Abstract
Multi-atlas labeling has come in wide spread use for whole brain labeling on magnetic resonance imaging. Recent challenges have shown that leading techniques are near (or at) human expert reproducibility for cortical gray matter labels. However, these approaches tend to treat white matter as essentially homogeneous (as white matter exhibits isointense signal on structural MRI). The state-of-the-art for white matter atlas is the single-subject Johns Hopkins Eve atlas. Numerous approaches have attempted to use tractography and/or orientation information to identify homologous white matter structures across subjects. Despite success with large tracts, these approaches have been plagued by difficulties in with subtle differences in course, low signal to noise, and complex structural relationships for smaller tracts. Here, we investigate use of atlas-based labeling to propagate the Eve atlas to unlabeled datasets. We evaluate single atlas labeling and multi-atlas labeling using synthetic atlases derived from the single manually labeled atlas. On 5 representative tracts for 10 subjects, we demonstrate that (1) single atlas labeling generally provides segmentations within 2mm mean surface distance, (2) morphologically constraining DTI labels within structural MRI white matter reduces variability, and (3) multi-atlas labeling did not improve accuracy. These efforts present a preliminary indication that single atlas labels with correction is reasonable, but caution should be applied. To purse multi-atlas labeling and more fully characterize overall performance, more labeled datasets would be necessary.
Keywords: Multi-Atlas Segmentation, Diffusion Tensor Imaging, White Matter
1. INTRODUCTION
Labeling white matter regions of interest poses interesting challenges relative to other anatomical structures[1, 2] due to the need to integrate both local and global information. The Eve white matter atlas has become a de facto atlas of the white matter labels. The Eve atlas provides labels along with fractional anisotropy (FA) maps, T1 weighted structural MRI, and multi-modal information. There are two primary barriers to use of Eve within atlas-based frameworks: (1) only one subject is labeled, and (2) the peripheral white matter regions are conservatively labeled (Figure 1). Here, we evaluate white-matter segmentation with single and multi-atlas approaches.
Figure 1.
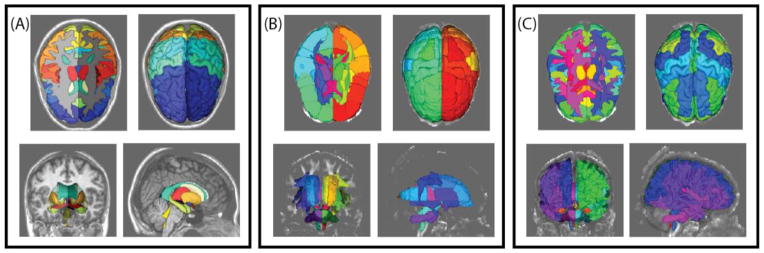
Example renderings of (A) BrainCOLOR cortical and sub-cortical labels, (B) Eve cortical and white matter labels and (C) rectified BrainCOLOR and Eve cortical and white matter labels.
2. METHODS
In this section, we describe the registration and segmentation procedures used for characterization of the white matter regions using the Eve atlas (Figure 2). Due to the unavailability of multiple labeled Eve atlases, several approaches were considered independently and jointly to segment the white matter. (1) First, we consider the registration framework used for registration of atlas images to target images by evaluating a standard single-modal registration of FA to FA in comparison to a joint cost multi-modal registration of T1+FA to T1+FA using the Advanced Normalization Toolkit (ANTs)[3]. (2) Second, we manually selected “atlases” from subjects visually verified to be well segmented by the single-atlas segmentation. We incorporated these atlases into a multi-atlas framework using Non-Local Spatial STAPLE (NLSS) [4, 5]. (3) Third, we incorporated the results of a cortical segmentation using the BrainCOLOR label set to serve as a guide to rectify the Eve labels to a well-defined white matter region.
Figure 2.
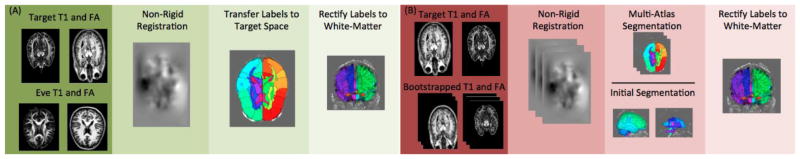
Flowcharts representing the approach for (A) single-atlas segmentation and (B) multi-atlas segmentation. The single-atlas segmentation uses non-rigidly registration of the Eve FA and T1 volumes to the target image and warping the labels to the target space. The resultant white matter labels are then rectified with the BrainCOLOR labels to form a unified segmentation. The multi-atlas segmentation uses a set of manually selected single-atlas samples as the input atlases and non-rigidly registers them to the target. The atlas labels are then fused through a label fusion approach and the resultant labels are rectified with the BrainCOLOR labels to form a unified segmentation.
2.1 Registrations
Each subject and atlas had both an FA map and structural T1 MRI. For each subject, the FA map was rigidly registered to the T1 volume to provide a consistent image space. Registration was performed two distinct ways using ANTs. First, registration considered only the FA map; second, the FA and T1 were used jointly and the cost functions for the multimodal registration were weighted equally. During translation, rotation, and affine registrations mutual information was maximized and locally normalized cross-correlation was maximized during non-rigid registration.
2.2 Single-Atlas Segmentation
Single-atlas segmentations were performed using both registration protocols. For the single-atlas single-modal (SASM) approach, the Eve FA map was non-rigidly registered to the target FA map using the previously defined registration protocol and the labels were transferred to target space with the resulting deformation field. For the single-atlas multimodal (SAMM) approach, the Eve FA map and structural T1 MRI were non-rigidly registered jointly to the target FA map and T1 image and the labels were transferred to target space with the resulting deformation field.
2.3 Multi-Atlas Segmentation
Since only one Eve subject was labeled and made available, multi-atlas segmentation is not feasible using the standard paradigm of register atlas images to the target, deform labels to the target image, and fuse the information between the registered labels and images. To get around this issue, we identified a set of “atlases” by segmenting subjects with the previously described SAMM protocol and manually identifying five subjects, which appeared to have better initial segmentations than others. The five new atlas-subjects are then registered to ten independent subjects using each of the previously defined single- and multi-modal registration procedures. The result of the two registration protocols were then independently segmented using Non-Local Spatial STAPLE (NLSS) to produce multi-atlas single-modal (MASM) and multi-atlas multi-modal (MAMM) segmentations.
2.4 Label Rectification
The Eve atlas was conservative in its labeling protocol and it left substantial space within the white matter either empty or defined as cortical surface. To overcome this, we segmented all of the subjects in the cohort with a multi-atlas segmentation with the BrainCOLOR labelset. The white-matter labels from the BrainCOLOR segmentation were then extracted. The white-matter labels from Eve were then grown to fill the space defined by the BrainCOLOR segmentation by finding voxels identified to be within the BrainCOLOR white-matter region neighboring at least on voxel identified as a white-matter label in Eve and selecting the most commonly occurring Eve white-matter label around it as its label. This process was performed iteratively until all of the space of the BrainCOLOR segmentation was filled with Eve labels or no more voxels could be reached. Rectification was incorporated with the four previous segmentation protocols producing four new segmentations, single-atlas single-modal rectified (SASMR), single-atlas multi-modal rectified (SAMMR), multi-atlas single-modal rectified (MASMR), and multi-atlas multi-modal rectified (MAMMR).
2.5 Methodological Comparison
To evaluate the results of the registration and segmentation protocols, a neuro-psychology doctoral scholar identified five tracts of interest (from the EVE dictionary) that were spread amongst the hemispheres of the brain. The selected tracts were the left external capsule, left middle cerebellar peduncle, left posterior thalamic radiation, right anterior corona radiata, and the corpus callosum. These tracts represent tracts in both hemispheres (identified by left and right), a corticospinal tract (anterior corona radiata), and a tract spanning the hemispheres (corpus callosum) and were selected as a representative sample of differing tracts throughout the brain. The doctoral scholar manually traced the tracts in ten randomly selected subjects to generate gold-standard segmentations for comparison with the automated transfer of Eve labels using FSL tools with the FA and principal Eigenvector maps. Mean surface distances and Hausdorff distances were calculated between each ground-truth and automated segmentation pair.
3. RESULTS
Multi-modal registration decreased the median Hausdorff distance by 1.02mm in the single-atlas non-rectified segmentation of the right anterior corona radiata (12.6% decrease) and of 0.936mm in the single-atlas non-rectified segmentation of the left posterior thalamic radiation (12.7% decrease) both of which are significant at significance level of p<0.05 (two sided t-test) (Figure 3). Figure 4 presents mean surface distance metrics for each tract. Multi-atlas segmentation decreased the median Hasudorff distance by 1.5mm compared to single-atlas single-modal in the right anterior corona radaita (18.8% decrease), and 0.55mm compared to single-atlas multi-modal in the right anterior corona radiate (7.8% decrease), but these results are marginally significant (p<0.1).
Figure 3.
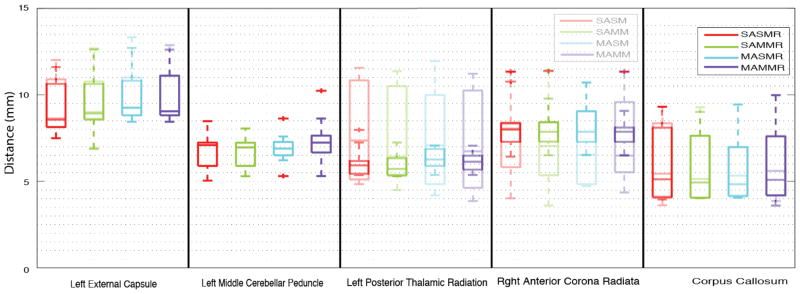
Hausdorff distances between segmentation protocols. The lighter bars indicate the results without rectification.
Figure 4.
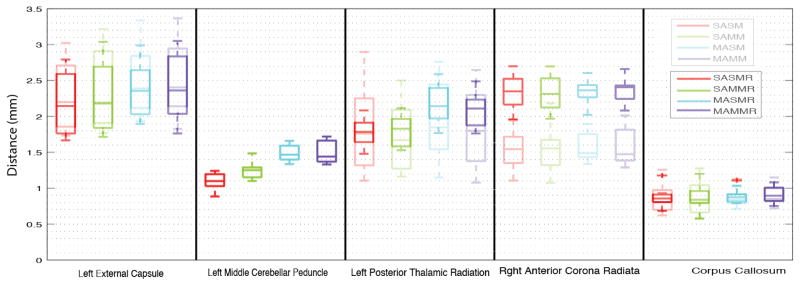
Mean surface distances between segmentation protocols. The lighter bars indicate the results without rectification.
Rectification provided the most consistent improvements of any segmentation technique. Within the left posterior thalamic radiation, label rectification reduced the median Hausdorff distance of single-atlas single-modal segmentation by 1.45mm (19.7% decrease), 0.72mm compared to single-atlas multi-modal segmentation (11.1% decrease), 0.84mm compared to multi-atlas single-modal segmentation (11.8% decrease), and 0.61mm compares the multi-atlas multi-modal segmentation (9.5% decrease). Within the right corpus callosum, label rectification reduced the median Hausdorff distance in single-atlas single-modal segmentation by 0.33mm (6.1% decrease), 0.22mm compares to single-atlas multimodal segmentation (4.2% decrease), 0.51mm compared to multi-atlas single-modal segmentation (9.6% decrease), and 0.53mm compares to multi-atlas multi-modal segmentation (9.5% decrease). The changes within the corpus callosum are statistically significant at a significance level of p<0.05.
No segmentation protocol produced consistently superior segmentation results. For the left external capsule the lowest median Hausdorff distance was achieved by single-atlas single-modal rectified segmentation (8.55mm) followed by single-atlas single-modal segmentation (8.65mm). For the left middle cerebellar peduncle the lowest median Hausdorff distance was achieved by multi-atlas multi-modal segmentation (6.89mm) and multi-atlas multi-modal segmentation produced the same value. For the left posterior thalamic radiation, the lowest median Hausdorff distance was achieved by single-atlas multi-modal rectified segmentation (5.71mm) followed by single-atlas single-modal rectified segmentation (5.91mm). For the right anterior corona radiata, the lowest median Hausdorff distance was achieved by multi-atlas multi-modal segmentation (6.49mm), followed by multi-atlas single-modal segmentation (6.55mm). For the corpus callosum, the lowest median Hausdorff distance was achieved by multi-atlas single-modal rectified segmentation (4.83mm), followed by single-atlas multi-modal rectified segmentation (4.93mm).
4. DISCUSSION
This work presents an initial study of atlas-based labeled with Eve. Conclusions should be treated as preliminary given the limited number of tracts that were manually labels and the difficulty of generalizing to all Eve white matter regions. In general, results were reassuring with mean surface distances of <2mm. However, Hausdorff distances were much greater (>10mm). Rectification with an automated multi-atlas approach focusing gray matter reduced outliers. Perhaps surprisingly, manually selected “good” single atlas results did not provide an effective alternative to multi-atlas labeling. Continued validation of Eve atlas propagation hinges upon the availability of additional subjects labeled with an equivalent protocol. As an additional benefit, true multi-atlas approaches would be possible with independently labeled subjects.
Acknowledgments
This work was supported in part by the Robert J. Kleberg, Jr. and Helen c. Kleberg Foundation. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. The project described was supported by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translation Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH,
References
- 1.Mori S, Oishi K, Jiang H, et al. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40(2):570–82. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oishi K, Faria A, Jiang H, et al. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer’s disease participants. Neuroimage. 2009;46(2):486–99. doi: 10.1016/j.neuroimage.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avants BB, Tustison NJ, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–44. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asman AJ, Landman BA. Formulating spatially varying performance in the statistical fusion framework. IEEE Trans Med Imaging. 2012;31(6):1326–36. doi: 10.1109/TMI.2012.2190992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asman AJ, Landman BA. Non-local statistical label fusion for multi-atlas segmentation. Med Image Anal. 2013;17(2):194–208. doi: 10.1016/j.media.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


