Abstract
Phagocytes are important players in host exposure to nanomaterials. Macrophages in particular are believed to be among the “first responders” and primary cell types that uptake and process nanoparticles, mediating host biological responses by subsequent interactions with inflammatory signaling pathways and immune cells. However, variations in local microenvironmental cues can significantly change the functional and phenotype of these cells, impacting nanoparticle uptake and overall physiological response. Herein we focus on describing the response of specific RAW 264.7 macrophage phenotypes (M1, INF-gamma/LPS induced and M2, IL-4 induced) to Stöber silica nanoparticle exposure in vitro and how this response might correlate with macrophage response to nanoparticles in vivo. It was observed that variations in macrophage phenotype produce significant differences in macrophage morphology, silica nanoparticle uptake and toxicity. High uptake was observed in M1, versus low uptake in M2 cells. M2 cells also displayed more susceptibility to concentration dependent proliferative effects, suggesting potential M1 involvement in in vivo uptake. Nanoparticles accumulated within liver and spleen tissues, with high association with macrophages within these tissues and an overall Th1 response in vivo. Both in vitro and in vivo studies are consistent in demonstrating that silica nanoparticles exhibit high macrophage sequestration, particularly those with Th1/M1 phenotype and in clearance organs. This sequestration and phenotypic response should be a primary consideration when designing new Stöber silica nanoparticle systems, as it might affect the overall efficacy.
Keywords: macrophage, nanoparticles, drug delivery, silica
Introduction
Macrophages play an important role in nanoparticle processing, and are believed to be primarily responsible for uptake and trafficking in vivo. Therapeutic capacity and clearance mechanisms in clinically relevant nanomedicines have been linked with macrophage activity[1, 2]. Successful nanoparticle clinical candidates should deliver high payloads to target sites, however in clinical applications, more than 95% of the total injected dose of drug ends up being cleared or residing in non-specific clearance organs[3]. Traditionally this has been attributed to the rapid association of these nanomedicines with elements of the mononuclear phagocytic system (MPS)[4]. To some extent this is a function of the opsonization that the particle undergoes when exposed to the blood and recognition of these opsins via the MPS[5], particularly Kupffer cells and splenic macrophages. If macrophages are indeed responsible for high clearance rates, reduced efficacy due to poor delivery of active drug payloads to specific targets and potential inflammatory mediated events are likely due to macrophage nanoparticle recognition and subsequent processing. If delivery vehicles were designed such that they avoided or harnessed this recognition system, payload delivery and subsequent efficacy could be significantly enhanced. However, in order to design appropriate systems one needs to understand nanoparticle macrophage interactions at a cellular level and how this might impact complete physiological responses.
Rational design of nanomedicines with specific macrophage interactions can be difficult, as local microenvironmental factors and cues in vivo can alter the phenotype and differentiation state of macrophages. This can drastically influence how they interact with the surrounding environment including nanomaterial processing and subsequent biological responses[6, 7]. We currently lack a full understanding of which macrophage phenotypes are important in the discovery and uptake of nanoparticle systems. A more complete understanding of this would be valuable in the design of new drug delivery systems, as this knowledge could guide nanomedicine design to achieve higher specificity towards desired targets.
The Th1/Th2 paradigm introduced decades ago[8, 9], illustrates different activation states and different macrophage phenotypes, which could help identify the macrophage phenotype which discovers and uptakes nanoparticles[10, 11]. M1 macrophages are generally considered janitorial cells, responsible for clearing foreign materials, pathogens and, potentially, nanomaterials. In vitro M1 cells are induced by IFN-gamma and are generally characterized by high iNOS, IL-12 release, and high expression of CD80. In general, M1s represent a Th1 response and, most likely, an inflammatory-mediated response[12]. In contrast, a Th2 state generally has an up-regulation of alternatively activated (or M2) cells. M2 cells are considered wound healing cells, inducing basement membrane breakdown, angiogenesis and general tissue repair. These cells are induced by IL-4 in vitro and are characterized by high arginase, IL-10 release and high expression of CD206[12]. The Th1/Th2 paradigm is a simplified immunological model system. Many macrophage phenotypes in vivo lie in between these two states and may even reside outside of them, making it extremely difficult to identify these cells in vivo[7]. However, the Th1/Th2 model system does represent the complex in vivo biological environment more accurately than traditional unpolarized macrophage models. This model may help explain whether a specific phenotype is responsible for nanomaterial processing and/or biological response and, if so, whether we can select for specific target phenotypes to harness therapeutic responses or reduce toxicity.
Evidence suggests variations in nanomaterial properties can alter macrophage uptake and initiate either Th1 or Th2 responses[13]. Clinically, environmental exposure to nanomaterials correlates directly to induced autoimmune disorders such as scleroderma and rheumatoid arthritis[14, 15]. These disease states are generally classified as a Th1 response, suggesting the involvement of M1 phenotypes[16]. In line with these findings, silica and titanium nanoparticles have been shown to induce M1 phenotypes in vivo, significantly up regulating inflammatory mechanisms[17, 18]. However, a recent study revealed that alternatively activated macrophage M2 phenotypes in vitro and in vivo took up 300nm particle replication in nonwetting templates (PRINT™) nanoparticles to a higher extent than M1 phenotypes[19]. In contrast, 200–600nm poly(lactic acid) particles induced a Th1 response while 2–8µm particles showed a Th2 response[20]. Poly(lactic-co-glycolic) acid (PLGA) nanoparticles showed a Th1 response even after priming in vivo for a Th2 response[21]. Interestingly, environmental exposure to crystalline silica has also been linked to silicosis, a disease characterized by fibrosis and a general Th2 response[22]. Injection of silica has also shown increased levels of IgG2a and IgE within serum indicating increased presence of antibodies[23], an M2 mediated adjuvant-response. However, research has shown an inability to induce an M2 activation state after macrophage incubation with superparamagnetic iron oxide particles and, to a limited degree, this effect is observed with silica[24]. In general, evidence suggests nanoparticle characteristics and testing environments can drastically affect the uptake and response within macrophage systems.
A correlation should be drawn between variations in nanomaterial properties and how these properties interact with the biological environment to induce either a Th1 or Th2 response and a basic understanding of which macrophage phenotypes could be responsible for these responses. We believe that Stöber silica nanoparticle systems interact with the biological environment and initiate a Th1 phenotypic response, as a function of M1 uptake in vitro and in vivo. Macrophage phenotype expression is dependent on disease state; understanding the phenotype that specific nanomaterial characteristics target will help to derive a better functional design platform for specific biomedical applications.
Materials and Methods
Particle synthesis and characterization
Spherical silica nanoparticles were prepared by previously reported modified Stöber methods[25, 26]. All particles were fluorescently labeled with fluorescein isothiocyanate (FITC) to assess cellular uptake. The constructs were sterilized by dry autoclaving. Transmission electron microscopy (TEM) images were taken on a Phillips TECHAI F2 (Hillsboro, OR) at an accelerating voltage of 80 kV. TEM samples were created by evaporating droplets of particles suspended in deionized water off copper grids. After micrograph collection, nanoconstruct size was measured utilizing Adobe Photoshop’s pixilation ruler measurement tool (Adobe, San Jose, CA). At minimum the sizes of 300 particles of each type were measured. Particle zeta potential of SNPs dispersed in DI water at a concentration of 1.0 mg/ml was measured using a Malvern Instruments Zetasizer Nano ZS (Westborough, MA). SNPs (50 or 25 mg/ml) were sonicated, vortexed and the final particle dispersions were prepared immediately before use from common stock in culture medium and vortexed before application to the culture cells. All particles were tested for endotoxin levels prior to cellular incubation; levels were below FDA recommended .05 EU/mL.
In vitro methods
Cell culture
RAW 264.7 murine macrophages were obtained from ATCC (Manassas, Virginia) and maintained in RPMI media supplemented with 10% FBS, at passage numbers 5–25. Cell cultures were incubated at 37°C in 5% CO2 and 95% humidified air and kept in logarithmic phase of growth throughout all experiments, never reaching full confluence.
Cell polarization and confirmation
Cells were seeded and allowed to adhere overnight at 37°C in 5% CO2. The following day the cells were treated with a M1 cocktail that consisted of LPS (100ng/mL) and IFN-gamma (300 units/mL), a M2 cocktail of IL-4 (10 units/mL) or an unpolarized cocktail with no additives (Sigma-Aldrich, St. Louis, MO). All cocktails were diluted in fresh media. The cells were incubated for 18 hours to obtain sufficient polarization. To confirm polarization, IL-10 and IL-12 were quantified via BD Cytokine Flow Cytometry protocol and colorimetrically for arginase and nitric oxide (described below).
Arginase and nitric oxide evaluation
Following cellular polarization in 96 well plates, cells were treated for 24 hours with a range of silica nanoparticle concentrations (5–250 ug/mL). Classen et al’s colorimetric protocols for the Griess reagent (detection of nitric oxide) and a urea assay (detection of arginase) were followed[27].
Cellular proliferation
Following polarization, cells were exposed to a range of concentrations (5–250 ug/mL) of silica nanoparticles for 72 hours in RPMI media supplemented with 10% FBS. Relative cell viability was assessed by utilizing a water-soluble tetrazolium salt, WST-8 [2–(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt], the key component in the Cell Counting Kit-8 from Dojindo Molecular Technologies, Inc. (Rockville, Maryland).
Cell uptake, visualization and quantification
The uptake of silica nanoparticles by cultured cells was visualized by confocal microscopy. Cells were grown on 24 well imaging plates at a density of ~9000 cells/cm2 polarized and incubated for 24 hours with 37.5 µg/ml FITC labeled silica nanoconstructs in RPMI media supplemented with 10% FBS. After incubation, cells were fixed with 4% formalin in PBS. Cell nuclei were stained with 2.5 µM 4′,6-diamidino-2-phenylindole (DAPI) according to the manufacturer's protocol. For CLSM (Olympus FluoView® FV1000, Olympus America Corp., Center Valley, PA), the intensity of the laser beam and the photodetector sensitivity were kept constant in order to compare the relative fluorescence intensities between experiments. Z stacks were collected and used for 3D reconstruction and visualization of intracellular particle localization. All image acquisitions and analyses were performed using FluoView 2.0 software.
Flow cytometry was used to quantify the amount of nanoparticle uptake. Cells were grown on 12 well plates at a density of ~15,000 cells/cm2 polarized and incubated with 37.5 µg/ml FITC labeled silica nanoconstructs in RPMI media supplemented with 10% FBS for 24 hours. Following incubation, cells were scraped to obtain a single cell suspension. Cells were suspended in PBS containing 1% BSA and analysis was performed on a FACSCalibur (BD Biosciences). Emitted light resulting from FITC-labeled nanoparticles was detected by the FL-2 detector. To calculate the background fluorescence of unlabeled cells, cells without any addition of nanoparticles were carried along as a negative control in every measurement. For whole-cell analysis 10,000 cells were counted. Data analysis was performed with BD CellQuest Pro (BD Biosciences).
Cellular autophagy
Coverslips were seeded at a density of ~9000 cells/cm2, polarized with polarization cocktails for 24 hours in RPMI media supplemented with 10% FBS. Following polarization cells were incubated for 24 hours with 37.5 µg/ml FITC labeled silica nanoconstructs in RPMI media supplemented with 10% FBS. After incubation cells were fixed, permeabilized via triton-X and stained intracellularly for LC3-II (Molecular Probes). Both colocalization and cellular internalization were visualized with confocal microscopy.
Cellular co-localization
6 well plates were seeded at a density of ~9000 cells/cm2 and polarized to either M1 or M2 phenotypes. Following polarization, wells were scraped and 1 well containing M1 polarized cells and 1 well containing M2 polarized cells were seeded on glass coverslips and allowed to adhere overnight. Prior to scraping M1 cells were preincubated with DiD to mark the phenotype while M2 cells were left unstained (Molecular Probes). Following adhesion these co-cultures were treated for 24 hours with 37.5 µg/ml FITC labeled silica nanoconstructs. After incubation cells were fixed, and stained with DAPI. Cellular internalization was visualized with confocal microscopy.
Transmission electron microscopy
The uptake of silica constructs by cultured cells was assessed by transmission electron microscopy. Cells were seeded on 6 well plates containing 1x1 cm ACLAR plastic at a density of ~9000 cells/cm2, polarized and after an overnight incubation, 37.5 µg/mL of silica nanoconstructs were added. Subsequently, cells were incubated for 24 hours after which they were washed with PBS and fixed with a 2.5% glutaraldehyde and 1% formaldehyde in 0.1M sodium cacodylate buffer with sucrose and calcium chloride. Cells were stained with uranyl acetate for 45 minutes at room temperature and TEM images were taken with a Phillips, TECHAI F2 TEM (Hillsboro, OR, USA) at an accelerating voltage of 80 kV.
In vivo methods
Animal handing
All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines of The University of Utah. Adult female CD-1 mouse injections were administered via the tail vein at 20 mg/kg of nanoparticle formulations with a max injection volume of 200 ul. Saline administration was used as a negative control. Mice were sacrificed at one and four week time points and organs analyzed. N=5 for all studies.
Biodistribution and clearance
At necropsy, organs were collected, isolated and weighed. Organs subsequently underwent acid digestion with aqua regia at 80 °C. Following complete organic digestion, the solution was suspended in 5% trace metal-grade nitric acid and HF was added to the solution to dissolve and stabilize the silica content. This solution was analyzed via inductively coupled plasma mass spectrometry (ICP-MS), and quantified against a silica and internal standard. Urine and fecal matter was pooled to assess the total silica content.
Histology
Following necropsy organs were fixed in 4% fresh paraformaldehyde. Samples were dehydrated, paraffin-embedded and cut into 4µm thick sections. Sections were stained with hematoxylin and eosin. Immunohistochemical staining was performed on air-dried room temperature sections, placed in a 60 °C oven to melt the paraffin. Sections were de-paraffinized with EZ Prep solution (Ventana Medical Systems). In the case of iNOS and Arginase sections were pretreated with CC1 (Cell Conditioner 1, pH 8.0) and F4/80 sections were digested with Protease 2. Primary antibodies were applied (iNOS: 1:50 for 2 hours, Arginase 1:800 for 1 hour, F4/80 1:200 for 2 hours) (Thermo Scientific and Abcam). Slides were detected using the IView DAB detection kit (Ventana Medical Systems), which is a Streptavidin-HRP system, utilizing DAB (3-3’ diaminobenzidine) as the chromogen and counterstained with hematoxylin. Sections were washed with DI water, dipped in iodine followed by sodium thiosulfate and dehydrated in graded alcohols cleared with xylene and coverslipped. Immunofluorescence was similarly performed with primary antibody incubation (C68 1:100 for 2hours, CD206 1:50 for 2 hours, LC3-II 1:100 for 1 hour) and subsequent secondary detection (Alexa Fluor 647) (Molecular Probes).
Immunofluorescence was imaged on an Olympus 1×50 inverted microscope. Quantification and visualization were performed utilizing a mean fluorescence intensity of collected tissue samples in ImageJ. Immunohistochemical staining was also visualized utilizing an Aperio slide scanner. Quantification and visualization were performed utilizing Aperio ScanScope software.
Transmission electron microscopy
The uptake of silica constructs within organs was assessed by transmission electron microscopy. Organs were fixed with a 2.5% glutaraldehyde and 1% formaldehyde in 0.1M sodium cacodylate buffer with sucrose and calcium chloride. Cells were stained with uranyl acetate for 45 minutes at room temperature and TEM images were taken with a Phillips, TECHAI F2 TEM (Hillsboro, OR, USA) at an accelerating voltage of 80 kV.
Results
Silica Nanoparticle Synthesis and Characterization
130nm FITC-labeled silica nanoparticles were synthesized and characterized via DLS, zeta potential, and IR (Supplemental Table 1). Characterization of particle stability in serum was assessed and confirmed that nanoparticles were in suspension. After verification of synthesis and stability, the particles were utilized to investigate the influence of uptake in M1 and M2 polarized macrophages in vitro and in vivo.
Polarization of M1 and M2 macrophages in vitro
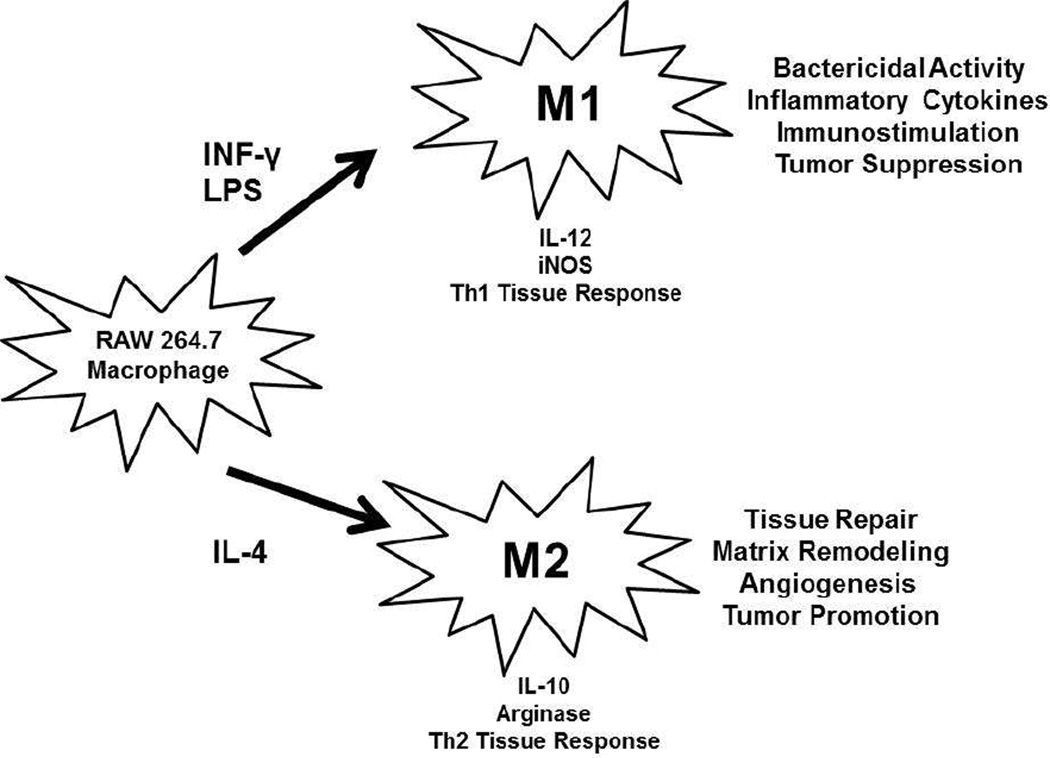
RAW 264.7 macrophages were polarized to M1 and M2 phenotypes utilizing traditional cytokine cocktails, IFN-gamma/LPS and IL-4, respectively (Diagram 1). M1 polarized macrophages exhibited expected morphological changes such as enhanced intracellular vacuoles and 5x increase in size when compared to M2 cells which exhibited very little to no vacuolization and spindle like morphology (Supplemental Figure 1). Additionally, iNOS and IL-12 were present in M1 polarization states and absent in M2. M2 states exhibited arginase and IL-10 but these cytokines were absent in M1 polarization (Supplemental Figure 1). The cytokine and morphological results confirmed RAW 264.7 polarization into M1 and M2 phenotypes. The confirmed polarized macrophages were utilized in in vitro testing.
Diagram 1.
Diagram depicting the in vitro macrophage polarization procedure. RAW 264.7 cells were polarized into M1 and M2 macrophages, by incubation for 24 hours with INF-γ and LPS or IL-4, respectively. These two polarization states have very different in vivo phenotypic functions, outlined above.
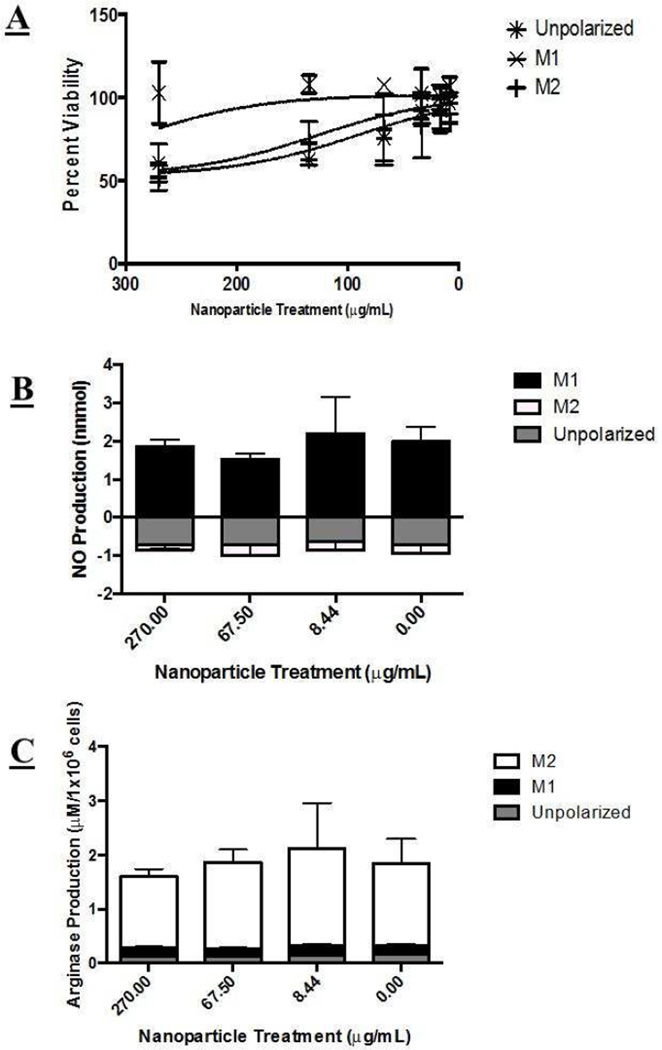
Nanoparticles were incubated with unpolarized RAW 264.7 cells, M1, and M2 polarization states for 72 hours (~2 population doubling times) to assess the relative influence of nanoparticle treatment on overall function and proliferation capacity. Nanoparticle solutions affected proliferation status in M2 and unpolarized cells, with higher concentrations exhibiting more pronounced toxicity (Figure 1A). The nanoparticle concentrations tested did not appear to affect M1 proliferation.
Figure 1.
Macrophage viability and cytokine excretion after nanoparticle treatment. A) Macrophage viability following nanoparticle treatment. Unpolarized and M2 cells showed a statistically significant decreased viability with increasing nanoparticle concentration. M1 cells however appeared to be statistically unaffected by nanoparticle treatment. B) iNOS did not appear to be significantly affected by any nanoparticle treatment. M1 cells exhibited a statistically significant higher native expression, as expected. C) Arginase activity was not significantly affected by nanoparticle treatment. M2 cells exhibited a statistically significant higher native expression, as expected. Statistical difference from control determined via paired t-test, p value < .05 indicated with a *, p values > .05 were determined insignificant.
We observed little to no effect (increase or decrease) on hallmark M1 and M2 markers, iNOS and arginase, after nanoparticle treatment at a variety of concentrations (Figure 1B and 1C).
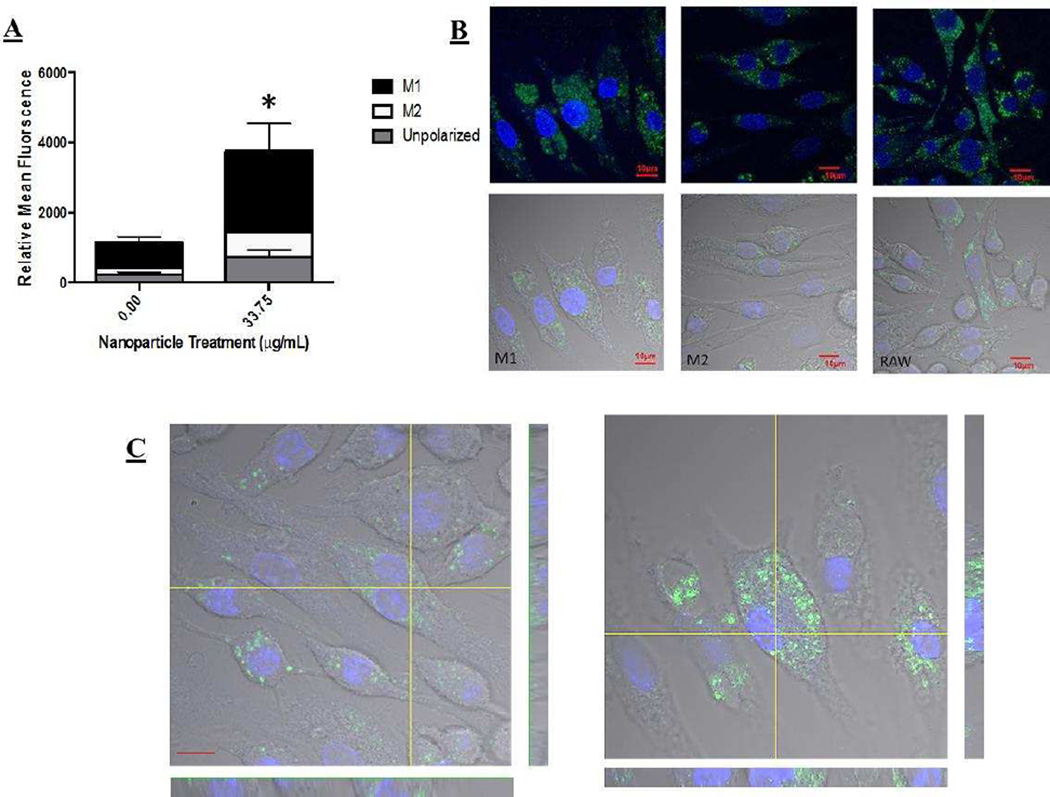
In an attempt to understand proliferative effects, we sought to investigate and quantify the degree of nanoparticle uptake within these polarization states. As such, nanoparticles were incubated with M1, M2, and unpolarized macrophages. The relative level of uptake was assessed and quantified via FACS and confirmed via confocal microscopy (Figure 2 and Supplemental Figure 2). Nanoparticles were taken up to a greater extent in M1 macrophages when compared to M2 and unpolarized macrophage phenotypes. In order to compensate for potential data abberations related to the significantly different cell size between M1 and M2 cells (40um vs. 10um, respectively), we normalized the FACS analysis to the size of each cell type, and still saw a statistical increase.
Figure 2.
A) Uptake of nanoparticles in different macrophage polarization states via FACS. M1 macrophages took up nanoparticles to the highest extent, statistically significant increase in uptake was shown. Statistical difference from control determined via paired t-test, p value < .05 indicated with a *, p values > .05 were determined insignificant. B) Confocal images confirming FACS analysis of nanoparticle uptake in polarized macrophages, with the highest degree of uptake in M1 cells. C) Z-stack images of cells showing internalization and not cellular surface association of particles in overall uptake.
The conflicting uptake and proliferation results led us to ask if internalization mechanisms within these polarization states were altered. To visualize nanoparticle localization within polarization states, TEM images were taken of macrophages incubated with nanoparticles (Supplemental Figure 3). A large accumulation of nanoparticles was observed in the vacuoles of M1 macrophages. Very few vacuoles were observed in M2 cells and the vacuoles displayed reduced nanoparticle association and vesicular definition. Similar results to M2 macrophages were observed for unpolarized cells.
Mechanisms of particle intracellular fate were investigated in macrophages. We previously identified intracellular fate consistent with autophagic activity after treatment with amine modified silica nanoparticles in unpolarized cells[28]. We investigated whether similar internalization was observed in M1 and M2 polarized cells. A high upregulation of autophagic activity in M2 and unpolarized macrophages was observed (Supplemental Figure 3 and 4). M1 cells exhibited little to no autophagy in any treatment group.
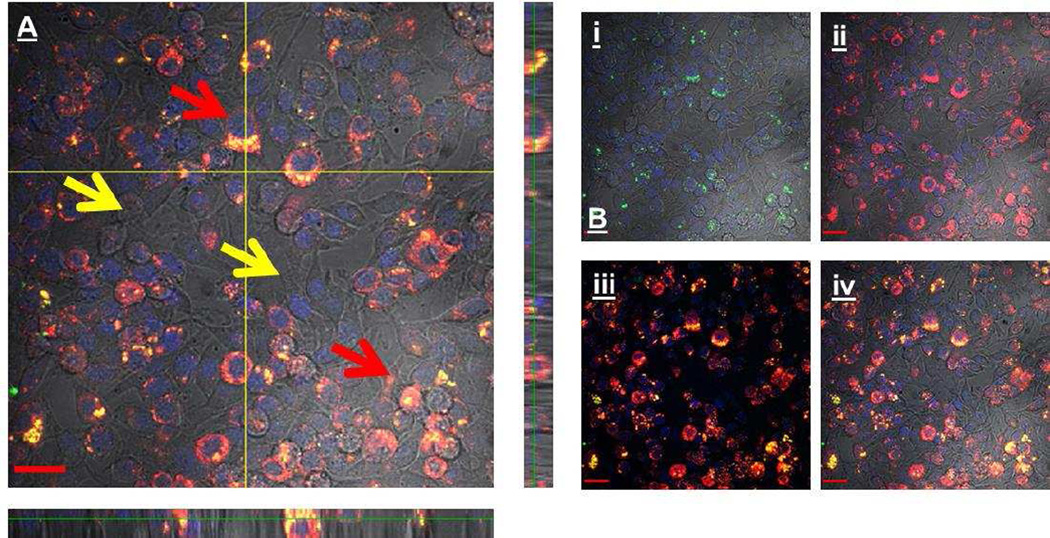
M1 cells have a higher sequestration of nanoparticles in isolated culture; however, in vivo polarization states are mixed. In order to more accurately mimic the in vivo system, both macrophage types were cultured together to determine if sequestration in one macrophage over the other would still be present. In this system, we observed an increase in particle accumulation in M1 macrophages (Figure 3).
Figure 3.
A) Z stack representing the uptake of nanoparticles within a co-culture of M1 and M2 cells. Red arrows: Nanoparticles (green) are nearly always co-localized (yellow) with M1 cells (red), providing evidence for M1 sequestration of nanoparticles. Yellow arrows: Cells that are unlabeled represent an M2 phenotype; these cells appear to avoid nanoparticle uptake. B) For clarification dyes are separated out to help visualization. i) Nanoparticle label ii) M1 macrophage label iii) colocalization of M1 and nanoparticle label without DIC iv) DIC and co-localization.
In vivo nanoparticle effect
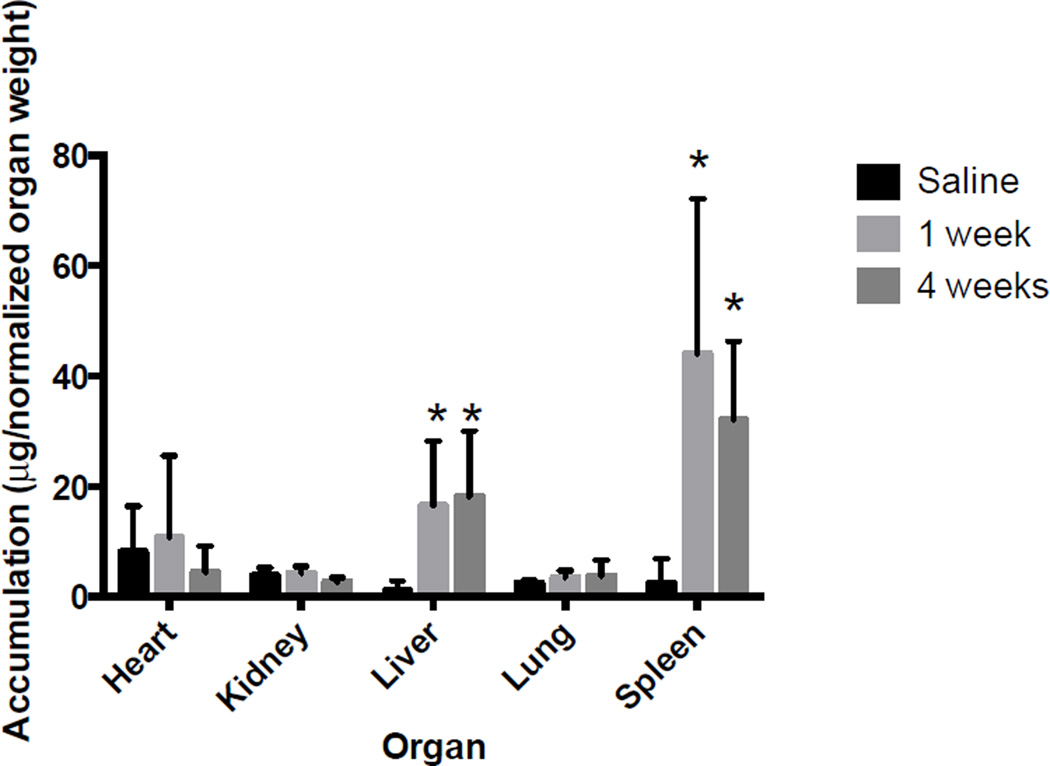
After observing an increase in nanoparticle uptake in M1 macrophages in both isolated and co-culture systems, we investigated if we could correlate this in vitro effect with an in vivo system. Adult CD-1 mice were injected with each nanoparticle formulation, and the effects of these systems after 1 and 4 weeks were observed. As expected with non-degradable nanoparticle systems, we observed high accumulation in the liver and spleen and very little association with other organs (Figure 4). While uptake and residence of nanoparticles were significantly increased within the liver and spleen, animals did not have any weight loss or gain over time compared to control, suggesting minimal toxic effects of these systems (Supplemental Figure 5).
Figure 4.
Silica nanoparticle organ accumulation, as determined via quantitative ICP-MS. Data has been normalized to organ weight to account for aberrations, and additionally normalized to brain weight; no differences were observed. We observed accumulation of silica nanoparticles primarily in splenic and hepatic tissues, with minimal accumulation in other organs. Silica was retained within these organs at statistically similar levels out to 4 week time points. Statistical difference from control determined via paired t-test, p value < .05 indicated with a *, p values > .05 were determined insignificant.
Following these observations, we wanted to see if accumulation and residence of these nanoparticles within the spleen or liver caused any great duress on the tissue. H&E staining did not show toxicity (Supplemental Figure 6); however, slight variations in cell content and morphology within the spleen and liver were observed. The spleen showed a slight but not significant increase in inflammatory cells, formation of multinucleated cells, and very slight destruction of lymphatic foci from 1 to 4 weeks (Figure 5). The liver showed a slight but not significant increase in hydropic degeneration or vacuolization and increased levels of inflammatory cells from 1 to 4 weeks (Figure 5). Overall, we observed little to no toxicity or induced inflammation in vivo after nanoparticle treatment.
Figure 5.
A) H&E images of spleen tissue of animals treated with silica nanoparticles at 1 and 4 weeks. Increased levels of macrophages and multinucleated cells are present in the spleen at both 1 and 4 weeks. However, overall inflammatory effects are minimal, as a function of silica nanoparticle treatment. B) H&E images of liver tissue of animals treated with silica nanoparticles at 1 and 4 weeks. Increased levels of vacuolization (increased white space) are present in the liver at both 1 and 4 weeks. However, overall inflammatory effects are minimal as a result of silica nanoparticle treatment.
The observation of a slight increase in inflammatory mediated cells in H&E staining led us to investigate the potential of increased levels of macrophages over time within these tissues. We observed a drastic increase in CD68 expression, a lysosomal marker within the liver from 1 to 4 weeks, suggesting the increase of macrophages with lysosomal-like compartments (Figure 6).
Figure 6.
Immunofluorescence of liver tissues stained with CD68. CD68 is a lysosomal marker that is useful in identification of increased levels of macrophages. This image suggests that we have a drastic increase or recruitment of macrophages within the liver at 4 weeks.
Confirmation of increased macrophage residence and identification of the phenotype of the up-regulated in vivo macrophage population was performed through tissue staining with F4/80 (a macrophage marker), iNOS (an M1 marker) and arginase (an M2 marker). We observed an upregulation of F4/80 and iNOS in the liver and spleen (Figure 7). These macrophage markers were patchy; staining was not uniform throughout the tissue. To quantify the level of upregulation a complete organ slide scan and colorimetric deconvolution were performed. Increased F4/80 and iNOS were observed in some areas of liver tissue however this was not statistically significant across the entire tissue (data not shown). Localized staining might be due to nanoparticule accumulation in only certain areas of the tissue. Local increases in these markers suggest that M1 macrophages may play an important role in the response to these nanoparticle systems. We observed no upregulation in arginase (Supplemental Figure 7) in any tissue. CD206 (M2 marker) and autophagic markers were also utilized, we observed little to no overall increase in staining (data not shown).
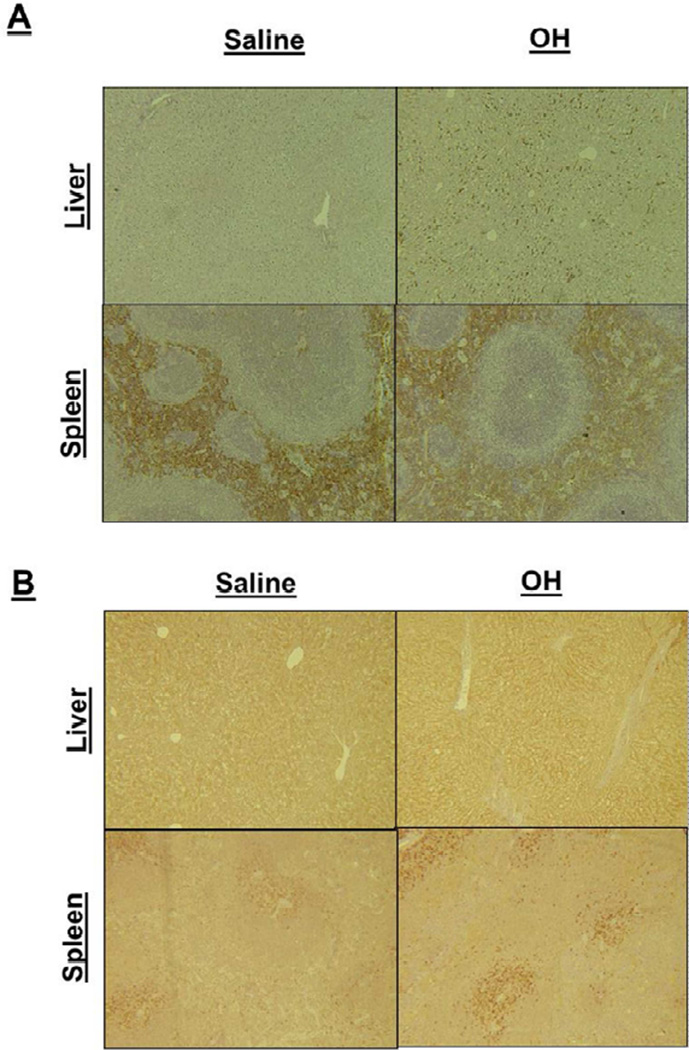
Figure 7.
A) Liver and splenic tissues stained for F4/80. An observable increase is shown in treatment groups of the F4/80 stain. This is a macrophage marker stain suggesting an increase in the local recruitment of macrophages within these tissues. 7B) Liver and splenic tissues stained for iNOS. An observable increase is shown in treatment groups. This is a marker for M1 macrophages, and may suggest increased levels of macrophages present in the tissue, as observed via CD68 and F4/80. However iNOS is present in other cell types and could be a function of their release, as well.
We sought to determine whether M1 macrophage upregulation was a result of recruitment as a reaction to nanoparticle residence or if proliferation of macrophages was a direct result of macrophage uptake of nanoparticles. We performed TEM on liver and spleen tissues (Figure 8) and observed macrophage nanoparticle uptake in both organs. Macrophages took up the majority of nanoparticles, however, nanoparticles were also observed in other cells capable of phagocytosis (neutrophils, endothelial cells, etc).
Figure 8.
TEM images of nanoparticles residing within macrophages of the liver and the spleen. These images support nanoparticle uptake within macrophage populations and help to support the increased macrophage response and recruitment is a direct result of internalization of nanoparticles within this cell type.
Discussion
Polarization of M1 and M2 macrophages in vitro
M1 cells appear to experience minimal toxic effects when treated with nanoparticles, even at very high concentrations. M1 cells also uptake nanoparticles to a greater degree than M2 cells, despite showing the lowest toxicity. This may suggest that polarizing cells to an M1 state induces a coping mechanism for foreign matter, which helps the cell to avoid interference with the rest of the cellular machinery and phenotypic state. This may be due to TLR4 induction as a result of treatment with LPS during the polarization protocol, as this is thought to help support cell survival and increase phagocytosis[29]. Globally, this behavior is not surprising, as characteristically the M1 phenotype is thought to remove foreign materials and pathogens from the local environment, which is likely to leave the cell, in most cases, unaffected.
Unlike M1 cells, M2 and unpolarized cells exhibit a toxic effect at higher nanoparticle treatment concentrations, despite reduction in overall uptake. The toxic concentrations in M2 and unpolarized cells are similar, which may suggest that they process the materials in a similar manner. However, the mechanism of toxicity remains to be studied.
Uptake of nanoparticles in M1 and M2 cells
Clear M1-specific uptake suggests that M1 cells have a higher sequestration of these nanoparticle systems. Some M1 surface receptors have been implicated in promiscuous poly anionic and silanol binding in vivo[30–32]. High sequestration in M1 cells of negatively charged terminal groups may help explain why these silanol-terminated particles were taken up to a greater degree in M1 cells in vitro.
Our results indicate that M1s clearly uptake more particles than M2 both in isolated cultures, as well as co-culture. Understanding this phenomenon might be useful when designing future in vitro assays and understanding nanoparticle processing. The results may help to correlate in vitro results to a greater extent with in vivo. Understanding that M1s are the primary phenotype which uptake these nanoparticle systems could help to explain downstream physiological effects, such as the induction of inflammation or the recruitment of other immunological cell types.
In vivo nanoparticle effect
We observed minimal toxic effects after nanoparticle administration to rodents by tail vein injection, which suggests that the cells which ingest nanoparticles in vivo may potentially be providing a cellular coping effect, helping to preserve the local tissue from further nanoparticle damage. We observed similar results with M1 systems in vitro. Increased levels of F4/80, iNOS and CD68 are indicative of macrophage involvement and may suggest primarily activated M1 macrophage involvement and induction of a Th1 response. Limited to no increase in arginase, CD206 or LC3-II suggests M2 macrophages play little to no role in the overall response to these nanoparticle systems.
Increased levels of inflammatory cells, destruction of lymphocytes, and increased vacuoles suggested the involvement of macrophages and potential induction of inflammatory mediated mechanisms. Hydropic degeneration is also a sign of mitochondrial dysfunction, a characteristic we have observed with nanoparticle exposure in vitro and can be detrimental in longer terms[33]. These studies were only carried out to 4 weeks, it will be essential in nanoparticle development to carry such studies out to at least a year to characteristically identify if inflammatory cell infiltration, tissue destruction and vacuolization persist and if further downstream issues are potentiated.
Drastic increases in lysosomes (CD68) in the liver may help to explain decreased levels of silica in some tissues and may help to support downstream inflammation. Silica may potentially be trafficked to this site, and cleared or stored in or around biliary tracts. It is important to note that CD68 is a lysosomal marker that is observed in cell types other than macrophages and one cannot discern between M1 and M2 polarization states with this marker. One can only assume that the macrophages or cells that have been stained are activated to a lysosomal or inflammatory like state, which could potentially be harmful to the animal in chronic situations.
Increased M1 and Th1 responses are not surprising, as M1 macrophages are traditionally involved in biological processing of nanoparticles. Crystalline silica has been linked as an adjuvant to induce autoimmune disorders[14] such as sclercoderma, a Th1 response. Anionic groups, like those found on the surface of silanol terminated silica nanoparticles, are also known to differentiate dendritic cells and upregulate CD80 and CD86 (F4/80) in vivo[34, 35]. Anionic group recognizing receptors overexpressed on M1 macrophages including Kupffer cells of the liver and within the red pulp macrophages of the spleen have also been shown in vivo to increase inflammatory M1 responses [36], and M1 phenotypes have shown to be overloaded with iron oxide particles in vivo [37]. Additionally, the terminal silanols seen within the nanoparticles investigated here induce complement mediated events which could facilitate specific uptake in M1 macrophages[38]. This may suggest that silica could induce a Th1 response as a result of nanoparticle uptake within these tissues.
Our TEM results show images that provide clear evidence for preferential uptake of nanoparticles in macrophages in vivo. We observe that CD68 and F4/80 expression coincide with this macrophage nanoparticle uptake within these organs, suggesting the response is a function of nanoparticle uptake and not simply because of nanoparticle residence. This may also suggest that nanoparticles maybe sequestered in M1 phenotypes, however future quantification is necessary to draw these specific conclusions. We did attempt to sort those macrophages that took up nanoparticles in vivo but were unable to convincingly obtain in vivo phenotypic data, a common problem in the macrophage field[39]. Quantification of both macrophage phenotypic uptake and macrophage versus other cell type uptake will be important in future studies.
General Implications
Changing the macrophage phenotype that takes up the nanoparticle could alter the physiological response. For example, if M2 macrophages instead took up silica nanoparticles to a higher extent, it would likely elicit a Th2 response and very different physiological patterns. It will be imperative in the future to pay attention to which particles are taken up by which phenotypes, as nanoparticle processing could initiate detrimental chronic responses dependent on the phenotypic uptake. Additionally, this could have therapeutic implications as differing disease states have variations in macrophage phenotypic responses.
Macrophage phenotype targeting and response could be useful therapeutically. The results of this study may suggest that silica nanoparticles could be utilized to target M1 macrophages in vivo. Such a strategy could potentially be utilized in the design of a treatment for neurodegenerative disorders, where an upregulation of M1 phenotypes is destructive to surrounding tissue. If one could target this phenotype and revert or change it to a M2 phenotype it may resolve the disease state. For example, in cases of spinal cord injury, an upregulation of M1 polarized macrophages is observed which could be preventing M2 cells from repairing the local environment [40]. M1 phenotypes have also been implicated in the development of multiple sclerosis, and M2 suppression has been implicated in relapse of MS suggesting targeting of these phenotypes maybe a viable therapeutic approach [41]. However, it is imperative even within disease states to understand the overall chronic implications of nanoparticle treatment and therefore the potential of these systems.
Conclusions
Our research demonstrates that Stöber silica nanoparticle systems, a commonly investigated nanomedical agent, interact with the biological environment and initiate an Th1 response in vivo, M1 phenotypic response in vitro and a general macrophage sequestration in vivo. We have shown that M1 macrophages have a higher sequestration of nanoparticles in vitro and may potentially regulate organ-level uptake and response in vivo. Uptake within M1 phenotypes may provide a protective effect or initiate long term chronic responses in vitro and in vivo. Coping and chronic response mechanisms will need further mechanistic evaluation. Surface chemistry of nanoparticles may potentially play a role in cellular uptake, increased circulation, and clearance. The increased levels of M1 uptake in vitro and increased macrophage response in vivo may also suggest that multiple macrophage phenotypic model systems must be used in vitro to understand potential effects of nanoparticles in vivo.
Supplementary Material
Acknowledgements
The authors would like to thank David Grainger for valuable discussions, and Nancy Chandler at the electron microscopy core of The University of Utah, Sheryl Tripp at ARUP Labs, and Cassandra Rice at The University of Utah with their help with TEM, IHC, and fluorescence quantification, respectively. We would also like to thank Ryan Robinson and Wiebke Gerlach with their help with animal handling, intravenous injection and necropsy. Support was provided by R01ES024681 from the NIH and a predoctoral fellowship (Heather Herd, W81XWH-11-1-0057) from the Department of Defense Breast Cancer Research Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Caron WP, Lay JC, Fong AM, La-Beck NM, Kumar P, Newman SE, et al. Translational studies of phenotypic probes for the mononuclear phagocyte system and liposomal pharmacology. J Pharmacol Exp Ther. 2013;347:599–606. doi: 10.1124/jpet.113.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caron WP, Song G, Kumar P, Rawal S, Zamboni WC. Interpatient pharmacokinetic and pharmacodynamic variability of carrier-mediated anticancer agents. Clinical pharmacology and therapeutics. 2012;91:802–812. doi: 10.1038/clpt.2012.12. [DOI] [PubMed] [Google Scholar]
- 3.Duncan R, Gaspar R. Nanomedicine(s) under the microscope. Mol Pharm. 2011;8:2101–2141. doi: 10.1021/mp200394t. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q, Jones SW, Parker CL, Zamboni WC, Bear JE, Lai SK. Evading immune cell uptake and clearance requires PEG grafting at densities substantially exceeding the minimum for brush conformation. Mol Pharm. 2014 doi: 10.1021/mp400703d. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 5.Mortimer GM, Butcher NJ, Musumeci AW, Deng ZJ, Martin DJ, Minchin RF. Cryptic epitopes of albumin determine mononuclear phagocyte system clearance of nanomaterials. ACS nano. 2014 doi: 10.1021/nn405830g. [DOI] [PubMed] [Google Scholar]
- 6.Cassetta L, Cassol E, Poli G. Macrophage polarization in health and disease. The Scientific World Journal. 2011;11:2391–2402. doi: 10.1100/2011/213962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. The Journal of experimental medicine. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Romagnani S. The Th1/Th2 paradigm. Immunology Today. 1997;18:263–266. doi: 10.1016/s0167-5699(97)80019-9. [DOI] [PubMed] [Google Scholar]
- 11.Gordon S. Alternative activation of macrophages. Nature reviews Immunology. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Phan N, Isely C, Bruene L, Bratlie KM. The Effect of Surface Modification and Macrophage Phenotype on Particle Internalization. Biomacromolecules. 2014 doi: 10.1021/bm5011382. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 14.Haustein U-F, Ziegler V, Herrmann K, Mehlhorn J, Schmidt C. Silica-induced scleroderma. Journal of the American Academy of Dermatology. 1990;22:444–448. doi: 10.1016/0190-9622(90)70062-m. [DOI] [PubMed] [Google Scholar]
- 15.Mohamed BM, Verma NK, Davies AM, McGowan A, Crosbie-Staunton K, Prina-Mello A, et al. Citrullination of proteins: a common post-translational modification pathway induced by different nanoparticles in vitro and in vivo. Nanomedicine (Lond) 2012;7:1181–1195. doi: 10.2217/nnm.11.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper GS, Miller FW, Germolec DR. Occupational exposures and autoimmune diseases. International Immunopharmacology. 2002;2:303–313. doi: 10.1016/s1567-5769(01)00181-3. [DOI] [PubMed] [Google Scholar]
- 17.Lucarelli M, Gatti AM, Savarino G, Quattroni P, Martinelli L, Monari E, et al. Innate defense functions of macrophages can be biased by nano-sized ceramic and metallic particles. European Cytokine Network. 2004;15:339–346. [PubMed] [Google Scholar]
- 18.Schanen BC, Das S, Reilly CM, Warren WL, Self WT, Seal S, et al. Immunomodulation and T helper TH(1)/TH(2) response polarization by CeO(2) and TiO(2) nanoparticles. PloS one. 2013;8:e62816. doi: 10.1371/journal.pone.0062816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones SW, Roberts RA, Robbins GR, Perry JL, Kai MP, Chen K, et al. Nanoparticle clearance is governed by Th1/Th2 immunity and strain background. The Journal of Clinical Investigation. 2013;123:3061–3073. doi: 10.1172/JCI66895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanchan V, Panda AK. Interactions of antigen-loaded polylactide particles with macrophages and their correlation with the immune response. Biomaterials. 2007;28:5344–5357. doi: 10.1016/j.biomaterials.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 21.Lutsiak M, Kwon GS, Samuel J. Biodegradable nanoparticle delivery of a Th2-biased peptide for induction of Th1 immune responses. Journal of Pharmacy and Pharmacology. 2006;58:739–747. doi: 10.1211/jpp.58.6.0004. [DOI] [PubMed] [Google Scholar]
- 22.Barbarin V, Xing Z, Delos M, Lison D, Huaux F. Pulmonary overexpression of IL-10 augments lung fibrosis and Th2 responses induced by silica particles. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2005;288 doi: 10.1152/ajplung.00329.2004. L841-L8. [DOI] [PubMed] [Google Scholar]
- 23.Granum B, Gaarder PI, Groeng E-C, Leikvold R-B, Namork E, Løvik M. Fine particles of widely different composition have an adjuvant effect on the production of allergen-specific antibodies. Toxicology Letters. 2001;118:171–181. doi: 10.1016/s0378-4274(00)00292-7. [DOI] [PubMed] [Google Scholar]
- 24.Kodali V, Littke MH, Tilton SC, Teeguarden JG, Shi L, Frevert CW, et al. Dysregulation of Macrophage Activation Profiles by Engineered Nanoparticles. ACS nano. 2013;7:6997–7010. doi: 10.1021/nn402145t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stöber W, Fink A, Bohn E. Controlled growth of monodisperse silica spheres in the micro size range. Journal of Colloids and Interface Science. 1968;26:62–69. [Google Scholar]
- 26.Blaaderen A, Vrji A. Synthesis and characterization of colloidal dispersions of fluorescent silica spheres. Langmuir. 1992;8:2921–2931. [Google Scholar]
- 27.Classen A, Lloberas J, Celada A. Macrophages and Dendritic Cells. Springer; 2009. Macrophage activation: classical vs. alternative; pp. 29–43. [DOI] [PubMed] [Google Scholar]
- 28.Herd HL, Malugin A, Ghandehari H. Silica nanoconstruct cellular toleration threshold in vitro. Journal of controlled release : official journal of the Controlled Release Society. 2011;153:40–48. doi: 10.1016/j.jconrel.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Platt N, Gordon S. Scavenger receptors: diverse activities and promiscuous binding of polyanionic ligands. Chemistry & Biology. 1998;5:R193–R203. doi: 10.1016/s1074-5521(98)90156-9. [DOI] [PubMed] [Google Scholar]
- 31.Chao SK, Hamilton RF, Pfau JC, Holian A. Cell surface regulation of silica-induced apoptosis by the SR-A scavenger receptor in a murine lung macrophage cell line (MH-S) Toxicology and Applied Pharmacology. 2001;174:10–16. doi: 10.1006/taap.2001.9190. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton RF, Jr, Thakur SA, Holian A. Silica binding and toxicity in alveolar macrophages. Free Radical Biology and Medicine. 2008;44:1246–1258. doi: 10.1016/j.freeradbiomed.2007.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malugin A, Ghandehari H. Caspase 3 independent cell death induced by amorphous silica nanoparticles. Nanoscience and Nanotechnology Letters. 2011;3:309–313. [Google Scholar]
- 34.Bartneck M, Keul HA, Wambach M, Bornemann J, Gbureck U, Chatain N, et al. Effects of nanoparticle surface-coupled peptides, functional endgroups, and charge on intracellular distribution and functionality of human primary reticuloendothelial cells. Nanomedicine: Nanotechnology, Biology and Medicine. 2012;8:1282–1292. doi: 10.1016/j.nano.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Yang D, Zhao Y, Guo H, Li Y, Tewary P, Xing G, et al. [Gd@ C82 (OH) 22] n Nanoparticles Induce Dendritic Cell Maturation and Activate Th1 Immune Responses. ACS nano. 2010;4:1178–1186. doi: 10.1021/nn901478z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Qian L, Zong G, Ma K, Zhu X, Zhang H, et al. Class A scavenger receptor promotes cerebral ischemic injury by pivoting microglia/macrophage polarization. Neuroscience. 2012;218:35–48. doi: 10.1016/j.neuroscience.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 37.Sindrilaru A, Peters T, Wieschalka S, Baican C, Baican A, Peter H, et al. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. The Journal of Clinical Investigation. 2011;121:985. doi: 10.1172/JCI44490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Governa M, Amati M, Fenoglio I, Valentino M, Coloccini S, Bolognini L, et al. Variability of biological effects of silicas: different degrees of activation of the fifth component of complement by amorphous silicas. Toxicology and applied pharmacology. 2005;208:68–77. doi: 10.1016/j.taap.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 39.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kigerl KA, Gensel JC, Ankeny DP, Alexander JK, Donnelly DJ, Popovich PG. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. The Journal of Neuroscience. 2009;29:13435–13444. doi: 10.1523/JNEUROSCI.3257-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikita J, Dubourdieu-Cassagno N, Deloire MS, Vekris A, Biran M, Raffard G, et al. Altered M1/M2 activation patterns of monocytes in severe relapsing experimental rat model of multiple sclerosis. Amelioration of clinical status by M2 activated monocyte administration. Multiple Sclerosis Journal. 2011;17:2–15. doi: 10.1177/1352458510379243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.