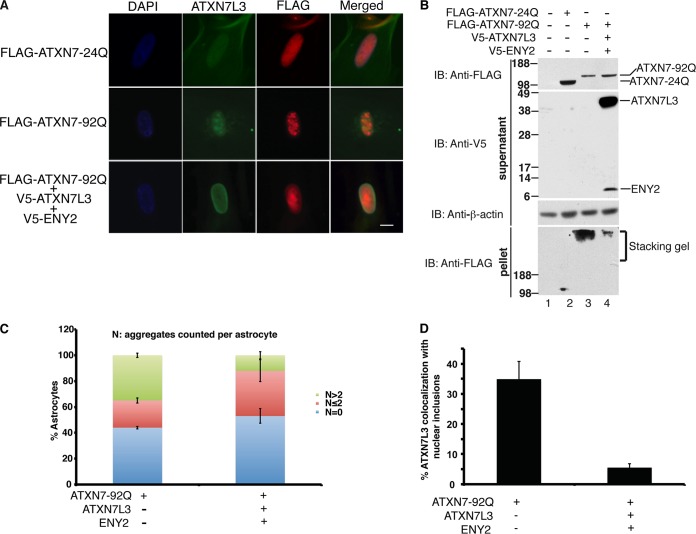
Aggregation of poly(Q) proteins is a hallmark of CAG expansion disorders (2). Aggregates associated with ATXN7-poly(Q) mutants also include transcriptional regulators, ubiquitin/proteasome proteins, cell death-associated proteins, chaperones, and ATXN7 protein partners (1). Previous work indicated that expression of ATXN7-92Q, but not that of ATXN7-24Q, leads to formation of aggregates that also contained Gcn5 in human astrocytes (27). We asked whether other DUBm components were also present within aggregates using double immunofluorescence staining with anti-ATXN7L3 antibodies and anti-Flag to detect ATXN7-92Q. We found that endogenous ATXN7L3 was present in nuclear inclusions in ∼35% of astrocytes containing ATXN7-92Q aggregates (Fig. 4A, middle row, and D [n = 86 aggregates]; see also Fig. S3 in the supplemental material). Since the solubility of ATXN7-92Q was improved in baculovirus upon coexpression of other DUBm subunits (Fig. 1D), we asked whether such coexpression would also increase solubility and decrease aggregation in mammalian cells. Indeed, immunoblot analyses of human astrocyte extracts showed that cooverexpression of ATXN7L3 and ENY2 greatly inhibited aggregation of ATXN7-92Q (Fig. 4B, compare lane 3 and 4). Cooverexpression of ATXN7L3 and ENY2 also clearly suppressed formation of ATXN7-92Q aggregates, as detected by immunofluorescence (Fig. 4A, bottom row; see also Fig. S3 in the supplemental material). We further quantified this effect by counting cells showing no aggregates (n = 0), 1 or 2 aggregates (n ≤ 2), or more than 2 aggregates (n > 2). Cooverexpression of ATXN7L3 and ENY2 greatly decreased the number of cells with more than 2 aggregates per nucleus (n > 2; ∼40% versus ∼10% in ATXN7-92Q alone) (Fig. 4C). Consistent with these results, colocalization of ATXN7L3 with nuclear inclusions also decreased upon cooverexpression of ATXN7L3 and ENY2 (Fig. 4A and B) (∼35% versus ∼5%; n = 86 aggregates). Overexpression of only ATXN7L3 or ENY2 with ATXN7-92Q did not cause any obvious decrease in aggregation of ATXN7-92Q (see Fig. S4A in the supplemental material; also data not shown). These results indicate that aggregates caused by ATXN7-poly(Q) mutants may be cytotoxic due to sequestration of important DUBm components and that cooverexpression of both ATXN7L3 and ENY2 may oppose this toxicity through suppression of aggregate formation.
FIG 4.
Overexpression of ATXN7L3 and ENY2 prevents sequestration of ATXN7L3 into aggregates with ATXN7-92Q in vivo. (A) Exogenous expression of ATXN7-92Q, but not that of ATXN7-24Q, causes aggregate formation in human astrocytes. ATXN7L3 colocalization with nuclear inclusions in ATXN7-92Q-expressing astrocytes was observed by immunofluorescence staining using anti-Flag and anti-ATXN7L3 antibodies. Cooverexpression of ATXN7L3 and ENY2 reduces this colocalization by inhibiting ATXN7-92Q aggregation. Nuclei were stained with DAPI (blue). Bar, 10 μm. (B) Upon cooverexpression of ATXN7L3 and ENY2 with ATXN7-92Q, the amount of soluble ATXN7-92Q in the supernatant fraction increased, whereas the amount of SDS-insoluble ATXN7-92Q aggregates in the pellet (trapped in the stacking gel) dramatically decreased compared to the amount obtained with overexpression of ATXN7-92Q alone. ATXN7-24Q is highly soluble. Anti-β-actin immunoblotting served as a loading control. ATXN7-24Q and ATXN7-92Q were tagged with Flag. ATXN7L3 and ENY2 were tagged with a V5 epitope. (C) Quantification of astrocytes containing nuclear inclusions. One hundred cells from multiple fields were counted and then sorted by the number of nuclear inclusions per cell (N). Three independent experiments were performed, and a two-tailed Student t test was used for statistical analysis; error bars represent ±SDs. (D) Quantification of astrocytes with endogenous ATXN7L3 colocalization with nuclear inclusions. Three independent experiments were performed, and a two-tailed Student t test was used for statistical analysis; error bars represent SDs.
Increased solubility of ATXN7-92Q rescues DUB activity in vivo.
Given that depletion of ATXN7L3 causes robust increases in global H2Bub levels (21), sequestration of ATXN7L3 into ATXN7-92Q aggregates might also affect global H2Bub levels. Consistent with previous work (27), we found that H2Bub levels were increased in ATXN7-92Q-overexpressing astrocytes (Fig. 5A). Interestingly, acetylation of H3K9 and K14 was not altered in ATXN7-92Q-overexpressing cells (Fig. 5A and B), even though Gcn5 is also associated with aggregates in ATXN7-poly(Q) cells (27). Importantly, cooverexpression of ATXN7L3 and ENY2 opposed the increase in global H2Bub levels (Fig. 5B), but overexpression of USP22 or ENY2 alone did not (see Fig. S4B in the supplemental material), suggesting that DUB activity compromised by aggregate formation can be rescued by cooverexpression of ATXN7L3 and ENY2. Together with our biochemical data that indicate that ATXN7L3 and ENY2 are required to activate USP22 activity, these findings suggest that aggregates caused by ATXN7-poly(Q) likely contribute to neuronal toxicity by impairing DUB activity through sequestering important DUBm components away from their substrates and that the increased solubility of the ATXN7-poly(Q) mutant by cooverexpression of DUBm components can counteract this toxicity.
FIG 5.
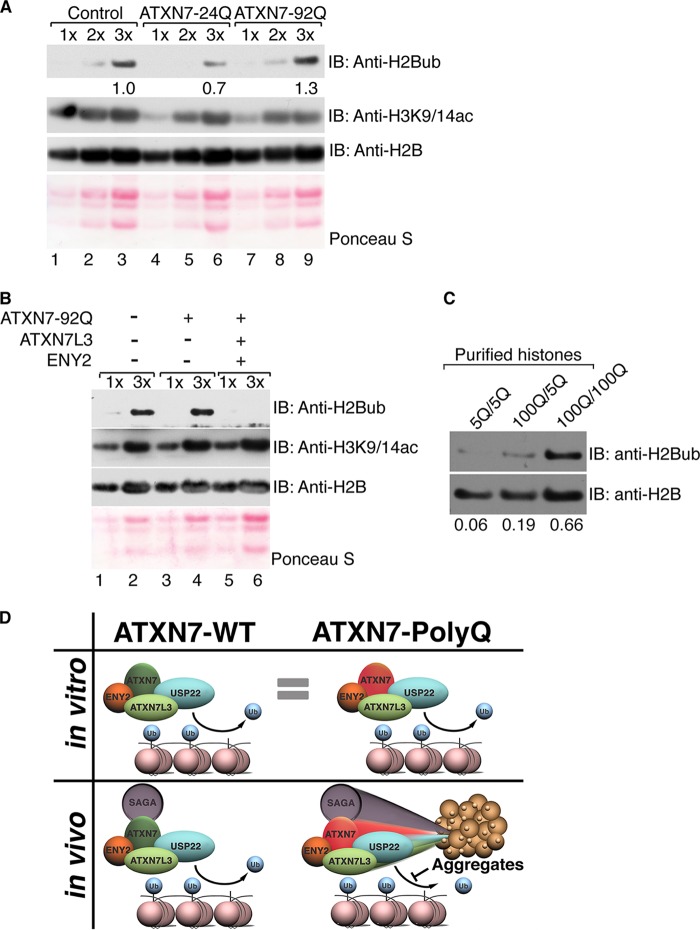
Solubility of ATXN7-92Q in vivo correlates with DUB activity. (A) Core histones were purified from human astrocytes infected with empty vector (lanes 1 to 3), ATXN7-24Q (lanes 4 to 6), or ATXN7-92Q (lanes 7 to 9) and were analyzed by electrophoresis of increasing sample amounts (1× to 3×), and the levels of H2Bub were monitored with antibodies specific for this modification. The levels of H2B ubiquitination increased in astrocytes expressing ATXN7-92Q, indicating impaired deubiquitination. Quantification of H2Bub levels was normalized to H2B levels using ImageJ software. (B) Cooverexpression of ATXN7L3 and ENY2 enhances global H2Bub deubiquitination in astrocytes expressing ATXN7-92Q, whereas H3K9/14ac levels are not altered. Different sample amounts (1× and 3×) were analyzed. H2B immunostaining or Ponceau S staining served as a loading control. (C) Core histones were purified from the cerebellums of 4-month-old mice bearing wild-type (5Q) or mutant (100Q) alleles of the ATXN7 gene. Both ATXN7100Q/100Q and ATXN7100Q/5Q mice exhibit SCA7 symptoms over time, but disease progression in ATXN7100Q/100Q mice was faster and more severe than that in ATXN7100Q/5Q mice. Immunoblot analysis was used to detect H2Bub levels with anti-H2Bub antibody. Samples from both ATXN7100Q/100Q and ATXN7100Q/5Q mice show increased H2B ubiquitination, with the effect being more severe in the samples from ATXN7100Q/100Q mice. H2Bub levels were normalized to H2B levels using ImageJ software. Representative results chosen from more than three independent experiments are shown. (D) Model of the contribution of ATXN7-poly(Q) to SCA7 disease in humans. In vitro, soluble DUBm with ATXN7-92Q exhibits DUB activity comparable to that of the DUBm with ATXN7-24Q. However, in vivo ATXN7-poly(Q) tends to form aggregates that sequester DUBm components away from their substrates. Incorporation into DUBm can help solubilize ATXN7-poly(Q) in vitro and in vivo, partially alleviating the formation of the poly(Q)-containing aggregates in astrocytes. Impaired DUB activity in the cerebellums of SCA7 patients expressing ATXN7-poly(Q) likely leads to increased global H2Bub levels, as well as the deregulation of other DUBm substrates, contributing to SCA7 disease.
To determine if the sequestration of DUBm components influences H2Bub in a mouse model of SCA7 (12, 34), we extracted histones from the cerebellums of mice bearing one or two alleles of the ATXN7100Q gene and compared H2Bub levels to those of histones extracted from age-matched wild-type ATXN75Q mice. We found a slight increase in H2Bub levels in histones isolated from mice heterozygous for the expanded ATXN7 allele (ATXN7100Q/5Q) and an even greater increase in H2Bub levels in ATXN7100Q homozygous mice (Fig. 5C). These results further indicate that DUB activity is indeed impaired by the ATXN7 poly(Q) expansions in a SCA7 mouse model.
DISCUSSION
The role of hallmark nuclear aggregates caused by poly(Q) expansions in the pathogenesis of neurodegenerative diseases is still controversial (2). Some studies suggest that nuclear inclusions are cytotoxic, whereas others indicate that they might be cytoprotective by acting to sequester the misfolded disease proteins (3, 35, 36). Our results demonstrate that although poly(Q) expansions in ATXN7 have no effect on the enzymatic activity or assembly of the SAGA DUBm in vitro, they promote aggregation in vivo, thereby sequestering crucial activators of USP22 activity away from H2Bub and other substrates. Our data, then, suggest that the nuclear aggregates lead to a loss of effective function for ATXN7-poly(Q) and associated proteins in vivo, favoring a cytotoxic rather than a cytoprotective role for the nuclear inclusions observed in SCA7 disease (Fig. 5D). Sequestration of crucial activators of USP22 away from H2Bub and other substrates likely impairs the efficient transcription of key genes relative to the development of SCA7, such as reelin (27).
Interestingly, we observed that the solubility of ATXN7-92Q in insect cells improved upon incorporation into the DUBm. Consistent with these findings, cooverexpression of ATXN7L3 and ENY2 in human astrocytes obviously suppressed ATXN7-92Q aggregation, indicating that imbalances in the levels of the poly(Q) protein relative to the levels of the other DUB components may contribute to disease development.
Based on the crystal structure of the yeast DUBm, Samara et al. (20) proposed that ATXN7 poly(Q) expansions either would disrupt interactions with the other DUBm subunits to compromise deubiquitinating activity or would cause ATXN7 to aggregate. Although the position of the Q tract in yeast Sgf73 is different from that in ATXN7, our experiments show that the presence of poly(Q) expansions does not affect the assembly or the activity of the DUBm in vitro. An intriguing interaction between the N-terminal portion of Sgf73, Sgf11, and Sus1 stabilizes module assembly and may explain how overexpression of the mammalian counterparts, ENY2 and ATXN7L3, promotes solubilization of mutant ATXN7 (19). The N-terminal tail of Sgf73 is not required for interaction with Ubp8 but is fundamental for generation of a three-protein junction between the N-terminal H1 helix of Sgf11, the C-terminal H5 helix of Sus1, and the N-terminal H1 helix of Sgf73 (19). Although structural information for the mammalian ATXN7 is lacking, the N-terminal tail of ATXN7 could nucleate similar interactions with ATXN7L3 and ENY2, and the subsequent aggregation caused by the poly(Q) proteins might be prevented when these associations are formed. The poly(Q) protein also has an extended half-life relative to that of nonexpanded ATXN7 (37), which could promote the accumulation and aggregation of ATXN7-poly(Q) over time. This process might explain the delayed onset of SCA7 and also the inverse relationship between poly(Q) length and age of disease onset. Limiting amounts of DUBm subunits in neural or retinal cells could also explain why these specific cell types selectively degenerate in SCA7.
We have previously shown that Gcn5 is present in aggregates formed by ATXN7-92Q (27) and that the loss of Gcn5 functions can contribute to SCA7 progression in mice (12). Our current analysis of global histone levels purified from human astrocytes confirms that H2Bub levels increase in the presence of ATXN7-92Q, while no changes in the levels of H3K9/K14 acetylation occur. Despite the fact that Gcn5 is also sequestered to nuclear foci, the unchanged levels of H3K9/K14 acetylation might be explained by functional compensation by other HAT modules present in human astrocytes, such as PCAF. Interestingly, trichostatin A (TSA) treatment of the human astrocytes rescues DUB activity but also increases the number of nuclear foci. McCullough et al. (27) suggested that TSA treatment induces the aggregation of mutant ATXN7 into nuclear foci, making room for the endogenous, wild-type form of ATXN7 to work more efficiently toward H2B deubiquitination. From our results, we now propose a different mechanism for the rescue of DUB activity. TSA might increase the expression levels of endogenous DUB subunits, allowing greater solubility and activity of ATXN7-poly(Q) within the DUBm.
Our current work and previous work suggest that sequestration of both enzymatic centers in SAGA upon ATXN7 poly(Q) expansion likely contributes to SCA7 development and progression. Our current findings indicate that therapies designed to improve the deubiquitinating or the acetyltransferase activity of SAGA may have limited success in reversing the symptoms of SCA7 disease, unless they also prevent aggregation of poly(Q)-expanded ATXN7.
Supplementary Material
ACKNOWLEDGMENTS
We thank Boyko Atanassov for many helpful discussions on this project, Didier Devys for ATXN7L3 and ENY2 cDNAs, and Huda Zoghbi for providing the SCA7 mouse model.
This work was supported by a research training award from the Cancer Prevention and Research Institute of Texas (CPRIT RP140106) to A.C.S. and by NIH grant 1R01-GM096472 to S.Y.R.D.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01454-14.
REFERENCES
- 1.Lebre AS, Brice A. 2003. Spinocerebellar ataxia 7 (SCA7). Cytogenet Genome Res 100:154–163. doi: 10.1159/000072850. [DOI] [PubMed] [Google Scholar]
- 2.Todd TW, Lim J. 2013. Aggregation formation in the polyglutamine diseases: protection at a cost? Mol Cells 36:185–194. doi: 10.1007/s10059-013-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 4.Giunti P, Stevanin G, Worth PF, David G, Brice A, Wood NW. 1999. Molecular and clinical study of 18 families with ADCA type II: evidence for genetic heterogeneity and de novo mutation. Am J Hum Genet 64:1594–1603. doi: 10.1086/302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koutelou E, Hirsch CL, Dent SY. 2010. Multiple faces of the SAGA complex. Curr Opin Cell Biol 22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samara NL, Wolberger C. 2011. A new chapter in the transcription SAGA. Curr Opin Struct Biol 21:767–774. doi: 10.1016/j.sbi.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Navarro S. 2009. Insights into SAGA function during gene expression. EMBO Rep 10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. 2005. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A 102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR III, Grant PA. 2005. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci U S A 102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, Picaud S, Zoghbi HY, Trottier Y, Tora L, Devys D. 2006. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol 4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke TL, Miller JL, Grant PA. 2013. Direct inhibition of Gcn5 catalytic activity by polyglutamine-expanded ataxin-7. J Biol Chem 288:34266–34275. doi: 10.1074/jbc.M113.487538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YC, Gatchel JR, Lewis RW, Mao CA, Grant PA, Zoghbi HY, Dent SY. 2012. Gcn5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model. Hum Mol Genet 21:394–405. doi: 10.1093/hmg/ddr474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YC, Grant PA, Dent SYR. 2012. SCA7: is the rub in the DUB? Trends Cell Mol Biol 7:73–79. [Google Scholar]
- 14.Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van-Dorsselaer A, Wurtz JM, Mandel JL, Tora L, Devys D. 2004. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet 13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. 2005. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol 25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. 2005. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol 25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. 2008. Yeast ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol 10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 19.Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. 2009. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell 35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson MA, Koutelou E, Hirsch C, Akdemir K, Schibler A, Barton MC, Dent SY. 2011. Ubp8 and SAGA regulate Snf1 AMP kinase activity. Mol Cell Biol 31:3126–3135. doi: 10.1128/MCB.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atanassov BS, Dent SY. 2011. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep 12:924–930. doi: 10.1038/embor.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. 2008. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol 10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 26.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, Hublarova P, Moyal L, Gana-Weisz M, Shiloh Y, Yarden Y, Johnsen SA, Vojtesek B, Berger SL, Oren M. 2008. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev 22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCullough SD, Xu X, Dent SY, Bekiranov S, Roeder RG, Grant PA. 2012. Reelin is a target of polyglutamine expanded ataxin-7 in human spinocerebellar ataxia type 7 (SCA7) astrocytes. Proc Natl Acad Sci U S A 109:21319–21324. doi: 10.1073/pnas.1218331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE. 2006. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol 8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- 29.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, Conaway JW. 2008. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell 31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohan RD, Dialynas G, Weake VM, Liu J, Martin-Brown S, Florens L, Washburn MP, Workman JL, Abmayr SM. 2014. Loss of Drosophila ataxin-7, a SAGA subunit, reduces H2B ubiquitination and leads to neural and retinal degeneration. Genes Dev 28:259–272. doi: 10.1101/gad.225151.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Case A, Stein RL. 2006. Mechanistic studies of ubiquitin C-terminal hydrolase L1. Biochemistry 45:2443–2452. doi: 10.1021/bi052135t. [DOI] [PubMed] [Google Scholar]
- 33.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. 2001. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J 20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo SY, Pennesi ME, Weeber EJ, Xu B, Atkinson R, Chen S, Armstrong DL, Wu SM, Sweatt JD, Zoghbi HY. 2003. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron 37:383–401. doi: 10.1016/S0896-6273(02)01190-X. [DOI] [PubMed] [Google Scholar]
- 35.Yang W, Dunlap JR, Andrews RB, Wetzel R. 2002. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum Mol Genet 11:2905–2917. doi: 10.1093/hmg/11.23.2905. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi T, Kikuchi S, Katada S, Nagai Y, Nishizawa M, Onodera O. 2008. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Hum Mol Genet 17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 37.Yvert G, Lindenberg KS, Devys D, Helmlinger D, Landwehrmeyer GB, Mandel JL. 2001. SCA7 mouse models show selective stabilization of mutant ataxin-7 and similar cellular responses in different neuronal cell types. Hum Mol Genet 10:1679–1692. doi: 10.1093/hmg/10.16.1679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.