Abstract
To better determine the history of modern birds, we performed a genome-scale phylogenetic analysis of 48 species representing all orders of Neoaves using phylogenomic methods created to handle genome-scale data. We recovered a highly resolved tree that confirms previously controversial sister or close relationships. We identified the first divergence in Neoaves, two groups we named Passerea and Columbea, representing independent lineages of diverse and convergently evolved land and water bird species. Among Passerea, we infer the common ancestor of core landbirds to have been an apex predator and confirm independent gains of vocal learning. Among Columbea, we identify pigeons and flamingoes as belonging to sister clades. Even with whole genomes, some of the earliest branches in Neoaves proved challenging to resolve, which was best explained by massive protein-coding sequence convergence and high levels of incomplete lineage sorting that occurred during a rapid radiation after the Cretaceous-Paleogene mass extinction event about 66 million years ago.
The diversification of species is not always gradual but can occur in rapid radiations, especially after major environmental changes (1,2). Paleobiological (3–7) and molecular (8) evidence suggests that such “big bang” radiations occurred for neoavian birds (e.g., songbirds, parrots, pigeons, and others) and placental mammals, representing 95% of extant avian and mammalian species, after the Cretaceous to Paleogene (K-Pg) mass extinction event about 66 million years ago (Ma). However, other nuclear (9–12) and mitochondrial (13,14) DNA studies propose an earlier, more gradual diversification, beginning within the Cretaceous 80 to 125 Ma. This debate is confounded by findings that different data sets (15–19) and analytical methods (20, 21) often yield contrasting species trees. Resolving such timing and phylogenetic relationships is important for comparative genomics, which can inform about human traits and diseases (22).
Recent avian studies based on fragments of 5 [~5000 base pairs (bp) (8)] and 19 [31,000 bp (17)] genes recovered some relationships inferred from morphological data (15, 23) and DNA-DNA hybridization (24), postulated new relationships, and contradicted many others. Consistent with most previous molecular and contemporary morphological studies (15), they divided modern birds (Neornithes) into Palaeognathae (tinamous and flightless ratites), Galloanseres [Galliformes (landfowl) and Anseriformes (waterfowl)], and Neoaves (all other extant birds). Within Neoaves, they proposed several large new clades, including a waterbird clade containing taxa such as penguins, pelicans, and loons, as well as a landbird clade containing woodpeckers, birds of prey, parrots, and songbirds. Despite these efforts, the relationships among the deepest branches within Neoaves; the positions of a number of chronically challenging taxa such as shorebirds, mousebirds, owls, and the enigmatic hoatzin; and the identification of the first divergence of Neoaves [proposed to have given rise to two equally large clades designated Metaves and Coronaves (25)] remain unresolved.
Although some of the findings of the initial multi-gene studies (8, 17) have since been corroborated with larger sequence (26–28) or transposable element (TE) insertion data sets (29), other proposed clades were not supported (27, 28). Furthermore, complete mitochondrial genome analyses recover different relationships (14, 18) and fail to support higher landbird monophyly [but see (30)]. Some of the differences among studies could arise from gene tree incongruence, possibly due to incomplete lineage sorting (ILS) of those genes (29, 31), nucleotide base composition biases (19), differences between data types (32, 33), or insufficient data (34, 35). Thus, it has been difficult to establish confidence in whether specific avian traits—such as vocal learning, predatory behavior, or adaptations to aquatic or terrestrial habitats—reflect single or multiple independent origins and under what ecological conditions these events have occurred.
A common assumption is that whole-genome data will improve phylogenetic reconstructions, due to the complete evolutionary record within each species’ genome and increased statistical power (34,35). We test this hypothesis through phylogenetic analysis on 48 avian genomes we collected or assembled, representing all commonly accepted extant neognath orders (36,37) and two palaeognaths, with several nonavian reptiles and human as outgroups.
Species choice, computational developments, and total evidence nucleotide data set
We chose species representing all neoavian orders according to different classifications [see supplementary materials section 1 (SM1)]. These include groups that have been challenging to place within the avian tree, such as the hoatzin, cuckoo-roller, nightjars, mousebirds, mesites, and seriemas (table S1). We also included species postulated to descend from deep nodes in their orders to break up potentially long branches, such as the kea for parrots (Psittaciformes) and the rifleman for songbirds (Passeriformes). We included vocal-learning species (oscine songbirds, hummingbirds, and parrots), used as models for spoken language in humans (38), and their proposed closest vocal-nonlearning relatives (suboscines, swifts, falcons, and/or cuckoos, depending on the tree) to help resolve differences in trees that lead to different conclusions on their independent gains (15, 17, 18, 26, 29, 38, 39). The resulting data set consisted of 45 avian genomes sequenced in part for this project [48 when including previously published species (40–42)] and three nonavian reptiles [American alligator, green sea turtle, and green anole lizard (43)] (table S1), with details reported in (44–52).
We were confronted with computational challenges not previously encountered in smaller-scale phylogenomic studies. Differently annotated genomes complicated the identification of orthologs, and the size of the data matrix made it impossible to use many standard phylogenetic tools. To address these challenges, we generated a uniform reannotation of the protein-coding genes for all avian genomes based on synteny in chicken and zebra finch (SM2). We found that the SATé iterative alignment program (53, 54) yielded more reliable alignments than other algorithms for large-scale data, and we developed alignment-filtering algorithms to remove unaligned and incorrectly overaligned sequences (SM3). We developed ExaML, a computationally more efficient version of the maximum likelihood program RAxML, for estimating species trees from genome-scale concatenated sequence alignments (SM4) (55–57). We also developed a statistical binning approach that improves multispecies coalescent analyses for handling gene trees with low phylogenetic signal to infer a species tree (SM5) (58). These computationally intensive analyses were conducted on more than 9 supercomputer centers and required the equivalent of >400 years of computing using a single processor (SM3 and SM4).
From these efforts, we identified a high-quality orthologous gene set across avian species, consisting of exons from 8251 syntenic protein-coding genes (~40% of the proteome), introns from 2516 of these genes, and a nonoverlapping set of 3769 ultraconserved elements (UCEs) with ~1000 bp of flanking sequences. This total evidence nucleotide data set comprised ~41.8 million bp (table S3 and SM4), representing ~3.5% of an average avian genome.
A genome-scale avian phylogeny
Total evidence nucleotide tree
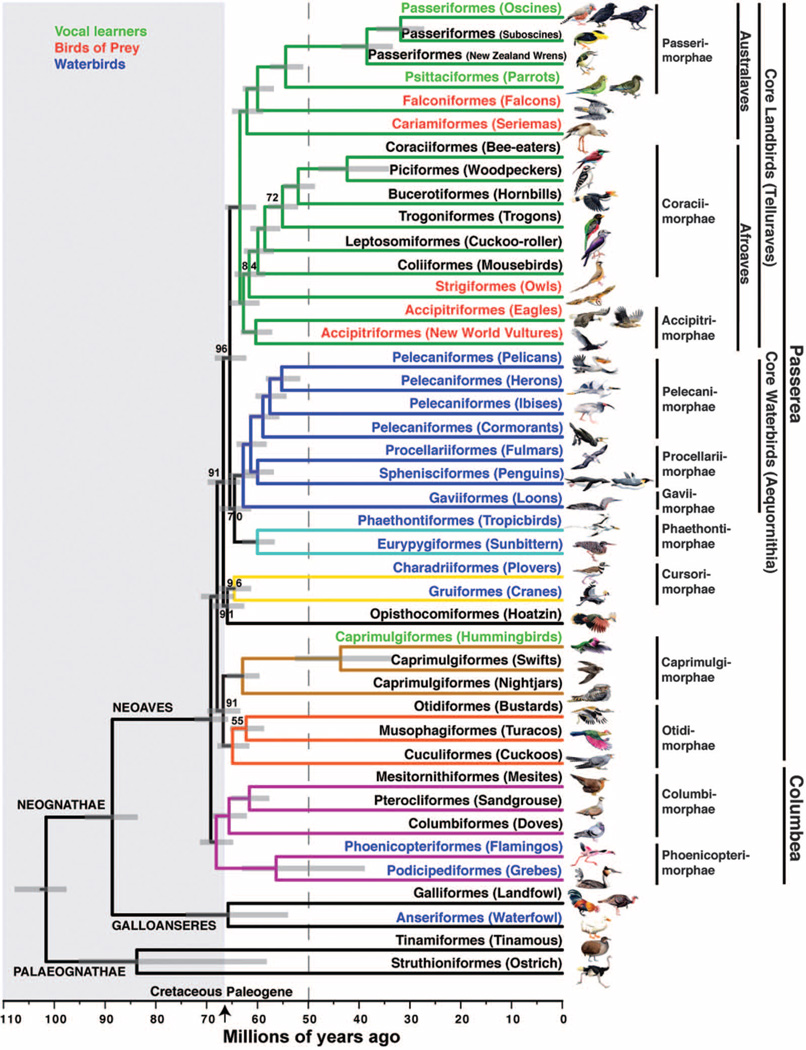
The total evidence nucleotide alignment partitioned by data type (introns, UCEs, and first and second exon codon positions; third positions excluded as described later) analyzed with ExaML under the GTR+GAMMA model of sequence evolution (SM4) resulted in a highly resolved total evidence nucleotide tree (TENT) (Fig. 1 and fig. S1). The three recognized major groupings within extant birds—Palaeognathae, Galloanseres, and Neoaves (the latter two united in the infraclass Neognathae)—were recovered with full (100%) bootstrap support (BS). The tree revealed the first divergence within extant Neoaves, resulting in two fully supported, reciprocally monophyletic sister clades that we named Passerea (after its most speciose group Passeriformes) and Columbea (after its most speciose group Columbiformes) (Fig. 1; see SM6 for rationale of clade names).
Fig. 1. Genome-scale phylogeny of birds.
The dated TENT inferred with ExaML. Branch colors denote well-supported clades in this and other analyses. All BS values are 100% except where noted. Names on branches denote orders (-iformes) and English group terms (in parentheses); drawings are of the specific species sequenced (names in table S1 and fig. S1). Order names are according to (36, 37) (SM6). To the right are superorder (-imorphae) and higher unranked names. In some groups, more than one species was sequenced, and these branches have been collapsed (noncollapsed version in fig. S1). Text color denotes groups of species with broadly shared traits, whether by homology or convergence.The arrow indicates the K-Pg boundary at 66 Ma, with the Cretaceous period shaded at left. The gray dashed line represents the approximate end time (50 Ma) by which nearly all neoavian orders diverged. Horizontal gray bars on each node indicate the 95% credible interval of divergence time in millions of years.
Within Passerea, the TENT strongly confirmed the monophyly of two large closely related clades that we refer to as core landbirds (Telluraves) and core waterbirds (Aequornithia) (8,16,17,27,36,59); we use the term “core” instead of “higher” to prevent interpretation that these groups are more advanced or more recently evolved than other birds. Within core landbirds, we found 100% BS for a previously more weakly supported clade (Australaves) containing seriemas (historically placed in Gruiformes), falcons (historically grouped with other diurnal birds of prey), parrots (historically difficult to place), and Passeriformes and a sister clade (Afroaves) containing Accipitrimorphae birds of prey, owls, mousebirds, woodpeckers, and bee eaters, among others (Fig. 1) (8, 17, 26, 29, 60). Core waterbirds were sister to a fully supported clade (Phaethontimorphae) containing tropicbirds and the sunbittern (Fig. 1) (27, 28). We did not include Phaethontimorphae in the core waterbirds because their relationship had relatively low 70% BS, although their aquatic (tropicbirds) and semiaquatic (sunbittern) lifestyles are consistent with a waterbird grouping, and multiple analyses presented below group them with 100% BS. The TENT also resolved at 100% BS taxa that were previously difficult to place, including uniting cuckoos, turacos, and bustards (Otidimorphae) and placement of the mousebird among core landbirds. The Columbea also had separate land-bird and waterbird groups. These results demonstrate that genome-scale data can help resolve difficult relationships in the tree of life.
Comparisons of TENT with previous studies
The TENT contradicted some relationships in avian phylogenies generated from morphological characters (15), DNA-DNA hybridization (24), and mitochondrial genomes (14,18) (Figs. 2, fig. S2, and Fig. 3A versus fig. S3, A to C). For example, our Falconiformes excluded the previously included eagles and New World vultures (now in Accipitrimorphae); our Coraciiformes was more narrowly delineated and excluded hornbills and cuckoo-rollers; our Pelecaniformes excluded tropicbirds; and our Gruiformes excluded seriemas, bustards, the sunbittern, and mesites. The TENT did not fully support the view based on one gene (β-fibrinogen) that the first divergence in Neoaves resulted in two equally large Metaves and Coronaves radiations (25). However, all Columbea species in the TENT were in the previously defined Metaves, supporting the hypothesis of two parallel radiations of birds with convergent adaptations (25).
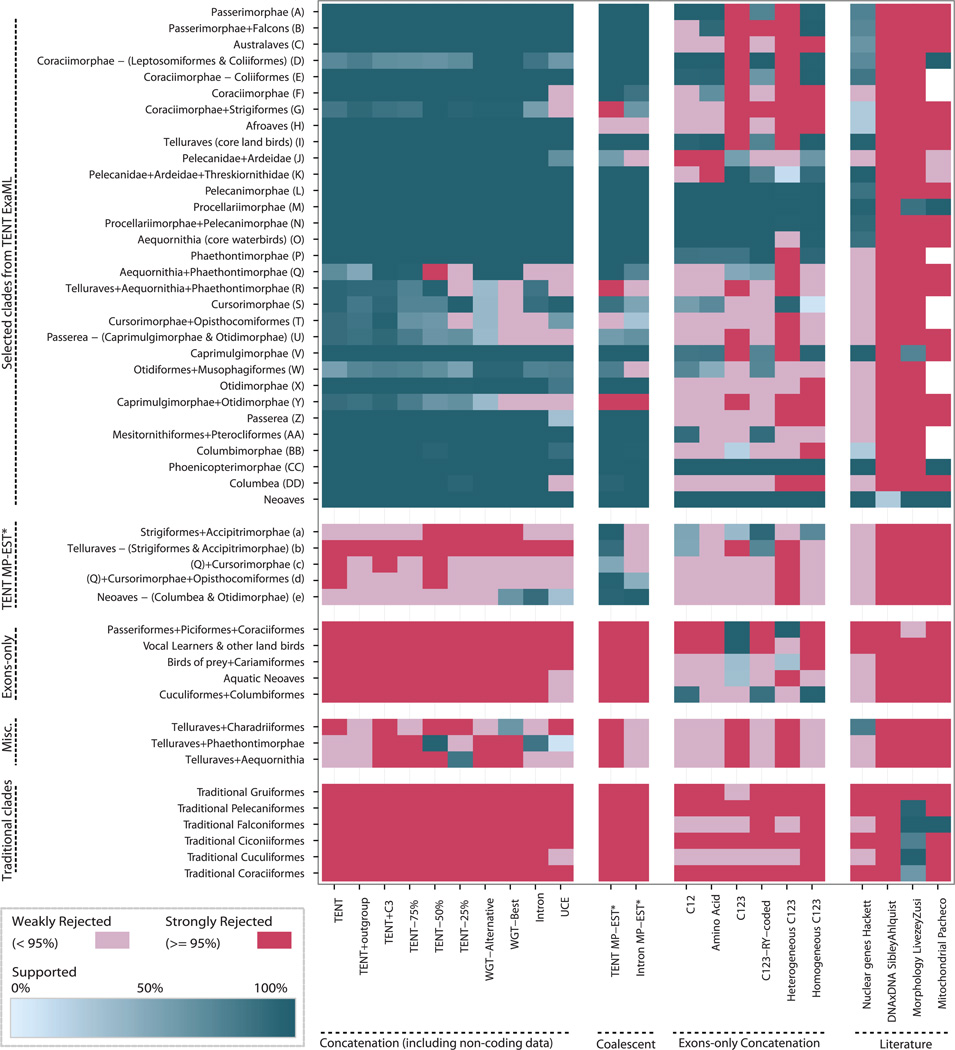
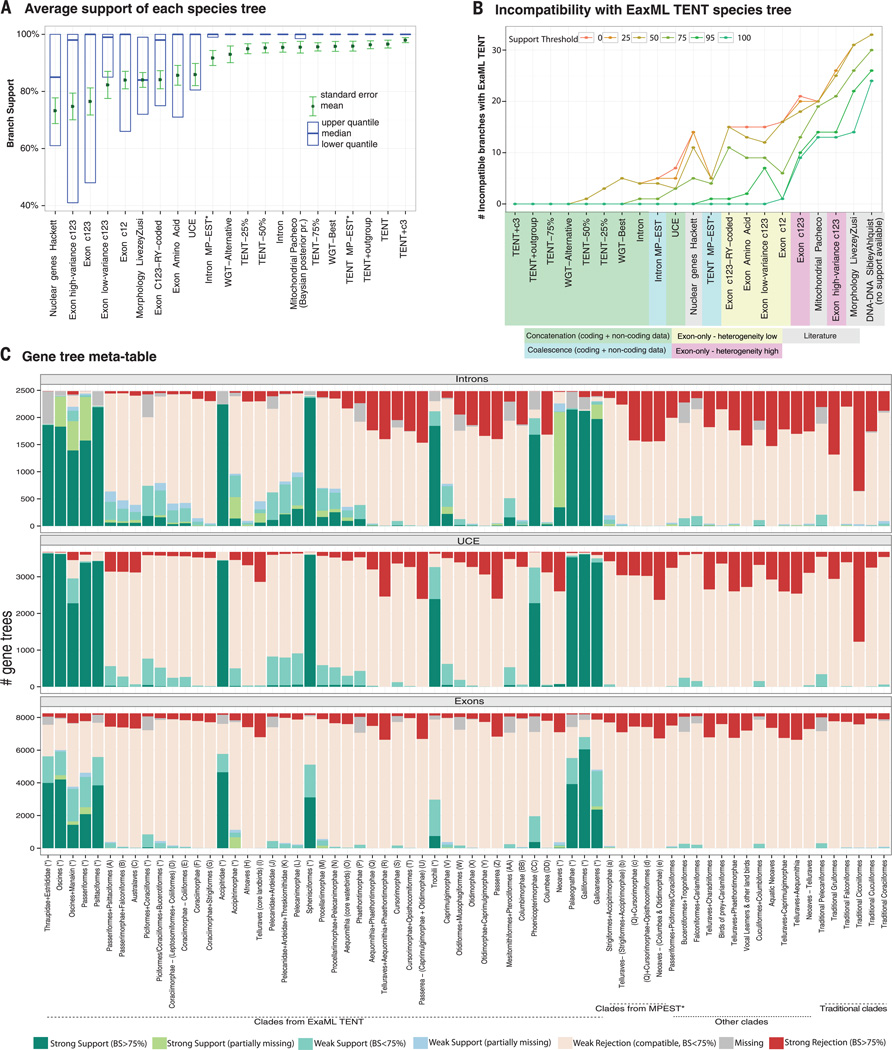
Fig. 2. Metatable analysis of species trees.
Results for different genomic partitions, methods, and data types are consistent with or contradict clades in our TENT ExaML, TENT MP-EST* and exon-only trees and previous studies of morphology (15), DNA-DNA hybridization (24), mitochondrial genes (14), and nuclear genes (17). Letters (A to DD and a to e) denote clade nodes highlighted in Fig. 3, A and B, of the ExaML and MP-EST* TENT trees. Each column represents a species tree; each row represents a potential clade. Blue-green signifies the monophyly of a clade, and shades show the level of its BS (0 to 100%). Red, rejection of a clade; white, missing data. We used a 95% cut-off (instead of a standard 75%) for strong rejection due to higher support values with genome-scale data. The threshold for the mitochondrial study was set to 99% due to Bayesian posterior probabilities yielding higher values than BS. An expanded metatable showing partitioned ExaML, unbinned MP-EST, and additional codon tree analyses is shown in fig. S2.
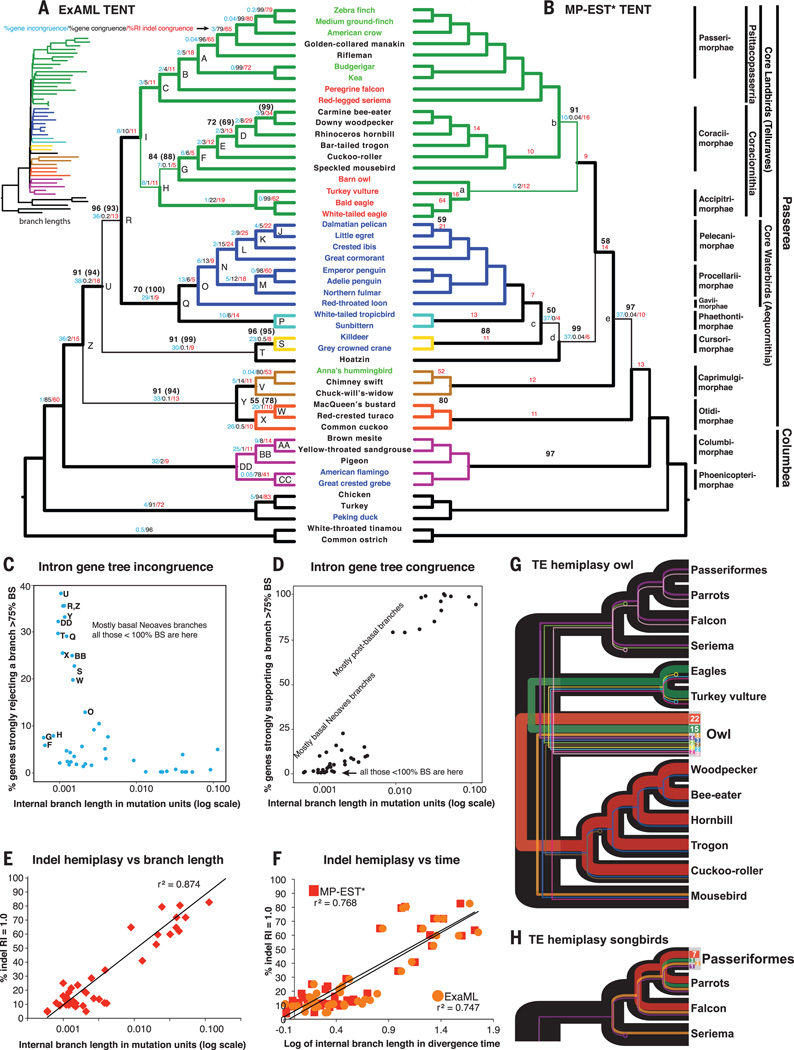
Fig. 3. Evidence of ILS.
(A) Cladogram of ExaML TENT avian species tree, annotated for nodes from Fig. 2 (letters), for branches with less than 100% BS without and with (parentheses) third codon positions, for strong (>75% BS) intron gene tree incongruence and congruence, and for indel congruence on all branches (except the root). Thin branch lines represent those not present in the MP-EST* TENT of (B). (Inset) ExaML branch lengths in substitution units (expanded view in fig. S7). Color coding of branches and species is as in Fig. 1. (B) Cladogram of MP-EST* TENT species tree, annotated similarly as in the ExaML TENT in (A). Thin branch lines represent those not present in the ExaML TENT of (A). (C) Percent of intron gene trees rejecting (≥75% BS) branches in the ExaML TENT species tree relative to branch lengths. Letters denote nodes in (A) that either have less than 100% support or are different from the MP-EST* TENT in (B). (D) Percent of intron gene trees supporting (≥75% BS) branches in the ExaML TENT species tree relative to branch lengths. (E) Indel hemiplasy [the inverse of percent of retention index (RI) = 1.0 indels that support the branch; see SM9] correlated with ExaML TENT branch length (log transformed). r2, correlation coefficient. (F) Indel hemiplasy correlated with ExaML and MP-EST TENT internal branch divergence times in millions of years (log transformed). Plotting with internal branch times was necessary to compare both trees (SM9). (G) TE hemiplasy with owls among the core landbirds. Line color, shared TE tree topology; line thickness, relative proportion of TEs that support a specific topology (total numbers shown in the owl node). Circles at end of lines indicate loss of the TE allele in that species after ILS, as the sequence assembly contains an empty TE insertion site (SM10). Only topologies with two or more TEs are shown. (H) TE hemiplasy with songbirds among the core landbirds.
The TENT was most congruent with past (8,17) and more recent (27,28) smaller-scale multilocus nuclear trees (Figs. 2 and 3A and fig. S3D), although most congruence was limited to the core landbirds and core waterbirds. Within the former, we recovered Australaves and Afroaves (60), although with a different branching ordering in our tree; our taxon sampling is insufficient to address the biogeographic justification of their names. The TENT recovered a number of groups not present in these previous trees, and even for those present, the TENT had higher BS (Fig. 2). Absence of nonavian outgroups in our TENT above was not responsible for variation with past studies because we recovered the same topology when including outgroups (Fig. 2 and fig. S4, A and B), despite the outgroups having only ~30% orthologous sequences in the TENT alignment (e.g., fig. S21; SM3).
More data are responsible for resolving early branches of the tree
Despite the many fully supported (100% BS) relationships in the TENT, lower support was obtained for 9 of the 45 internal branches (although still within the high 70 to 96% BS range). Almost all were at deep divergences within the Neoaves, after the Columbea and Passerea divergence and before the ordinal divergences (Fig. 1 and fig. S1). The monophyly of each of the superorders, however, had 100% BS. The presence of these lower BS values is in contrast to the expectation that genome-scale alignments would result in complete phylogenetic resolution (34, 35, 61).
However, consistent with this hypothesis, we found that most relationships that had less than 100% BS with the full TENT data exhibited a steady increase in support with an increase in random subsets of the TENT data (Fig. 2 and fig. S5). The placement of the Phaethontimorphae (sunbittern and tropicbirds) and hoatzin changed when smaller (25 to 50%) amounts of data were analyzed. Further exploring data amount, we used the assembled ~1.1-billion-bp chicken genome (40) as a reference to generate a 322-million-bp MULTIZ alignment of putatively orthologous genome regions across all species, comprising ~30% of an average assembled avian genome and corresponding to the maximal orthologous sequence obtainable across all orders under our homology criteria (SM3). We ran ExaML on the alignment for ~42 CPU years, with 20 maximum likelihood searches on distinct starting trees and 50 bootstrap replicates before reaching our convergence criterion (SM4) on a whole-genome tree (WGT). Notably, all runs resulted in one of two trees: one identical to the TENT topology (fig. S4C) and a second almost identical to the TENT (fig. S4D). This latter tree differed from the TENT by local shifts in five branches, all clades that had less than 100% BS in the TENT (fig. S4, A and D). Given the relatively minor differences between the second WGT and the TENT, together they corroborate the majority of relationships in the avian tree of life. Although the WGT has more data (table S3), the orthology (SM2) and alignment (SM3) qualities are higher for the TENT, and thus we consider the TENT more reliable.
Noncoding data contribute more to the TENT topology
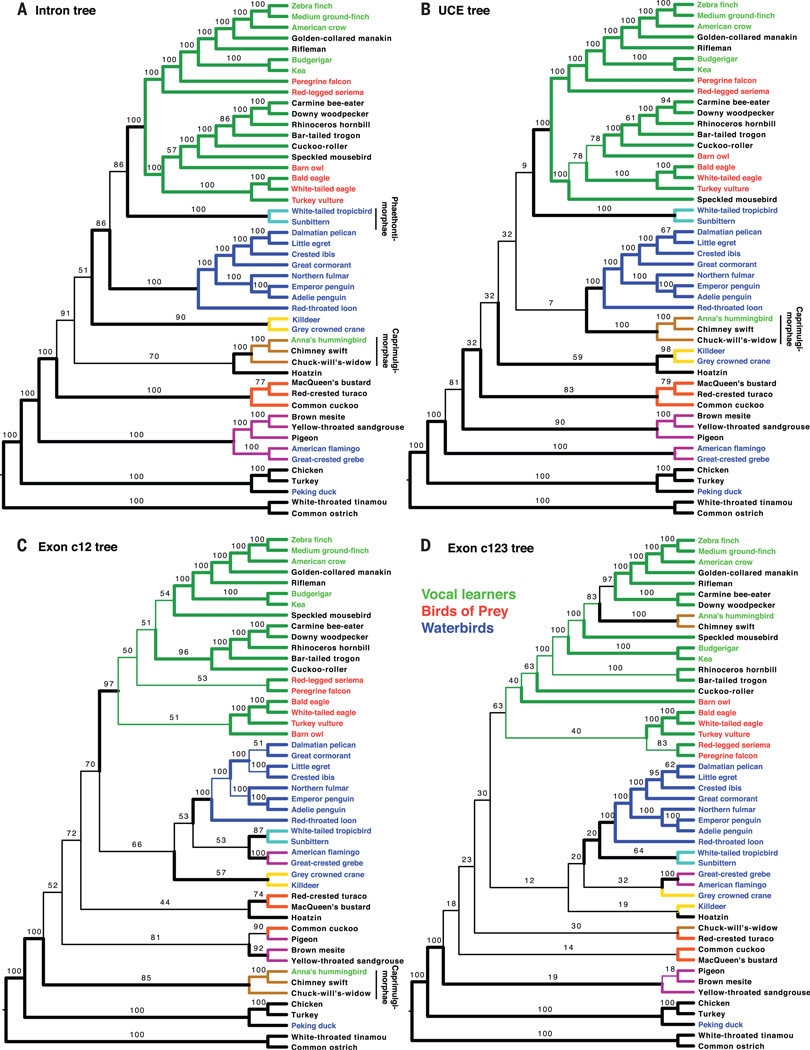
We sought to determine if different genomic partitions contribute differently to the TENT and found that ExaML trees using only introns or UCEs from the TENT data were largely congruent with the TENT and WGTs for branches that had strong support (BS > 75%) in the intron and UCE trees (Figs. 2; 4, A and B; and 5B). However, the intron tree, and even more so the UCE tree, had lower resolution than the TENT (Fig. 5A), mostly on deep branches (Fig. 4, A and B), consistent with fewer data leading to lower resolution on deeper branches. For the intron tree, some lower-resolution branches had local shifts, but they matched those found in the second WGT or the 25 to 75% data subsets of the TENT; an exception was Phaethontimorphae, which moved from being sister to core waterbirds with 70% BS in the TENT (but 100% BS in the WGTs) to sister to core landbirds with 86% BS in the intron tree. For the UCE tree, the lower-resolution, deep branches had more distant shifts. Trees created from analysis of the first and second codon positions (exon c12) of the TENT data also had lower levels of BS (~39 to 64%) but with more topological differences on the deep branches (Figs. 2, 4C, and 5A), yet all but one of the fully resolved relationships (local difference in egret + ibis + pelican) were congruent with the TENT (Fig. 5B).
Fig. 4. Species trees inferred from concatenation of different genomic partitions.
(A) Intron tree. (B) UCE tree. (C) Exon c12 tree. (D) Exon c123 tree. The tree with the highest likelihood for each ExaML analysis is shown. Color coding of branches and species is as in Fig. 1 and fig. S1. Thick branches denote those present in the ExaML TENT. Numbers give the percent of BS.
Fig. 5. Comparisons of total support among species trees and gene trees.
(A) Average BS across all branches of species trees from varying input data as in Fig. 2, ordered left to right from lowest to highest values. (B) Numbers of incompatible branches (out of 45 internal), at different support thresholds, with the ExaML TENT tree, ordered left to right from most to least compatible (expanded analysis in fig. S6). (C) Analyses of intron, exon, and UCE gene tree congruence and incongruence with nodes in the ExaML TENT, MP-EST* TENT, and other species trees. Names and letters for clades are as in Figs. 2 and 3. “Missing” denotes the case in which an ortholog is not present for any taxa or is present for only one taxon, and hence monophyly cannot be determined. “Partially missing” indicates the case in which some taxa are missing but at least two of the taxa are present, and thus we can still categorize it as either monophyletic or not. For further details, see SM7.
These findings demonstrate that noncoding intron sequences lend greater support for the TENT than the protein-coding and UCE sequences, consistent with intron sequences having a higher rate of evolution (SM4) and thus greater phylogenetic signal. These differences are not merely due to shorter alignments of the exon and UCE sequences, because each accounted for ~25% of the TENT data, similar in sequence length to the random 25% subset of the TENT with introns (table S3) that produced a tree with a higher average BS and a topology closer to the full TENT (Fig. 5A and fig. S5D).
Incomplete lineage sorting and impact on deep branches
Deeper branches exhibit higher gene tree incongruence
We next investigated ILS, a population-level process that results in incongruence between gene trees and the species tree (62). Consistent with conditions that could lead to ILS (63), the TENT had a wall of many (25 of 45; ~55%) very short internal branches (0.0006 to 0.002 substitutions per site), almost all at deep divergences within Neoaves (Fig. 3A, inset, and fig. S7). Indeed, all nine branches with <100% BS were among the shortest in the TENT (fig. S8), many in succession, suggesting that reduced BS could be related to conflict among gene trees.
To test this hypothesis, we compared the distribution of gene trees that have strong conflict (>75% BS) with branches of the ExaML TENT. We focused on introns because they had greater gene tree resolution (higher average BS) than exons or UCEs (fig. S24 and SM4). The 2485 introns with orthologs available in the two outgroup Palaeognathae species ranged from exhibiting no conflict to exhibiting considerable conflict (up to 950 genes or 38%) for some branches of the TENT (Fig. 3A, blue numbers, and Fig. 5C). The percentage of gene tree conflict was successively higher for the shorter and deeper branches of the TENT (Fig. 3A), particularly those with <100% BS (e.g., branches R, U, and Z in Fig. 3, A and C). Conversely, these short branches had fewer (0 to 20%) intron gene trees supporting them at high (>75%) BS levels (Fig. 3A, black numbers, and Fig. 3D). These findings suggest that ILS could have affected the inferred relationships of some of the deep branches of Neoaves in the concatenated tree analysis.
Multispecies coalescent approach infers a species tree similar to the TENT
To determine if ILS affected the concatenated tree analyses, we explored whether a multispecies coalescent model leads to a different tree topology. Multispecies coalescent methods estimate the species tree from a set of gene trees and are statistically consistent when discordance among gene trees results from ILS (64,65). However, the inferred species tree can have low resolution (BS) and be less topologically accurate when the input gene trees are poorly resolved (33, 66), a problem that many of our genes faced (SM4). Thus, we developed a statistical binning technique that first groups genes into sets based on phylogenetic similarities, from each set estimates a supergene tree, and uses them in the maximum pseudolikelihood estimation of the species tree (MP-EST) multispecies coalescent approach (67) to infer a species tree (SM5) (58).
This approach produced more accurate estimated species trees compared with MP-EST applied to unbinned gene data sets that have low phylogenetic signal (i.e., figs. S2 and S9; SM7) (58). It produced a highly resolved binned MP-EST (MP-EST*) TENT tree that was highly congruent with the ExaML TENT (Fig. 3, A and B). There were only local shifts of five clades, nearly all on lower-support (<100% BS) branches of the ExaML and MP-EST* TENTs (Fig. 3, A and B). The monophyly of Afroaves was the only case of 100% BS in the ExaML TENT that conflicted with the MP-EST* TENT tree and involved a local shift in the owl with mousebirds and Accipitrimorphae birds of prey. Two branches with <100% BS in the ExaML TENT increased to 100% in the MP-EST* TENT, including Phaethontimorphae with core waterbirds. The intron trees supported some branches more in the ExaML and some more in the MP-EST* TENT (Fig. 3, A and B). Nevertheless, the overall topology of both trees was very similar, including the basal Columbea and Passerea divergence.
All estimates of gene trees differ from our candidate species trees
No single intron, exon, or UCE locus from our TENT data set had an estimated topology identical to the ExaML TENT or MP-EST* TENT (fig. S10, A and B). The top three loci (all introns) with the closest inferred topologies differed from the two versions of the TENT on more than 20 to 30% of their branches. Average topological distance with the ExaML species tree was 63% for the introns, 66% for the UCEs, and 80% for the exons. To test whether our total evidence data set missed some genes with the TENT topologies, we constructed a more comprehensive collection of genes trees with phylomeDB, which assigns orthology using maximum likelihood analyses (http://phylomedb.org) [see SM8 and (68)]. For ~13,000 (low-coverage genomes) to ~18,000 (high-coverage genomes) annotated genes across avian species (44), phylomeDB inferred orthologs for 94.58% of them and these agreed with the synteny-based orthology of the 8251 proteincoding genes of the TENT by 93%. This more complete set of protein-coding genes still did not have a single estimated gene tree that was fully congruent with the ExaML or MP-EST* TENT trees (fig. S10, C and D), and there was overall low congruence with the species trees (http://tol.cgenomics.org/birds_v1) (fig. S11, A and B). The conflicting nodes largely reflected branches with low statistical support (approximate likelihood ratio test < 0.95), which primarily corresponded to the short successive deep branches of Neoaves. These findings can be explained by both a low amount of phylogenetic signal in individual loci (figs. S24 to S26 and SM4) and a high amount of ILS during the neoavian radiation.
Indels suggest a high degree of ILS at the earliest branches of the Neoaves tree
We further assessed ILS using insertions and deletions (indels) (69), because they have less homoplasy (convergence) than single nucleotides (SM9), and unlike gene trees, indels can be examined as discrete characters mapped to a reference tree without the added inference of constructing trees from them. We scored 5.7 million indels from the TENT alignment, of which 24% were shared by two or more taxa (table S3). We found indel incongruence on all branches of the ExaML TENT, as measured inversely by the percent of the indel characters uniquely defining each branch (Fig. 3A, red numbers; SM9). Like the gene trees, there appeared to be a successive decrease in the percentage of indels that supported deeper branches of each major clade (Fig. 3A). Most branches with the highest levels of indel incongruence belonged to the shortest and deepest ones that made local shifts in analyses, with the two branches joining mousebirds and owls exhibiting the highest indel incongruence and the shortest internal branch lengths in the ExaML TENT (Fig. 3A and fig. S7). Consistent with these findings, indel incongruence was inversely correlated with internal branch length, and branch length explained 87% (r2) of the variation in the percentage of nonhomoplasious indels defining each branch (Fig. 3E). The correlation of indel incongruence versus branch time was similar for both ExaML and MP-EST* TENT trees (Fig. 3F).
Indel incongruence is not due to the indels supporting another species tree, as applying ExaML on indels from the total evidence alignment as binary data produced a total evidence indel tree that was largely congruent with the ExaML TENT and MP-EST* TENT for all but one node with a local shift of pigeon within Columbea (fig. S12). Homoplasy due to convergence is thought to be positively correlated with branch length [i.e., long branch attraction (70)]. The only known source of incongruence that is inversely correlated with internal branch length is hemiplasy (differential inheritance of polymorphic alleles) (64, 71). Because hemiplasy is a hallmark of ILS and 87% of the variation in indel incongruence is explained by branch length, our indel findings suggest high levels of ILS during the basal radiation of Neoaves, with comparable support for the ExaML or MP-EST* versions of the TENT.
Transposable elements with higher ILS in the deepest branch of core landbirds with owls
We tested for a signature of ILS in TE insertions, which have extremely low homoplasy because independent insertions into the same location in a genome are rare (SM10) (72, 73). We focused on the owl because its position exhibited one of the strongest incongruences among the species tree results. Of 3671 barn owl long terminal repeat TE insertion loci orthologous in all of the bird genomes, 61 were informative for owls among core landbirds and showed two dominant exclusive TE topologies: (i) an owl + Accipitrimorphae topology, as seen in the MP-EST* TENT; and (ii) an owl + Coraciimorphae topology that excludes mousebird, as seen in the UCE tree (Fig. 3G compared to Figs. 3B and 4B). Nine other topologies had fewer markers supporting them. In contrast, for 25 informative TEs of Neoaves in (29), 13 were informative for Australaves, and of these, 3 were exclusive for Passeriformes + parrots, 7 for Passeriformes + parrots + falcons, and 2 for the latter group plus seriemas, with no alternative topologies for the first two groups (Fig. 3H). If the passeriform TE insertions exhibited a similar mixture of alternative distributions as for the owl, just 10 markers would result in conflicting distributions (4 with one, 3 with another, and 3 for the remaining topologies) instead of a conflict-free topology. Although this analysis is limited to specific taxa, it suggests higher ILS near the deepest branches of Afroaves involving the owl, consistent with the branch length, gene tree, and indel findings.
Overall, these results reveal considerable ILS during the neoavian radiation and that, even with genome-scale data, ILS may affect the inference of small local relationships in the deep branches of the species tree that have long been more challenging to resolve. However, ILS does not affect the majority of other phylogenetic relationships we found using genome-scale data.
Protein-coding data resolve avian phylogeny poorly but reflect life history traits
Codon positions of protein-coding genes and life history relationships
We investigated sources of lower resolution and incongruence for the tree based on proteincoding sequences (Fig. 4C). This is crucial for phylogenomic inference, as many studies [including transcriptome analyses (19, 74)] use only protein-coding genes to infer species trees. We found that ExaML analyses with either all (c123; Fig. 4D) or individual codon positions (c1, c2, c3; fig. S13, A to C) produced trees with lower BS (Fig. 5A) and greater differences in topologies (Fig. 2 and fig. S2) compared with noncoding data and coding + noncoding combined. The differences between coding versus noncoding trees were not solely due to shorter sequence length of the coding data, because the full coding data set (13.3 million bp for c123) produced a tree with fully supported (100% BS) relationships that were incongruent with those fully supported in the intron (19.3 million bp), TENT (37.4 million bp without the third codon position), and WGT (322.1 million bp) (Figs. 2 and 5B, and table S3). Surprisingly, the c123 topology associated species more with life history traits than the TENT topology. This included a strongly supported clade (100% BS on most branches) that comprised the three groups of vocal learners (parrots, songbirds, and hummingbirds) and most of the nonpredatory core landbirds, a monophyletic clade of diurnal birds of prey and seriemas (albeit with low 40% BS), and a monophyletic clade of all aquatic and semiaquatic species of Passerea and Columbea (also with low 20% BS) (Fig. 4D). Partitioning the data to account for possible differences in evolutionary rates among genes (SM4) did not result in a tree more similar to the TENT, but instead in a tree with increased support for monophyletic groupings of species with these broadly shared traits (fig. S14C). The c1, c2, and amino acid tree topologies (fig. S13, A, B, and D) were more congruent with the c12 tree (Figs. 2 and 4C), consistent with these two codon positions largely specifying amino acid identity. In contrast, the c3 tree was very similar to the c123 tree but with higher BS (63 to 82%) for similar trait groupings; it moreover brought all basal neoavian landbirds together as sister to all neoavian aquatic/semiaquatic species (figs. S2 and S13C). Most individual gene trees show weak to strong rejection of these relationships (Fig. 5C).
As expected (19), the third codon position exhibited greater base composition variation among species than the other codon positions and even other genomic partitions (fig. S15A). Although all codon positions violated the stationarity assumption in the GTR + GAMMA model of sequence evolution, the third codon position exhibited a much stronger violation (fig. S15B). Reducing this variation by RY recoding of purines (R) and pyrimidines (Y) on the third codon position (SM4) made the c123 tree topology more similar to the c12 topology (Fig. 2 and fig. S14D). These results demonstrate that the third codon position exerts a strong influence on the protein-coding–tree topology, overriding signals from the first and second codon positions. They also suggest that a signal in the third codon position could also be associated with convergent life history traits.
Heterogeneous protein-coding genes associated with life history traits
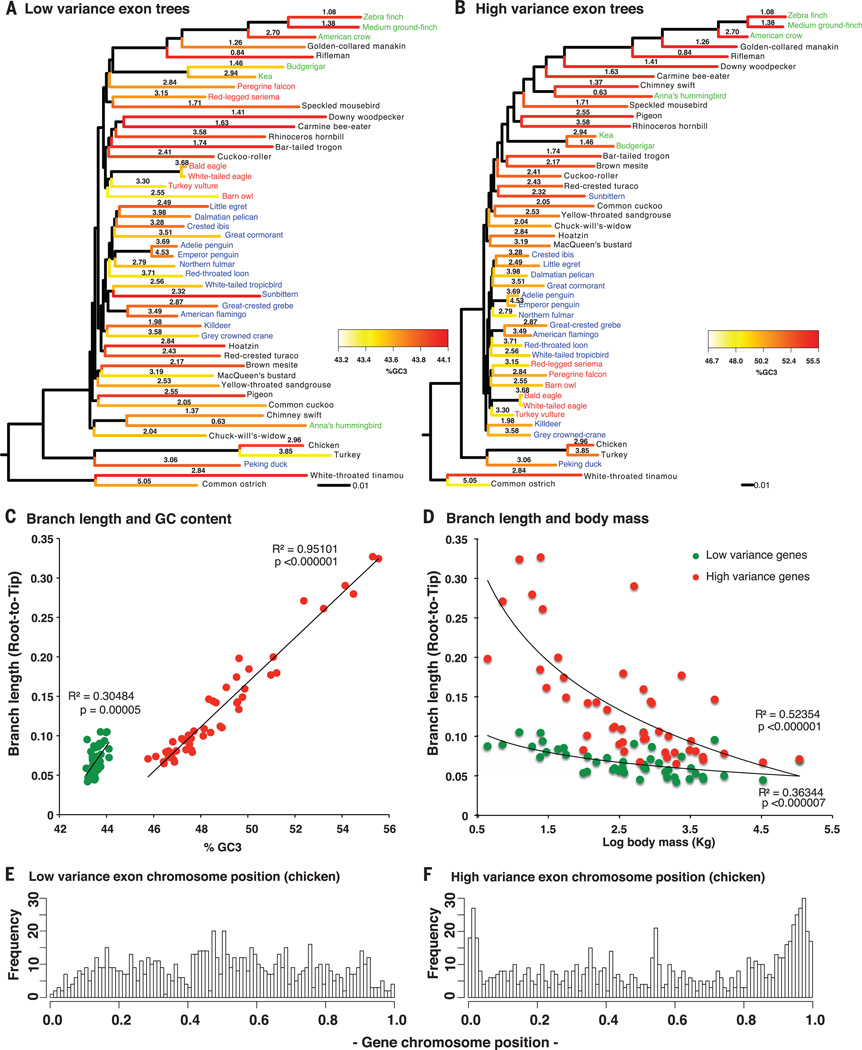
We further investigated the source of the conflict in the protein-coding genes (SM11) and found that trees using all codon positions from the 10% most compositionally homogeneous (low-variance) exons (n = 830) were most congruent with the c12 tree and, thus, more similar to the TENT than to the c123 tree (Figs. 2 and 6A; cladograms in fig. S16, A to C). Conversely, trees using all codon positions from the 10% most compositionally heterogeneous (high-variance) genes (n = 830) were more congruent with the exon c123 and c3 trees (Figs. 2 and 6B and fig. S16, B and D). The branch lengths of the high-variance exon tree showed a strong positive correlation with GC content and a negative correlation with the average body mass of species, seen at a much lesser magnitude in the low-variance exon tree (Fig. 6, A to D). The correlations for the high-variance genes were also strongest on the third codon position (fig. S17, A and B) (75, 76). In addition, the genomic positions of the high-variance genes were skewed toward the ends of the chromosomes, whereas the positions of the low-variance genes were skewed toward the center (Fig. 6, E and F, and fig. S17, C and D). Although the available introns of these genes had significant correlations among GC content and body mass and among GC content and chromosome position, they exhibited less heterogeneity overall (fig. S17, A to D) and yielded trees that were much more congruent with each other and with the TENT (figs. S2 and S17, E and F). An ExaML TENT tree that included the third codon position (TENT + c3) was identical in topology to the ExaML TENT without the third codon position and had increased support for six of the nine branches that had less than 100% BS (fig. S1 versus fig. S18, also Figs. 3A and 5A).
Fig. 6. Life history incongruence in protein-coding trees.
(A) Species tree inferred from low-base composition variance exons (n = 830 genes) graphed with branch length, third codon position GC (GC3) content (heatmap), and log of body mass (numbers on branches). (B) Species tree inferred from high-base composition variance exons (n = 830 genes), graphed similarly as in (A). The %GC3 scale is higher and ~10 times wider for the high-variance genes, and the branch lengths are ~3 times longer [black scales at the bottom of (A) and (B)]. Color coding of species’ names is as in Fig. 1. Cladograms of trees in (A) and (B) are in figs. S16, A and B. (C and D) Correlations of branch length with GC content (C) and body mass (D) of the low-variance and high-variance exons. Correlations were still significant after independent contrast analyses for phylogenetic relationships (SM11). (E and F) Relative chromosome positions of the low-variance (E) and high-variance (F) exons normalized between 0 and 1 for all chicken chromosomes and separated into 100 bins (bars). The height of each bar represents the number of genes in that specific relative location. The two distributions in (E) and (F) are significantly different (P < 2.2 × 10−16, Wilcoxon rank sum test on grouped values). For further details, see SM11.
These results suggest that in the context of protein-coding data only, high-base compositional heterogeneity and life history have a strong impact on incongruence with the species tree, and thus are not suitable for generating a highly resolved phylogeny. However, in the context of large amounts of noncoding genomic data, the phylogenomic signal in the exon data adds support to the species tree.
Dating the radiation of Neoaves
The generation of a well-resolved avian phylogeny allowed us to address the timing of avian diversification. To estimate the avian timetree with genomic-scale data, we used first and second codon positions from 1156 clock-like exon genes (which do not strongly exhibit the above protein-coding compositional bias), calibrated with 19 conservatively chosen avian fossils (plus nonavian outgroups) as minimum bounds for lineage ages (with a maximum-bound age constraint of 99.6 Ma for Neornithes), in a Bayesian autocorrelated relaxed clock method using MCMCTREE (77) on the fixed ExaML TENT topology (SM12).
Our results suggest that after the Palaeogna-thae and Neognathae divergence about 100 Ma in the Late Cretaceous, the Palaeognathae diverged into their two stem lineages [ostrich and tinamous (11,78)] about 84 Ma, and the Neognathae diverged into their stem lineages (Galloanseres and Neoaves) about 88 Ma (Fig. 1). Although the 95% credibility interval for the ostrich-tinamou divergence is broad, its lower bound is consistent with the fossil record (79). In contrast, both the earliest divergence within Galloanseres and an explosive diversification within Neoaves were dated to occur around the K-Pg boundary, with 95% credibility intervals spanning 6.5 million years, on average. In particular, the most basal divergences within Neoaves (Columbea, Passerea, and two more) occurred before the K-Pg transition (67 to 69 Ma) and all others after, with nearly all ordinal divergences completed by 50 Ma (Fig. 1, dashed line). The estimated age for the basal split of Passeriformes, representing ~60% of all living ~10,400 avian species, was around 39 Ma These divergence times conflict with some previous studies based on nuclear (9–12) and mitochondrial (13,14) DNA but are consistent with the fossil record (80), including the identification of Vegavis iaai, a very Late Cretaceous (66 to 68 Ma) stem-anseriform fossil (80, 81), and the dearth of verifiable Neoaves fossils in the Late Cretaceous (5). These findings were similar regardless of the specific tree from this study we dated or whether we used a later minimum age (86.5 Ma) for Neornithes (table S16; more discussion on dating in SM12).
Discussion
Our study is an example of the extraordinary amount of genomic sequence data required to produce a highly supported phylogeny spanning a rapid radiation. The conflict we observe with other data types (14,15, 24) can no longer be considered to be due to error from smaller amounts of sequence data (8,17) nor to differences in concatenation versus coalescence methods (27, 28). The absence of a single gene tree identical to the avian species tree is consistent with studies in yeast (82), indicating that phylogenetic studies based on one or several genes, especially for rapid radiations, will probably be insufficient. The major sources of the gene tree incongruence we find are low-resolution gene trees and substantial ILS during the rapid radiation. It is possible that some of the deep branches of the species tree are in the anomaly zone (63), although the gene tree support is not high enough to confidently test this idea. It is also possible that some gene and local species tree incongruence could reflect ancient hybridization during the radiation, but distinguishing between this and other sources of hemiplasy (83) would require more complete assemblies, genes without missing data across species, and development of new methods (84). Finally, it is also possible that insufficient taxon sampling contributed to the local species tree incongruence, known to lead to long-branch attraction (70). We did seek to break up some long branches, specifically within core land-birds and core waterbirds. However, the very large-scale data collection for this study made it necessary to prioritize species for specific parts of the tree. Moreover, the potential to add taxa that will break up long branches is limited for a number of groups because the species either are extinct or there are no more major lineages to sample, suggesting that further study of analytical methods for whole genomes will prove to be as important as additional taxa.
Genomic-scale amounts of protein-coding sequence data were not only insufficient but were also misleading for generating an accurate avian phylogeny due to convergence. One possible explanation is convergent GC-biased gene conversion in exons, where AT-GC mismatches are corrected by DNA repair molecules in a biased manner to produce more gametes with the GC allele (85). GC-biased gene conversion correlates with recombination rate (86), and new GC alleles reach fixation more easily in species with larger population sizes, which tend to also have smaller body sizes (87). Recombination also tends to be higher toward the ends of chromosomes (88), where we found higher GC-rich high-variance exons. An alternative possibility is that the associations of ecology and/or life history are related to convergent exon-coding mutations for those traits in avian genomes (89, 90).
With a well-resolved tree, it becomes possible to more confidently infer evolution of convergent traits. Our tree lends support for either three independent gains of vocal learning (38, 91) or two gains (hummingbirds and the common ancestor of parrots and oscine songbirds) followed by two losses (in New Zealand wrens and suboscines) (29, 39). However, a single origin for parrots and oscines followed by two losses (three events) is not much less parsimonious than independent origins in parrots and oscines (two events). In addition, the suboscine Procnias bellbirds have recently been shown to be vocal learners (92, 93), suggesting that there could have been a fourth gain or a regain after a loss of vocal learning in other suboscines. The non-monophyly of the birds of prey at the deepest branches of the Australaves and Afroaves radiations suggests that the common ancestor of core landbirds may have been an apex predator, followed by two losses of the raptorial trait. Seriema at the deepest branch of Australaves could be considered to belong to a raptorial taxon because they kill vertebrate prey (94) and are the sole living relatives of the extinct giant “terror birds,” apex predators during the Paleogene (95, 96). The deepest branches after Accipitriformes and owl among the Afroaves, the mousebirds and cuckoo-roller, have Eocene relatives with raptor-like feet (97), and the cuckoo-roller specializes on chameleon prey (98). This suggests that losses of the predatory phenotype were gradual across successive divergences of each of the two core landbird radiations. More broadly, the Columbea and Passerea clades appear to have many ecologically driven convergent traits that have led previous studies to group them into supposed monophyletic taxa (8,17,25). These convergences include the footpropelled diving trait of grebes in Columbea with loons and cormorants (15) in Passerea, the wading-feeding trait of flamingos in Columbea with ibises and egrets (24,99) in Passerea, and pigeons and sandgrouse in Columbea with shorebirds (killdeer) in Passerea (24). These long-known trait and morphological alliances suggest that some of the traditional nongenomic trait classifications are based on polyphyletic assemblages.
In conclusion, our genome-scale analysis supports the hypothesis of a rapid radiation of diverse species occurring within a relatively short period of time (36 lineages within 10 to 15 million years; Fig. 1) during the K-Pg transition, with many interordinal divergences in the 1- to 3-million-year range. This rate of divergence is consistent with modern speciation rates, but it is notable that so many lineages from a single stem lineage survived extinction. Subsequent ecological diversification of surviving lineages is consistent with a proliferation of the earliest fossil stem representatives of most modern orders by the latest Paleocene to Eocene. Our finding is broadly consistent with recent estimates for placental mammals [(100), but see SM12 (101)] and thus supports the hypothesis that the K-Pg transition was associated with a rapid species radiation caused by a release of ecological niches following the environmental destruction and species extinctions linked to an asteroid impact (2, 4, 5,102).
Supplementary Material
ACKNOWLEDGMENTS
Genome assemblies, annotations, alignments, tree files, and other data sets used or generated in this study are available at GigaScience, the National Center for Biotechnology Information (NCBI), ENSEMBL, CoGe, UCSC, and other sources listed in SM13 (table S17). We thank S. Edmunds at GigaScience, K. Pruit at NCBI, and P. Flicek at ENSEMBL for making this possible. The majority of genome sequencing and annotation was supported by internal funding from BGI. Additional major support is from the coordinators of the project: E.D.J. from the HHMI and NIH Directors Pioneer Award DP1OD000448; S.M. from an HHMI International Student Fellowship; G.Z. from Marie Curie International Incoming Fellowship grant (300837); T.W. from NSF DEB 0733029, NSF DBI 1062335, and NSF IR/D program; and M.T.P.G. from a Danish National Research Foundation grant (DNRF94) and a Lundbeck Foundation grant (R52-A5062). J. Fjeldså generated the bird drawings used in the figures. O.A.R. acknowledges a uniform biological material transfer agreement between San Diego Zoo Global and BGI used for some tissue samples. Additional acknowledgements are listed in the supplementary materials. We thank the following for allowing us to conduct the computationally intensive analyses for this study: Heidelberg Institute for Theoretical Studies; San Diego Supercomputer Center, with support by an NSF grant; SuperMUC Petascale System at the Leibniz Supercomputing Center; Technical University of Denmark; Texas Advanced Computing Center; Georgia Advanced Computing Resource Center, a partnership between the University of Georgia’s Office of the Vice President for Research and Office of the Vice President for Information Technology; Amazon Web Services; BGI; the Nautilus supercomputer at the National Institute for Computational Sciences of the University of Tennessee and Smithsonian; and Duke University Institute for Genome Sciences and Policy.
Footnotes
R.E.G. declares that he is President of Dovetail Genomics, with no conflicts of interest.
www.sciencemag.org/content/346/6215/1320/suppl/DC1
Supplementary Text SM1 to SM13
Figs. S1 to S29
Tables S1 to S17
References (103–277)
Appendices
REFERENCES AND NOTES
- 1.Venditti C, Meade A, Pagel M. Nature. 2010;463:349–352. doi: 10.1038/nature08630. [DOI] [PubMed] [Google Scholar]
- 2.Yoder JB, et al. J. Evol. Biol. 2010;23:1581–1596. doi: 10.1111/j.1420-9101.2010.02029.x. [DOI] [PubMed] [Google Scholar]
- 3.Feduccia A. Science. 1995;267:637–638. doi: 10.1126/science.267.5198.637. [DOI] [PubMed] [Google Scholar]
- 4.Schulte P, et al. Science. 2010;327:1214–1218. doi: 10.1126/science.1177265. [DOI] [PubMed] [Google Scholar]
- 5.Longrich NR, Tokaryk T, Field DJ. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15253–15257. doi: 10.1073/pnas.1110395108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksepka DT, Boyd CA. Paleobiology. 2012;38:112–125. [Google Scholar]
- 7.Cohen KM, Finney S, Gibbard PL. International Chronostratigraphic Chart v2013/01. International Commission on Stratigraphy. 2013 www.stratigraphy.org/ICSchart/ChronostratChart2013-01.jpg.
- 8.Ericson PG, et al. Biol. Lett. 2006;2:543–547. doi: 10.1098/rsbl.2006.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meredith RW, et al. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 10.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. Nature. 2012;491:444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 11.Haddrath O, Baker AJ. Proc. Biol. Sci. 2012;279:4617–4625. doi: 10.1098/rspb.2012.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee MS, Cau A, Naish D, Dyke GJ. Syst. Biol. 2014;63:442–449. doi: 10.1093/sysbio/syt110. [DOI] [PubMed] [Google Scholar]
- 13.Brown JW, Rest JS, García-Moreno J, Sorenson MD, Mindell DP. BMC Biol. 2008;6:6. doi: 10.1186/1741-7007-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pacheco MA, et al. Mol. Biol. Evol. 2011;28:1927–1942. doi: 10.1093/molbev/msr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livezey BC, Zusi RL. Zool. J. Linn. Soc. 2007;149:1–95. doi: 10.1111/j.1096-3642.2006.00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayr G. J. Zool. Syst. Evol. Res. 2011;49:58–76. [Google Scholar]
- 17.Hackett SJ, et al. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- 18.Pratt RC, et al. Mol. Biol. Evol. 2009;26:313–326. doi: 10.1093/molbev/msn248. [DOI] [PubMed] [Google Scholar]
- 19.Nabholz B, Künstner A, Wang R, Jarvis ED, Ellegren H. Mol. Biol. Evol. 2011;28:2197–2210. doi: 10.1093/molbev/msr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Song S, Liu L, Edwards SV, Wu S. Proc. Natl. Acad. Sci. U.S.A. 2012;109:14942–14947. doi: 10.1073/pnas.1211733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alföldi J, Lindblad-Toh K. Genome Res. 2013;23:1063–1068. doi: 10.1101/gr.157503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters JL. In: Check-List of Birds of the World. Mayr E, Paynter RA, Traylor MA, Greenway JC, editors. Cambridge, MA: Harvard Univ. Press; pp. 1937–1987. [Google Scholar]
- 24.Sibley CG, Ahlquist JE. Phylogeny and Classification of Birds: A Study in Molecular Evolution. New Haven, CT: Yale Univ. Press; 1990. [Google Scholar]
- 25.Fain MG, Houde P. Evolution. 2004;58:2558–2573. doi: 10.1111/j.0014-3820.2004.tb00884.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang N, Braun EL, Kimball RT. Mol. Biol. Evol. 2012;29:737–750. doi: 10.1093/molbev/msr230. [DOI] [PubMed] [Google Scholar]
- 27.Kimball RT, Wang N, Heimer-McGinn V, Ferguson C, Braun EL. Mol. Phylogenet. Evol. 2013;69:1021–1032. doi: 10.1016/j.ympev.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 28.McCormack JE, et al. PLOS ONE. 2013;8:e54848. doi: 10.1371/journal.pone.0054848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suh A, et al. Nat. Commun. 2011;2:443. doi: 10.1038/ncomms1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahmood MT, McLenachan PA, Gibb GC, Penny D. Genome Biol. Evol. 2014;6:326–332. doi: 10.1093/gbe/evu016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matzke A, et al. Mol. Biol. Evol. 2012;29:1497–1501. doi: 10.1093/molbev/msr319. [DOI] [PubMed] [Google Scholar]
- 32.Chojnowski JL, Kimball RT, Braun EL. Gene. 2008;410:89–96. doi: 10.1016/j.gene.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Patel S, Kimball RT, Braun EL. J. Phylogenetics Evol. Biol. 2013;1:9–10. [Google Scholar]
- 34.Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Trends Genet. 2002;18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- 35.Rokas A, Williams BL, King N, Carroll SB. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- 36.Cracraft J. In: The Howard and Moore Complete Checklist of the Birds of the World. Dickinson EC, Remsen JV, editors. Aves Press; Eastbourne, UK: 2013. pp. xxi–xliii. [Google Scholar]
- 37.Dickinson EC, Remsen JV, editors. The Howard and Moore Complete Checklist of Birds of the World. Eastbourne, UK: Aves Press; 2013. [Google Scholar]
- 38.Jarvis ED. Ann. N.Y. Acad. Sci. 2004:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clayton DF, Balakrishnan CN, London SE. Curr. Biol. 2009;19:R865–R873. doi: 10.1016/j.cub.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hillier LW, et al. Nature. 2004;432:695–716. [Google Scholar]
- 41.Dalloul RA, et al. PLOS Biol. 2010;8:e1000475. [Google Scholar]
- 42.Warren WC, et al. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alföldi J, et al. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang G, et al. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro MD, et al. Science. 2013;339:1063–1067. doi: 10.1126/science.1230422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan X, et al. Nat. Genet. 2013;45:563–566. doi: 10.1038/ng.2588. [DOI] [PubMed] [Google Scholar]
- 47.Huang Y, et al. Nat. Genet. 2013;45:776–783. doi: 10.1038/ng.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, et al. Nat. Genet. 2013;45:701–706. doi: 10.1038/ng.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganapathy G, et al. Gigascience. 2014;3:11. doi: 10.1186/2047-217X-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S, et al. Genome Biol. 2014 10.1186/s13059-014-0557-1. [Google Scholar]
- 51.Li C, et al. GigaScience. 2014;3:27. doi: 10.1186/2047-217X-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Green RE, et al. Science. 2014;346:1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu K, Raghavan S, Nelesen S, Linder CR, Warnow T. Science. 2009;324:1561–1564. doi: 10.1126/science.1171243. [DOI] [PubMed] [Google Scholar]
- 54.Liu K, et al. Syst. Biol. 2012;61:90–106. doi: 10.1093/sysbio/syr095. [DOI] [PubMed] [Google Scholar]
- 55.Stamatakis A, et al. Bioinformatics. 2012;28:2064–2066. doi: 10.1093/bioinformatics/bts309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Stamatakis A. “The multi-processor scheduling problem in phylogenetics”. IEEE 26th International Parallel and Distributed Processing Symposium. :691–698. Shanghai, 21 to 25 May 2012. [Google Scholar]
- 57.Stamatakis A, Aberer AJ. “Novel parallelization schemes for large-scale likelihood-based phylogenetic inference”. IEEE27th International Symposium on Parallel and Distributed Processing. :1195–1204. Boston, 20 to 24 May 2013. [Google Scholar]
- 58.Mirarab S, Bayzid MS, Boussau B, Warnow T. Science. 2014;346:1250463. doi: 10.1126/science.1250463. [DOI] [PubMed] [Google Scholar]
- 59.Yuri T, et al. Biology (Basel) 2013;2:419–444. doi: 10.3390/biology2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ericson PG. J. Biogeogr. 2012;39:813–824. [Google Scholar]
- 61.Kim J. Mol. Phylogenet. Evol. 2000;17:58–75. doi: 10.1006/mpev.2000.0816. [DOI] [PubMed] [Google Scholar]
- 62.Maddison WP. Syst. Biol. 1997;46:523–536. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- 63.Rosenberg NA. Mol. Biol. Evol. 2013;30:2709–2713. doi: 10.1093/molbev/mst160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Degnan JH, Rosenberg NA. Trends Ecol. Evol. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 65.Edwards AW. Genetics. 2009;183:5–12. doi: 10.1534/genetics.109.107847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bayzid MS, Warnow T. Bioinformatics. 2013;29:2277–2284. doi: 10.1093/bioinformatics/btt394. [DOI] [PubMed] [Google Scholar]
- 67.Liu L, Yu L, Edwards SV. BMC Evol. Biol. 2010;10:302. doi: 10.1186/1471-2148-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. Nucleic Acids Res. 2014;42:D897–D902. doi: 10.1093/nar/gkt1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmons MP, Ochoterena H. Syst. Biol. 2000;49:369–381. [PubMed] [Google Scholar]
- 70.Felsenstein J. Inferring Phylogenies. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- 71.Avise JC, Robinson TJ. Syst. Biol. 2008;57:503–507. doi: 10.1080/10635150802164587. [DOI] [PubMed] [Google Scholar]
- 72.Ray DA, Xing J, Salem AH, Batzer MA. Syst. Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- 73.Han KL, et al. Syst. Biol. 2011;60:375–386. doi: 10.1093/sysbio/syq100. [DOI] [PubMed] [Google Scholar]
- 74.Lemmon EM, Lemmon AR. Annu. Rev. Ecol. Evol. 2013;44:99–121. [Google Scholar]
- 75.Weber CC, Boussau B, Romiguier J, Jarvis ED, Ellegren H. Genome Biol. 2014;15:549. doi: 10.1186/s13059-014-0549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weber CC, Nabholz B, Romiguier J, Ellegren H. Genome Biol. 2014;15:542. doi: 10.1186/s13059-014-0542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.dos Reis M, Yang Z. Mol. Biol. Evol. 2011;28:2161–2172. doi: 10.1093/molbev/msr045. [DOI] [PubMed] [Google Scholar]
- 78.Smith JV, Braun EL, Kimball RT. Syst. Biol. 2013;62:35–49. doi: 10.1093/sysbio/sys067. [DOI] [PubMed] [Google Scholar]
- 79.Houde P. Nature. 1986;324:563–565. doi: 10.1038/324563a0. [DOI] [PubMed] [Google Scholar]
- 80.Mayr G. Syst. Biodivers. 2013;11:7–13. [Google Scholar]
- 81.Clarke JA, Tambussi CP, Noriega JI, Erickson GM, Ketcham RA. Nature. 2005;433:305–308. doi: 10.1038/nature03150. [DOI] [PubMed] [Google Scholar]
- 82.Salichos L, Rokas A. Nature. 2013;497:327–331. doi: 10.1038/nature12130. [DOI] [PubMed] [Google Scholar]
- 83.Salichos L, Stamatakis A, Rokas A. Mol. Biol. Evol. 2014;31:1261–1271. doi: 10.1093/molbev/msu061. [DOI] [PubMed] [Google Scholar]
- 84.Twyford AD, Ennos RA. Heredity (Edinb.) 2012;108:179–189. doi: 10.1038/hdy.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duret L, Galtier N. Annu. Rev. Genomics Hum. Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 86.Mugal CF, Arndt PF, Ellegren H. Mol. Biol. Evol. 2013;30:1700–1712. doi: 10.1093/molbev/mst067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Romiguier J, Ranwez V, Douzery EJ, Galtier N. Genome Res. 2010;20:1001–1009. doi: 10.1101/gr.104372.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rudd MK, et al. PLOS Genet. 2007;3:e32. doi: 10.1371/journal.pgen.0030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parker J, et al. Nature. 2013;502:228–231. doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Proteome-wide analyses among the 48 bird genomes were conducted in (44) where it was found that vocal-learning bird species have convergent amino acid changes in a set of genes expressed in the song-learning nuclei [Google Scholar]
- 91.Nottebohm F. Am. Nat. 1972;106:116–140. [Google Scholar]
- 92.Saranathan V, Hamilton D, Powell GV, Kroodsma DE, Prum RO. Mol. Ecol. 2007;16:3689–3702. doi: 10.1111/j.1365-294X.2007.03415.x. [DOI] [PubMed] [Google Scholar]
- 93.Kroodsma D, et al. Wilson J. Ornithol. 2013;125:1–14. [Google Scholar]
- 94.Redford KH, Peters G. J. Field Ornithol. 1986;57:261–269. [Google Scholar]
- 95.Alvarenga HMF, Hofling E. Papeis Avulsos Zool. 2003;43:55–91. [Google Scholar]
- 96.Mayr G. Paleogene Fossil Birds. Berlin, Heidelberg: Springer; 2009. pp. 139–152. [Google Scholar]
- 97.Houde P, Olson SL. Natural History Museum of Los Angeles County Science Series. Vol. 36. Los Angeles: Natural History Museum of Los Angeles County; 1992. pp. 137–160. [Google Scholar]
- 98.Forbes-Watson AD. Ibis. 1967;109:425–430. [Google Scholar]
- 99.Olson SL, Feduccia A. Smithson. Contrib. Zool. 1980;316:1–73. [Google Scholar]
- 100.O’Leary MA, et al. Science. 2013;339:662–667. doi: 10.1126/science.1229237. [DOI] [PubMed] [Google Scholar]
- 101.dos Reis M, Donoghue PC, Yang Z. Biol. Lett. 2014;1020131003 doi: 10.1098/rsbl.2013.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Benton MJ. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:3667–3679. doi: 10.1098/rstb.2010.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.