Abstract
Human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) provide a potential source of cells to repair injured ventricular myocardium. CM differentiation cultures contain non-cardiac cells and CMs of both nodal and working subtypes. Direct application of such cultures in clinical studies could induce arrhythmias; thus, further purification of working-type CMs from heterogeneous cultures is desirable. Here, we designed 10 molecular beacons (MBs) targeting NPPA mRNA, a marker associated with working-type CMs and highly up-regulated during differentiation. We examined these MBs by solution assays and established their specificity using NPPA-overexpressing CHO cells as well as hPSC-CMs. We selected one MB for subsequent CM subtype isolation using fluorescence-activated cell sorting because the signal-to-background ratio was the highest for this MB in solution assays and a linear correlation was observed between MB signals and the CM purity in differentiation cultures. Compared with cells with low MB signals, cells positively selected based on MB signal had higher expression levels of genes associated with working-type CMs and lower expression levels of genes associated with nodal-type CMs. Therefore, the MB-based method is capable of separating working-type CMs from nodal-type CMs with high specificity and throughput, potentially providing working-type CMs for biomedical applications.
Keywords: Cardiomyocyte, Flow cytometry, Gene expression, Molecular imaging, Stem cell
1. Introduction
Human cardiac tissues have limited regenerative capacity. Damage in cardiac tissues from ischemia or other pathological conditions can result in heart failure, the leading cause of death in the United States. Human pluripotent stem cells (hPSCs), including both human embryonic stem cells (hESCs) [1] and induced pluripotent stem cells (iPSCs) [2, 3], hold great promise for cardiac cell therapy due to their ability to generate patient-matched functional cardiomyocytes (CMs) [4–9]. In preclinical studies, hESC-CMs survived in infarcted rat heart following transplantation and prevented progression of heart failure [10, 11]. Extensive engraftment in infarcted hearts was also achieved in pigtail macaques following transplantation of hESC-CMs at a clinically relevant dose; however, arrhythmias were observed in these animals [12].
CM differentiation cultures contain a mixture of nodal and working (atrial and ventricular) cardiac subtypes [4–9]. These subtypes are suitable for different applications: enriched working-type CMs for repairing injured ventricular myocardium, and enriched nodal-type CMs for developing a biological pacemaker. Direct application of the CM mixture could lead to arrhythmias because working-type and nodal-type CMs have distinct electrophysiological properties and functions [4–8]. Therefore, from both efficacy and safety points of view, highly homogeneous populations of a specific CM type may be required for cardiac cell therapy. To meet this requirement, it is necessary to develop methods for controlling subtype specification during stem cell differentiation and/or isolating individual CM subtypes from differentiation cultures of hPSCs.
While significant improvement has been made in increasing the efficiency of CM differentiation from hPSCs [10, 13–15], none of the existing methods can generate homogeneous cell populations for a specific subtype. Further, methods for the isolation and enrichment of each CM subtype have not been established due to the lack of well-defined surface markers. It is desirable to have a new platform that can distinguish and enrich cardiac subtypes with high specificity and throughput.
Molecular beacons (MBs) have the potential to achieve this goal. MBs are stem-loop (hairpin) oligonucleotide probes with a fluorophore at one end and a quencher at the other end [16]. When a MB is in the hairpin conformation, fluorescence of the fluorophore is quenched. Upon proper excitation, fluorescence signals are emitted when MBs are hybridized to their complementary RNA target sequences. Because of these characteristics, MBs targeting specific genes can be used to detect the presence, level and localization of mRNA expression in living cells [16, 17]. They have also been used to identify and isolate stem cells or their derivatives based on specific mRNA expression. For example, MBs specific for stem cell marker Oct4 can distinguish undifferentiated mouse embryonic stem cells from differentiated cells [18]. MBs targeting Sox2, a marker for undifferentiated stem cells and neural stem cells, have been used to sort live undifferentiated stem cells from retinoic acid-treated mouse embryonic stem cells [19]. MBs targeting the specific CM marker myosin heavy chain beta (MYH7) mRNA have recently been used to enrich CMs derived from mouse and human embryonic stem cells [20].
To date, the use of MBs to target specific mRNAs for isolating human CM subtypes has not been demonstrated. Here we report the development of an MB-based method that was used successfully to detect and separate working-type CMs from hPSC differentiation cultures. Based on the expression of putative subtype-associated genes during CM differentiation, we designed 10 MBs targeting NPPA mRNA and examined the specificity of these MBs using respectively engineered NPPA-overexpressing CHO cells and stem cell-derived CMs. We selected one NPPA–targeting MB that has the highest signal-to-background ratio and delivered this MB via nucleofection into the CM differentiation cultures. We then separated the cells by FACS into two populations based on fluorescence signal from the MBs, one was with high MB signal and positively selected (putative working-type), and the other with low MB signal. Comparison of the expression of specific genes in these two populations of cells confirmed that the MB-based method is capable of separating working-type CMs from nodal-type CMs with high specificity.
2. Materials and Methods
2.1. hPSC culture and CM differentiation
iPSC line IMR90 [3] and the NIH-approved hESC line WA07 [1] (H7, NIHhESC-10-0061, obtained from the WiCell Research Institute, Madison, WI) were maintained in mouse embryonic fibroblast conditioned medium (MEF-CM) supplemented with basic fibroblast growth factor (bFGF) (8 ng/ml) [21]. Cells were passaged for cardiac differentiation 6 days after seeding or until colonies occupied approximately ~80% of the well surface area. Cells were rinsed with 2 ml DPBS and incubated with 2 ml versene (EDTA) for 10 min at 37°C; then versene was aspirated and replaced with 1 ml/well of MEF-CM containing bFGF (8 ng/ml). Cells were dissociated by gentle flowing of MEF-CM all over the wells and triturating (~15 times) to generate single cell suspension. Further, cells were seeded at a density of 4×105 cells in 1 ml of MEF-CM for each well of 24-well Matrigel-coated plates. Cells were fed daily by replacing MEF-CM supplemented with bFGF (8 ng/ml) until compactly covering the wells. Usually, 2 to 4 days after seeding, cells were ready for induction. At the day of induction (day 0), MEF-CM was replaced with 1 ml warm medium (RPMI 1640 + B27 without Insulin) supplemented with 100 ng/ml activin A. After 24 hr (day 1), activin A-containing medium was replaced with 1 ml warm medium (RPMI 1640 + B27 without Insulin) supplemented with 10 ng/ml bone morphogenetic protein 4 (BMP-4). Then, induced cultures were incubated for additional 4 days without any medium change. From day 5 onwards, BMP-4 containing medium was replaced with 1 ml warm RPMI 1640 + B27 without any growth factors and the medium was replaced on alternate days. Cells were observed under microscope daily for beating cells, which typically started after day 8, and cells were harvested at day 14 to determine CM yield.
2.2. MB design
The mRNA sequence of the target gene NPPA was retrieved from NCBI and the secondary structure of the NPPA mRNA was predicted using the online mFOLD server (http://mfold.rna.albany.edu/?q=mfold). Multiple regions predicted to be single stranded and with 30–70% GC content were selected as target sequences. Oligonucleotides of 17–25 bases long complementary to these target sequences were selected to form the loop region of the MB (Table 1), and two short (4–7 bases) complementary sequences were added respectively to the 5′ and 3′ ends of loop sequence to form the stem region of the MB (Table 1 and Fig. 1B). Different stem designs (length and sequence) were compared based on the predicted hairpin structures using the qwikfold server (http://mfold.rna.albany.edu/?q=DINAMelt/Quickfold), and stems that gave desired melting temperatures were selected to form MBs [22]. To minimize the potential quenching of the dye, special care was taken to avoid a G within 3 bps from the 5′ end where the fluorophore is conjugated. To achieve high specificity, the target sequences of MBs were analyzed using BLAST to screen the Homo sapiens reference mRNA database; only MBs returning e-values less than 1 for the target gene and more than 1 for other genes were selected for synthesis. A fluorophore (FAM or Cy3) and a quencher (Black Hole Quencher 1 or 2) were added to the 5′- and 3′-end of the stem-loop hairpin respectively to form an MB. All HPLC purified beacons were ordered from IDT (Integrated DNA Technologies, Coralville, IA). An oligonucleotide perfectly complementary to the MB loop sequence was ordered as a positive control, and a second oligonucleotide with 6-base mismatches compared with the target sequence was used as a negative control.
Table 1.
Molecular beacon designs for detecting NPPA mRNA
| Name | Sequencea | Fluorophore | Quencher | Complex Melting Tm | Solution Assay Normalized SBRb | CHO-NPPA | CHO | hPSC-CMs | hPSCs |
|---|---|---|---|---|---|---|---|---|---|
| NPPA-targeting Molecular Beacons | |||||||||
| NPPA_A | CCTCACCCTGCTTGTCCTCCCTGGCTGTTATGTGAGG | FAM | BHQ1 | 71.0 | 6.0 | Negc | Neg | Neg | N/A |
| NPPA_B | ACCTCTTGCAGTCTGTCCCTAGGAGGT | FAM | BHQ1 | 66.3 | 6.8 | Neg | Neg | Neg | N/A |
| NPPA_C | CATCGAATCCATCAGGTCTGCGTTGGACGATG | Cy3 | BHQ2 | 59.4 | 4.7 | Neg | Neg | Neg | N/A |
| NPPA_D | CCTCGATCTCTCTGGGCTGGGCTGACTTCGAGG | Cy3 | BHQ2 | 62.4 | 1.0 | N/A | N/A | N/A | N/A |
| NPPA_E | CTAGCCGGGCACGACCTCATCTTCTAAAGGCTAG | Cy3 | BHQ2 | 61.3 | 0.8 | N/A | N/A | N/A | N/A |
| NPPA_F | CATAGCTTCTTCATTCGGCTCACTGAGCTATG | Cy3 | BHQ2 | 60.2 | 1.0 | N/A | N/A | N/A | N/A |
| NPPA_G | CACAGTGTTGACAGGAAGCTGCAGCTGTG | Cy3 | BHQ2 | 66.7 | 14.1 | Posd | Neg | Pos | Neg |
| NPPA_H | CTGCCAATGCATGGGGTGGGAGAGGCAG | Cy3 | BHQ2 | 69.9 | 0.8 | Neg | Neg | Pos | Pos |
| NPPA_I | CACGCAAATTCTTGAAATCCATCAGCGTG | Cy3 | BHQ2 | 58.7 | 8.2 | Pos | Neg | Pos | Pos |
| NPPA_J | ATCCCTCAGCTTGCTTTTTAGGAT | Cy3 | BHQ2 | 61.5 | 4.1 | Neg | Neg | Pos | Pos |
| Control Molecular Beacons | |||||||||
| Random-Fam | ACGACGCGACAAGCGCACCGATACGTCGT | 2x FAM | None | N/A | N/A | Pos | Pos | Pos | Pos |
| MHC1 | CCTCCATCTTCTTCTTCACGGAGG | Cy3 | BHQ2 | 57.5 | 6.8 | N/A | N/A | Pos | Neg |
Underlined sequences indicate stem sequences.
SBR: signal-to-background ratio.
Neg: low MB signal.
Pos: high MB signal.
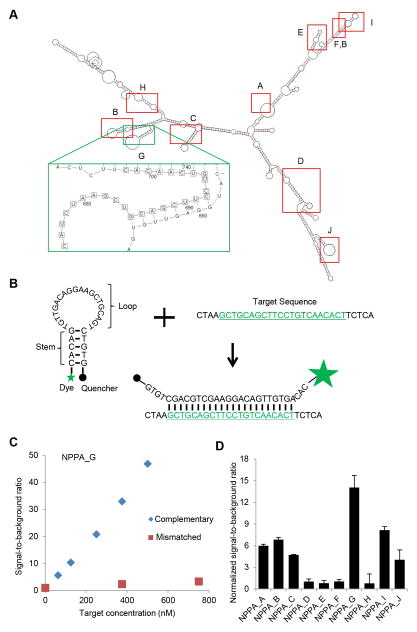
Fig. 1.
Gene expression during CM differentiation from hPSCs. A) CM differentiation procedure. B) Purity of CMs as detected by flow cytometry analysis of α-actinin. Isotype was used as a negative control to establish the gating parameters. C) Expression of genes associated with different stages of differentiation. D) Expression of genes associated with working-type CMs during differentiation. E) Expression of genes associated with nodal-type CMs during differentiation.
2.3. Solution assays
The quality of synthesized MBs was determined by measuring the fluorescence of MBs (with a fixed concentration) hybridized to positive and negative control oligonucleotides with varied concentrations. MBs were suspended in Tris-EDTA buffer, pH 8 (100 μM solution) and added to wells in an opaque 384-well plate at a final beacon concentration of 500 nM/well. Increasing concentrations (DNA, 0–500 nM) of 20–30 bp positive-control (complementary) oligonucleotides were then added to the wells and the resulting fluorescence signals were measured. MB Fluorescence was also measured when hybridized to the negative control nucleotides (with 6-bp mismatched) at concentrations of 750 nM and 375 nM. Sample plates were incubated at 37°C for 10 min, and fluorescence was read with a plate reader (Safire, Ex: 490, Em: 525 or Ex: 540, Em: 565 all bandpass filters at 7 nm, Tecan) every 10 min for a total of 30 min. The signal-to-background ratio was defined as the ratio of the fluorescence of an MB in the presence of its target to the fluorescence of the MB in the absence of its target. Normalized signal-to-background ratio was defined as the ratio of the fluorescence of an MB in the presence of its complementary target at the highest concentration to the fluorescence of the MB in the presence of a mismatched target at the highest concentration.
2.4. MB delivery
Cells were incubated with accutase for 10–15 min till rounded in shape and then gently pipeted and collected in DMEM containing 10% FBS. Cells were washed once with PBS by centrifuging at 100 g for 5 min. 1×106 cells were suspended in 100 μl of Amaxa SF cell line nucleofection solution containing 1 μM molecular beacons. Cell suspension was immediately transferred to nucleofector cuvette and nucleofection was performed in Amaxa Nucleofector II using A033 program. Nucleofected cells were immediately transferred to a 6-well plate containing 1 ml RPMI 1640 medium containing B27 supplement and further incubated at 37°C for 10 min. Cells were collected in a 5 ml tube and spun at 200 g for 5 min and resuspended in 100 μl PBS. Cells were acquired using FACS Canto (FITC channel for FAM and PE channel for Cy3) within an hour of nucleofection.
2.5. Establishment of NPPA expressing cell lines
Stable cell-lines (CHO-NPPA cells) were created by expressing NPPA for initial screening of MBs before testing them on CMs. Cells (2.5×105 cells/well) were seeded in a volume of 2 ml complete growth medium per well in a 6-well plate, 18–24 hr before transfection at the density of ~80% confluent. The transfection was performed according to the manufacturer’s instructions using 9 μl Lipofectamine 2000 reagent (Life Technologies) in 100 μl serum-reduced Opti-MEM I medium (Life Technologies) containing 3 μg of pReceiver-M68 vector containing the NPPA gene (GeneCopoeia, Inc). After treatment, the cells were incubated for 24 hr in the culture media. For the generation of stable cell lines, the titration of the selective antibiotic puromycin (Life Technologies) was carried out to generate a killing curve. Different puromycin concentrations were applied (0, 2, 4, 6, 10, 15, 20 μg/ml) onto 1×106 CHO cells seeded in the wells of a 12-well plate. For selection of positive cells after transfection, the lowest concentration of puromycin was used in which no cell survived after 7 days. Transfected CHO cells were selected initially at a concentration of 15 μg/ml puromycin in the complete medium. Cells not expressing the construct were killed by puromycin and the concentration of puromycin was lowered to 7.5 μg/ml to maintain the cell line.
2.6. Flow cytometry analysis of α-actinin
Differentiation of hPSCs to cardiac cells was analyzed by intracellular staining of α-actinin using flow cytometry. Differentiated cultures at days 14 to 45 were harvested by incubating with 0.25% trypsin-EDTA at 37°C for 10 min and subsequently neutralized by 10% FBS in DMEM. Cells were counted and 0.5×106 cells each for α-actinin and isotype control along with compensation controls (unstained, EMA only and α-actinin only) were aliquoted in 15 ml tubes. Then cells were washed once with PBS (5 ml/tube) and resuspended in 0.5 ml staining buffer (SB) (PBS containing 2% heat-inactivated FBS) containing 1 μg/ml ethidium monoazide (EMA) to distinguish live and dead cells and incubated in dark on ice for 15 min. Further, cells were pelleted, resuspended in 1 ml PBS and exposed to bright light for 10 min by placing tubes horizontally on ice bed under a desk lamp. Cells were washed once with 2 ml of PBS and fixed in 2% paraformaldehyde (PFA) at room temperature for 15 min by adding 4% PFA (0.5 ml) in equal volumes of cell suspension. Cells were washed again and resuspended in 100 μl of PBS and then permeabilized by adding 900 μl of ice cold absolute methanol and incubated on ice for 30 min. Cells were washed once and blocked with blocking solution (BS) (20% goat serum (Invitrogen) in SB) at room temperature for 30 min. After blocking, cells were washed and incubated for 20 min at room temperature with anti-human α-actinin mouse IgG1 primary antibody (0.5 μg/test) or mouse IgG1 isotype control (0.5 μg/test) in 100 μl of SB. Cells were washed two times and incubated for 15 min at room temperature in dark with secondary antibody, goat anti-mouse IgG1 conjugated with Alexa-488 (0.5 μg/test). Cells were then again washed three times and resuspended in 200 μl of SB. Stained cells were analyzed by BD FACS Canto II by adjusting voltage and compensation using appropriate excitation and detection channels; FITC and PerCP Cy5.5 for α-actinin and EMA respectively. Forward versus side scatter quadrants were defined and at least 10,000 EMA negative (live) events were acquired. Finally, dot plots were generated upon data analysis using FlowJo software to display the frequency of α-actinin positive cells.
2.7. Flow cytometry analysis of MB signals and cell sorting
Negative control cells were nucleofected as aforementioned without any MBs. These cells were used to establish gating parameters for live cells in the forward and side scatter (FSC-SSC) dot plots. Negative control cells in the live cell gate were then used to establish the gating parameters for FITC and Cy3 fluorescence (MB signal), with the maximum negative control representing the minimum threshold. Samples containing MBs were analyzed using the same gates, and only events in the live cell gate and with fluorescence higher than the FITC or Cy3 fluorescence thresholds were counted as MB positive cells. This procedure was followed on Accuri C6, the BD FACS Canto, and the BD FACS Aria flow cytometry systems.
FACS Aria was used for sorting NPPA MBhi (with high MB signal) and NPPA MBlo (with low MB signal) cells. Initial gate was applied in the FSC-SSC dot plot to exclude dead cells and debris. Further, only single cells were included in the analysis based on gating cells under width vs. height. On the Aria, the 488 nm and 561 nm laser powers were both decreased to 25 mW to reduce photobleaching of Cy3 observed with the higher intensity illumination in the Aria system. As analyzed by BD FACSDiva software, after NPPA MB delivery, the cell population with Cy3 fluorescence (MB signal) higher than that in the negative control cells was collected as NPPA MBhi cells and the cell population with Cy3 fluorescence signal comparable to that in the negative control was collected as NPPA MBlo/neg cells.
2.8. qRT-PCR
Total RNA was extracted at different time-points during the course of differentiation according to the recommendations of the manufacturer using Aurum total RNA mini kit (Bio-Rad). Briefly, cells were rinsed once with 1 ml PBS in the culture plate and replaced with 350 μl lysis solution supplemented with 1% β-mercaptoethanol, then pipeted up and down several times to lyse the cells thoroughly. Lysed cells were collected in RNAse- and DNAse-free 1.5 ml tubes and kept at −80°C until RNA isolation. Individual RNA sample (1 μg) was reverse transcribed by adding 100 U of Superscript III enzyme and random primers in 20 μl reaction mixture containing Vilo reaction buffer as per manufacturer’s instruction in SuperScript® VILO™ cDNA Synthesis Kit (Life Technologies). Reaction mixture was then incubated at three different temperature cycles: 25°C for 10 min, 42°C for 2 hr and 25°C for 5 min in a Bio-Rad thermal cycler. One reaction without superscript III (minus RT) was also used as a negative control. Further, reaction mixture was diluted 15-times to 300 μl and 2 μl cDNA was used for real-time PCR. Human specific PCR primers (Table S1) for different genes were retrieved from open access websites (http://primerdepot.nci.nih.gov/ or http://pga.mgh.harvard.edu/primerbank/). Real-time PCR reaction was performed in 20 μl mixture containing 2x iTaq SyBr green master mix, 2 μl of cDNA mix, and 200 nM of forward and reverse primers. Reaction cycle was initial denaturation step at 95°C for 10 min, 40 cycles of two-step PCR with 15 s of denaturation at 95°C followed by 1 min of annealing/extension at 60°C. A melt curve was performed for all reactions to check for product integrity and primer-dimer formation as default settings in Applied Biosystems 7500 real-time PCR systems. In this method, mRNA levels for each sample were normalized to the GAPDH or ACTB (β-actin) mRNA levels and expressed as a relative increase or decrease compared with levels in control samples. Data are presented as mean±STD.
2.9. Immunocytochemical analysis
CHO cells or CHO-NPPA cells were grown to confluency in LabTek coverslip bottom dishes. Cell media was aspirated and the dish was rinsed with Hank’s Balanced Salt Solution. Samples were then incubated with cold 4% PFA in PBS for 10 min. PFA was aspirated, and the dish was rinsed with PBS, with 5 min of incubation time between rinses. Cells were permeabilized using 0.1% v/v Tween 20 in PBS for 10 min at room temperature and then blocked by incubating with 5% goat serum and 0.1% Tween 20 for 1 hr at room temperature. The blocking solution was removed and cells were incubated with mouse monoclonal NPPA antibody (Millipore) at a dilution of 1 to 1000 in PBS with 0.1% Tween 20 overnight at 4°C. Cells were then rinsed 3 times with PBS and 0.1% Tween 20 before incubating with goat-anti-mouse secondary antibody conjugated to Alexa 594 (Life Technologies) at a dilution of 1 to 1000 for 1 hr at room temperature. The secondary antibody solution was then aspirated and cells were incubated for 5 min at room temperature with 1 μg/mL Hoechst stain. Cells were then rinsed another 3 times with PBS before imaging using a Deltavision microscope (Applied Precision) with a 60X oil-immersion lens (NA 1.4).
3. Results
3.1. Identification of optimal subtype-specific mRNAs as MB targets
Previously, we showed that both working- and nodal-type CMs were generated from hPSCs following growth factor-induced differentiation [11]. To facilitate the selection of appropriate subtype-specific mRNAs for MB targeting, we examined the expression of a number of genes during CM differentiation that were reported to be associated with CM subtypes. Monolayer cultures of H7 cells were sequentially treated with activin A and BMP4 for CM induction [10] (Fig. 1A). Typically, beating cells were observed at days 8–10 and increased in number overtime. Flow cytometry analysis indicated that ~75% CMs were generated at day 14 (Fig. 1B). A time-course expression analysis was performed using RNA samples of cell cultures collected from days 0 to 14 during differentiation. The qRT-PCR analysis showed expected gene expression patterns for CM differentiation: stem cell markers OCT4 were down-regulated soon after the initiation of differentiation; mesoderm marker T up-regulated and reached the highest level at day 2 and gradually down-regulated afterwards; and subsequently, expression of cardiac progenitor marker NKX2-5 and CM marker cTnT increased (Fig. 1C). Among cardiac subtype-associated genes, several genes were up-regulated dramatically upon differentiation, including putative working-type CM-associated genes IRX4 [23], NPPA [24, 25], NPPB [26], MYL2 [27], and MYL7 [28] (Fig. 1D) as well as nodal-type CM-associated genes HCN4 [29], SHOX2 [30], TBX3 [31] and TBX18 [32] (Fig. 1E). However, several other putative subtype-associated genes were detected at similar or decreased levels in differentiation cultures compared with undifferentiated cultures, including SCN5A [26] and GJA1 (connexin 43 or Cx43) [33–35] for working-type CMs and GJA3 (Cx30.2) for nodal-type CMs [35, 36] (Fig. 1D). This outcome is likely due to the fact that some of subtype-associated genes are also expressed by other cell types in stem cell cultures. For example, connexin 43 is highly expressed by undifferentiated hPSCs [37] although it is differentially expressed by working-type and nodal-type CMs [33–35]. Based on these results, we selected NPPA as a target gene for MB design because NPPA showed the largest change in the expression level upon CM differentiation from hPSCs. NPPA, a gene encoding natriuretic peptide A, is a specific marker for the early working-type cells of atria and ventricles in the developing heart [24, 25], but it is undetectable in pacemaker cells such as sinus nodes and atroventricular nodes.
3.2. Design and validation of MBs for targeting NPPA mRNAs
Based on the predicted secondary structure of NPPA mRNA (Fig. 2A), we designed 10 MBs using fluorophores FAM or Cy3 at one end of the stem-loop structure with DNA backbone and quenchers BHQ1 or BHQ2 at the other end (Table 1, and a representative MB design in Fig. 2B). To examine the quality of synthesized MBs targeting the NPPA mRNA, we quantified fluorescence signals when MBs were hybridized to perfectly complementary or mismatched synthetic targets using a solution assay. All NPPA MBs except NPPA_D and NPPA_H showed relatively low levels of the signal-to-background ratio (<10) when hybridized to mismatched targets (Fig. S1). The fluorescence signal of all NPPA MBs displayed a linear response to increasing concentrations of complementary targets (Fig. 2C and Fig. S2A where representative results are shown). Among all MBs, NPPA_G MB showed the highest normalized signal-to-background ratio (Fig. 2D). Three MBs (NPPA_D, NPPA_E, and NPPA_F) showing signal-to-background ratios of less than 4 were eliminated from further testing (Table 1).
Fig. 2.
Solution assays of MBs targeting mRNAs associated with cardiac subtypes. A) Predicted secondary structure of NPPA mRNAs and locations of NPPA MBs. B) structure of NPPA_G MB. MB is an oligonucleotide hybridization probe, hairpin shaped molecule with an internally quenched fluorophore whose fluorescence is restored when it binds to a target NPPA mRNA sequence. C) Solution assay of NPPA_G MB. D) Summary of normalized signal-to-background ratio of MBs in the solution assays (mean±STD, N=3 reactions for each MB).
3.3. Evaluation of MBs on NPPA-overexpressing CHO cells
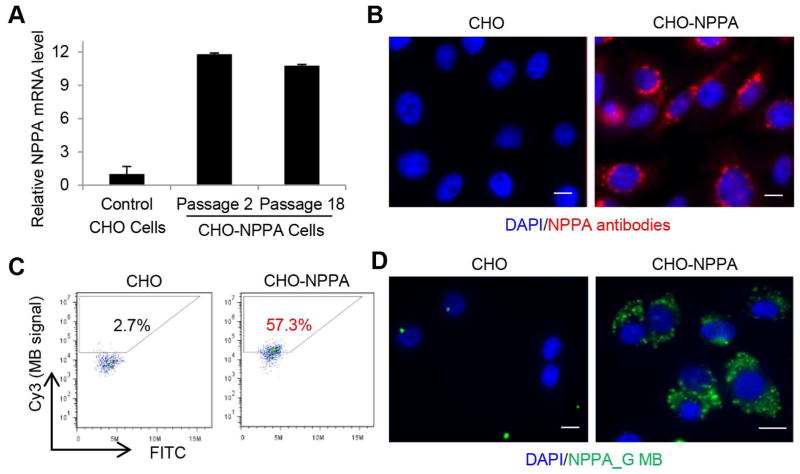
To determine whether the MBs can detect NPPA expression in cell culture, we first constructed a positive control cell line by transfecting CHO cells with a plasmid expressing both NPPA and puromycin resistance genes. After puromycin selection, cells were clonally expanded and verified for expression of NPPA by immunocytochemistry and qRT-PCR (Figs. 3A and 3B). We then delivered NPPA-targeting MBs into CHO-NPPA and control CHO cells respectively using the Amaxa Nucleofector. Of the 7 NPPA MBs examined, NPPA_G gave higher fluorescence signal in CHO-NPPA cells than in control CHO cells according to flow cytometry (Fig. 3C). Under fluorescence microscopy, we observed that the NPPA_G signal originated from cytoplasmic but not nuclear loci in CHO-NPPA cells and was undetectable on control CHO cells (Fig. 3D), suggesting that this MB can detect NPPA expression in CHO-NPPA cells. NPPA_I also produced fluorescence signal in CHO-NPPA cells but not in control CHO cells while other MBs had undetectable signal in CHO-NPPA cells (Table 1). NPPA_G displayed a higher signal in CHO-NPPA cells than NPPA_I did. Fifty-seven percent of the cells were identified as NPPA positive with NPPA_G (Fig. 3C) while only 15% of the cells were identified as NPPA positive with NPPA_I (Fig. S2B). This observation is consistent with the results shown in Fig. 2D where MBs NPPA_G had higher normalized signal-to-background ratio than NPPA_I.
Fig. 3.
Generation of CHO cells overexpressing NPPA and evaluation of NPPA MBs. A) qRT-PCR demonstrating overexpression of NPPA in CHO-NPPA cells. B) Immunocytochemical analysis of NPPA expression in CHO control cells and CHO-NPPA cells. C) Flow cytometry analysis of NPPA_G MB in CHO control cells and CHO-NPPA cells. D) Imaging of cells with beacons demonstrating cytoplasmic localization in CHO-NPPA cells. CHO control cells were used as a negative control. Scale bars, 10 μm.
3.4. Specificity of NPPA MBs in stem cell-derived CMs
To determine whether the MBs can detect NPPA expression in hPSC-CMs, we evaluated them using CM differentiation cultures as positive cells and undifferentiated cells as a negative control. To ascertain that MBs can be delivered into these cells using nucleofection, we first transfected a positive control MB in which both arms of the stem were labeled with a fluorophore FAM without a quencher. After MB delivery, positive signals were detected in both CM differentiated cells and undifferentiated cells. We then similarly examined the NPPA MBs (Table 1) and found that only NPPA_G MB resulted in fluorescence signal in CM differentiated cells but no signal in undifferentiated stem cells. NPPA_H, NPPA_I and NPPA_J MBs showed signals in both hPSC-CMs and undifferentiated cells, while NPPA_A, NPPA_B and NPPA_C MBs did not generate any detectable signal in hPSC-CMs (Table 1).
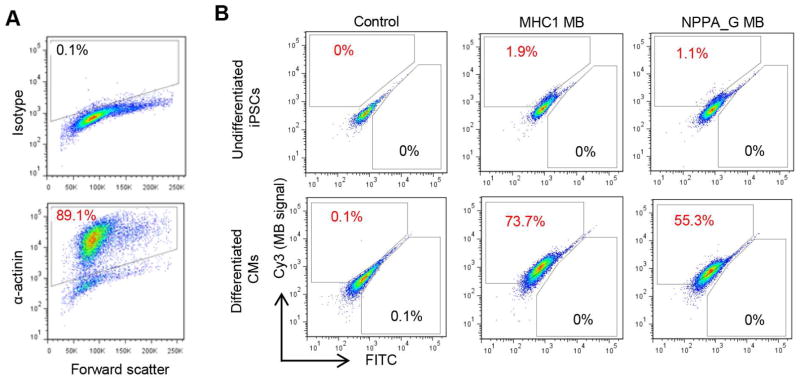
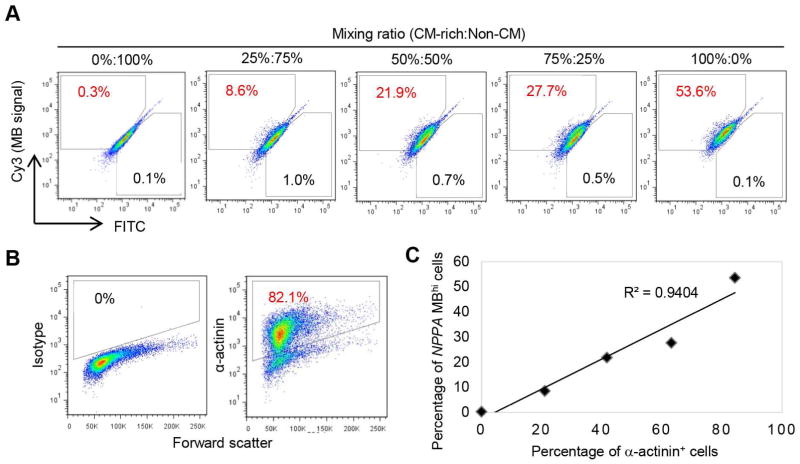
To examine the specificity of NPPA_G MB with hPSC-CM differentiation cultures, we first compared the NPPA_G MB with MHC1 MB, an MB targeting myosin heavy chain beta (MYH7), which was used successfully to detect total CMs (both working-type and nodal-type CMs) [20]. The two MBs were delivered respectively into a differentiation culture that contained ~89% α-actinin (a CM marker) positive cells (Fig. 4A). As expected, MB signals were very low in undifferentiated hPSCs, and there were fewer NPPA_MBhi cells (cells with high fluorescence signal from NPPA_G MB) than MHC1 MBhi cells (~55% vs. ~74%, Fig. 4B) in the differentiation culture. These results suggest that the NPPA_G MB is capable of detecting a subset of the hPSC-CMs. To further establish the specificity of NPPA_G MB, we next prepared cell mixtures containing various ratios of the CM differentiated cells (CM-rich) and cells from a mock differentiation culture without growth factor induction (Non-CM) (Fig. 5A). The CM differentiation culture contained 84% α-actinin positive cells (Fig. 5B), and the mock differentiation culture contained non-beating cells with a different morphology. The 5 cell mixtures therefore contained 0% to 84% CMs, with ratios of CM-rich:Non-CM equal to 0%:100%, 25%:75%, 50%:50%, 75%:25%, and 100%:0%. When detected by NPPA_G MB, these cell mixtures showed 0.3% to 53.6% NPPA MBhi cells (Fig. 5A). The percentage of NPPA MBhi cells correlated very well with the percentage of CMs (α-actinin positive cells) in the cell mixtures (R2=0.9404; Fig. 5C), suggesting that the NPPA_G MB was able to specifically detect a subset of total CMs in the hPSC-CM differentiation cultures.
Fig. 4.
Evaluation of NPPA MBs in iPSC-derived CMs. A) Purity of CMs for cells used in (B) as detected by flow cytometry analysis of α-actinin. Isotype was used as a negative control to establish the gating parameters. B) MB signals in undifferentiated cells vs. iPSC-derived CMs. Cells that were nucleofected without any MBs were used as negative control cells to establish the gating parameters for FITC and Cy3 fluorescence. MB signals were detected by the Cy3 fluorescence.
Fig. 5.
Examination of sensitivity and specificity of NPPA_G MB by mixing CM differentiated cells (CM-rich) and non-CM. A) Detection of NPPA MBhi cells in cell mixtures using different ratios of CM-rich and non-CM. B) Purity of CMs for cells used in (A) as detected by flow cytometry analysis of α-actinin. Isotype was used as a negative control to establish the gating parameters. C) Correlation between MB signal and purity of CMs.
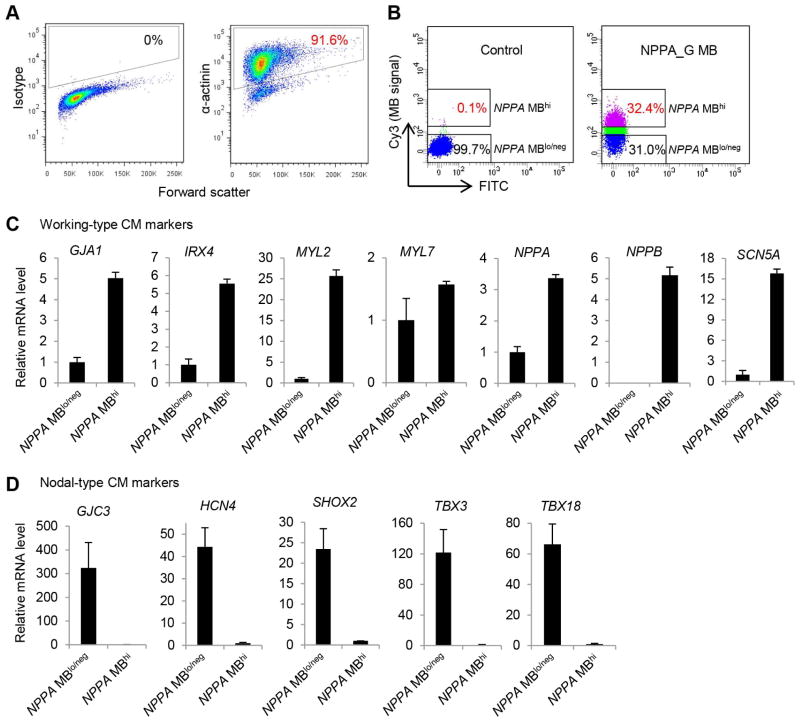
3.5. Cell sorting using NPPA_G MB
We performed live cell sorting of the cells identified based on NPPA_G MB signal using FACS. A differentiation culture containing 91.6% α-actinin positive cells (Fig. 6A) was dissociated and delivered with the NPPA_G MB using nucleofection. Cells with high signal from NPPA_G MB (NPPA MBhi) and those with low MB signal (NPPA MBlo/neg) were collected following the FACS sorting (Fig. 6B). To characterize molecular features of these cells, we examined expression of putative markers for CM subtypes by qRT-PCR (Fig. 6C). Compared with the NPPA MBlo/neg cells, the NPPA MBhi cells showed increased expression of ~3- to 26-fold for 6 genes (out of 7 genes examined) associated with working-type CMs, including GJA1, IRX4, MYL2, NPPA, NPPB, and SCN5A. In addition, compared with the NPPA MBhi cells, the NPPA MBlo/neg cells had ~25- to 300-fold higher expression levels for 5 genes (out of 5 genes examined) associated with nodal-type CMs, including GJC3, HCN4, TBX3, TBX18, and SHOX2, indicating that the NPPA MBlo/neg population contained enriched cells expressing nodal-associated genes. These results strongly suggest that the MB-based approach has the potential as a simple assay to distinguish and separate CM subtypes derived from hPSCs.
Fig. 6.
Expression of genes associated with CM subtypes in NPPA_G MB-isolated cells. A) Purity of CMs used for sorting of NPPA MBhi and NPPA MBlo/neg cells. Differentiation culture was dissociated and analyzed for CM purity by flow cytometry analysis of α-actinin. Isotype was used as a negative control to establish the gating parameters. B) Sorting of live NPPA MBhi and NPPA MBlo/neg cell populations on the basis of MB fluorescence intensity as compared to control cells that were nucleofected without any MBs using FACS Aria and BD FACSDiva software. C) Relative expression levels of genes associated with working-type CMs in NPPA MBhi and NPPA MBlo/neg cells detected by qRT-PCR. D) Relative expression levels of genes associated with nodal-type CMs in NPPA MBhi and NPPA MBlo/neg cells detected by qRT-PCR.
4. Discussion
In this study, we have designed and validated MBs targeting NPPA mRNA a marker known to be associated with human working-type CMs at the early stage. The selection of NPPA as a target for the MB-based approach was also based on our evaluation of relative expression levels of genes associated with CM subtypes during hPSC differentiation. The specificity of the NPPA-targeting MBs was evaluated using different assays: (1) in solution assay when MBs were mixed with perfectly complementary or mismatched synthetic targets, NPPA_G MB showed the highest signal-to-background ratio; (2) using CHO cell lines, NPPA_G MB produced high fluorescence signal in CHO cells overexpressing NPPA but not in the control CHO cells with very low NPPA expression; (3) NPPA_G MB signal enabled the detection of a subset of CMs in hPSC-CMs but not in undifferentiated hPSCs; (4) in hPSC differentiation culture, the percentage of cells detected based on signals from NPPA_G MB was correlated well with the amount of CMs in the mixed cell population; and (5) importantly, two cell populations from hPSC differentiation culture with high and low MB signal separated by FACS showed a gene expression pattern specific for working-type and nodal-type CMs, respectively. Taken together, these results clearly demonstrate the ability of NPPA_G MB to isolate working-type CMs with high specificity.
Our study underscores that the MB-based cell isolation technique requires careful design and evaluation of multiple MBs in the initial screening stage for their sensitivity and specificity. An improved MB technique, such as ratiometric bimolecular beacons (RBMBs), may overcome some of the limitations of conventional MBs for detecting mRNA in live cells [38]. The RBMBs contain a unique structure conjugated with a reference dye, which can significantly reduce false-positive signals and achieve higher sensitivity and specificity.
Overall, the MB-based method can provide a powerful tool for isolating, enriching, and characterizing specific cell types. For the detection of cardiac subtypes, the conventional method is to measure intracellular action potentials from single beating cells using the patch clamp technique, a labor-intensive and time consuming process that requires highly trained electrophysiologists to carry out. Compared with the electrophysiology, the cardiac subtype-specific MBs can be used in a much higher throughput manner. In addition, isolating cells with MBs is a versatile and robust procedure that can be readily applied to many cell types. The NPPA_G MB developed in this study has the potential to be used as a simple and rapid cell-based assay to identify working-type CMs in mixed cell populations and to evaluate the efficiency of CM differentiation into working subtype lineage from hPSCs. A high-throughput and simple assay for the detection of cardiac subtypes will facilitate not only the characterization of cell preparations for therapy or other applications but also basic research on developmental biology in understanding the regulation of subtype specification. Although an electrophysiology approach may still be useful in confirming the specificity of NPPA MBs, the gene expression profiles of the NPPA MBhi cells in comparison with the NPPA MBlo/neg cells strongly suggest that the NPPA_G MB can specifically isolate working-type CMs.
Although much progress has been made in controlling subtype specification in differentiating stem cells [39–41], the generation of pure population of cardiac subtypes is still challenging and a better understanding of molecular regulation of cardiac subtypes is needed in order to generate cells that can meet high standards for clinical and pharmaceutical applications. For example, a homogeneous population of working-type CMs without contamination of nodal-type CMs may be required in order to prevent arrhythmias for cardiac repair. Generation of specific cardiac subtypes will also be advantageous for studying disease mechanisms using patient-specific iPSCs and for improving the accuracy of drug testing using hPSC-CMs. Although cardiac subtypes can also be examined using a reporter gene under the control of a cardiac subtype-specific promoter in genetically modified hPSCs, genetic modification can be a safety hazard in therapeutic applications [42]. In contrast, the MB-based method has the advantage of isolating live cardiac subtypes without genetic modification. Importantly, the resulting cardiac subtypes generated by MB-based sorting can be a great cell source for further investigation of gene expression profiles associated with specific cardiac subtypes, which can in turn lead to the identification of novel gene signatures associated with cardiac subtypes. These MB-isolated cells can also facilitate the study of the specification of cardiac subtypes from hPSCs, leading to the identification of molecular mechanisms governing cardiac subtype specification. Such novel genes and pathways may have implications in cardiac diseases involving these subtypes and can also be exploited to efficiently produce homogeneous cardiac subtypes. For example, identification of a cell surface marker specific to one of the cardiac subtypes may provide a better approach to enrich specific subtype for clinical studies and applications.
5. Conclusion
We have developed an MB-based approach to isolate human working-type CMs from differentiation cultures of hPSCs by targeting NPPA mRNA, and confirmed the specificity of the MB with different assays. The MB-based method provides opportunities for developing high-throughput assays for detecting cardiac subtypes, and can help accelerate studies to understand the regulation of cardiac subtype specification from hPSCs, therefore facilitating future applications of hPSCs in cardiac cell therapy and other applications.
Supplementary Material
Figure S1: Signal-to-background ratio of MBs when MBs were hybridized with mismatched targets in the solution assay.
Figure S2: Evaluation of NPPA_I MB. A) Signal-to-background ratio of NPPA_I MB when the NPPA_I MB was hybridized with its complementary target or mismatched target in the solution assay. B) Detection of NPPA mRNA in CHO control cells and CHO-NPPA cells using NPPA_I MB by flow cytometry.
Table S1. Primers for qRT-PCR
Acknowledgments
This study was supported in part by an NIH-NHLBI grant R21HL118454 (C.X. and G.B.) and in part by the NIH-NHLBI Program of Excellence in Nanotechnology award under Contract No. HHSN268201000043C (G.B. and C.X.). We thank Aaron Rae at the Emory Children’s Pediatric Research Flow Cytometry Core for assistance with flow cytometry analysis and cell sorting.
Appendix
Supplementary Information includes 1 table and 2 supplementary figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–14. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–8. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 6.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93:32–9. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 7.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den Brink S, Hassink R, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–40. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circulation Research. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zwi L, Caspi O, Arbel G, Huber I, Gepstein A, Park IH, et al. Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation. 2009;120:1513–23. doi: 10.1161/CIRCULATIONAHA.109.868885. [DOI] [PubMed] [Google Scholar]
- 10.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–24. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, Police S, Hassanipour M, Li Y, Chen Y, Priest C, et al. Efficient generation and cryopreservation of cardiomyocytes derived from human embryonic stem cells. Regen Med. 2011;6:53–66. doi: 10.2217/rme.10.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–7. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111:1125–36. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1848–57. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–60. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santangelo P, Nitin N, Bao G. Nanostructured probes for RNA detection in living cells. Ann Biomed Eng. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 17.Lennon FE, Hermann CD, Olivares-Navarrete R, Rhee WJ, Schwartz Z, Bao G, et al. Use of molecular beacons to image effects of titanium surface microstructure on beta1 integrin expression in live osteoblast-like cells. Biomaterials. 2010;31:7640–7. doi: 10.1016/j.biomaterials.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee WJ, Bao G. Simultaneous detection of mRNA and protein stem cell markers in live cells. BMC Biotechnol. 2009;9:30. doi: 10.1186/1472-6750-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsson HM, Lee ST, Roccio M, Velluto D, Lutolf MP, Frey P, et al. Sorting live stem cells based on Sox2 mRNA expression. PLoS One. 2012;7:e49874. doi: 10.1371/journal.pone.0049874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ban K, Wile B, Kim S, Park HJ, Byun J, Cho KW, et al. Purification of cardiomyocytes from differentiating pluripotent stem cells using molecular beacons that target cardiomyocyte-specific mRNA. Circulation. 2013;128:1897–909. doi: 10.1161/CIRCULATIONAHA.113.004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nature Biotech. 2001;19:971–4. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 22.Tsourkas A, Behlke MA, Bao G. Structure-function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res. 2002;30:4208–15. doi: 10.1093/nar/gkf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–4. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- 24.Houweling AC, Somi S, Van Den Hoff MJ, Moorman AF, Christoffels VM. Developmental pattern of ANF gene expression reveals a strict localization of cardiac chamber formation in chicken. Anat Rec. 2002;266:93–102. doi: 10.1002/ar.10042. [DOI] [PubMed] [Google Scholar]
- 25.Houweling AC, van Borren MM, Moorman AF, Christoffels VM. Expression and regulation of the atrial natriuretic factor encoding gene Nppa during development and disease. Cardiovasc Res. 2005;67:583–93. doi: 10.1016/j.cardiores.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Horsthuis T, Buermans HP, Brons JF, Verkerk AO, Bakker ML, Wakker V, et al. Gene expression profiling of the forming atrioventricular node using a novel tbx3-based node-specific transgenic reporter. Circ Res. 2009;105:61–9. doi: 10.1161/CIRCRESAHA.108.192443. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Kubalak SW, Minamisawa S, Price RL, Becker KD, Hickey R, et al. Selective requirement of myosin light chain 2v in embryonic heart function. J Biol Chem. 1998;273:1252–6. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- 28.Kubalak SW, Miller-Hance WC, O’Brien TX, Dyson E, Chien KR. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–70. [PubMed] [Google Scholar]
- 29.Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr Patterns. 2003;3:777–83. doi: 10.1016/s1567-133x(03)00125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–8. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 31.Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–99. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 32.Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–97. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 33.Verheijck EE, van Kempen MJ, Veereschild M, Lurvink J, Jongsma HJ, Bouman LN. Electrophysiological features of the mouse sinoatrial node in relation to connexin distribution. Cardiovasc Res. 2001;52:40–50. doi: 10.1016/s0008-6363(01)00364-9. [DOI] [PubMed] [Google Scholar]
- 34.Gros D, Dupays L, Alcolea S, Meysen S, Miquerol L, Theveniau-Ruissy M. Genetically modified mice: tools to decode the functions of connexins in the heart-new models for cardiovascular research. Cardiovasc Res. 2004;62:299–308. doi: 10.1016/j.cardiores.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Kreuzberg MM, Willecke K, Bukauskas FF. Connexin-mediated cardiac impulse propagation: connexin 30.2 slows atrioventricular conduction in mouse heart. Trends Cardiovasc Med. 2006;16:266–72. doi: 10.1016/j.tcm.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gros D, Theveniau-Ruissy M, Bernard M, Calmels T, Kober F, Sohl G, et al. Connexin 30 is expressed in the mouse sino-atrial node and modulates heart rate. Cardiovasc Res. 2010;85:45–55. doi: 10.1093/cvr/cvp280. [DOI] [PubMed] [Google Scholar]
- 37.Xu C, Jiang J, Sottile V, McWhir J, Lebkowski J, Carpenter MK. Immortalized fibroblast-like cells derived from human embryonic stem cells support undifferentiated cell growth. Stem Cells. 2004;22:972–80. doi: 10.1634/stemcells.22-6-972. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Song Y, Shah AY, Lekova V, Raj A, Huang L, et al. Quantitative assessment of ratiometric bimolecular beacons as a tool for imaging single engineered RNA transcripts and measuring gene expression in living cells. Nucleic Acids Res. 2013;41:e152. doi: 10.1093/nar/gkt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA. Neuregulin/ErbB Signaling Regulates Cardiac Subtype Specification in Differentiating Human Embryonic Stem Cells. Circ Res. 2010;107:776–86. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanwar V, Bylund JB, Hu J, Yan J, Walthall JM, Mukherjee A, et al. Gremlin 2 promotes differentiation of embryonic stem cells to atrial fate by activation of the JNK signaling pathway. Stem Cells. 2014;32:1774–88. doi: 10.1002/stem.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–87. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–9. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Signal-to-background ratio of MBs when MBs were hybridized with mismatched targets in the solution assay.
Figure S2: Evaluation of NPPA_I MB. A) Signal-to-background ratio of NPPA_I MB when the NPPA_I MB was hybridized with its complementary target or mismatched target in the solution assay. B) Detection of NPPA mRNA in CHO control cells and CHO-NPPA cells using NPPA_I MB by flow cytometry.
Table S1. Primers for qRT-PCR