Abstract Abstract
The expression of long noncoding RNAs (lncRNAs) in human heart failure (HF) has not been widely studied. Using RNA sequencing (RNA-Seq), we compared lncRNA expression in 22 explanted human HF hearts with lncRNA expression in 5 unused donor human hearts. We used Cufflinks to identify isoforms and DESeq to identify differentially expressed genes. We identified the noncoding RNAs by cross-reference to Ensembl release 73 (Genome Reference Consortium human genome build 37) and explored possible functional roles using a variety of online tools. In HF hearts, RNA-Seq identified 84,793 total messenger RNA coding and noncoding different transcripts, including 13,019 protein-coding genes, 2,085 total lncRNA genes, and 1,064 pseudogenes. By Ensembl noncoding RNA categories, there were 48 lncRNAs, 27 pseudogenes, and 30 antisense RNAs for a total of 105 differentially expressed lncRNAs in HF hearts. Compared with donor hearts, HF hearts exhibited differential expression of 7.7% of protein-coding genes, 3.7% of lncRNAs (including pseudogenes), and 2.5% of pseudogenes. There were not consistent correlations between antisense lncRNAs and parent genes and between pseudogenes and parent genes, implying differential regulation of expression. Exploratory in silico functional analyses using online tools suggested a variety of possible lncRNA regulatory roles. By providing a comprehensive profile of right ventricular polyadenylated messenger RNA transcriptome in HF, RNA-Seq provides an inventory of differentially expressed lncRNAs, including antisense transcripts and pseudogenes, for future mechanistic study.
Keywords: long noncoding RNA, right ventricle, human heart failure, gene expression, transcriptional profiling
The results reported from the Encyclopedia of DNA Elements (ENCODE) project are transforming our nascent understanding of integrated human genomics.1 Fully 75% of the human genome is transcribed into some type of RNA,2 although only 1.22% of the genome encodes the exons that comprise the 20,687 known protein-coding genes.1 The vast majority of the human genome, therefore, is transcribed into non–protein-coding RNAs, including the 8,801 small RNAs, 9,640 long noncoding RNAs (lncRNAs; defined as nontranslated RNA <200 base pairs [bp] in length) and 11,224 pseudogenes identified to date.1 Evolving research has identified diverse epigenetic regulatory roles for lncRNAs in development, homeostasis, and disease.3 lncRNAs are likely to play important regulatory roles in human heart failure as well.4
Via high-throughput sequencing, RNA sequencing (RNA-Seq) provides a comprehensive profile of polyadenylated protein-coding messenger RNA (mRNA) and non–protein-coding lncRNA transcripts.5 We used RNA-Seq to generate the lncRNA transcriptional profile of the right ventricle in end-stage human heart failure. Human right ventricular (RV) tissue may be obtained at either endomyocardial biopsy or surgery. Diagnostic endomyocardial biopsy, given its attendant risks, is rarely performed for patients with pulmonary arterial hypertension or isolated RV heart failure and yields small amounts of tissue. Explanted RV human myocardium, by contrast, is readily obtainable at cardiac transplantation and yields abundant tissue collectable under strictly controlled conditions to prevent RNA degradation. We thus examined the human RV transcriptome in hearts explanted at the time of transplantation for end-stage left ventricular (LV) failure.
Methods
Clinical characterization of study subjects
De-identified clinical data were abstracted for all study subjects. RV contractile function was classified as normal or dysfunctional on the basis of the presence of mild or greater RV hypokinesis by visual inspection on the echocardiogram performed most closely to explantation. RV size and function were also quantified by tricuspid annular plane systolic excursion, the index of myocardial performance, and fractional area change according to American Society of Echocardiography guidelines. Pulmonary hypertension was defined as a mean pulmonary artery pressure greater than 25 mmHg by the right heart catheterization performed most closely to explantation.
RNA preparation
The myocardial samples were free of macroscopic infarction or fibrosis and were collected from approximately the same cross-sectional region of the unused donor and heart failure RVs. Ninety to 120 mg of tissue per study subject was used for RNA extraction. Total RNA was isolated using miRNeasy Mini kit (Qiagen) following the manufacturer’s instructions.
RNA-Seq
We first performed a quality check of the input total RNA by running an aliquot on the Agilent Bioanalyzer to confirm integrity, and a Qubit RNA fluorometry assay was used to measure concentration. For each library, 100 ng of total RNA underwent enrichment for poly(A)-containing mRNA using poly(T) oligo-attached magnetic beads. Following purification, eluted poly(A) RNA was cleaved into small fragments of 120–210 bp (divalent cations, elevated temperature). The cleaved RNA fragments were copied into first-strand complementary DNA (cDNA) using SuperScript II reverse transcriptase and random primers. This was followed by second-strand cDNA synthesis using DNA polymerase I and RNase H. The cDNA fragments then underwent an end-repair process, the addition of a single “A” base, and ligation of the Illumina multiplexing adapters. The products were purified and enriched with polymerase chain reaction (PCR) to create the final cDNA sequencing library. The cDNA library underwent quality control check via an Agilent Bioanalyzer HS DNA assay to confirm final library size and via Agilent Mx3005P quantitative PCR (qPCR) machine using the KAPA Illumina library quantification kit to determine concentration. From a 2-nM stock, samples were pooled by molarity for multiplexing. From the pool, 12 pM was loaded into each well for the flow cell on the Illumina cBot for cluster generation. The flow cell was loaded onto the Illumina HiSeq 2500 using v3 chemistry and HTA 1.8 and sequenced at paired-end 50 bp with a target of 30 million pass filter reads per library. The raw sequencing reads in BCL format were processed through CASAVA-1.8.2 for FASTQ conversion and demultiplexing. The RTA chastity filter was used, and only the pass filter reads were retained for any further analysis.
Analysis of gene and transcript expression
We first performed quality control on raw sequence-derived data to identify potential outliers before doing any advanced analysis by applying tools such as Fastx Toolkit and FastQC. RNA read alignment and mapping were performed by Bowtie/TopHat, and transcriptome reconstruction was performed by Cufflinks for both mRNA and lncRNA.6 For Cufflinks, a minimum RPKM (reads per kilobase of exon model per million mapped reads) value of ≥1 was required for further analysis.7 Cufflinks was also used to detect and quantitate alterative spliced transcripts and isoforms.8
For differential expression quantification of mRNA and lncRNA genes,9 we used three read-count-based negative binomial methods, DESeq,10 edgeR,11 and baySeq12 to explore the consistency of differential expression quantification across methods. Each method aligned then adjusted the unadjusted read-count tables before analysis. For each platform, inflated type I error due to multiple comparisons was adjusted, and all adjusted tests were significant at the two-sided significance level of P ≤ 0.05. To examine parsimony and exclude potential bias across the three methods, we also compiled a sum of ranks from the three read-count-based methods to identify differentially expressed genes (DEGs) consistently selected across the three platforms. Following inspection of this ranking across the three platforms, for final selection of DEGs, we elected to use the results afforded by DESeq. DESeq afforded the best compromise between permissive (edgeR) and restrictive (baySeq) identification of DEGs and afforded the best reconciliation with the sum-of-ranks approach. In reporting adjusted total read counts for individual genes, we divided the unadjusted total gene read counts in a given library by the total read counts for all genes in same library and then multiplied this by 1 × 106.
Exploration of lncRNA, pseudogene, and antisense function
Exploratory characterization of selected lncRNAs and antisense RNAs was provided by the online lncRNA search engine and bioinformatics and annotation tool LNCipedia (http://www.lncipedia.org),13 a database for human lncRNA transcripts and genes that provides basic transcript information, gene structure and sequence from the genome sequence assembly tool Ensembl, secondary structure from RNAfold image, protein-coding potential from Coding Potential Calculator (CPC; used to assess protein-coding potential of a given RNA transcript) and HMMER, Proteomics Identification (PRIDE) database search, and microRNA (miRNA; nontranslated RNA usually 18–24 bp in length) binding site predictions from MiRTarget2 (an online miRNA target search engine and bioinformatics tool). Exploratory functional annotation of selected pseudogenes was provided by the on-line resource tool pseudoMap (http://pseudomap.mbc.nctu.edu.tw/).14 The pseudoMap tool maps a known pseudogene to both (1) the predicted endogenous short interfering RNAs (esiRNAs, which may silence mRNA translation via RNA interference) produced from that pseudogene and (2) the predicted miRNA targets for that pseudogene (by binding miRNA, a pseudogene may act as an “miRNA decoy,” providing competitive endogenous RNA-mediated regulation of mRNA translation). In selected instances, either the DIANA-LncBase (http://www.microrna.gr/LncBase)15 or Segal Lab of Computational Biology at the Weismann Institute of Science online tools (http://genie.weizmann.ac.il)16 was used for computational prediction of miRNA targets on lncRNAs or parent genes. For exploring noncoding RNA-protein interactions, RNA-Protein Interaction Prediction (RPISeq), which uses both random forest (RF) and support vector machine (SVM) classifiers to predict RNA-protein interactions using only sequence information (http://pridb.gdcb.iastate.edu/RPISeq),17 and catRAPID, an online in silico RNA and protein binding prediction tool that uses sequence information to evaluate the interaction propensities of polypeptide and nucleotide chains using physicochemical properties (http://s.tartagilalab.com/catrapid/omics),18 were used.
Results
Subjects
The clinical and echocardiographic characteristics of the subjects who underwent transplant appear in Table 1.
Table 1.
Clinical and echocardiographic characteristics of study subjects
| Characteristic | Nonischemic HF (n = 11) | Ischemic HF (n = 11) |
|---|---|---|
| Clinical | ||
| Age, years | 49 ± 14 | 54 ± 9 |
| Sex (M/F) | 7/4 | 9/2 |
| No. of subjects with diabetes | 5 | 8 |
| Serum creatinine level, mg/dL | 1.6 ± 0.7 | 1.6 ± 0.7 |
| Hematocrit, % | 34 ± 5 | 35 ± 6 |
| Therapy, no. of subjects | ||
| ACEI/ARB | 7 | 7 |
| Beta-blocker | 10 | 9 |
| Aldosterone inhibitor | 9 | 9 |
| PDE V inhibitor | 4 | 1 |
| Inotropic therapy | 10 | 10 |
| Intra-aortic balloon pump* | 0 | 5 |
| Ventricular assist device | 1 | 0 |
| Hemodynamic | ||
| Right atrial pressure, mmHg | 9 ± 6 | 8 ± 6 |
| Mean pulmonary artery pressure, mmHg | 26 ± 8 | 27 ± 10 |
| Pulmonary capillary wedge pressure, mmHg | 16 ± 8 | 17 ± 7 |
| Pulmonary vascular resistance, Woods units | 2.2 ± 0.6 | 2.6 ± 1.7 |
| Cardiac index, L/min/m2 | 2.2 ± 0.4 | 2.1 ± 0.4 |
| Echocardiographic | ||
| RV basal, cm | 4.6 ± 0.5 | 4.5 ± 0.8 |
| RV long axis, cm | 8.3 ± 1.1 | 7.3 ± 0.6 |
| TAPSE, mm | 14.3 ± 4.9 | 11.7 ± 3.9 |
| RIMP | 54.7 ± 25.6 | 49.6 ± 10.7 |
| FAC, % | 29.2 ± 20.7 | 33.2 ± 11.4 |
| RAP, estimated mmHg | 8.7 ± 4.7 | 6.3 ± 5.1 |
| Moderate or severe dysfunction, no. of subjects | 5 | 6 |
Data are mean value ± standard deviation, unless otherwise indicated. Asterisk indicates P < 0.05. ACEI: angiotensin-converting enzyme inhibitor; ARB: angiotensin receptor blocker; FAC: right ventricular fractional area change; HF: heart failure; PDE V: phosphodiesterase type V; RAP, right atrial pressure; RIMP: RV index of myocardial performance; RV: right ventricular; TAPSE: tricuspid annular plane systolic excursion.
Heart failure RV transcriptome
By the read count methods, there were 791,427,072 total paired-end, Ensembl-annotated unadjusted reads summed across the 32 cDNA libraries. The average number of aligned, Ensembl-annotated unadjusted reads per library was 24,732,096, with a range of reads per library from 18,607,371 to 35,024,568. There were 29,501,480, 25,357,404, and 23,506,029 average unadjusted reads in the unused donor LV, unused donor RV, and heart failure RV libraries, respectively.
There were 77,768 and 83,793 total mRNA coding and noncoding transcripts identified in the unused donor RVs and heart failure RVs, respectively (Table 2). The heart failure RVs expressed 13,019 protein-coding genes, 2,085 lncRNAs, and 1,064 pseudogenes, and all types of transcripts were more abundant in the heart failure RVs than unused donor RVs.
Table 2.
Cumulative number of genes and transcripts expressed compared with GENCODE
| Variable | Total in ENCODE (GRch37_63, GENCODE version 16 lncRNA) | Expressed in normal RV (% ENCODE) (n = 5) | Expressed in normal LV (% ENCODE) (n = 5) | Expressed in failing RV (% ENCODE) (n = 22) |
|---|---|---|---|---|
| Genes expressed | ||||
| Total number of genes | 56,340 | 15,169 (27) | 15,083 (27) | 17,247 (31) |
| Protein-coding genes | 20,007 | 12,086 (60) | 12,001 (60) | 13,019 (65) |
| lncRNA genes | 13,220 | 1,474 (11) | 1,428 (11) | 2,085 (16) |
| Pseudogenes (processed and unprocessed) | 13,196 | 793 (6) | 819 (6) | 1,064 (8) |
| Transcripts expressed | ||||
| Total number | 174,930 | 77,768 (44) | 77,046 (44) | 83,793 (48) |
| Protein coding (full and partial length) | 125,376 | 71,792 (57) | 71,204 (57) | 76,033 (61) |
| lncRNA transcripts | 22,444 | 3,905 (17) | 3,723 (17) | 5,102 (23) |
For this study, a gene was considered expressed if the value for fragments per kilobase of exon was ≥1. ENCODE: Encyclopedia of DNA Elements; lncRNA: long noncoding RNA. LV: left ventricle; RV: right ventricle.
lncRNAs
There were 2,085 and 1,474 lncRNAs expressed in heart failure RVs and unused RVs, respectively (Table 3). In heart failure RVs versus unused donor RVs, 78 lncRNAs (48 lncRNAs and 30 natural antisense transcripts), representing 3.7% of the total 2,085 lncRNAs, were differentially expressed genes. The 48 lncRNA differentially expressed genes included intergenic, intronic, and processed transcripts. Among the 48 lncRNA heart failure RV differentially expressed genes, 35 were decreased in expression and 13 were increased in expression compared with unused donor RVs (Tables 3, 4). As a rule, lncRNAs exhibited a wide range of expression as denoted by the mean (± standard deviation) and maximum-minimum range (Table 4). The most abundantly expressed lncRNA in the heart failure RV group was RP11-206l10.11 (ENSG00000228794.3), a novel lncRNA with no predicted miRNA targets (Table 3).
Table 3.
Differentially expressed long noncoding RNAs (lncRNAs)
| Ensembl ID | Gene ID | L2F change | Adjusted P value | Chrom | Trans | lncRNA length, base pairs |
|---|---|---|---|---|---|---|
| ENSG00000255414.1 | AP000783.2 | −5.3 | 7.40E−14 | 11 | 1 | 2,471 |
| ENSG00000259383.1 | RP11-403B2.6 | Inf | 3.70E−13 | 15 | 1 | 748 |
| ENSG00000265542.1 | RP11-60A24.3 | −2.6 | 1.87E−07 | 17 | 2 | 505, 472 |
| ENSG00000268913.1 | AC026806.2 | 2.2 | 3.06E−07 | 19 | 1 | 443 |
| ENSG00000236423.1 | RP13-15E13.1 | −3.1 | 1.26E−06 | 1 | 3 | 634, 1,940, 3,059 |
| ENSG00000267653.1 | RP1-193H18.3 | −3.1 | 2.96E−05 | 17 | 1 | 584 |
| ENSG00000224260.2 | RP1-272L16.1 | −6.6 | 3.93E−05 | 1 | 2 | 641, 863 |
| ENSG00000253270.1 | RP11-1105014.1 | −3.2 | 6.00E−05 | 8 | 1 | 467 |
| ENSG00000260711.1 | RP11-747H7.3 | −2.6 | 7.77E−05 | 14 | 1 | 3,040 |
| ENSG00000228058.1 | RP11-552D4.1 | −4.9 | 8.76E−05 | 1 | 1 | 654 |
| ENSG00000270132.1 | CTC-458A3.8 | −3.3 | 1.30E−04 | 8 | 1 | 642 |
| ENSG00000261606.1 | AP000783.2 | −1.9 | 2.50E−04 | 11 | 1 | 2,471 |
| ENSG00000246375.2 | RP11-10L7.1 | 1.8 | 3.60E−04 | 4 | 1 | 1,654 |
| ENSG00000249816.2 | LINC00964 | −2.1 | 6.20E−04 | 8 | 9 | 443–2,203 |
| ENSG00000228794.3 | RP11-206l10.11 | −1.03 | 8.70E−04 | 1 | 7 | 612–6,606 |
| ENSG00000246328 | AC020926.1 | −1.9 | 1.30E−03 | 16 | 1 | 3,309 |
| ENSG00000260391.1 | RP11-71H17.7 | −1.3 | 1.50E−03 | 3 | 1 | 2,538 |
| ENSG00000230102.2 | RP11-407B7.1 | 2.2 | 2.50E−03 | 3 | 15 | 203–2,666 |
| ENSG00000235387.1 | LINC0096 | −1.4 | 3.10E−03 | 9 | 1 | 1,612 |
| ENSG00000245651.2 | RP11-620J15.2 | 2.2 | 3.10E−03 | 12 | 2 | 549, 3,507 |
| ENSG00000263860.1 | RP11-218M11.3 | −4.7 | 3.40E−03 | 17 | 1 | 280 |
| ENSG00000259176.1 | RP11-69H14.6 | −5.8 | 5.20E−03 | 15 | 14 | 346–2,317 |
| ENSG00000250763.1 | RP11-974F13.6 | −1.5 | 6.30E−03 | 5 | 2 | 689, 3,763 |
| ENSG00000255328.1 | RP11-326C3.12 | −2.2 | 7.60E−03 | 11 | 3 | 457, 587, 827 |
| ENSG00000227403.1 | AC009299.3 | Inf | 9.30E−03 | 2 | 2 | 765, 2,333 |
| ENSG00000260186.1 | RP11-481J2.2 | −2.2 | 9.60E−03 | 16 | 3 | 538, 644, 930 |
| ENSG00000258819.1 | RP11-7F17.3 | −1.8 | 9.80E−03 | 14 | 3 | 424, 861, 1,817 |
| ENSG00000249460.1 | RP11-665C14.2 | 3.9 | 1.00E−02 | 4 | 1 | 493 |
| ENSG00000266743.1 | RP11-94B19.6 | 3.7 | 1.10E−02 | 18 | 1 | 464 |
| ENSG00000244227.1 | RP11-298021.5 | 1.1 | 1.40E−02 | 3 | 7 | 436–2,350 |
| ENSG00000261480.1 | RP11-578F21.6 | −2.5 | 1.60E−02 | 15 | 1 | 557 |
| ENSG00000261116.1 | RP3-523K23.2 | −2.7 | 1.60E−02 | 6 | 1 | 1,933 |
| ENSG00000259366.1 | CTD-2647L4.4 | −2 | 1.70E−02 | 8 | 1 | 552 |
| ENSG00000270074.1 | RP11-351l21.11 | 1.3 | 1.70E−02 | 8 | 1 | 1,547 |
| ENSG00000225882.1 | RP3-410B.11 | Inf | 1.70E−02 | X | 1 | 708 |
| ENSG00000223617.1 | RP11-54H7.2 | −3.6 | 1.80E−02 | 13 | 1 | 698 |
| ENSG00000260512.1 | RP13-259F12.2 | −0.9 | 2.10E−02 | 11 | 1 | 5,329 |
| ENSG00000232104.2 | RP11-380J14.1 | −1.8 | 2.20E−02 | 1 | 1 | 4,676 |
| ENSG00000260025.1 | RP11-509J21.1 | −1.9 | 2.50E−02 | 9 | 2 | 496, 552 |
| ENSG00000249868.4 | RP11-490M8.1 | 1.5 | 2.50E−02 | 2 | 1 | 366 |
| ENSG00000232044.3 | RP11-63E5.6 | −1.6 | 2.60E−02 | 8 | 1 | 880 |
| ENSG00000233013.4 | AC073479.1 | −1.7 | 3.00E−02 | 2 | 5 | 580–3,329 |
| ENSG00000182021.5 | FAM157B | −2.8 | 3.10E−02 | 9 | 2 | 1,278, 1,795 |
| ENSG00000264920.1 | RP11-38107.3 | −1.1 | 4.00E−02 | 9 | 5 | 834–2,566 |
| ENSG00000248017 | AC026449.1 | −3.1 | 4.30E−02 | 5 | 1 | 818 |
| ENSG00000219159.3 | AC011298.2 | Inf | 4.50E−02 | 2 | 2 | 949, 3,398 |
| ENSG00000250392.1 | RP11-700N1.1 | 0.9 | 4.50E−02 | 4 | 1 | 642 |
| ENSG00000256910.1 | AL034397.1 | −1.3 | 4.80E−02 | X | 2 | 1,690, 1,815 |
Chrom: chromosome; Ensembl: Ensembl release 74; inf: value cannot be calculated; L2F: log2-fold change; Trans: transcripts.
Table 4.
Long noncoding RNAs (lncRNAs): unused donor (DON) right ventricle (RV) versus heart failure (HF) RV read counts and LNCipedia notes
| Gene ID | DON RV MEAN | DON RV SD | DON RV MIN | DON RV MAX | HF RV MEAN | HF RV SD | HF RV MIN | HF RV MAX | L2F change | L2F change P value | LNCipedia notesa |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AP000783.2 | 99.6 | 143.0 | 2 | 346 | 2.3 | 2.5 | 0 | 11 | −5.3 | 7.40E−14 | Novel lncRNA, 2 exons; miR targets 4760-3p, 942, 4659-3p |
| RP11-403B2.6 | 0 | 0.0 | 0 | 0 | 16.1 | 62.9 | 0 | 292 | Inf | 3.70E−13 | No miR targets identified |
| RP11-60A24.3 | 70.4 | 33.2 | 16 | 98 | 8.8 | 6.8 | 0 | 21 | −2.6 | 1.87E−07 | lnc-MRPS23-1:1; no miR targets identified |
| AC026806.2 | 34.6 | 12.3 | 26 | 55 | 137.7 | 70.2 | 38 | 372 | 2.2 | 3.06E−07 | No entry |
| RP13-15E13.1 | 50.4 | 45.9 | 5 | 127 | 5.4 | 3.1 | 0 | 13 | −3.1 | 1.26E−06 | lnc-DFFB; miR targets 1913, 324-3p, 187-5p (634 bp) and 149 (3,059 bp) |
| RP1-193H18.3 | 146 | 79.4 | 35 | 239 | 12.5 | 13.1 | 0 | 48 | −3.1 | 2.96E−05 | No entry |
| RP1-272L16.1 | 7.8 | 12.5 | 1 | 30 | 0.2 | 0.7 | 0 | 3 | −6.6 | 3.93E−05 | No entry |
| RP11-1105014.1 | 29.6 | 41.5 | 3 | 102 | 2.8 | 3.5 | 0 | 16 | −3.2 | 6.00E−05 | lnc-INTS10-2; miR targets 508-5p, 765, 4255, 4760-3p |
| RP11-747H7.3 | 53.4 | 43.7 | 10 | 125 | 11.0 | 7.7 | 1 | 28 | −2.6 | 7.77E−05 | lnc-TC2N-1; no miR targets identified |
| RP11-552D4.1 | 6.6 | 2.6 | 3 | 9 | 0.3 | 0.6 | 0 | 2 | −4.9 | 8.76E−05 | lnc-GALNT2-2:2; miR targets 4540, 4680-3p, 148a-3p, 148b-3p, 2861, 5006-3p |
| CTC-458A3.8 | 24.6 | 17.3 | 14 | 55 | 2.0 | 2.3 | 0 | 10 | −3.3 | 1.30E−04 | No entry |
| AP000783.2 | 304.4 | 84.3 | 179 | 407 | 66.8 | 60.2 | 2 | 241 | −1.9 | 2.50E−04 | miR targets 4760-3p, 942, 4659a-3p |
| RP11-10L7.1 | 43.8 | 16.1 | 17 | 60 | 155.9 | 174.3 | 8 | 535 | 1.8 | 3.60E−04 | lnc-HERC6-2; miR targets 5011-5p, 7-5p |
| LINC00964 | 83.8 | 51.9 | 38 | 161 | 14.5 | 10.3 | 1 | 38 | −2.1 | 6.20E−04 | lnc-ZNF572-1; miR targets 586, 3126-3p |
| RP11-206l10.11 | 3,581.8 | 497.7 | 2,923 | 4,200 | 1,625.1 | 417.8 | 1,009 | 2,385 | −1.03 | 8.70E−04 | lnc-AL669831.1-5; no miR targets identified |
| AC020926.1 | 412.8 | 245.5 | 137 | 750 | 94.5 | 58.8 | 31 | 240 | −1.9 | 1.30E−03 | miR targets 449a, 34a-5p |
| RP11-71H17.7 | 220 | 75.2 | 116 | 324 | 86.1 | 47.1 | 21 | 224 | −1.3 | 1.50E−03 | lnc-KALRN-1; no miR targets identified |
| RP11-407B7.1 | 7.4 | 3.0 | 4 | 11 | 32.9 | 13.9 | 11 | 56 | 2.2 | 2.50E−03 | lnc-CPN2-4; miR targets 641, 3609, 3617 |
| LINC0096 | 130.6 | 35.9 | 77 | 167 | 44.3 | 18.2 | 13 | 74 | −1.4 | 3.10E−03 | lnc-HRCT-1; miR targets 4652-3p, 584-3p, 4695-5p, 30d-3p |
| RP11-620J15.2 | 5.6 | 2.1 | 4 | 9 | 22.6 | 10.1 | 8 | 41 | 2.2 | 3.10E−03 | lnc-CTDSP2-1; miR targets 4498, 4713-5p, 4674, 4685-3p, 4287, 3664-3p |
| RP11-218M11.3 | 3 | 3.2 | 0 | 8 | 0.0 | 0.2 | 0 | 1 | −4.7 | 3.40E−03 | No miR targets identified |
| RP11-69H14.6 | 6.6 | 8.3 | 0 | 20 | 0.1 | 0.3 | 0 | 1 | −5.8 | 5.20E−03 | lnc-OR4N4-1; no miR targets identified |
| RP11-974F13.6 | 139 | 71.9 | 82 | 260 | 48.5 | 30.7 | 14 | 114 | −1.5 | 6.30E−03 | lnc-SERF1B-1; no miR targets identified |
| RP11-326C3.12 | 14.8 | 4.8 | 10 | 20 | 2.8 | 1.5 | 0 | 6 | −2.2 | 7.60E−03 | lnc-IFITM2-3; miR targets (12 total) 29a-5p, 888-3p, 524-5p, 520d-5p, 1229, 124-3p, |
| AC009299.3 | 0 | 0.0 | 0 | 0 | 2.8 | 2.7 | 0 | 10 | Inf | 9.30E−03 | lnc-TANK1; miR targets 4282, 194-5p, 5003-5p, 4799-5p, 216a, 20a-3p, 376b, 376a-3p |
| RP11-481J2.2 | 219.4 | 122.1 | 77 | 381 | 46.0 | 36.2 | 9 | 139 | −2.2 | 9.60E−03 | lnc-NDRG4-1; no miR targets identified |
| RP11-7F17.3 | 26.8 | 8.7 | 18 | 38 | 6.4 | 4.1 | 2 | 19 | −1.8 | 9.80E−03 | lnc-IRF2BPL-1; miR targets 626, 4753-3p |
| RP11-665C14.2 | 6.2 | 6.6 | 1 | 16 | 68.8 | 66.5 | 15 | 272 | 3.9 | 1.00E−02 | lnc-DCTD-6:2; miR targets 4679, 27b-5p |
| RP11-94B19.6 | 1 | 1.2 | 0 | 3 | 12.5 | 10.6 | 0 | 40 | 3.7 | 1.10E−02 | lnc-ZNF516-6; no miR targets identified |
| RP11-298021.5 | 119.2 | 32.6 | 87 | 158 | 213.2 | 63.4 | 108 | 344 | 1.1 | 1.40E−02 | lnc-SERPINI1-4; miR targets 4511, 345-3p, 1305, 2052, 4699-5p, 203, 544b, 5590-5p |
| RP11-578F21.6 | 9.4 | 4.0 | 5 | 15 | 1.5 | 1.2 | 0 | 4 | −2.5 | 1.60E−02 | No miR targets identified |
| RP3-523K23.2 | 22.6 | 13.5 | 8 | 44 | 2.5 | 4.7 | 0 | 21 | −2.7 | 1.60E−02 | lnc-FAM83B-1:1; no miR targets identified |
| CTD-2647L4.4 | 18.2 | 11.5 | 4 | 31 | 4.2 | 3.2 | 0 | 11 | −2 | 1.70E−02 | lnc-EXTL3-5; no miR targets identified |
| RP11-351l21.11 | 42 | 11.7 | 30 | 56 | 93.8 | 32.5 | 36 | 168 | 1.3 | 1.70E−02 | No entry |
| RP3-410B.11 | 2.8 | 6.3 | 0 | 14 | 0.8 | 2.2 | 0 | 10 | Inf | 1.70E−02 | lnc-BEND2-1; miR targets (13 total) 548aa, 548t-3p, 4482-3p, 600, 3545-5p, 29b-1-5p |
| RP11-54H7.2 | 5.2 | 6.3 | 0 | 16 | 0.6 | 0.8 | 0 | 2 | −3.6 | 1.80E−02 | lnc-MYO16-1; miR targets (22 total) 5011-3p, 525-5p, 629-3p, 502-5p, 374b-5p, 473b |
| RP13-259F12.2 | 283.8 | 75.1 | 213 | 375 | 128.4 | 54.0 | 55 | 267 | −0.9 | 2.10E−02 | lnc-GUCY1A2-1; no miR targets identified |
| RP11-509J21.1 | 13.4 | 7.7 | 5 | 23 | 2.9 | 1.8 | 0 | 7 | −1.8 | 2.20E−02 | lnc-KCNV2-3; miR targets 5706, 4782-5p, 46777-5p, 4768-3p, 5004-3p, 765, 548a |
| RP11-490M8.1 | 31.8 | 14.3 | 19 | 54 | 70.8 | 40.1 | 18 | 174 | −1.9 | 2.50E−02 | No miR targets identified |
| RP11-63E5.6 | 31.4 | 11.5 | 16 | 41 | 9.4 | 4.8 | 1 | 20 | 1.5 | 2.50E−02 | No miR targets identified |
| AC073479.1 | 69 | 37.5 | 34 | 129 | 15.1 | 15.0 | 0 | 52 | −1.6 | 2.60E−02 | lnc-SOX11-3; miR targets 452-5p, 330-3p, 5699 |
| FAM157B | 5.6 | 3.1 | 3 | 9 | 1.6 | 1.9 | 0 | 6 | −1.7 | 3.00E−02 | lnc-CACNA1B-1; no miR targets identified |
| RP11-38107.3 | 179.2 | 66.3 | 107 | 285 | 67.9 | 27.5 | 31 | 144 | −2.8 | 3.10E−02 | No entry |
| RP11-420A6.2 | 563.4 | 112.9 | 458 | 707 | 836.1 | 200.6 | 364 | 1,285 | −1.1 | 4.00E−02 | lnc-SP6-2; no miR targets identified |
| AC026449.1 | 3.8 | 2.6 | 1 | 7 | 0.2 | 0.4 | 0 | 1 | −3.1 | 4.30E−02 | lnc-AP351-1; miR targets 4495, 1273g-3p, 3185, 4303, 5006, 362-5p, 3943 |
| AC011298.2 | 0 | 0.0 | 0 | 0 | 2.0 | 1.8 | 0 | 5 | Inf | 4.50E−02 | No miR targets identified |
| RP11-700N1.1 | 337 | 111.4 | 142 | 424 | 543.7 | 127.8 | 293 | 778 | 0.9 | 4.50E−02 | No miR targets identified |
| AL034397.1 | 53.4 | 23.3 | 33 | 84 | 19.4 | 7.6 | 8 | 34 | −1.3 | 4.80E−02 | lnc-HEPH-1 miR targets (37 total) 4700-3p, 143-3p, 4770, 122-5p, 202-5p, 760, 4501 |
bp: base pairs; lnc: long noncoding; MAX: read count maximum; MEAN: read count mean; MIN: read count minimum; miRNA: microRNA; SD: read count standard deviation; L2F: log2-fold change.
LNCipedia is available at http://www.lncipedia.org.13
Exploratory in silico functional analyses using online tools suggested a range of possible lncRNA functions. For example, the most differentially expressed lncRNA AP000783.2 (ENSG00000255414) is a 2,471-bp, 2-exon novel lncRNA transcribed from an intergenic region on ch11:123,325,106–123,331,118 forward strand. Although its function is unknown, AP000783.2 possesses 81 DIANA-predicted miRNA target sites with miRNA targeted gene (miTG) scores >0.7 (the higher the score, the higher the probability of miRNA binding), including target sites for miRNA-942 (score, 0.981), miRNA-580 (score 0.961), and miRNA-4760-3p (score, 0.946), which suggests that AP000783.2 may serve as an miRNA “decoy” for these or other miRNAs. The second most differentially expressed lncRNA, RP11-403B2.6, is a 748-bp, 2-exon transcript from ch15:20,964,640–20,980,123 reverse strand. Interestingly, it was not expressed in any of the five unused donor RVs or LVs and was expressed in only 4 of the 22 heart failure RVs with up to 60 and 292 read counts when expressed. It has no DIANA-predicted miRNA target sites, and its function is unknown. The third most differentially expressed lncRNA, RP11-60A24.3 (ENSG00000265542), is a novel lncRNA transcribed from ch17:55,849,307–55,912,110 reverse strand with 2 transcripts (a 2-exon, 472-bp transcript and a 3-exon, 505-bp transcript) due to alternative splicing. Interestingly, the lncRNA RP11-60A24.3 intronic region includes the sequence for a separate lncRNA gene, RN7SKP94 (ENSG00000222976, lncRNA 7SK small nuclear pseudogene 94). RP11-60A24.3 was decreased 2.5 log2-fold (P value 1.9E−07) in heart failure RVs, expressed in all unused donor RVs, and not expressed in one heart failure RV. Its function is unknown, but it possesses 13 DIANA-predicted miRNA binding sites with miTG scores >0.7, including target sites for miRNA-654-5p (score, 0.937), miRNA-1207-3p (score, 0.936), and miRNA-541-3p (0.864), which suggests that RP11-60A24.3 may play a role as miRNA “decoy.”
As an example of exploratory in silico functional analysis using online tools of a somewhat better characterized lncRNA, RP11-298021.5 (ENSG00000244277.1) is a 7-exon, 2,350-bp transcript with 7 alternatively spliced isoforms transcribed from chr3:167,613,736–167,645,799, an intergenic region conserved in mice but not fish between the SERPINI1 and GOLIM4 protein-coding genes. Although its function is unknown, based upon a predicted CPC score of 2.1, RP11-298021.5 possesses modest protein-coding potential. There are no known RP11-298021.5 associations with proteins based upon the absence of hits in the PRIDE database. MirTarget2 prediction identified 8 miR targets with scores >80, denoting the possibility of interaction with these miRs. These miR targets do not overlap with the MirTarget2-predicted miRs interacting with SERPINI1 or GOLIM4, suggesting that, if lnc-SERPINI1–4 acts as a miR decoy, it may not serve as a decoy for nearby SERPINI1 or GOLIM4 transcripts.
Antisense transcripts
There were 30 natural antisense transcript differentially expressed genes in heart failure RVs versus unused donor RVs (Table 5). Of these 30, 12 were increased in expression and 18 were decreased in expression in heart failure RVs. Interestingly, RP11-524H19.s (parent gene unknown) and RP11-597M17.1 (parent gene SCARA5) natural antisense transcripts were not expressed in any heart failure RV, and RP3-467K16.7 (parent gene FHAD1) natural antisense transcript was not expressed in any unused donor RV.
Table 5.
Differentially expressed natural antisense transcripts
| Ensembl ID | Gene ID | L2F change | L2F adj P value | Chrom | Trans | Length, base pairs | Parent sense gene | LNCipedia notesa | EZH2 binding |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000249307.1 | RP11-438E5.1 | −2.24 | 0.000004 | 4 | 11 | 525–973 | BMP2K | lnc-BMP2K-1; possible coding; no miR targets identified | 0.75/0.33 |
| ENSG00000242349.1 | NPPA-AS1 | 2.30 | 0.0003 | 1 | 2 | 936, 1,032 | NPPA | lnc-CLCN6-1; low coding potential; no miR targets identified; NPPA exonic transcript | 0.65/0.91 |
| ENSG00000232110.3 | RP11-149123.3 | 2.47 | 0.0005 | 10 | 3 | 495, 603, 232 | LIPA | lnc-IFIT-2; low coding potential; no miR targets identified | 0.75/0.81 |
| ENSG00000232480.1 | RP11-224019.2 | 2.65 | 0.0014 | 1 | 1 | 557 | Unknown (close to TGFb2) | lnc-GPATCH2-2; probable noncoding; no miR targets identified | 0.80/0.94 |
| ENSG00000268157.1 | AC010524 | −2.34 | 0.0019 | 19 | 1 | 494 | TEAD2 | No entry | 0.75/0.40 |
| ENSG00000269256.1 | RP11-325D15.2 | −4.23 | 0.0035 | 10 | 1 | 592 | C10orf11 | No entry | 0.85/0.94 |
| ENSG00000233251.3 | AC007743 | −1.27 | 0.0047 | 2 | 3 | 1,623, 1,657, 3,075 | CCDC85A | lnc-EFEMP1-3; probable coding; miR targets (29 total) 5581-3p, 432-5p, 2467-5p, 3187-3p, 1236, 4683, 3916, 561-5p, 3190-5p, 1184 | 0.80/0.92 |
| ENSG00000203593.3 | RP5-1096D14.6 | 2.18 | 0.0050 | 12 | 1 | 629 | CACNA1C | lnc-LRTM2-1; possible coding; no miR targets identified | 0.45/0.91 |
| ENSG00000256377.1 | RP11-1060J15.4 | −2.00 | 0.0055 | 12 | 3 | 672, 691, 1,110 | REP15 | lnc-MANSC4-3; possible coding; no miR targets identified | 0.85/0.98 |
| ENSG00000259065.1 | RP5-1021l20.1 | 1.38 | 0.0062 | 14 | 2 | 538, 573 | C14orf43 | lnc-PTGR2-1; possible coding; no miR targets identified | 0.55/0.52 |
| ENSG00000261616.1 | RP11-602.3 | −1.29 | 0.0072 | 15 | 1 | 2,677 | TTC23 | lnc-SYNM-2; possible coding; no miR targets identified | 0.75/0.97 |
| ENSG00000177133.5 | LINC00982 | −1.11 | 0.0077 | 1 | 7 | 280–2,917 | PRDM16 | lnc-TTC34-3; probable coding; no miR targets identified | 0.70/0.61 |
| ENSG00000237686.1 | RP5-1120P11.1 | 1.72 | 0.0082 | 6 | 2 | 556, 2,232 | C6orf223 | lnc-MRPL14-2; noncoding; miR targets (29 total) 519-3p, 145-5p, 1322, 4459, 4298, 547-5p, 1302, 526-5p, 654-3p, 3659 | 0.75/0.93 |
| ENSG00000259291.1 | RP11-617F23.1 | 1.11 | 0.0097 | 15 | 1 | 1,168 | ZNF710 | lnc-IDH2-1; noncoding; no miR targets | 0.80/0.77 |
| ENSG00000260000.2 | RP3-467N11.11 | 1.47 | 0.0112 | 6 | 1 | 1,517 | ASCC3 | No entry | 0.80/0.95 |
| ENSG00000264235.1 | RP13-270P17.1 | 2.23 | 0.0186 | 18 | 2 | 723, 1,035 | MYL12A | lnc-MYOM1-1; noncoding; no miR targets | 0.80/0.98 |
| ENSG00000224251.1 | RP11-49907.7 | −2.17 | 0.0244 | 10 | 2 | 302, 425 | AKR1C2 | lnc-AKR1C1-2; noncoding; no miR targets | 0.45/0.98 |
| ENSG00000231329.2 | RP1-225E12.2 | −2.00 | 0.0275 | 6 | 14 | 367–829 | HECA | lnc-TXLND-3; noncoding; no miR targets | 0.65/0.85 |
| ENSG00000253858.1 | CTB-147C13.1 | 2.89 | 0.0332 | 5 | 1 | 567 | KCNlP1 | lnc-KCNMB1-1; noncoding; no miR targets | 0.60/0.83 |
| ENSG00000253397.1 | RP11-597M17.1 | N/C | 0.0337 | 8 | 1 | 359 | SCARA5 | lnc-ESCO2-2; noncoding; no miR targets; expressed in 3/5 DON and 0/22 HF hearts | 0.65/0.83 |
| ENSG00000244300.2 | RP11-475N22.4 | −1.26 | 0.0348 | 3 | 3 | 722, 1,163, 1,578 | GATA2 | lnc-EEFSEC-3; coding (long transcripts); 4722-3p, 4652-3p, 4341-5p, 5589-5p, 4668-5p, 4762-3p, 3688-3p, 3148, 4699-5p, 1182 | 0.70/0.66 |
| ENSG00000246451 | RP11-894P9.1 | −1.90 | 0.0379 | 14 | 1 | 1,404 | KLC1 | lnc-XRCC3-1; coding; miR targets (14 total) 570-3p, 542-3p, 363-5p, 24-3p, 155-5p, 2110, 1587, 374b-3p, 501-5p, 4776-3p | 0.80/0.95 |
| ENSG00000260618.1 | RP11-23N2.4 | −1.78 | 0.0404 | 15 | 2 | 612, 635 | KIAA1370 | lnc-MAPK6-9; noncoding; no miR targets | 0.60/0.96 |
| ENSG00000263220.1 | RP11-420A6.2 | −3.44 | 0.0419 | 17 | 1 | 512 | RABEP1 | lnc-NUP88-1; noncoding; no miR targets | 0.75/0.96 |
| ENSG00000234779.1 | RP11-62F24.2 | −1.82 | 0.0427 | 9 | 1 | 428 | BNC2 | lnc-CNTLN-2; coding; no miR targets | 0.55/0.68 |
| ENSG00000236498.1 | AC107081 | 2.39 | 0.0442 | 2 | 1 | 802 | CCT4 | lnc-COMMD1-1; noncoding; no miR targets | 0.70/0.99 |
| ENSG00000261295.1 | RP11-524D16_A.3 | −1.47 | 0.0442 | X | 1 | 652 | SYTL4 | lnc-SRPX2-1; noncoding; no miR targets | 0.75/0.95 |
| ENSG00000244998.1 | CTD-3064M3.4 | −1.71 | 0.0445 | 8 | 1 | 1,380 | PTP4A3 | lnc-GPR20-2; coding; miR targets (18 total) 5571-3p, 4794, 326, 384, 645, 3674, 4665-5p, 5196-5p, 1275, 3911 | 0.75/0.76 |
| ENSG00000224984.1 | RP11-524H19.2 | Inf | 0.0448 | 6 | 1 | 586 | Unknown | lnc-HMGCLL1-1; noncoding; no miR targets; expressed in 3/5 DON and 0/22 HF hearts | 0.70/0.96 |
| ENSG00000236045.1 | RP3-467K16.7 | Inf | 0.0464 | 1 | 1 | 727 | FHAD1 | lnc-CASP9-2; noncoding; no miR targets; expressed in 0/5 DON and 19/22 HF hearts | 0.75/0.91 |
adj: adjusted; Chrom: chromosome; EHZ2: predicted binding to EZH2 component of polycomb 2 repressive complex; Ensembl: Ensembl release 74; L2F change: log2-fold change; lncRNA: long noncoding RNA; Trans: transcripts.
LNCipedia is available at http://www.lncipedia.org.13
The relationship between parent gene and natural antisense transcription pair expression in heart failure RV versus unused donor RVs was complex (Table 6). Parent and natural antisense transcript were both decreased in 8 pairs, parent and natural antisense transcript were both increased in 6 pairs, parent increased and natural antisense transcript decreased in 7 pairs, parent decreased and natural antisense transcript increased in 3 pairs, parent unchanged with natural antisense transcript increased in 2 pairs, parent unchanged with natural antisense transcript decreased in 1 pair, and status unknown in 3 pairs (two parent genes unknown, KIAA1370 parent of RP11-23N2.4 not expressed) in heart failure RVs. Unused donor versus heart failure RV parent/natural antisense transcript ratios were significantly increased for 7 parent∶natural antisense transcript pairs and significantly decreased in 4 parent∶natural antisense transcript pairs (Table 6).
Table 6.
Differentially expressed natural antisense transcripts and parent sense genes
| Ensembl ID | Gene ID | DON RV MEAN | DON RV SD | DON RV CORR | HF RV MEAN | HF RV SD | HF RV CORR | Sense∶antisense ratio P value |
|---|---|---|---|---|---|---|---|---|
| ENSG00000156269 | BMP2K | 65.4 | 18.6 | 0.69 | 45.3 | 16.9 | −0.3 | 0.03 |
| ENSG00000249307.1 | RP11-438E5.1 | 69.6 | 24.9 | 11.3 | 6.4 | |||
| Ratio sense/antisense | 0.02 | 0.03 | 0.02 | 0.1 | ||||
| ENSG00000175206 | NPPA | 1,583.2 | 1,857.6 | 0.7 | 42,372.7 | 59,245.0 | 1.0 | 0.003 |
| ENSG00000242349.1 | NPPA-AS1 | 11.6 | 3.4 | 70.4 | 90.7 | |||
| Ratio sense/antisense | 118.3 | 124.8 | 506.9 | 256.7 | ||||
| ENSG00000107798 | LIPA | 430.2 | 62.2 | −0.5 | 280.3 | 86.3 | 0.1 | 0.00001 |
| ENSG00000232110.3 | PR11-149123.3 | 7.0 | 4.4 | 35.1 | 16.7 | |||
| Ratio sense/antisense | 61.5 | 58.3 | 11.8 | 14.3 | ||||
| Unknown | Unknown | N/C | N/C | N/C | N/C | N/C | N/C | N/C |
| ENSG00000232480.1 | RP11-224019.2 | 3.0 | 2.0 | 18.9 | 15.6 | |||
| Ratio sense/antisense | N/C | N/C | N/C | N/C | ||||
| ENSG00000074219 | TEAD2 | 1,312.4 | 201.9 | −0.5 | 749.0 | 154.1 | −0.4 | 0.1 |
| ENSG00000268157.1 | AC010524 | 18.6 | 6.7 | 4.1 | 2.7 | |||
| Ratio sense/antisense | 70.6 | 36.2 | 296.0 | 300.9 | ||||
| ENSG00000148655 | C10orf11 | 307.4 | 143.4 | −0.7 | 109.8 | 34.4 | 0.2 | 0.2 |
| ENSG00000269256.1 | RP11-325D15.2 | 4.4 | 2.7 | 0.4 | 0.7 | |||
| Ratio sense/antisense | 67.3 | 22.2 | 104.7 | 50.8 | ||||
| ENSG00000055813 | CCDC85A | 179.4 | 50.6 | −0.2 | 93.4 | 31.8 | 0.6 | 0.5 |
| ENSG00000233251.3 | AC007743 | 271.0 | 123.1 | 112.1 | 51.8 | |||
| Ratio sense/antisense | 0.8 | 0.5 | 1.0 | 0.4 | ||||
| ENSG00000151067 | CACNA1C | 2,699.0 | 381.4 | 0.03 | 2,640.7 | 549.2 | 0.04 | 0.00001 |
| ENSG00000203593.3 | RP5-1096D14.6 | 6.8 | 3.8 | 21.3 | 10.8 | |||
| Ratio sense/antisense | 493.6 | 247.9 | 157.2 | 78.7 | ||||
| ENSG00000174236 | REP15 | 27.0 | 9.2 | 0.3 | 17.1 | 7.3 | −0.1 | 0.1 |
| ENSG00000256377.1 | RP11-1060J15.4 | 26.6 | 13.4 | 8.3 | 8.5 | |||
| Ratio sense/antisense | 1.3 | 1.0 | 4.0 | 3.8 | ||||
| ENSG00000156030 | C14orf43 | 351.6 | 92.5 | −0.9 | 262.0 | 102.3 | 0.1 | 0.00005 |
| ENSG00000232480.1 | RP5-1021l20.1 | 3.0 | 2.0 | 18.9 | 15.6 | |||
| Ratio sense/antisense | 191.7 | 167.2 | 23.9 | 19.5 | ||||
| ENSG00000103852 | TTC23 | 331.2 | 116.1 | 1.0 | 257.8 | 51.9 | 0.7 | 0.006 |
| ENSG00000261616.1 | RP11-602.3 | 205.2 | 126.5 | 87.0 | 35.9 | |||
| Ratio sense/antisense | 1.8 | 0.4 | 3.3 | 1.1 | ||||
| ENSG00000142611 | PRDM16 | 645.8 | 243.1 | 1.0 | 288.4 | 108.2 | 0.8 | 0.9 |
| ENSG00000177133.5 | LINC00982 | 697.6 | 400.6 | 292.5 | 108.9 | |||
| Ratio sense/antisense | 1.0 | 0.2 | 1.0 | 0.3 | ||||
| ENSG00000181577 | C6orf223 | 1.8 | 1.5 | 0.1 | 4.0 | 2.4 | 0.1 | 0.4 |
| ENSG00000237686.1 | RP5-1120P11.1 | 34.4 | 17.2 | 99.6 | 36.9 | |||
| Ratio sense/antisense | 0.1 | 0.1 | 0.0 | 0.0 | ||||
| ENSG00000140548 | ZNF710 | 119.0 | 88.0 | −1.0 | 84.8 | 50.0 | −0.1 | 0.006 |
| ENSG00000259291.1 | RP11-617F23.1 | 406.8 | 120.5 | 742.5 | 201.5 | |||
| Ratio sense/antisense | 0.4 | 0.4 | 0.1 | 0.1 | ||||
| ENSG00000112249 | ASCC3 | 329.6 | 122.7 | 0.1 | 316.9 | 106.3 | 0.6 | 0.000001 |
| ENSG00000260000.2 | RP3-467N11.11 | 28.4 | 3.0 | 79.8 | 45.1 | |||
| Ratio sense/antisense | 11.7 | 4.1 | 4.6 | 1.7 | ||||
| ENSG00000101608 | MYL12A | 34,160.4 | 17,356.8 | 0.6 | 73,047.6 | 39,647.2 | 0.2 | 0.1 |
| ENSG00000264235.1 | RP13-270P17.1 | 76.0 | 59.4 | 310.6 | 174.7 | |||
| Ratio sense/antisense | 541.4 | 343.2 | 315.7 | 258.6 | ||||
| ENSG00000151632 | AKR1C2 | 516.6 | 201.0 | 0.9 | 157.7 | 61.1 | 0.6 | 0.3 |
| ENSG00000224251.1 | RP11-49907.7 | 10.4 | 6.7 | 2.6 | 2.1 | |||
| Ratio sense/antisense | 58.0 | 17.3 | 81.3 | 47.6 | ||||
| ENSG00000112406 | HECA | 1,374.2 | 430.4 | 0.1 | 1,155.0 | 186.5 | 0.3 | 0.2 |
| ENSG00000231329.2 | RP1-225E12.2 | 24.8 | 24.3 | 6.6 | 4.4 | |||
| Ratio sense/antisense | 103.1 | 94.0 | 352.5 | 385.2 | ||||
| ENSG00000182132 | KCNIP1 | 3.2 | 4.1 | 0.3 | 13.4 | 7.1 | 0.7 | 0.4 |
| ENSG00000253858.1 | CTB-147C13.1 | 1.0 | 1.0 | 6.1 | 5.3 | |||
| Ratio sense/antisense | 2.0 | 2.6 | 3.0 | 2.1 | ||||
| ENSG00000168079 | SCARA5 | 982.4 | 370.4 | 0.7 | 226.5 | 158.6 | N/C | N/C |
| ENSG00000253397.1 | RP11-597M17.1 | 2.2 | 2.9 | 0 | 0 | |||
| Ratio sense/antisense | 596.9 | 535.1 | N/C | N/C | ||||
| ENSG00000179348 | GATA2 | 395.2 | 78.9 | 0.4 | 182.4 | 53.0 | 0.2 | 0.3 |
| ENSG00000244300.2 | RP11-475N22.4 | 58.8 | 23.9 | 20.9 | 7.6 | |||
| Ratio sense/antisense | 7.5 | 2.7 | 9.9 | 4.3 | ||||
| ENSG00000126214 | KLC1 | 1,262.4 | 170.1 | −0.9 | 1,269.0 | 205.6 | −0.1 | 0.1 |
| ENSG00000246451 | RP11-894P9.1 | 104.6 | 94.8 | 30.6 | 27.7 | |||
| Ratio sense/antisense | 26.4 | 21.2 | 86.8 | 78.3 | ||||
| KIAA1370 (not found) | KIAA1370 (not found) | N/C | N/C | N/C | N/C | N/C | N/C | N/C |
| ENSG00000260618.1 | RP11-23N2.4 | 45.4 | 28.9 | 13.5 | 14.0 | |||
| Ratio sense/antisense | N/C | N/C | N/C | N/C | ||||
| ENSG00000029725 | RABEP1 | 1,645.0 | 445.5 | −0.1 | 1,472.0 | 342.3 | 0.3 | 0.03 |
| ENSG00000263220.1 | RP11-420A6.2 | 3.4 | 1.9 | 0.9 | 1.1 | |||
| Ratio sense/antisense | 405.0 | 150.0 | 1,115.2 | 575.1 | ||||
| ENSG00000173068 | BNC2 | 35.6 | 13.2 | 0.3 | 19.7 | 10.2 | 0.2 | 0.3 |
| ENSG00000234779.1 | RP11-62F24.2 | 19.4 | 15.5 | 5.4 | 2.9 | |||
| Ratio sense/antisense | 2.4 | 1.3 | 5.3 | 5.5 | ||||
| ENSG00000115484 | CCT4 | 1,816.6 | 508.1 | 0.2 | 1,774.5 | 372.2 | 0.0 | 0.001 |
| ENSG00000236498.1 | AC107081 | 2.0 | 1.2 | 7.0 | 3.4 | |||
| Ratio sense/antisense | 1,144.4 | 708.0 | 371.7 | 305.4 | ||||
| ENSG00000102362 | SYTL4 | 331.2 | 71.7 | 0.4 | 151.5 | 57.4 | 0.6 | 0.4 |
| ENSG00000261295.1 | RP11-524D16_A.3 | 25.0 | 16.6 | 8.0 | 6.3 | |||
| Ratio sense/antisense | 18.9 | 12.5 | 23.2 | 9.5 | ||||
| ENSG00000184489 | PTP4A3 | 6,209.2 | 1,847.1 | −0.2 | 9,032.6 | 2,230.3 | 0.2 | 0.01 |
| ENSG00000244998.1 | CTD-3064M3.4 | 17.4 | 11.3 | 4.3 | 2.5 | |||
| Ratio sense/antisense | 464.0 | 214.3 | 2,903.2 | 1,950.1 | ||||
| Unknown | Unknown | N/C | N/C | N/C | N/C | N/C | N/C | N/C |
| ENSG00000224984.1 | RP11-524H19.2 | 2.4 | 4.3 | 0 | 0 | |||
| Ratio sense/antisense | N/C | N/C | N/C | N/C | ||||
| ENSG00000142621 | FHAD1 | 2.2 | 1.3 | N/C | 9.9 | 7.5 | 0.7 | N/C |
| ENSG00000236045.1 | RP3-467K16.7 | 0 | 0 | 2.9 | 2.5 | |||
| Ratio sense/antisense | N/C | N/C | 3.7 | 2.1 |
CORR: read count correlation; DON RV: unused donor right ventricle; Ensembl: Ensembl release 73; HF RV: heart failure right ventricle; MEAN: read count mean; N/C: not calculated due to absence of sufficient read counts; P value: sense∶antisense ratio P value by t test; SD: read count standard deviation.
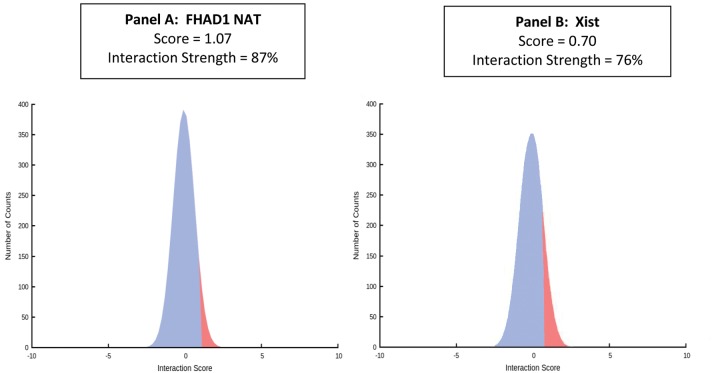
Exploratory in silico analyses using online tools suggested multiple possible functional roles of natural antisense transcripts, as previously reported.19 Of the 27 transcripts with entries in LNCipedia, 13 possessed protein-coding potential (no identified or known protein transcripts), and 5 possessed high-likelihood miRNA target sequences. For example, the GATA2 parent gene and its natural antisense transcript were both targets of miR 4722 (MiRTarget scores 0.77 and 0.82, respectively), and CCDC85A parent gene and its natural antisense transcript were both targets of miR 1184 (MiRTarget scores 0.76 and 0.64, respectively), suggesting that some natural antisense transcripts may serve as miR decoys. By RPISeq, 7 natural antisense transcripts were predicted with probability ≥0.80 by both RF and SVM classifiers to be binding partners for the polycomb repressive complex 2 (PRC2) EZH2 subunit. By the catRAPID algorithm, the FHAD1 natural antisense transcript had a greater strength of RNA∶protein interaction with EZH2 than did Xist (Fig. 1), a natural antisense transcript previously reported to bind to and regulate PRC2 EZH2.19 Interestingly, an atrial natriuretic peptide (NPPA) natural antisense transcript was among the natural antisense transcript differentially expressed genes, and NPPA natural antisense transcripts have been previously reported to modulate the expression of NPPA splicing isoforms.20
Figure 1.
Interaction strength with EZH2. By the catRAPID binding prediction algorithm, the FHAD1 NAT (A) has a greater strength of RNA∶protein interaction with the PRC2 component EZH2 than does Xist (B), a long noncoding RNA previously reported to bind to and regulate the H3K27me3 trimethylation activity of EZH2.
Pseudogenes
There were 1,064 and 793 transcribed pseudogenes expressed in heart failure RVs and unused donor RVs, respectively (Table 2). There were 27 pseudogenes, with differentially expressed genes representing 2.5% of all pseudogenes in heart failure RVs versus unused donor RVs (Table 2). These included 14 processed, 10 unprocessed, 2 unitary, and 1 unknown type pseudogenes. Of the 27 total pseudogenes, 10 were transcribed in antisense, and 17 were transcribed in sense. The parental coding genes included 2 unknown genes, 2 zing finger protein pseudogenes (i.e., 2 pseudogenes had parental pseudogenes), and known parental protein-coding genes with a variety of disparate, functional proteins.
As previously reported, in general, the expression of the parental coding gene was more abundant than the expression of the corresponding pseudogenes (Table 7).21 There were exceptions to this pattern, however, because the expression of the FBX043 pseudogenes (ENSG00000236155, RP11-231P20.2), for example, was considerably more abundant than the expression of the parental FBX043 gene. The ratio of parental gene read count number to pseudogene read count number was highly variable between individual subjects and across different parental gene∶pseudogene pairings and significantly different for several parental gene∶pseudogene pairings between unused donor and heart failure RVs (Table 8).
Table 7.
Differentially expressed pseudogenes
| Ensembl gene | Gene name | L2F change | L2F adj P value | Chrom | Trans | Pseudogene type | Length, base pairs | Parental gene | PseudoMap query results: esiRNA products, miRNA targets |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000230916 | RP11-10B2.1 | 5.53 | 9.07E−11 | X | 1 | Processed, forward strand | 699 | PTGS1 | No entry |
| ENSG00000177359 | RP11-551L14.1 | −4.22 | 7.00E−10 | 12 | 6 | Unprocessed, reserve strand | 520–4,427 | STARD9 | esiRNAs 58366, 54458; no miR regulators |
| ENSG00000171658 | RP11-443P15.2 | −4.56 | 8.78E−10 | 3 | 7 | Unprocessed, forward strand | 330–1,789 | NMRAL1 | miRNA regulators 324-3p, 1913 |
| ENSG00000224163 | RP11-309L24.6 | 3.03 | 1.00E−05 | 7 | 2 | Unprocessed, reverse strand | 543–560 | FLNC | No entry |
| ENSG00000236155 | RP11-231P20.2 | 1.82 | 0.00013 | 1 | 4 | Unprocessed, forward strand | 334–2,108 | FBXO43 | esiRNAs 7335, 419, 7397, 7411, 7329, 7318, 54243, 49311, 56320 |
| ENSG00000227671 | RP11-488L18.4 | −1.78 | 0.00061 | 1 | 4 | Unprocessed, reverse strand | 430–1,465 | ZFP TPG (ENSG00000215198) | esiRNA18005 |
| ENSG00000225415 | RP3-509I19.1 | −3.46 | 0.00070 | 6 | 2 | Processed, forward strand | 825–1,053 | CCRL1 | esiRNAs 15275, 15276, 15277 |
| ENSG00000256356 | RP11-320N7.3 | 3.14 | 0.00153 | 1 | 1 | Processed, forward strand | 2,069 | HSPA8 | No entry |
| ENSG00000249780 | RP11-352E6.2 | −3.34 | 0.00379 | 4 | 1 | Processed, reverse strand | 340 | MT-CO3 | No entry |
| ENSG00000235695 | RP11-395P16.1 | 4.06 | 0.01058 | 3 | 1 | Processed, forward strand | 309 | HIGD2A | No entry |
| ENSG00000183458 | RP11-958N24.1 | −1.92 | 0.01718 | 16 | 3 | Unprocessed, forward strand | 514, 2,519, 6,830 | PKD1 | esiRNAs 30192, 30161, 56802, 30137, 30122 |
| ENSG00000227827 | RP11-958N24.2 | −2.66 | 0.01786 | 16 | 1 | Unprocessed, forward strand | 9,932 | PKD1 | No entry |
| ENSG00000214558 | RP11-74E24.2 | −2.80 | 0.02258 | 6 | 2 | Processed, forward strand | 2,436, 3,237 | ZC3H11B (ZF CCCH-type TPG) | >30 esiRNAs including esiRNA 11710, 10424, 10422, 11749, 1731, 10414, 10409, 10412, 10425, 11725 |
| ENSG00000254398 | RP11-578F21.2 | −2.20 | 0.02530 | 15 | 1 | Unprocessed, reverse strand | 546 | HERC2 | No entry |
| ENSG00000227141 | RP11-545A16.3 | −4.16 | 0.03084 | 1 | 1 | Processed, forward strand | 2,471 | CENTG2 | No entry |
| ENSG00000232546 | RP11-458F8.1 | −3.33 | 0.03754 | 7 | 1 | Unprocessed, forward strand | 298 | GTF2I | No entry |
| ENSG00000205898 | RP11-3B12.4 | −2.58 | 0.04785 | 7 | 1 | Processed, reverse strand | 574 | C11orf58 | No entry |
| ENSG00000232403 | RP11-511I11.1 | −1.97 | 0.04855 | 2 | 1 | Processed, reverse strand | 312 | RPL27A | No entry |
| ENSG00000225400 | RP11-1114A5.5 | 1.36 | 0.05243 | X | 1 | Processed, forward strand | 651 | RAB28 | 14 esiRNAs including eisRNAs 12075, 12079, 12078, 11235, 12076, 11239, 11248, 12077, 12073, 12074 |
| ENSG00000215452 | RP5-981L23.1 | −2.35 | 0.05378 | 20 | 2 | Unitary, processed, reverse strand | 1,499, 2,722 | ZNF248 | eisRNAs 39275, 48721, 45063 |
| ENSG00000240831.1 | AC112777.1 | 1.02 | 0.03339 | 12 | 1 | Processed, forward strand | 165 | Novel rRNA | No entry |
| ENSG00000249214 | AC022558.1 | 4.04 | 0.0003 | 15 | 1 | Forward strand | 132 | Unknown/novel | No entry |
| ENSG00000226121 | AHCTF1P1 | −2.16 | 0.0010 | 2 | 2 | Processed, reverse strand | 1,809, 6,821 | AHCTF1 | No entry |
| ENSG00000225898 | AC002075.3 | −3.90 | 0.0069 | 7 | 1 | Processed, forward strand | 676 | TSN | No entry |
| ENSG00000236732 | AC094019.4 | −3.66 | 0.0120 | 3 | 1 | Unprocessed, reverse strand | 472 | RPL21 | esiRNAs 19973, 29053 |
| ENSG00000226469 | AC003029.4 | −1.35 | 0.0452 | 12 | 1 | Unitary, forward strand | 2,000 | ADAM21 | No entry |
| ENSG00000233870 | AC007881.4 | −3.01 | 0.0550 | 2 | 1 | Unprocessed, forward strand | 187 | NAGK | No entry |
Chrom: chromosome; Ensembl: Ensemble release 73; esiRNA: endogenous silencing RNA; L2F adj: log2-fold change adjusted; miRNA: microRNA; Trans: transcripts.
Table 8.
Differentially expressed pseudogene and parent gene read counts
| Ensemble ID | Gene ID | DON RV MIN | DON RV MAX | DON RV MEAN | DON RV SD | DON RV CORR | HF RV MIN | HF RV MAX | HF RV MEAN | HF RV SD | HF RV CORR | Parent∶TPG ratio P value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000095303 | PTGS1 | 49 | 142 | 100.0 | 33.2 | −0.36 | 18 | 156 | 71.1 | 33.0 | 0.66 | 0.46 |
| ENSG00000230916 | RP11-10B2.1 | 11 | 48 | 23.6 | 15.8 | 6 | 9,631 | 1,028.0 | 2,381.2 | |||
| Ratio | 2 | 12 | 6.3 | 4.5 | 0 | 16 | 4.6 | 4.5 | ||||
| ENSG00000159433 | STARD9 | 464 | 915 | 666.4 | 177.4 | 0.61 | 187 | 606 | 360.5 | 113.3 | 0.12 | 0.02 |
| ENSG00000177359 | RP11-551L14.1 | 17 | 411 | 197.6 | 193.0 | 0 | 21 | 6.8 | 6.1 | |||
| Ratio | 1 | 34 | 12.4 | 13.8 | 12 | 245 | 99.2 | 75.5 | ||||
| ENSG00000153406 | NMRAL1 | 243 | 457 | 350.6 | 79.0 | −0.61 | 163 | 703 | 374.3 | 132.4 | 0.59 | 0.01 |
| ENSG00000171658 | RP11-443P15.2 | 0 | 70 | 35.4 | 30.2 | 0 | 8 | 1.5 | 1.8 | |||
| Ratio | 5 | 35 | 13.2 | 14.8 | 37 | 521 | 239.0 | 148.9 | ||||
| ENSG00000128591 | FLNC | 7,405 | 68,685 | 29,113.8 | 23,179.7 | 0.82 | 12,874 | 85,574 | 34,519.5 | 17,945.1 | 0.00 | 0.15 |
| ENSG00000224163 | RP11-309L24.6 | 3 | 9 | 5.4 | 2.6 | 0 | 256 | 25.2 | 63.9 | |||
| Ratio | 2,468 | 8,265 | 5,194.6 | 2,604.5 | 237 | 38,914 | 12,647.5 | 10,832.6 | ||||
| ENSG00000156509 | FBX043 | 1 | 6 | 3.6 | 2.3 | 0.57 | 0 | 12 | 3.6 | 3.1 | 0.93 | 0.00 |
| ENSG00000236155 | RP11-231P20.2 | 29 | 47 | 37.4 | 6.7 | 70 | 188 | 112.5 | 29.4 | |||
| Ratio | 0 | 0 | 0.1 | 0.1 | 0 | 0 | 0.032 | 0.0 | ||||
| ENSG000000215198 | RP11-182N22.7 | 0 | 2 | 0.6 | 0.9 | 0.83 | 0 | 2 | 0.2 | 0.5 | 0.97 | 0.89 |
| ENSG00000227671 | RP11-488L18.4 | 116 | 318 | 216.4 | 196.1 | 15 | 154 | 62.9 | 34.6 | |||
| Ratio | 0 | 0 | 0.0 | 0.0 | 0 | 0 | 0.004 | 0.0 | ||||
| ENSG00000129048 | CCRL1 | 37 | 676 | 278.4 | 260.0 | 0.77 | 5 | 90 | 44.0 | 23.3 | −0.27 | 0.24 |
| ENSG00000225415 | RP3-509I19.1 | 16 | 397 | 183.0 | 163.8 | 0 | 75 | 17.4 | 18.7 | |||
| Ratio | 1 | 4 | 1.9 | 1.1 | 1 | 27 | 5.6 | 6.7 | ||||
| ENSG00000170606 | HSPA4 | 1,125 | 2,748 | 2,159.8 | 661.6 | 0.81 | 1,512 | 4,215 | 2,394.8 | 710.1 | 0.21 | 0.04 |
| ENSG00000256356 | RP11-320N7.3 | 0 | 3 | 1.6 | 1.1 | 0 | 30 | 9.3 | 8.6 | |||
| Ratio | 916 | 2,379 | 1,392.1 | 682.2 | 88 | 2,422 | 601.2 | 677.1 | ||||
| ENSG00000198938 | MT-CO3 | 412,866 | 1,123,986 | 758,553.2 | 316,252.7 | −0.59 | 524,558 | 1,216,500 | 769,377.6 | 171,110.3 | 0.48 | 0.59 |
| ENSG00000249780 | RP11-352E6.2 | 1 | 64 | 14.0 | 28.0 | 0 | 4 | 1.1 | 1.1 | |||
| Ratio | 6,451 | 1,064,846 | 527,405.4 | 385,001.9 | 228,661 | 1,216,500 | 616,128.4 | 289,694.0 | ||||
| ENSG00000146066 | HIGD2A | 1,670 | 3,298 | 2,358.8 | 660.6 | 0.17 | 1,706 | 4,695 | 2,605.8 | 806.0 | −0.51 | 0.09 |
| ENSG00000235695 | RP11-395P16.1 | 0 | 1 | 0.4 | 0.5 | 0 | 32 | 4.4 | 9.6 | |||
| Ratio | 1,670 | 3,298 | 2,484.0 | 1,151.2 | 110 | 2,778 | 732.4 | 1,042.9 | ||||
| ENSG00000008710 | PKD1 | 2,192 | 2,864 | 2,495.2 | 278.1 | 0.68 | 1,555 | 3,434 | 2,371.0 | 473.1 | 0.01 | 0.20 |
| ENSG00000183458 | RP11-958N24.1 | 18 | 87 | 45.6 | 33.1 | 1 | 80 | 16.1 | 17.2 | |||
| Ratio | 33 | 122 | 79.6 | 43.3 | 36 | 2,461 | 408.9 | 549.1 | ||||
| ENSG00000008710 | PKD1 | 2,192 | 2,864 | 2,495.2 | 278.1 | −0.51 | 1,555 | 3,434 | 2,371.0 | 473.1 | 0.09 | 0.13 |
| ENSG00000227827 | RP11-958N24.2 | 20 | 99 | 45.8 | 32.6 | 0 | 24 | 8.5 | 7.0 | |||
| Ratio | 22 | 127 | 77.8 | 44.1 | 78 | 2,399 | 454.9 | 525.6 | ||||
| ENSG00000215817 | ZC3H11B | 2 | 20 | 10.2 | 8.6 | 0.96 | 0 | 6 | 2.5 | 2.3 | 0.35 | 0.51 |
| ENSG00000214558 | RP11-74E24.2 | 6 | 28 | 14.2 | 10.1 | 0 | 10 | 3.0 | 3.2 | |||
| Ratio | 0 | 1 | 0.7 | 0.3 | 0 | 2.5 | 0.9 | 0.7 | ||||
| ENSG00000206149 | HERC2 | 1,061 | 1,675 | 1,371.8 | 232.5 | 0.80 | 546 | 1,161 | 857.1 | 140.9 | −0.41 | 0.13 |
| ENSG00000254398 | RP11-578F21.2 | 18 | 99 | 52.4 | 40.5 | 0 | 35 | 10.1 | 8.7 | |||
| Ratio | 16 | 64 | 39.7 | 22.9 | 33 | 738 | 180.8 | 199.9 | ||||
| ENSG00000157985 | AGAP1 | 133 | 198 | 164.6 | 24.8 | −0.05 | 53 | 132 | 87.9 | 21.3 | N/C | N/C |
| ENSG00000227141 | RP11-545A16.3 | 0 | 5 | 2.0 | 2.1 | 0 | 1 | 0.0 | 0.2 | |||
| Ratio | 31 | 67 | 54.5 | 20.4 | 31 | 65 | 65.0 | N/C | ||||
| ENSG00000077809 | GTF2I | 2,846 | 3,963 | 3,320.2 | 435.7 | 0.25 | 2,064 | 3,218 | 2,719.5 | 343.2 | 0.51 | 0.16 |
| ENSG00000232546 | RP11-458F8.1 | 1 | 15 | 6.0 | 5.3 | 0 | 3 | 0.9 | 1.1 | |||
| Ratio | 225 | 2,993 | 1,081.5 | 1,091.4 | 909 | 3,191 | ||||||
| ENSG00000110696 | C11orf58 | 1,317 | 3,311 | 2,215.4 | 851.8 | 0.45 | 810 | 2,602 | 1,661.6 | 426.5 | 0.38 | 0.02 |
| ENSG00000205898 | RP11-3B12.4 | 0 | 14 | 7.6 | 5.7 | 0 | 5 | 1.3 | 1.6 | |||
| Ratio | 110 | 478 | 296.6 | 158.8 | 308 | 2,593 | 1,236.6 | 719.5 | ||||
| ENSG00000131469 | RPL27 | 8,100 | 11,516 | 9,989.4 | 1,605.8 | 0.65 | 5,944 | 12,985 | 9,044.7 | 1,764.4 | 1.00 | 0.03 |
| ENSG00000232403 | RP11-511I11.1 | 0 | 4 | 1.8 | 1.5 | 0 | 1 | 0.2 | 0.4 | |||
| Ratio | 2,646 | 8,100 | 5,537.7 | 2,235.0 | 7,579 | 10,848 | 8,804.0 | 1,466.4 | ||||
| ENSG00000147274 | RBMX | 1,515 | 2,375 | 1,907.6 | 383.7 | 0.95 | 1,241 | 2,443 | 1,911.7 | 277.4 | −0.03 | 0.00 |
| ENSG00000225400 | RP11-1114A5.5 | 15 | 36 | 25.6 | 8.4 | 25 | 79 | 50.5 | 16.2 | |||
| Ratio | 66 | 101 | 77.9 | 14.7 | 17 | 73 | 41.4 | 12.7 | ||||
| ENSG00000198105 | ZNF248 | 379 | 615 | 510.6 | 489.7 | −0.09 | 227 | 624 | 425.0 | 93.7 | −0.01 | 0.13 |
| ENSG00000215452 | RP5-981L23.1 | 2 | 35 | 12.2 | 13.4 | 0 | 9 | 2.9 | 2.8 | |||
| Ratio | 14 | 205 | 102.3 | 88.3 | 56 | 484 | 208.1 | 142.6 | ||||
| Unknown | Novel rRNA | 0 | N/C | N/C | N/C | N/C | N/C | N/C | N/C | |||
| ENSG00000240831.1 | AC112777.1 | 2,366 | 5,431 | 3,743.8 | 3,406.4 | 2,502 | 25,096 | 7,494.8 | 6,133.6 | |||
| Ratio | 0 | N/C | N/C | N/C | N/C | |||||||
| Unknown | Unknown/novel | 0 | N/C | N/C | N/C | N/C | N/C | N/C | N/C | |||
| ENSG00000249214 | AC022558.1 | 0 | 1 | 0.8 | 0.4 | 0 | 95 | 5.0 | 20.1 | |||
| Ratio | 0 | N/C | N/C | N/C | N/C | |||||||
| ENSG00000153207 | AHCTF1 | 420 | 1,075 | 724.8 | 260.6 | 0.82 | 237 | 602 | 376.8 | 96.2 | −0.34 | 0.13 |
| ENSG00000226121 | AC009487.6 | 14 | 80 | 40.8 | 29.3 | 1 | 20 | 9.0 | 5.2 | |||
| Ratio | 11 | 47 | 23.8 | 13.8 | 21 | 267 | 61.0 | 52.5 | ||||
| ENSG00000211460 | TSN | 638 | 1,179 | 943.2 | 216.8 | −0.66 | 695 | 1,340 | 989.3 | 165.3 | 1.00 | 0.39 |
| ENSG00000225898 | AC002075.3 | 0 | 9 | 3.0 | 3.7 | 0 | 1 | 0.1 | 0.3 | |||
| Ratio | 92 | 1,100 | 581.1 | 529.2 | 0 | 965 | 961.0 | 5.7 | ||||
| ENSG00000122026 | RPL21 | 266 | 448 | 367.0 | 76.0 | −0.06 | 261 | 516 | 332.4 | 67.7 | 0.79 | 0.02 |
| ENSG00000236732 | AC094019.4 | 3 | 19 | 7.0 | 6.8 | 0 | 7 | 0.9 | 1.6 | |||
| Ratio | 20 | 149 | 85.6 | 57.3 | 38 | 516 | 266.8 | 138.4 | ||||
| ENSG00000139985 | ADAM21 | 2 | 11 | 6.0 | 3.6 | 0.13 | 1 | 11 | 5.0 | 2.9 | 0.66 | 0.18 |
| ENSG00000226469 | AC003029.4 | 18 | 106 | 58.8 | 37.4 | 6 | 39 | 19.9 | 8.5 | |||
| Ratio | 0 | 0 | 0.1 | 0.2 | 0 | 1 | 0.3 | 0.2 | ||||
| ENSG00000124357 | NAGK | 603 | 870 | 695.4 | 103.6 | −0.79 | 538 | 890 | 681.5 | 90.7 | 0.62 | 0.01 |
| ENSG00000233870 | AC007881.4 | 1 | 8 | 4.0 | 2.7 | 0 | 2 | 0.5 | 0.6 | |||
| Ratio | 75 | 870 | 316.4 | 324.4 | 351 | 890 | 690.7 | 147.1 |
CORR: read count correlation; DON RV: unused donor right ventricle; Ensembl: Ensembl release 73; MAX: read count maximum; MEAN: read count mean; MIN: read count minimum; SD: read count standard deviation; HF RV: heart failure right ventricle; N/C: not calculated due to absence of sufficient read counts; P value: parent∶pseudogene (TPG) ratio P value by t test.
Functional annotation provided by the online resource tool “pseudoMap” yielded a variety of both predicted pseudogene esiRNA products and pseudogene miRNA binding targets (Table 7). From the available results in pseudoMap, the predicted pseudogene esiRNA products did not typically target the parent gene mRNAs, and the pseudogene miRNA binding partners did not typically overlap with the parent gene mRNA or miRNA binding partners. There were exceptions, however. For example, both the RPL27A pseudogene and RPL27A parent gene mRNA 3′ end uracil-rich terminal repeat (3′-UTR; the most common mRNA binding site for micro-RNAs) were predicted targets for miRNA 612, suggesting that the RPL27A pseudogene could serve as a miRNA decoy for miRNA 612, and both the parent gene NMRAL1 and the NMRAL1 pseudogenes were targets for miR 342-3p and miR 1913. The esiRNAs produced from the FBXO43 pseudogenes were not predicted to target FBXO43 transcripts but may target FBXO43-associated transcripts, including MAPK1, MDM2, OSMR, and IRAK3 transcripts (for each pseudoMap target score >200, minimum fold energy for RNA sequence ≤50).
Patterns of parent gene and pseudogene expression also suggested possible regulatory interactions. For example, the parent gene NMRAL1 was not differentially expressed in heart failure versus unused donor RVs (374 ± 132 vs. 351 ± 79 read counts). However, the NMRAL1 pseudogene was significantly reduced in heart failure versus unused donor RVs (2 ± 2 vs. 13 ± 15 read counts; Table 8). pseudoMap predicted miR 342-3p and miR 1913 as regulators of both the parent gene and pseudogene. If the NMRAL1 pseudogene served as an miRNA decoy, then reduction in NMRAL1 pseudogene expression without parallel reduction in NMRAL1 expression might permit miR 324-3p and/or miR 1913 repression of NMRAL1 translation. NMRAL1 (HSCARG) is a redox sensor protein.22 Reduced levels of NMRAL1 by RNA interference increases nitric oxide production and reduces cell viability, whereas overexpression of NMRAL1 increases cell viability.
Discussion
This study is notable for the high quality of isolated RNA, robust RNA-Seq count numbers, and abundant expression of both protein-coding and non–protein-coding poly(A) mRNAs. Transcripts in the failing RVs (as annotated in GENCODE, version 16, a tabulation of all known genes based on known human genome sequence) included 17,247 total genes, including 13,019 protein-coding genes (65% of all known protein-coding genes), 2,085 lncRNAs (16% of all known lncRNAs), and 1,064 pseudogenes (8% of all known pseudogenes). These findings are in general agreement with earlier RNA-Seq studies in mice, albeit with slightly larger numbers of transcripts.23-25 The heart failure RVs expressed a larger number of RNA transcripts in all categories, including lncRNAs and pseudogenes, compared with the unused donor RVs. Overall, 3.7% of lncRNAs and 2.5% of pseudogenes were differentially expressed in the heart failure RV versus unused donor RVs. Exploratory in silico analyses using online tools suggested multiple roles for lncRNAs, particularly roles as (1) potential miRNA decoys and (2) high-affinity binding partners for chromatin modifiers, such the EZH2 component of PRC2.
lncRNA regulatory roles
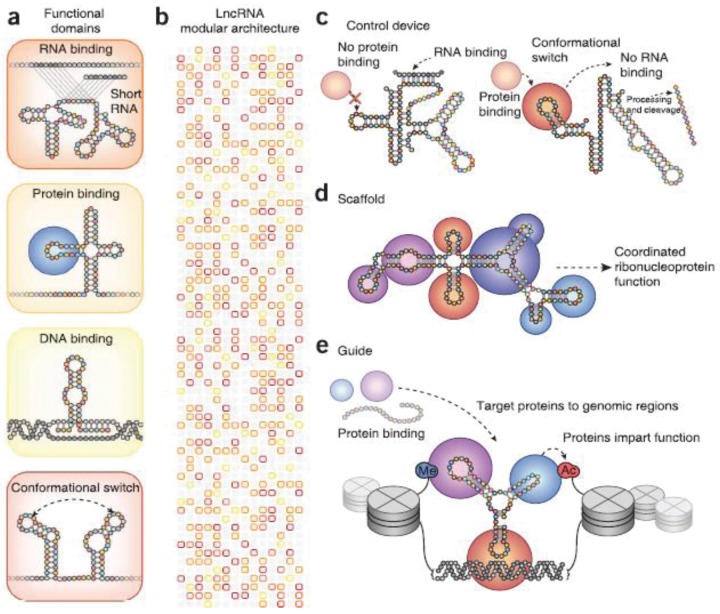
lncRNAs play a variety of defined critical regulatory roles, including X-inactivation and imprinting via interaction with DNA and/or DNA binding proteins,26 gene repression or enhancement via interaction with PRC1 and PRC2,27 and gene expression enhancement during development and differentiation in part via interaction with the mediator complex.28 Additional myriad lncRNA regulatory mechanisms reported to date include (1) transcriptional silencing or augmentation by interaction with transcriptional machinery or chromatin-altering binding partners, (2) interaction with actively transcribing mRNA, (3) direct interaction with DNA itself, (4) nuclear esiRNA generation via lncRNA “parent” fragmentation, (5) cytoplasmic miRNA decoy via the competing endogenous RNA hypothesis of miRNA regulation, and (6) interference with cytoplasmic mRNA stability by competition for mRNA stabilization factors.29 This functional diversity devolves from the wide array of protein, RNA and DNA binding domains, flexible scaffolding capabilities, modular secondary structures (Fig. 2), embedded esiRNAs, alternative transcripts, and alternatively spliced isoforms characteristic of lncRNAs.30
Figure 2.
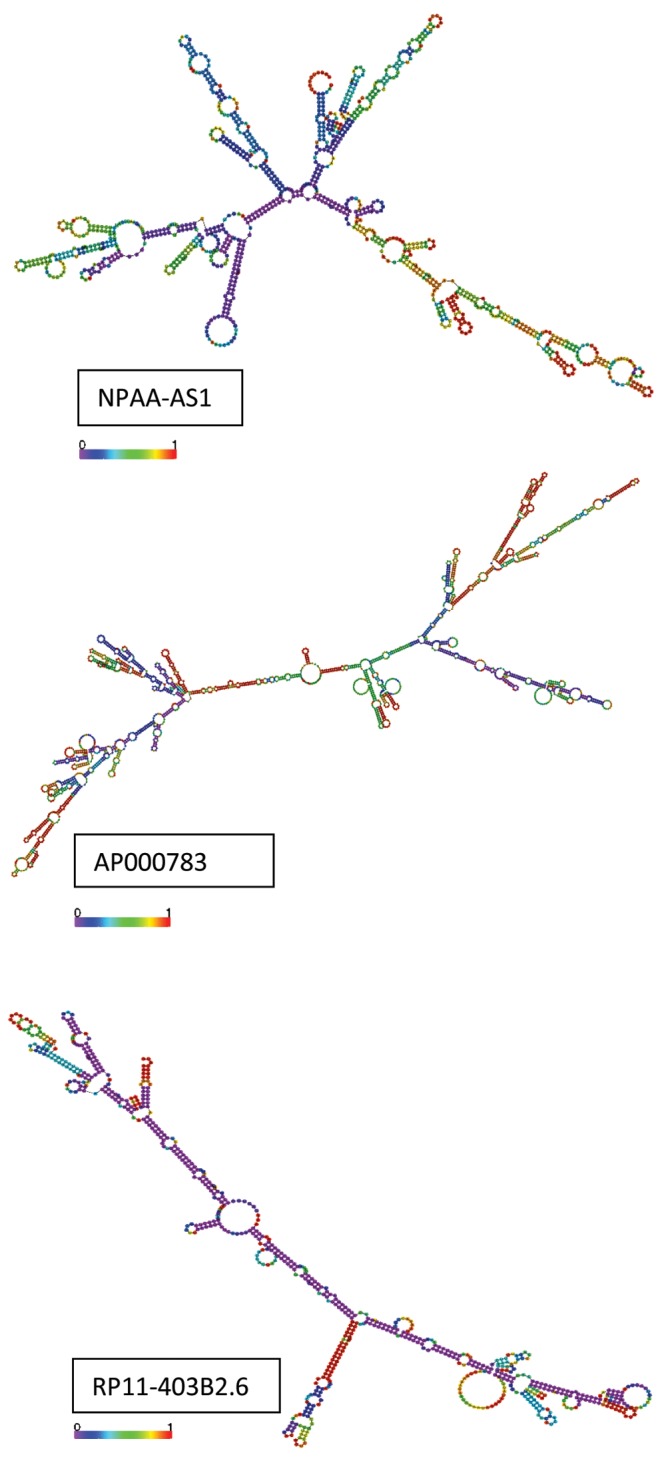
Three representative examples of long noncoding RNA (lncRNA) secondary structure. Examples of the diverse lncRNA RNAfold proposed secondary structures for three of the differentially expressed lncRNAs. Notable are the unique secondary conformations coupled with preserved modular features, including loops. The RNAfold proposed secondary structures were downloaded from http://www.lncipedia.org.13
lncRNAs in cardiac disease: evidence to date
Although the role of lncRNAs in epigenetic regulation in development, differentiation, and cancer is well established3 and will likely provide opportunities for novel targeted therapies,31 the role of lncRNAs in cardiac development and disease has not been widely studied.32 In a recent study, the lncRNA “Braveheart” was required for both progression of nascent mesoderm toward a cardiac fate and maintenance of cardiac fate in neonatal mouse cardiomyocytes in part via interaction with the SUZ12 component of PRC2.33 lncRNA NAT transcripts34 have been shown to regulate expression of troponin I,35 myosin heavy and light chains,36 and atrial natriuretic peptide.20 A higher antisense/sense mRNA ALC-1 ratio was associated with lower ALC-1 protein expression in hypertrophied human myocardium.37 The DMPK 3′-UTR CTG triplet repeat, when expanded 50–2,000 times, as in myotonic dystrophy, leads to “RNA toxicity” by sequestration of RNA-binding proteins, downregulation of connexins 40 and 43, induction of Nkx2–5 expression, and promulgation of heart block.38 The SRA1 gene yields both (1) lncRNAs termed “steroid receptor RNA activators” (SRAs), which are able to coactivate steroid nuclear receptors, and (2) mRNAs translated into steroid receptor RNA activator proteins (SRAPs).39 SRAPs may bind SRAs, and the SRAP/SRA transcripts serve as a “bi-faceted” genetic system with important roles in myogenic differentiation40 and human dilated cardiomyopathies.41 In separate genome-wide association studies, single-nucleotide polymorphism (SNP) variants in the novel lncRNA MIAT conferred increased risk of myocardial infarction,42 and SNPs in the lncRNA ANRIL locus carried the strongest genetic susceptibility for coronary artery disease.43 As more mechanistic examples, the lncRNA ANRIL, a natural antisense transcript of the protein-coding INK4b/ARF/INK4a complex, binds chromobox 7 of PRC1, thereby regulating the PRC1-mediated methylation of histone H3 lysine 27 and effecting the transcriptional silencing of INK4a in cis.44 The lncRNA cardiac hypertrophy related factor in mice acts as an endogenous “sponge” for mi489, thereby relieving the mi489-mediated repression of hypertrophy-inducing Myd88.45 esiRNA silencing of the lncRNA MALAT1, which under normal conditions is highly expressed in endothelial cells of different origin, resulted in endothelial-dependent reduced vascular growth both in vitro and in vivo.46
Speculative roles of lncRNAs in the heart
Little is presently known regarding the targets of the lncRNAs expressed in the heart.47 We could not find published literature identifying the targets of the differentially expressed lncRNAs identified in this study. Thus, we used available online tools for lncRNA in silico preliminary functional analyses. Future efforts, including our own, will be directed toward elucidating lncRNA targets using RNA silencing (esiRNA) approaches in animal and cellular models. Elucidation of the roles of specific lncRNAs in the heart will prove challenging given the large number of lncRNAs, the large number of potential lncRNA targets and interaction partners, and the known mechanistic diversity of lncRNAs themselves.47 Four broad functional roles as signals, decoys, guide, and scaffolds have been proposed for lncRNAs (Fig. 3).48 First, lncRNAs may serve as signals themselves once transcribed and/or modulate existing signals by interacting with chromatin-binding protein complexes or other RNA species.49 Second, lcnRNAs may serve as decoys for both RNAs and proteins. The “endogenous competing RNA hypothesis” proposes that multiple RNA species (mRNAs, lncRNAs, small nucleolar RNAs, and miRNAs) all “compete” for specific regulatory miRNA binding partners in a process of combinatorial regulation based on relative binding affinities and expression levels.50 Third, lncRNAs often possess high-affinity binding sites for proteins and may serve as decoys. By binding to DNA and forming heteroduplexes or heterotriplexes, lcnRNAs may also guide chromatin-binding proteins to regulatory cis and transgenomic regions.48 Fourth, lncRNAs, due to their modular structure, may serve as scaffolds for protein complex and RNA-binding partner and assist or direct the assembly of larger chromatin regulatory complexes.51 lncRNAs thus likely play key roles in the epigenetic regulation of gene expression during adaptive and maladaptive ventricular remodeling and the transition to failure.47,49
Figure 3.
Domain architecture and function of long noncoding RNAs (lncRNAs). A, lncRNAs structural domains can sense or bind other RNAs via complementary base pair interactions, proteins, and DNA that can induce allosteric conformations changes to other structures in the lncRNA. B, Alternative splicing can combine these structural domains into lncRNA modular architecture. C, Coupling sensing and actuator domains permit lncRNAs to function as “control” devices. In the example on the left, binding of RNA (gray) induced a conformational change that prevents protein binding. On the right, the protein can bind in the absence of RNA, inducing the formation of a stem-loop secondary structure that can be processed and cleaved to generate RNA output. D, lncRNAs such as HOTAIR act as molecular scaffolds, binding multiple proteins to form complex ribonucleoprotein structures. E, lncRNAs such as Cist can target the catalytic function of proteins to specific genome sites. lncRNAs can recruit chromatin-modifying proteins (purple, blue) to target sites by association with a DNA-binding protein such as YY1 (red). The chromatin modifiers then modify local histones to influence the expression of adjacent genes.
lncRNAs in the RV
Animal models have demonstrated significant differences in RV mRNA expression transcriptional profiles in response to experimental pressure52-56 and volume overload.57,58 Most of these studies, however, examined only the expression of mRNA by earlier-generation microarrays, which did not include probes for lncRNAs and thus provided an incomplete readout of the comprehensive RV transcriptome.5,6 lncRNA differential expression by RNA-Seq in RV pressure versus volume overload experimental models has not been reported to date. Based upon the dynamic nature of RV mRNA expression in pressure versus volume overload models, however, it is likely that lncRNA expression differences exist in such models and play key regulatory roles in the epigenetic regulation of the differential mRNA transcription profiles reported. Differential expression of lncRNAs and associated regulatory pathways is likely a conserved feature of RV remodeling independent of the type of RV remodeling stimulus. We would anticipate that at least some of the differentially expressed lncRNAs and associated pathways identified in this study would play roles in RV adaptive and maladaptive remodeling and the transition to RV failure in pulmonary arterial hypertension.
lncRNA in the RV versus LV
In the mouse, the normal RV and normal LV are epigenetically distinguishable based on differential expression patterns of epigenetic regulators of histone acetylation and methylation.59 Differences in RV versus LV mRNA transcription have been reported in the normal mammalian heart,60 in some61,62 but not all63 pressure overload experimental models, and in human congenital heart disease.64 Although the RV versus LV may invoke similar regulatory pathways in response to pressure overload, the RV mRNA transcriptional response appears generally more vigorous than the LV response.65 Based upon the aggregate microarray data to date, however, it is not yet clear whether targeting pathways specific to the stressed or failing RV, targeting pathways common to both the RV and LV, or targeting the mechanical interactions between the RV and LV will afford novel opportunities to attenuate RV adverse remodeling and the transition to RV failure.65,66
Pathobiology of RV failure in LV failure and future potential applications of lncRNA transcriptional profiling
The pathobiology of RV failure in the setting of LV failure is likely distinct based upon the etiology of LV failure.65 Given the relative resistance of the RV to irreversible ischemic injury, RV failure in ischemic LV failure most likely devolves from the chronic elevation of pulmonary vascular impedance resulting from progressive LV diastolic hypertension.67 In impedance overload, the RV remodels by concentric hypertrophy and adopts over time with unrelieved overload many of the geometric and cardiodynamic features of the LV. In nonischemic LV failure, by contrast, RV failure may result from the same heritable structural or contractile protein mutations or infectious/toxic injuries affecting the LV.67 Heritable protein mutations or injuries delimit the RV remodeling repertoire to any additional insult or overload imposed by LV failure. Hence, RV failure pathobiology due to heritable protein mutations or injuries diverges sharply from the RV failure pathobiology of pulmonary vascular impedance overload alone. Beyond these broad etiologic factors, other modifying genetic and environmental factors—including common comorbid conditions that alter pulmonary hemodynamics independent of the heart (e.g., obstructive sleep apnea and chronic obstructive pulmonary disease), general medical comorbidities (e.g., diabetes mellitus), and specific heart failure pharmacologic therapies—introduce higher orders of heterogeneity into the pathobiology of RV failure in the setting of LV failure.65,66 To some degree, the RV transcriptome in end-stage LV failure would be expected to reflect the different etiologies of LV failure, the variable pulmonary vascular impedance conditions dependent on LV function, and the integration of these and other diverse genetic and environmental modifying factors. Collectively, these pathobiologic considerations in end-stage human LV heart failure suggest that (1) the RV transcriptome is not likely to mirror directly the LV transcriptome and (2) the RV transcriptome may exhibit substantial patient-to-patient heterogeneity. Such RV transcriptome heterogeneity was supported by the striking variability in subject-to-subject expression of many differentially expressed lncRNAs identified in this study and may afford a potential future opportunity to personalize therapies for RV failure.
Beyond providing insight into fundamental mechanisms of RV myocardial development and remodeling as well as affording an opportunity to personalize therapies, lncRNA transcriptional profiling may yield diagnostic clinical assays and prognostic biomarkers in human heart failure. A recent study of explanted human LVs reported that the LV lncRNA transcriptional profile discriminated the etiology of ischemic versus nonischemic heart failure with greater precision than did the LV mRNA or miRNA transcriptional profiling.68 This study also showed that lncRNA expression profiles, but not mRNA or miRNAs, were markedly altered in response to the reverse remodeling observed with LV mechanical support.68 lncRNA profiling and therapeutic targeting may complement the evolving roles of miRNA profiling and therapeutic targeting in the LV69 and in RV dysfunction.70 In a recent study, the mitochondrial lncRNA LIPCAR was upregulated in the plasma of patients who developed adverse ventricular remodeling and heart failure after myocardial infarction and was shown to be an independent predictor of survival in patients with heart failure.71
Findings of additional speculative interest
Several of the findings in the present study are of additional speculative interest. First, we found expression of approximately 3,000 lncRNAs (including pseudogenes) representing 16% of known lncRNAs and 8% of known pseudogenes in RV myocardium. Approximately 6% overall of these lncRNAs were differentially expressed in heart failure versus unused donor RVs. These percentages of overall lncRNA expression and differential lncRNA are both modest. Most lncRNA expression, like mRNA expression, appears conserved in the failing RV, which is perhaps not surprising given the dependence of working myocardium (failing or not) on the highly expressed, highly conserved mitochondrial energetic and sarcomeric contractile protein components that dominate the myocardial transcriptome.68 Second, there were a larger number of transcripts expressed overall in the heart failure versus unused donor RVs. This may have arisen from a variety of factors. The heart failure subjects had a greater number of comorbidities and medications, both of which may have induced different and richer expression patterns. Although the explantation techniques, cardio-preservation protocols, and tissue-preparation processes were identical in the unused donor and heart failure subjects, the unused donors and heart failure subjects were in markedly different physiologic states at the time of explanation. The unused donor subjects were brain dead, hormonally resuscitated, anesthetized without induction, were not supported on cardiopulmonary bypass, and had multiple vital organs explanted before cardiac explantation. Conversely, the heart failure subjects had intact brain function, were anesthetized with careful induction, and were supported by cardiopulmonary bypass with fully intact vital organ function at the time of explantation. Collectively, these differences in physiologic state may have contributed to the gene expression differences found.72
Limitations
Our study was limited to explanted end-stage human myocardium. Although the RNA submitted for sequencing from the homogenized myocardial tissue was of high quality, we cannot exclude selective enrichment of RNAs due to sample-to-sample differences in tissue characteristics or composition, myocardial perfusion, or blood admixture. Although our tissue collection and preservation protocol was designed to minimize RNA degradation, the sequential processes of harvesting, transport, frozen storage, thawing, tissue homogenization, and RNA extraction cannot preserve all tissue RNA.
By design, we analyzed only RV tissues. Thus, we cannot compare differential expression of LV lncRNA versus RV lncRNA. The earlier study of LV lncRNA differential expression, cited above, did not provide a tabulation of differentially expressed LV lncRNAs to permit comparison with the tabulation of differentially expressed RV lcnRNAs in our study.68 All patients were treated with medications, including inotropes, before transplantation, which likely altered gene expression in the RV. RNA-Seq has inherent potential limitations owing to 5′ bias and dependence on library-preparation methodology.7 As demonstrated, substantial biological variation was found in read counts between subjects. The pooling of samples with such read count variation challenges statistical inference based on an assumed distribution of read counts. Statistical significance, as in all such studies, is accepted as a “proxy” for biological significance, realizing that meaningful biological variation may or may not result in statistical differences in expression if there are threshold, graduated, or nonlinear expression effects on translation.
We did not perform confirmatory qPCR or other assays for the differentially expressed genes reported. Multiple earlier studies have demonstrated equivalent precision of RNA-Seq and qPCR.5,6,73,74 Given the limitations of coverage, copy readout, and specificity of microarray probes, confirmatory qPCR is required to validate microarray findings. RNA-Seq, however, is not limited by coverage, copy readout, and specificity, and confirmatory qPCR is not currently recommended by experts for well-performed RNA-Seq.5,6,73,74 For this study, RNA-Seq was performed in the Vanderbilt Genomics Core laboratory, a facility with highly trained and experienced personnel and state-of-the-art equipment.
We compared RVs from subjects with end-stage LV failure with the RVs of unused human donors. Unused human donors do not have “normal” RV myocardium because of the state of brain death before harvest. Earlier studies have reported heterogeneity in gene expression in unused donor hearts attributable to clinical factors and harvest/storage factors.72 Finally, all such studies are inherently limited by the lack of understanding of fundamental aspects of gene expression, including the relationship of transcribed protein-coding or non–protein-coding mRNA to translated protein.75
Conclusion
RNA-Seq provides a comprehensive profile of the polyadenylated lncRNA transcriptome in human heart failure. Over 100 polyadenylated lncRNAs are differentially expressed in heart failure RVs versus unused donor RVs. Differentially expressed lncRNAs, including natural antisense transcripts and pseudogenes, likely possess a variety of regulatory functions.
Despite the likely critical roles of lncRNAs in myocardial pathobiology, myocardial lncRNA research is in its infancy. The current study was descriptive and exploratory by design. It describes a reasonably large number of differentially expressed lncRNAs in human heart failure and explores their possible functional roles via accessible, existing online resources and databases. It is hoped that these results provide investigators with an inventory of differentially expressed candidate lncRNAs, natural antisense transcripts, and pseudogenes for future mechanistic studies in human heart failure.
Source of Support: This study was supported by CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Conflict of Interest: None declared.
References
- 1.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed]
- 2.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, et al. Landscape of transcription in human cells. Nature 2012;489:101–108. [DOI] [PMC free article] [PubMed]
- 3.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861–874. [DOI] [PubMed]
- 4.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation 2010;121:1022–1032. [DOI] [PMC free article] [PubMed]
- 5.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. [DOI] [PMC free article] [PubMed]
- 6.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-Seq experiments with Tophat and Cufflinks. Nat Protoc 2012;7:562–578. [DOI] [PMC free article] [PubMed]
- 7.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5:621–628. [DOI] [PubMed]
- 8.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech 2010;28:511–515. [DOI] [PMC free article] [PubMed]
- 9.Berger B, Peng J, Singh M. Computational solutions for omics data. Nat Rev Genet 2013;14:333–346. [DOI] [PMC free article] [PubMed]
- 10.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11:R106. [DOI] [PMC free article] [PubMed]
- 11.Robinson MD, McCarthy DJ, Smyth GK. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–140. [DOI] [PMC free article] [PubMed]
- 12.Hardcastle TJ, Kelly KA. Bayseq: empirical bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 2010;11:422. [DOI] [PMC free article] [PubMed]
- 13.Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res 2013;41:D246–D251. [DOI] [PMC free article] [PubMed]
- 14.Chan WL, Yang WK, Huang HD, Chang JG. Pseudomap: an innovative and comprehensive resource for identification of siRNA-mediated mechanisms in human transcribed pseudogenes. Database (Oxford) 2013;2013:bat001. [DOI] [PMC free article] [PubMed]
- 15.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG. Diana-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res 2013;41:D239–D245. [DOI] [PMC free article] [PubMed]
- 16.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet 2007;39:1278–1284. [DOI] [PubMed]
- 17.Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics 2011;12:489. [DOI] [PMC free article] [PubMed]
- 18.Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics 2013;29:2928–2930. [DOI] [PMC free article] [PubMed]
- 19.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet 2013;14:880–893. [DOI] [PubMed]
- 20.Annilo T, Kepp K, Laan M. Natural antisense transcript of natriuretic peptide precursor a (NPPA): structural organization and modulation of NPPA expression. BMC Mol Biol 2009;10:81. [DOI] [PMC free article] [PubMed]
- 21.Poliseno L. Pseudogenes: newly discovered players in human cancer. Sci Signal 2012;5:re5. [DOI] [PubMed]
- 22.Zhao Y, Zhang J, Li H, Li Y, Ren J, Luo M, Zheng X. An NADPH sensor protein (HSCARG) down-regulates nitric oxide synthesis by association with argininosuccinate synthetase and is essential for epithelial cell viability. J Biol Chem 2008;283:11004–11013. [DOI] [PubMed]
- 23.Lee JH, Gao C, Peng G, Greer C, Ren S, Wang Y, Xiao X. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res 2011;109:1332–1341. [DOI] [PMC free article] [PubMed]
- 24.Sakabe NJ, Aneas I, Shen T, Shokri L, Park SY, Bulyk ML, Evans SM, Nobrega MA. Dual transcriptional activator and repressor roles of tbx20 regulate adult cardiac structure and function. Hum Mol Genet 2012;21:2194–2204. [DOI] [PMC free article] [PubMed]
- 25.Song HK, Hong SE, Kim T, Kim do H. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PloS ONE 2012;7:e35552. [DOI] [PMC free article] [PubMed]
- 26.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013;152:1308–1323. [DOI] [PubMed]
- 27.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, et al. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011;477:295–300. [DOI] [PMC free article] [PubMed]
- 28.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 2013;494:497–501. [DOI] [PMC free article] [PubMed]
- 29.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300–307. [DOI] [PubMed]
- 30.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature 2012;482:339–346. [DOI] [PMC free article] [PubMed]
- 31.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 2013;12:433–446. [DOI] [PubMed]
- 32.Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res 2012;111:1349–1362. [DOI] [PubMed]
- 33.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013;152:570–583. [DOI] [PMC free article] [PubMed]
- 34.Luther HP. Role of endogenous antisense RNA in cardiac gene regulation. J Mol Med (Berlin) 2005;83:26–32. [DOI] [PubMed]
- 35.Voigtsberger S, Bartsch H, Baumann G, Luther HP. Cell type-specific expression of endogenous cardiac troponin I antisense RNA in the neonatal rat heart. Mol Cell Biochem 2009;324:1–11. [DOI] [PubMed]
- 36.Haddad F, Qin AX, Bodell PW, Jiang W, Giger JM, Baldwin KM. Intergenic transcription and developmental regulation of cardiac myosin heavy chain genes. Am J Phisiol Heart Circ Physiol 2008;294:H29–H40. [DOI] [PubMed]
- 37.Ritter O, Haase H, Schulte HD, Lange PE, Morano I. Remodeling of the hypertrophied human myocardium by cardiac BHLH transcription factors. J Cell Biochem 1999;74:551–561. [DOI] [PubMed]
- 38.Yadava RS, Frenzel-McCardell CD, Yu Q, Srinivasan V, Tucker AL, Puymirat J, Thornton CA, Prall OW, Harvey RP, Mahadevan MS. RNA toxicity in myotonic muscular dystrophy induces NKX2–5 expression. Nat Genet 2008;40:61–68. [DOI] [PMC free article] [PubMed]
- 39.Cooper C, Vincett D, Yan Y, Hamedani MK, Myal Y, Leygue E. Steroid receptor RNA activator bi-faceted genetic system: heads or tails? Biochimie 2011;93:1973–1980. [DOI] [PubMed]
- 40.Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell 2006;11:547–560. [DOI] [PubMed]
- 41.Friedrichs F, Zugck C, Rauch GJ, Ivandic B, Weichenhan D, Muller-Bardorff M, Meder B, et al. HBEGF, SRA1, and IK: three cosegregating genes as determinants of cardiomyopathy. Genome Res 2009;19:395–403. [DOI] [PMC free article] [PubMed]
- 42.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 2006;51:1087–1099. [DOI] [PubMed]
- 43.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007;316:1491–1493. [DOI] [PubMed]
- 44.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010;38:662–674. [DOI] [PMC free article] [PubMed]
- 45.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting mir-489. Circ Res 2014;114:1377–1388. [DOI] [PubMed]
- 46.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 2014;114:1389–1397. [DOI] [PubMed]
- 47.Peters T, Schroen B. Missing links in cardiology: long non-coding RNAs enter the arena. Pflugers Arch 2014;466:1177–1187. [DOI] [PubMed]
- 48.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904–914. [DOI] [PMC free article] [PubMed]
- 49.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298–1307. [DOI] [PMC free article] [PubMed]
- 50.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 2011;146:353–358. [DOI] [PMC free article] [PubMed]
- 51.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by polycomb repressive complex 2. Nat Struct Mol Biol 2013;20:1250–1257. [DOI] [PMC free article] [PubMed]
- 52.Archer SL, Fang YH, Ryan JJ, Piao L. Metabolism and bioenergetics in the right ventricle and pulmonary vasculature in pulmonary hypertension. Pulm Circ 2013;3:144–152. [DOI] [PMC free article] [PubMed]
- 53.Drake JI, Bogaard HJ, Mizuno S, Clifton B, Xie B, Gao Y, Dumur CI, Fawcett P, Voelkel NF, Natarajan R. Molecular signature of a right heart failure program in chronic severe pulmonary hypertension. Am J Resp Cell Mol Biol 2011;45:1239–1247. [DOI] [PMC free article] [PubMed]
- 54.Gomez-Arroyo J, Mizuno S, Szczepanek K, Van Tassell B, Natarajan R, dos Remedios CG, Drake JI, et al. Metabolic gene remodeling and mitochondrial dysfunction in failing right ventricular hypertrophy secondary to pulmonary arterial hypertension. Circ Heart Fail 2013;6:136–144. [DOI] [PMC free article] [PubMed]
- 55.Reddy S, Zhao M, Hu DQ, Fajardo G, Hu S, Ghosh Z, Rajagopalan V, Wu JC, Bernstein D. Dynamic microRNA expression during the transition from right ventricular hypertrophy to failure. Physiol Genomics 2012;44:562–575. [DOI] [PMC free article] [PubMed]
- 56.Vonk-Noordegraaf A, Haddad F, Chin KM, Forfia PR, Kawut SM, Lumens J, Naeije R, et al. Right heart adaptation to pulmonary arterial hypertension: physiology and pathobiology. J Am Coll Cardiol 2013;62:D22–D33. [DOI] [PubMed]
- 57.Bartelds B, Borgdorff MA, Smit-van Oosten A, Takens J, Boersma B, Nederhoff MG, Elzenga NJ, van Gilst WH, De Windt LJ, Berger RM. Differential responses of the right ventricle to abnormal loading conditions in mice: pressure vs. volume load. Eur J Heart Fail 2011;13:1275–1282. [DOI] [PubMed]
- 58.Reddy S, Zhao M, Hu DQ, Fajardo G, Katznelson E, Punn R, Spin JM, Chan FP, Bernstein D. Physiologic and molecular characterization of a murine model of right ventricular volume overload. Am J Physiol Heart Circ Physiol 2013;304:H1314–H1327. [DOI] [PMC free article] [PubMed]
- 59.Mathiyalagan P, Chang L, Du XJ, El-Osta A. Cardiac ventricular chambers are epigenetically distinguishable. Cell Cycle 2010;9:612–617. [DOI] [PubMed]
- 60.Tabibiazar R, Wagner RA, Liao A, Quertermous T. Transcriptional profiling of the heart reveals chamber-specific gene expression patterns. Circ Res 2003;93:1193–1201. [DOI] [PubMed]
- 61.Friehs I, Cowan DB, Choi YH, Black KM, Barnett R, Bhasin MK, Daly C, et al. Pressure-overload hypertrophy of the developing heart reveals activation of divergent gene and protein pathways in the left and right ventricular myocardium. Am J Physiol Heart Circ Physiol 2013;304:H697–H708. [DOI] [PMC free article] [PubMed]
- 62.Apitz C, Honjo O, Humpl T, Li J, Assad RS, Cho MY, Hong J, Friedberg MK, Redington AN. Biventricular structural and functional responses to aortic constriction in a rabbit model of chronic right ventricular pressure overload. J Thorac Cardiovasc Surg 2012;144:1494–1501. [DOI] [PubMed]
- 63.Kreymborg K, Uchida S, Gellert P, Schneider A, Boettger T, Voswinckel R, Wietelmann A, et al. Identification of right heart-enriched genes in a murine model of chronic outflow tract obstruction. J Mol Cell Cardiol 2010;49:598–605. [DOI] [PubMed]
- 64.Kaufman BD, Desai M, Reddy S, Osorio JC, Chen JM, Mosca RS, Ferrante AW, Mital S. Genomic profiling of left and right ventricular hypertrophy in congenital heart disease. J Cardiac Fail 2008;14:760–767. [DOI] [PubMed]
- 65.Friedberg MK, Redington AN. Right versus left ventricular failure: differences, similarities, and interactions. Circulation 2014;129:1033–1044. [DOI] [PubMed]
- 66.Simon MA. Assessment and treatment of right ventricular failure. Nature Rev Cardiol 2013;10:204–218. [DOI] [PubMed]
- 67.Dell’Italia LJ. Anatomy and physiology of the right ventricle. Cardiol Clin 2012;30:167–187. [DOI] [PubMed]
- 68.Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNA in failing human heart and remodeling with mechanical circulatory support. Circulation 2014;129:1009–1021. [DOI] [PMC free article] [PubMed]
- 69.Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Circ Res 2013;113:676–689. [DOI] [PubMed]
- 70.Thum T, Batkai S. MicroRNAs in right ventricular (dys)function (2013 Grover conference series). Pulm Circ 2014;4:185–190. [DOI] [PMC free article] [PubMed]
- 71.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 2014;114:1569–1575. [DOI] [PubMed]
- 72.Barth AS, Kumordzie A, Frangakis C, Margulies KB, Cappola TP, Tomaselli GF. Reciprocal transcriptional regulation of metabolic and signaling pathways correlates with disease severity in heart failure. Circ Cardiovasc Genet 2011;4:475–483. [DOI] [PMC free article] [PubMed]
- 73.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 2011;12:87–98. [DOI] [PMC free article] [PubMed]
- 74.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-Seq. Nat Biotech 2013;31:46–53. [DOI] [PMC free article] [PubMed]
- 75.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature 2011;473:337–342. [DOI] [PubMed]