Abstract
Xerostomia (dry mouth), resulting from radiation damage to the parotid glands, is one of the most common and distressing side effects of head-and-neck cancer radiotherapy. Recent MRI studies have demonstrated that the volume reduction of parotid glands is an important indicator for radiation damage and xerostomia. In the clinic, parotid-volume evaluation is exclusively based on physicians’ manual contours. However, manual contouring is time-consuming and prone to inter-observer and intra-observer variability. Here, we report a fully automated multi-atlas-based registration method for parotid-gland delineation in 3D head-and-neck MR images. The multi-atlas segmentation utilizes a hybrid deformable image registration to map the target subject to multiple patients’ images, applies the transformation to the corresponding segmented parotid glands, and subsequently uses the multiple patient-specific pairs (head-and-neck MR image and transformed parotid-gland mask) to train support vector machine (SVM) to reach consensus to segment the parotid gland of the target subject. This segmentation algorithm was tested with head-and-neck MRIs of 5 patients following radiotherapy for the nasopharyngeal cancer. The average parotid-gland volume overlapped 85% between the automatic segmentations and the physicians’ manual contours. In conclusion, we have demonstrated the feasibility of an automatic multi-atlas based segmentation algorithm to segment parotid glands in head-and-neck MR images.
Keywords: Image registration, support vector machine, segmentation, MRI, parotid gland, head-and-neck cancer, radiation toxicity, xerostomia
1. INTRODUCTION
Radiation therapy is an important treatment modality in head-and-neck cancer. Worldwide head-and-neck cancer statistics indicate that there are about 640,000 new cases diagnosed each year [1]. The incidence of head and neck cancer varies widely across the world [2]. For example, nasopharyngeal cancer is one of the most prevalent cancers in China, than in the rest of the world [3]. Many patients with head-and-neck cancers receive radiation therapy as part of their treatment, which frequently causes considerable morbidity. Xerostomia (dry mouth), resulting from radiation damage to the parotid glands, is one of the most common and distressing side effects of head-and-neck cancer radiotherapy [4, 5]. Recent MRI studies have demonstrated volume reduction of parotid glands is an important indicator of radiation damage and dry mouth. In the clinic, the parotid-gland volume evaluation is exclusively based on physicians’ manual contours. However, manual contouring is time-consuming and prone to intra-observer and inter-observer variability [6]. Tools for automated parotid-gland segmentation are thus helpful for physicians in evaluating radiation-induced parotid damage.
We have developed a fully automatic multi-atlas-based segmentation method to delineate the parotid glands in head-and- neck MR images. An atlas, in the context of this work, is considered as two image volumes: one 3D head-and-neck MR scan and its corresponding parotid-gland mask (manually contoured by physician). Given an atlas, segmentation for a target subject can be estimated using image registration. A multi-atlas-based segmentation process is normally consisted of three steps: a registration step where the target image is registered to the multiple atlases, a fusion step where the labels are transferred from the atlas to the target, and a final segmentation step in which transferred labels are used to segment the target image.
A critical assumption of atlas-based segmentation is that a deformation exists to align the atlas with the target so that the objects of interest line up perfectly. However, insufficient similarity between the atlas and the target image often results in local mismatches, which in turn leads to segmentation errors [7]. Several studies have shown that multi-atlas segmentation outperforms methods that use a single atlas [8, 9]. This multi-atlas approach to segmentation reduces the effect of errors associated with individual propagated atlases. For example, a registration error for a particular propagated atlas is less likely to affect the final segmentation when combined with other atlases [8]. van Rikxoort and collaborators employed an atlas selection strategy to choose a registered atlas image most similar to the target image, and the subsequent segmentation is the same as the single atlas-based method [10]. van der Lijn et. al. created a probabilistic atlas based on registered atlases, defined an energy function, and minimized this function to get segmentation results [11]. In a prostate ultrasound study, Yang et. al. utilized a support vector machine (SVM) to combine multiple atlases to provide prostate segmentation [12, 13]. In this paper, we employ a SVM to combine multiple subject-specific atlases to reach a consensus segmentation estimate for the target parotid glands. The key concept of SVM is the use of hyperplanes to define decision boundaries separating between data points of different classes.
Our multi-atlas-based segmentation method was tested with 5 patients post radiotherapy for their head-and-neck malignancies. The detailed steps of our segmentation method and its evaluation results are reported in the following sections.
2. METHODS
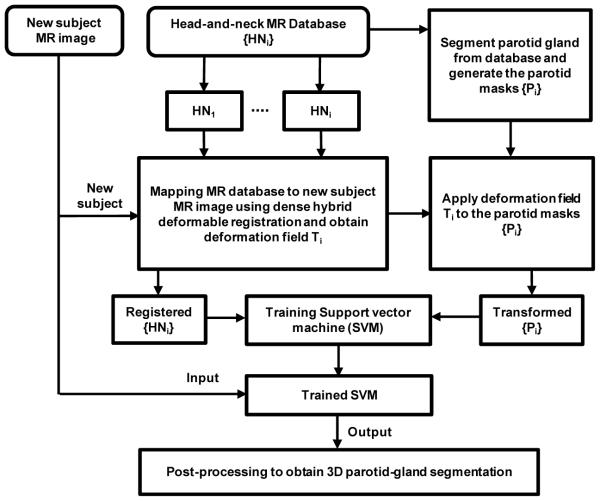
Our segmentation method consists of two major components: (1) atlas-based registration and (2) SVM training and parotid segmentation. Figure 1 shows a schematic flow chart of the proposed algorithm.
Figure 1.
Flow chart of multi-atlas based segmentation algorithm of the parotid gland.
2.1 Hybrid Deformable Registration
We built a patient MRI database for the proposed project. The database included the head-and-neck MR images and parotid-gland masks segmented by the physician. We registered the MR images in the database to the new MR images. A hybrid deformable registration method was used to obtain the spatial deformation field between the target MR image and the images in the database. The same transformation was then applied to the segmented parotid mask in the database.
Due to the fact that the sizes of the parotid glands may vary among patients, and head-and-neck MR images may be acquired at various positions and orientation, the non-rigid registration (translations, rotations, scaling, and deformation) is needed to normalize the image with respect to the template, and thus build the multiple-atlas database. In order to handle intensity contrast and inhomogeneity changes [14, 15] that often exist between MRI images, we applied a hybrid image matching metric [16]. This hybrid metric is a combination of the popular mutual information (MI) metric and a normalized sum-of-squared-differences (NSSD) metric:
| (1) |
| (2) |
| (3) |
where I1 and I2 denote the atlas and the target images respectively; H(I1) and H(I2) denote the marginal entropies of I1 and I2; and H(I1, I2) denotes their joint. α and β are the relative weighting of the two terms (α = β = 0.5). σI = Gs ∗ I denotes the local intensity mean, and al = Gs ∗ (I − uI) denotes the local intensity variation of image I. Gσ denotes a Gaussian filter with kernel size s (the kernel size s is chosen to be three times the image voxel size). This hybrid similarity measure provides a better image alignment than using the MI metric alone since the NSSD-term is an edge-based alignment metric, and it cannot take into account the local image contrast changes. Thus, this metric tends to improve the segmentation accuracy for all structures comparing to the MI-only method.
2.2 Support Vector Machine Training and Segmentation
SVM are supervised machine learning models with associated learning algorithms that analyze data and recognize patterns, used for classification and regression analysis. The idea behind SVMs is to map the original data points from the input space to a high-dimensional, feature space such that the classification problem becomes simpler in the feature space. Given a set of training examples, each marked as belonging to one of two categories, an SVM training algorithm builds a model that assigns target subject into one category or the other. An SVM model is a representation of the examples as points in space, mapped so that the examples of the separate categories are divided by a clear gap that is as wide as possible. The target subject is then mapped into that same space and predicted to belong to a category based on the side of the gap they fall on.
There are many hyperplanes that might classify the data. One reasonable choice as the best hyperplane is the one that represents the largest separation, or margin, between the two classes (parotid glands and non-parotid glands). So the hyperplane is chosen so that the distance from it to the nearest data point on each side is maximized. If such a hyperplane exists, it is known as the maximum-margin hyperplane. A nonlinear classifier was created by applying the kernel trick to maximum-margin hyperplanes [17]. The resulting algorithm is similar to a linear classifier, except that every dot product is replaced by a nonlinear kernel function. This allows the algorithm to fit the maximum-margin hyperplane in a high dimensional transformed feature space [18]. In this paper a Gaussian radial basis function (k(xi, xj) = exp(−r xi − xj||2)) is used as the kernel of SVM.
Our method uses multiple subject-specific atlas pairs to train the kernel SVM. The registered MR images are the input of a training pairs for SVM [12, 13]. The corresponding transformed parotids mask is used as the output of the training pairs. We use trained SVM to segment the target subject. The trained SVM is then applied to the target MR image in order to segment the parotid glands. The output of trained kernel SVM after inputting the target MR image is a binary image (volume) consisting of many “0” and “1” points. We performed some post-processing such as smoothing to obtain 3D segmented parotid glands (left and right).
Segmentation Evolution: Our segmentation method was tested with T2-weighted MR images (voxel size: 0.4688×0.4688×5.0 mm3) of 5 patients, 3 males and 2 females, following radiotherapy for nasopharyngeal cancer. The data were made available by China-Japan Union Hospital of Jilin University, Jilin, China. All parotid glands were contoured by an experience radiation oncologist (NW). We used leave-one-out cross-validation method to evaluate the proposed segmentation algorithm. In other words, we used the 4 training datasets as the atlases and applied the proposed method to process the remaining subject. To evaluate the performance of our segmentation method, we compared the parotid gland volumes between the automatic and the physician’s contours with the Dice overlap ratio. The Dice overlap ratio is defined as follows:
| (4) |
where Vol1 and Vol2 are binary parotid segmented volumes.
3. RESULTS
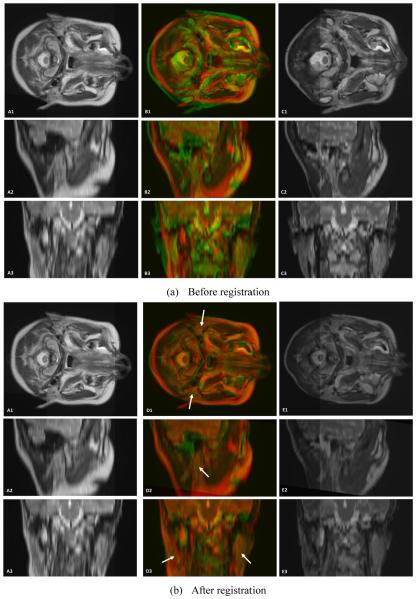
Parotid-gland segmentation was successfully applied to the T2-weighted MR images of all 5 patients. Figure 2 shows the deformable registration results between a target patient (require to be segmented) and the floating head-and-neck MR images (with parotid-gland segmented). The atlas MR image is registered to the target image, yielding a transformation which allows the atlas segmentation to be transformed and treated as a segmentation estimate for the target subject. As demonstrated in Fig. 3, the proposed automatic segmentation method worked well for the parotid glands, and achieved similar results as compared to physician’s manual segmentation.
Figure 2.
Visual assessment of the registration of 3D head-and-neck MR images. A1, A2 and A3 are head-and-neck MR reference images in transverse, sagittal and coronal directions. C1, C2 and C3 are head-and-neck MR floating images in three directions. B1, B2 and B3 are the fused images between the reference and floating MR images. E1, E2 and E3 are the 3D registered floating MR images. D1, D2 and D3 are the fused images between the reference MR and registered floating MR images (arrows indicate the parotid glands).
Figure 3.
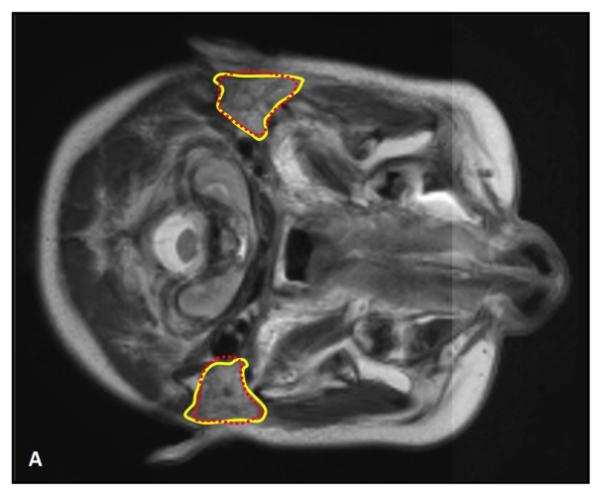
Comparison between the automatic and manual segmentations of the parotid glands. Manual segmentations are shown in solid yellow lines, while the automatic segmentations are shown in dotted red line. Close matching between our automatic segmentations and the physicians’ manual contours indicates the robustness the proposed method.
Table 1 provides quantitative evaluation of our segmentation algorithm using leave-one-out cross-validation method. The Dice overlap ranged from 81.8% to 89.1% with a mean of 85.3%.
Table 1.
Dice overlap between automatic and manual segmented parotid-gland volume
| Patients | No. 1 | No. 2 | No. 3 | No. 4 | No. 5 | Mean ± STD | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Parotid Gland | Left | Right | Left | Right | Left | Right | Left | Right | Left | Right | |
| Dice Overlap (%) | 89.1 | 85.9 | 86.5 | 83.9 | 84.5 | 87.7 | 82.6 | 81.8 | 84.2 | 86.3 | 85.3 ± 2.3 |
4. CONCLUSION
We propose an automatic algorithm for parotid-gland segmentation in head-and-neck MR images based on multi-atlas-based registration and machine learning. In multi-atlas methods, atlases within a database can be registered to a target image; their segmentations can be transformed and subsequently combined to provide a consensus segmentation estimate for the target subject. Although atlas-based segmentation method has been used to delineate parotid glands in head-and-neck CT images [16, 19-21], its application in MR images is new. In addition, we incorporated SVM to combine multiple subject-specific atlases, and improved the segmentation accuracy. Overall, automatic segmentation of parotid gland following head-and-neck radiotherapy to assess radiation-induced parotid damage is novel, and its clinical implication is significant. In vivo clinical results of 5 head-and-neck MRI datasets demonstrated the feasibility of our segmentation algorithm.
ACKNOWLEDGEMENTS
This research was supported by the National Natural Science Foundation of China (81201737), the Natural Science Foundation of Jilin Province (20090458 & 201015183), the Basic Scientific Research Foundation of Jilin University (2009 Young Teachers Innovation Program 421010043430 & 2011 Scientific Frontier and Interdisciplinary Innovation Program 450060445247) and the Young Scholars Rese rch Foundation of China-Japan Union Hospital (2009).
REFERENCES
- [1].Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- [2].Boyle P, Levin B. World Cancer Report 2008. 2008 [Google Scholar]
- [3].Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- [4].Yang X, Tridandapani S, Beitler J, et al. Ultrasound GLCM texture analysis of radiation-induced parotid-gland injury in head-and-neck cancer radiotherapy: An in vivo study of late toxicity. Medical Physics. 2012;39(9):5732. doi: 10.1118/1.4747526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yang X, Tridandapani S, Beitler JJ, et al. Ultrasound Histogram Assessment of Parotid Gland Injury Following Head-and-Neck Radiotherapy: A Feasibility Study. Ultrasound in Medicine and Biology. 2012;38(9):1514–1521. doi: 10.1016/j.ultrasmedbio.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hoogeman MS, Han X, Teguh D, et al. Atlas-based auto-segmentation of CT images in head and neck cancer: What is the best approach? International Journal of Radiation Oncology Biology Physics. 2008;72(1):S591–S591. [Google Scholar]
- [7].Crum WR, Griffin LD, Hill DLG, et al. Zen and the art of medical image registration: correspondence, homology, and quality. Neuroimage. 2003;20(3):1425–1437. doi: 10.1016/j.neuroimage.2003.07.014. [DOI] [PubMed] [Google Scholar]
- [8].Aljabar P, Heckemann RA, Hammers A, et al. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage. 2009;46(3):726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- [9].Heckemann RA, Hajnal JV, Aljabar P, et al. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33(1):115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- [10].van Rikxoort EM, Isgum I, Arzhaeva Y, et al. Adaptive local multi-atlas segmentation: Application to the heart and the caudate nucleus. Medical Image Analysis. 2010;14(1):39–49. doi: 10.1016/j.media.2009.10.001. [DOI] [PubMed] [Google Scholar]
- [11].van der Lijn F, den Heijer T, Breteler MMB, et al. Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts. Neuroimage. 2008;43(4):708–720. doi: 10.1016/j.neuroimage.2008.07.058. [DOI] [PubMed] [Google Scholar]
- [12].Yang X, Fei B. 3D prostate segmentation of ultrasound images combining longitudinal image registration and machine learning. Proc. SPIE. 2012;8316:83162O. doi: 10.1117/12.912188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang X, Schuster D, Master V, et al. Automatic 3D segmentation of ultrasound images using atlas registration and statistical texture prior. Proc. SPIE. 2011;7964:796432. doi: 10.1117/12.877888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang X, Fei B. A wavelet multiscale denoising algorithm for magnetic resonance (MR) images. Measurement Science and Technology. 2011;22:025803. doi: 10.1088/0957-0233/22/2/025803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yang X, Fei B. A multiscale and multiblock fuzzy C-means classification method for brain MR images. Medical Physics. 2011;38(6):2879–2891. doi: 10.1118/1.3584199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Han X, Hibbard L, O'Connell N, Willcut V. Automatic Segmentation of Parotids in Head and Neck CT Images using Multi-atlas Fusion. MICCAI 2010 Grand Challenges in Medical Image Analysis: Head & Neck Autosegmentation Challenge, Beijing. 2010 [Google Scholar]
- [17].Boser BE, Guyon IM, Vapnik VN. A Training Algorithm for Optimal Margin Classifiers. COLT '92 Proceedings of the fifth annual workshop on Computational learning theory. 1992:144–152. [Google Scholar]
- [18].Meyer D, Leisch F, Hornik K. The support vector machine under test. Neurocomputing. 2003;55(1-2):169–186. [Google Scholar]
- [19].Han X, Hoogeman MS, Levendag PC, et al. Atlas-Based Auto-segmentation of Head and Neck CT Images. Medical Image Computing and Computer-Assisted Intervention - MICCAI. 2008;5242:434–441. doi: 10.1007/978-3-540-85990-1_52. [DOI] [PubMed] [Google Scholar]
- [20].Yang J, Zhang Y, Zhang L, Dong L. Automatic Segmentation of Parotids from CT Scans Using Multiple Atlases. Medical Image Computing and Computer-Assisted Intervention - MICCAI. 2010:323–330. [Google Scholar]
- [21].Hollensen C, Hansen MF, Hojgaard L, Specht L, Larsen R. Segmenting the Parotid Gland using Registration and Level Set Methods. MICCAI: Grand Challenges in Medical Image Analysis: Head & Neck Autosegmentation Challenge, Beijing. 2010 [Google Scholar]