Abstract
Background & objectives:
Tuberculosis is a major health problem in India, and the emergence of multidrug resistant (MDR) and extensively drug resistant (XDR) strains of Mycobacterium tuberculosis (Mtb) has further complicated the situation. Though several studies characterizing drug sensitive and drug resistant strains are available in literature, almost all studies are done on unrelated strains. Therefore, the objective of this study was to compare the proteomic data of four sequential isolates of Mtb from a single patient who developed MDR-TB during the course of anti-tuberculosis therapy (ATT).
Methods:
In this study, using two-dimensional (2D) gel electrophoresis and MALDI-TOF mass spectrometry, we compared and analyzed the cell lysate proteins of Mtb sequential clinical isolates from a patient undergoing anti-TB treatment. The mRNA expression levels of selected identified proteins were determined by quantitative real-time polymerase chain reaction (qRT-PCR).
Results:
The genotypes of all four isolates remained homologous, indicating no re-infection. The initial isolate (before treatment) was sensitive to all first-line drugs, but the consecutive isolates were found to be resistant to isoniazid (INH) and rifampicin (RIF) and developed mutations in the katG, inhA and rpoB. The intensities of 27 protein spots were found to be consistently overexpressed in INH and RIF resistant isolates. The most prominent and overexpressed proteins found during the development of drug resistance were GarA (Rv1827), wag31 (Rv2145c), Rv1437 and Rv2970c.
Interpretation & conclusions:
This preliminary proteomic study provides an insight about the proteins that are upregulated during drug resistance development. These upregulated proteins, identified here, could prove useful as immunodiagnostic and possibly drug resistant markers in future. However, more studies are required to confirm these findings.
Keywords: 2D gel electrophoresis, tuberculosis, MDR-TB, MALDI-TOF, proteomics
Tuberculosis (TB) is a global emergency with an estimated nine million new cases and more than 1.5 million deaths occurring annually1. The situation has worsened after AIDS epidemic and with the emergence of multidrug resistant (MDR) forms of the causative agent Mycobacterium tuberculosis (Mtb). These drug resistant strains are more infectious by virtue of their high transmissibility in the population. Therefore, identification of the reliable diagnostic, prognostic and drug resistance markers is an urgent research priority. Various in vitro and in vivo studies have identified chromosomal mutations as determinants of drug resistance2,3,4. For example, mutation (s) in rpoB allele confers rifampicin (RIF) resistance (RIFr) in 90-95 per cent isolates2, while isoniazid (INH)-resistance (INHr) is attributed to mutation (s) in one or more alleles viz., katG, inhA, ahpC and ndh. However, in about 20 per cent of INHr isolates, none of these known mutations are found, suggesting the possibility of unknown mutations or mechanisms2,3. On the other hand, the genetic mutations may not necessarily correlate with phenotypic resistance; further suggesting that other factors such as drug impermeability, drug-efflux pumps, formation of survivable “persister cells” under drug pressure and several other host factors could be involved in the outcome of treatment2.
Hence, for unraveling the mechanism(s) of drug resistance, understanding the mode of action of anti-TB drugs is very crucial. Many studies have elucidated the mode of action of various anti-TB drugs using genetic analysis, mRNA expression and DNA microarray analysis4,5. Several groups have also explored the proteome of Mtb and provided comprehensive details about the subcellular localization and confirmed the genomic annotation6,7,8,9. In these studies two-dimensional (2D) gel electrophoresis followed by mass spectrometry (MS) identification of the differentially regulated proteins substantially helped in identifying the complex pathways and their regulatory enzymes. These studies also elucidated modes of action of various drugs and discovered new antigens that could be potential candidates for developing vaccines and diagnostics6,7,9,10. However, only a few studies are available which show differential expression of specific proteins in the drug resistant but not in drug susceptible cells7,8,9,11. Further, in all these studies, either the non-pathogenic mycobacteria or laboratory collections of drug sensitive and drug resistant strains of Mtb from different patients have been used. In the present study, protein profile of sequentially collected four clinical isolates of Mtb was analyzed using 2D gel electrophoresis and the differentially expressed proteins were identified by MALDI-TOF-MS analysis. All isolates were from the same patient, who developed MDR-TB during the course of chemotherapy.
Material & Methods
The study was conducted between January 2006 and June 2010 at the TB Laboratory, Division of Clinical Microbiology and Molecular Medicine, Department of Laboratory Medicine, All India Institute of Medical Sciences (AIIMS), New Delhi, India. This study was approved by the Institutional Ethics Committee of AIIMS and written informed consent was obtained from the patient. The patient was being treated at the designated microscopy and DOTS (directly observed treatment-short course) centres of Shahpurjat, New Delhi. This patient (22 yr old male) was diagnosed as having pulmonary TB on the basis of clinical and radiological findings and sputum smear microscopy. He was prescribed with anti-TB treatment (ATT) under the DOTS programme. The thrice a week treatment regimen comprised isoniazid, rifampicin, pyrazinamide (PZA) and ethambutol (EMB) (category I treatment) in intensive phase for two months followed by four month treatment with two (isoniazid and rifampicin) drugs regimen. Pre-treatment sputum specimen was used for isolation of Mycobacterium sp. by BACTEC MGIT-960 (Becton Dickinson, Sparks, MD, USA), which was positive. The isolate was identified as Mtb by conventional phenotypic and in-house PCR method12. This culture was labelled as isolate A, and was subjected to 16sRNA gene sequencing. The patient though took full six months course of treatment but became irregular in taking drugs after initial improvement in his clinical symptoms. After three months of cessation of treatment (6+3= 9 month13, his condition again deteriorated and his sputum culture was again positive for Mtb. We labelled this second culture as isolate B. He was re-treated with isoniazid, rifampicin, pyrazinamide, ethambutol and streptomycin (SM) (category II regimen). Within two months his clinical condition improved but he again defaulted. After an asymptomatic period of about four months his symptoms reappeared. His sputum was again culture positive and this culture was labelled as isolate C. The patient was again prescribed with the same treatment for 12 months after counselling but he stopped treatment after six months. His condition further deteriorated and he died of multisystem failure. The fourth sample was received just before his death and the isolate from this sputum sample was labelled as isolate D.
All the four clinical isolates (A, B, C & D) were identified as Mtb using standard protocols12,13. The anti-mycobacterial drug susceptibility testing was performed on all the isolates by both BACTEC™ MGIT-960 (Becton Dickinson, Sparks, MD, USA) and proportional method using Middlebrook 7H10 (Difco, USA) agar plates containing first-line anti-TB drugs (SM 2.0 µg/ml, INH 0.2 µg/ml, RIF 1.0 µg/ml, EMB 6.0 µg/ml)13,14. All four isolates were also genotyped by spoligotyping and identified using SITVIT-WEB database15. The rpoB, inhA and katG gene targets were sequenced using the primers as described elsewhere13.
Preparation of mycobacterial whole cell lysate: All Mtb isolates were grown without shaking in Middlebrook 7H9 medium supplemented with 0.2 per cent (v/v) glycerol, 10 per cent oleic acid, albumin-dextrose and catalase (OADC, Difco, USA) at 37°C for two weeks. Whole cell lysate was prepared according to protocol of Sharma et al11. Cells were washed three times with normal saline and then suspended in sonication buffer [50 mM tris-HCl containing 10 mM MgCl2, 0.1% sodium azide, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1mM ethylene glycol tetra acetic acid (EGTA); PH 7.4] at a concentration of 1g wet cell mass per 5ml, and then broken by intermittent sonication for 15 min at 4°C using sonicator (Sonics & Materials Inc, USA). The homogenate was centrifuged at 12,000×g for 20 min at 4°C. The pellets were discarded and supernatant was stored at -70°C until further use.
Protein precipitation with sodium dodecyl sulphate (SDS)-trichloroacetic acid (TCA)-acetone: The cell lysates were treated with 1 per cent SDS and then subjected to TCA-acetone precipitation procedure9. The protein pellet was suspended in appropriate volume of two-dimensional rehydration buffer (Bio-Rad, USA), and the protein concentration was estimated using the Bradford method16.
Two-dimensional gel electrophoresis: Isoelectric focusing (IEF) was done using the in-gel rehydration method (Bio-Rad, USA).
2D gels were analysed using PDQuest Advanced software (version 8.0) (Bio-Rad, USA). After acquisition, the images were analyzed using step-wise spot detection and spot matching followed by differential expression analysis. The quantity of each spot was normalized by total valid spot intensity. The expression differences for all four mycobacterial isolates were compared using the same software. Images for sensitive and resistant isolates were manually checked for artifactual spots, merged spots and missed spots, and spots with more isolate-specific variability were omitted in the downstream processing. Equal amount of protein was loaded in all gels and experiments were repeated three times with three independent biological replicates.
In-gel digestion of protein spots with trypsin: Protein spots of interest were excised from the coomassie brilliant blue R250 stained 2D gels using spot picker Investigator ProPic (Genomic Solutions Ltd., Huntingdon, UK). Digestion of proteins and spotting of peptides on matrix assisted LASER desorption/ionization-time of flight (MALDI-TOF) target plate was carried out using protein digester investigator ProPrep (Genomic Solutions, Huntingdon, UK).
For protein digestion, method of Shevchenko et al17 was followed with slight modifications. In brief, the gel plugs were de-stained and dehydrated by washing three times (10 min) with 25 mM NH4HCO3-50 per cent acetonitrile (ACN) (1:1v/v) solution and treated with freshly prepared 10 mM dithiothreitol (DTT) in 50 mM NH4HCO3 for 45 min at 56°C. After incubation, DTT was replaced with freshly prepared 55 mM iodoacetamide for 30 min and then dehydrated with 100 per cent ACN. The dried gel pieces were incubated for 12 h at 37 °C with 25 mM NH4HCO3 containing 0.02 μg/μl of mass spectrometry grade trypsin (Promega, USA). The resulting peptides were extracted twice from the gel pieces, using peptide extraction buffer [1:1v/v mixture of 70% ACN and 0.1% trifluoroacetic acid (TFA)].
Mass spectrometric analysis: Mass spectrometry (MS) was carried out as described earlier9. The digested samples were desalted and concentrated on C-18 ZipTips (Millipore, USA) using the manufacturer's protocol before mass spectrometric analysis. ZipTips were eluted on MTP 384 target plate with 2 μl of α-cyano-4-hydroxycinnamic acid (HCCA) (Sigma-Aldrich, USA) saturated solution dissolved in 50 per cent ACN and 0.2 per cent TFA. Mass spectra of digested proteins were acquired using Autoflex II TOF/TOF 50 (Bruker GmbH, Leipzig, Germany) in positive reflectron mode. AnchorChip target plate was placed in sample inlet of the instrument, controlled by flexControl 2.4 software (Bruker, Germany). The instrument was equipped with a 337 nm nitrogen LASER, delayed extraction electronics, and a 50Hz digitizer and percentage of LASER energy was maintained at 30-40 per cent. The pulse energy was 105 µJ and pulse duration was 1.3 nano sec. Final mass spectra were produced by averaging 1500-2500 LASER shots taken at different positions within each spot. The spectra were acquired in positive reflection mode in the mass range of 500-3000 m/z. Calibration was performed using peptide calibration standard II (Bruker, Germany). The proteolytic masses obtained, were processed through FlexAnalysis v.2.4 programme (Bruker, Germany) for peak detection. Initially, the spectra acquired were processed for baseline subtraction with 80 per cent baseline flatness followed by smoothening with threshold of signal-to-noise ratio (S/N) >5. The contaminant m/z peaks originating from human keratin, trypsin auto-digestion and matrix were removed from the spectra to generate the peptide mass list for the database search. Finally, the proteolytic masses obtained were evaluated using MASCOT, a peptide mass fingerprinting (PMF) tool (Matrix Sciences, UK). Peak detection in MALDI spectra and peak lists were submitted to the UniProtKB/Swiss-Prot database using the MASCOT search engine (http://www.matrixscience.com ) to identify the proteins from the annotated Mtb chromosome (strain H37Rv, EMBL/GenBank/DDBJ entry AL123456). Peptide mass tolerance was set in range of 50-100 ppm, with carbamidomethyl-cystein set as fixed modification, oxidation of methionine as variable modification and only 0 or 1 missed cleavage site was allowed. Further, matched precursor ions of identified proteins were selected for subsequent fragmentation using post source decay (PSD) for MS/MS. Lift_ATT method was performed in flex control software; parent peak mass spectrum was acquired by hitting LASER for 400-550 shots followed by acquisition of fragments of selected precursor ions for the same number of shots. Both parent and fragment spectra were pooled to generate MS/MS spectrum of a particular peptide. MS/MS spectrum was submitted to database using MASCOT wizard (Matrix Sciences, UK). The same parameters were used for MS/MS search in addition to the fragment mass tolerance from 0.5 to 2.0 Da.
Glycogen estimation: Logarithmic and stationary phase growth of Mtb sensitive (isolate A) and MDR (isolates B,C,D) isolates were collected by centrifugation (3 min at 5000 x g and at 4°C), and the pellet (15-20 mg wet weight) was re-suspended in 0.25M Na2CO3 and incubated at 95°C for 4 h. The glycogen content was estimated by following the procedure of Schulze et al18.
Isolation of Mtb total RNA and real-time quantitative PCR (qRT-PCR): Mtb H37Rv was grown in Middlebrook 7H9 broth containing 10 per cent OADC, and was treated with INH (0.1µg/ml), RIF (1.0µg/ml), EMB (5µg/ml) and INH (0.1µg/ml) +RIF (1.0µg/ml). Total RNA was isolated using a TRI reagent (Sigma, USA) following manufacturer's instructions. To analyze mRNA expression, cDNA was synthesized from 1 µg of total RNA by using Superscript III (Invitrogen Life Technologies, USA) and random primers (Invitrogen, Life Technologies, USA), followed by amplification of the gene(s) by gene-specific primers, using master mix SYBR green (Applied Biological Materials Inc., Canada). The expression is represented in fold increase. 16sRNA was used as internal control for mRNA expression analysis of ahpC, Rv1827, pknA, pknB, pknG and wag31.
Each reaction was repeated thrice with three independent RNA samples in a smart cycler Cepheid machine (Cepheid, USA). RT-PCR conditions were as follows: an initial denaturation step of 10 min, followed by 40 amplification cycles of 30 sec at 95°C, 30 sec at 60°C and 30 sec at 72°C. Melting curve analysis was carried out to confirm the specificity of the amplified product. After baseline corrections and determination of threshold settings, calculation and statistical analyses were carried out using the 2-∆∆CT Method19. The results are shown as fold increase in expression profile.
Results
Drug resistance pattern and mutations in katG, inhA and rpoB genes of the isolates: The four sequential culture isolates were identified as Mtb by conventional phenotypic and in-house PCR method. Genotyping was done by spoligotyping and the results confirmed that all the isolates belonged to the Central Asian Strain Delhi (CAS1_Delhi, ST26) genotype. The drug susceptibility test results showed that the initial isolate (isolate A) was sensitive to all the four first-line drugs (SM, INH, RIF and EMB), but the consecutive isolates (isolates B, C and D) became resistant to three drugs; INH, RIF and EMB. The minimum inhibitory concentration (MIC) of isolate B increased as compared to isolate A against INH, RIF and EMB. However, it was still sensitive to kanamycin. The isolates C and D became resistant not only to INH, RIF and EMB but also to kanamycin. The sequencing of the rpoB (RIFr), katG and inhA (INHr) regions revealed mutated alleles associated with resistance to the respective drugs13. Morphologically the resistant isolates were stunted, thicker and cocco-bacillary in shape.
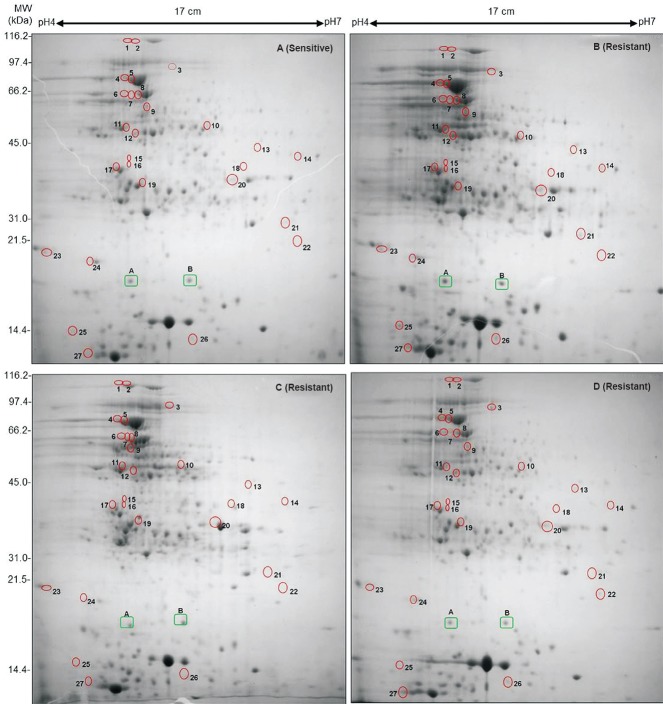
Differentially expressed proteins in drug sensitive and resistant isolates: The cell lysate proteins of four Mtb isolates were analyzed by 2D gel electrophoresis, which showed 430 protein spots in isolates A and 495, 556 and 395 spots in isolates B, C and D, respectively (Figs. 1 & 2). Quantitative analysis of 2D gel spots was carried out using PDQuest software which revealed 27 spots upregulated in MDR isolates (Table I). The spots showing more than 2-fold upregulation were further identified by MALDI-TOF/TOF MS (Table II). To rule out possibility of any artifact, proteins showing equal intensity were taken as internal control (represented as square in Fig. 1). Upregulated proteins were functionally classified according to TubercuList web server which showed that most of the identified proteins belonged to the functional group 0, 1, 2, 3, 5, 7 and 9; corresponding to virulence, detoxification and adaptation (18.5%), lipid metabolism (11.11%), information pathway (14.81%), cell wall and cell process (3.7%), insertion sequence and phages (18.51%), and intermediary metabolism and information (29.62%); respectively (Table III). The magnified regions of upregulated proteins are shown in Fig. 3. Of the 27 upregulated proteins, eight were hypothetical protein (Rv2004c), probable glutamyl-tRNA (GLN) amidotransferase A gatA (Rv3011c), possible phosphoserine aminotransferase SerC (Rv0884c), probable lipase/esterase LipN (Rv2970c), probable phosphoglycerate kinase Pgk (Rv1437), conserved hypothetical protein with FHA domain, GarA (Rv1827), bacterioferritin (Rv1876) and conserved hypothetical protein (Rv0543) and were not found in 2D-PAGE database system accessible at http://www.mpiib-berlin.mpg.de/2D-PAGE, whereas three proteins probable iron-regulated aconitate hydratase Acn (Rv1475c), probable chaperone protein DnaK (Rv0350) and 60 kDa chaperonin 2 groEL2 (Rv0440), were found in two spots. Six proteins (gatA, serC, fbd, garA, Rv2204c and Rv0543c) in isolate B, 10 proteins (Rv685c, Rv3457c, Rv1479, Rv2970c, Rv1437, qor, and two spots each of Rv1475c and groEL2 family) in isolate C, three proteins (fadB, fabG4 and rrf) in isolate D, three proteins (Rv3075c, Rv1436 and GroES) in isolates C as well as in D were found upregulated. Only five proteins were consistently upregulated in all the three resistant isolates and these were identified as, chaperonin protein dnaK HSP70 (spots 4 and 5), hypothetical protein (Rv2004, spot 8), antigen 84 (wag31, spot 19) and bfrA (spot 24) (Fig. 1).
Fig. 1.
2D gel profiles of whole cell lysate proteins of Mtb clinical isolates collected sequentially from a single patient. The upregulated proteins are highlighted by circles. (A) First Mtb isolate (before treatment) sensitive to all 4 drugs, (B) Second isolate (during treatment) acquires MDR, (C) Third isolate (after 15 months of treatment) acquires drug resistance to yet another drug kanamycin and (D) Fourth MDR isolate after 27 months. Two Proteins, A (Rv1080c) and B (Rv2140c) marked in green rectangles, were selected for observing expression variation in all four samples and showed similar level of expression in all gels.
Fig. 2.
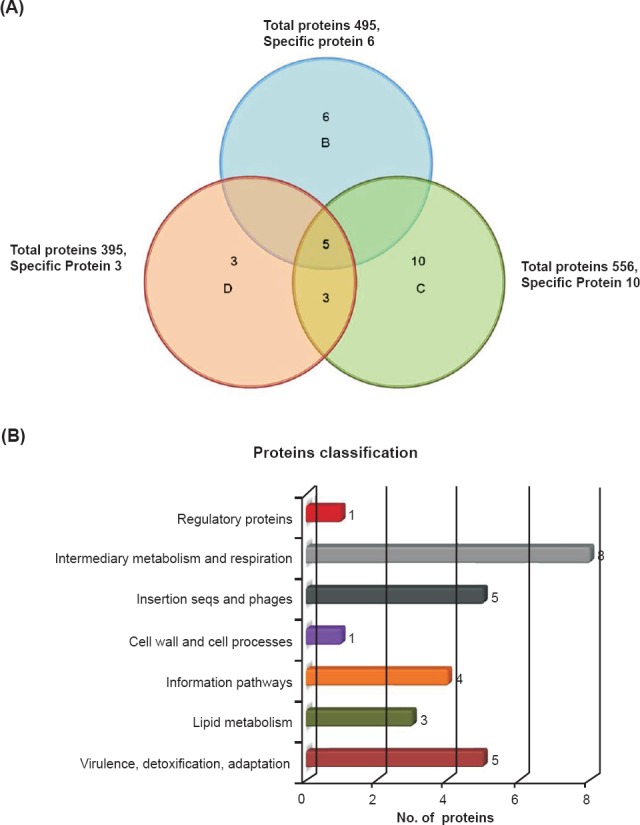
Upregulated proteins, their distributions, amount of overlap and functional classification in Mtb clinical isolates: (A) Upregulated proteins distribution and amount of overlap in clinical drug resistant isolates. (B) Functional classification of differentially expressed proteins according to the TubercuList Server.
Table I.
Details of upregulated proteins identified by mass spectrometry in drug resistant isolates
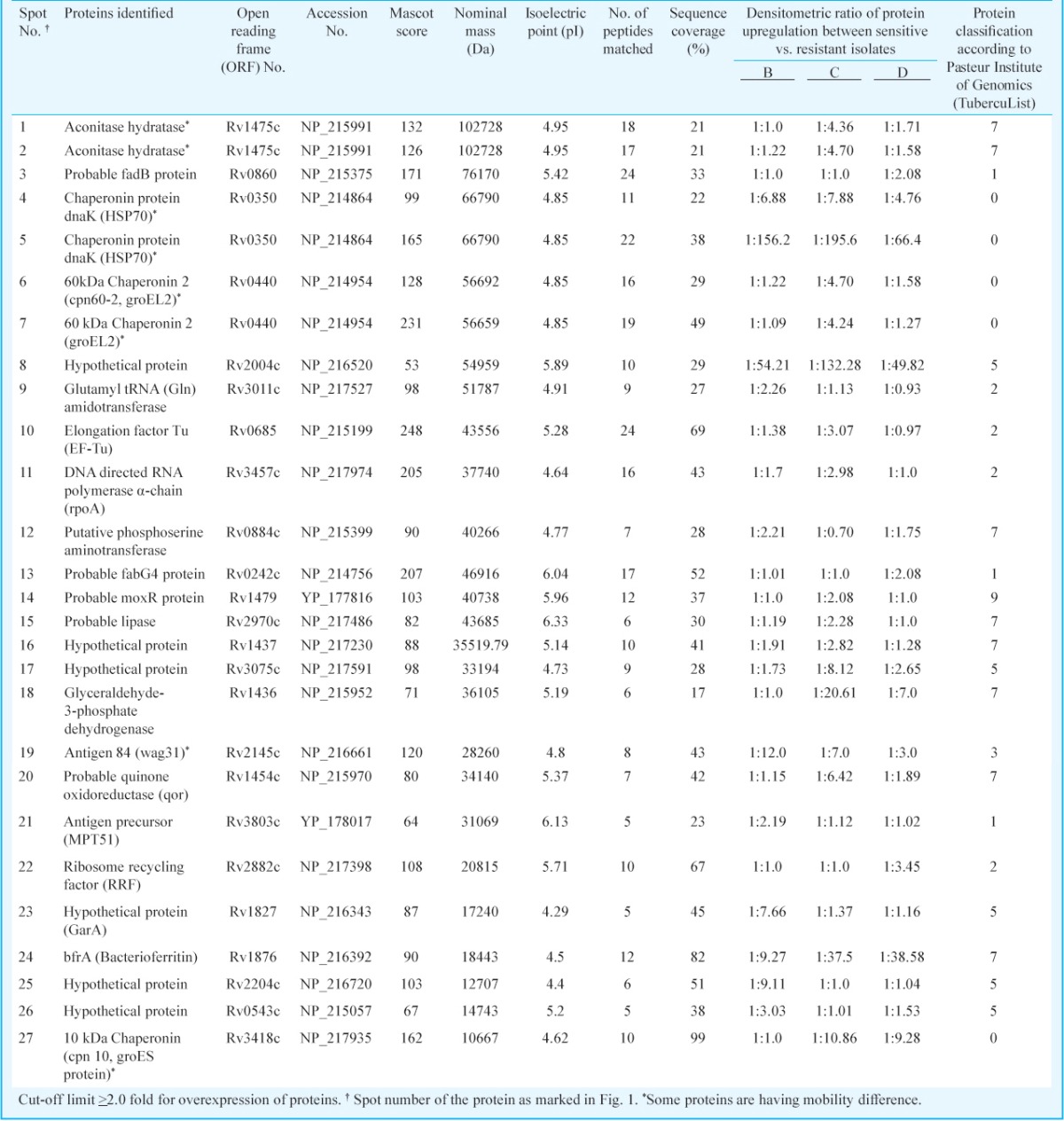
Table II.
MALDI-TOF/TOF (MS/MS) analysis of overexpressed proteins in MDR-TB clinical isolates
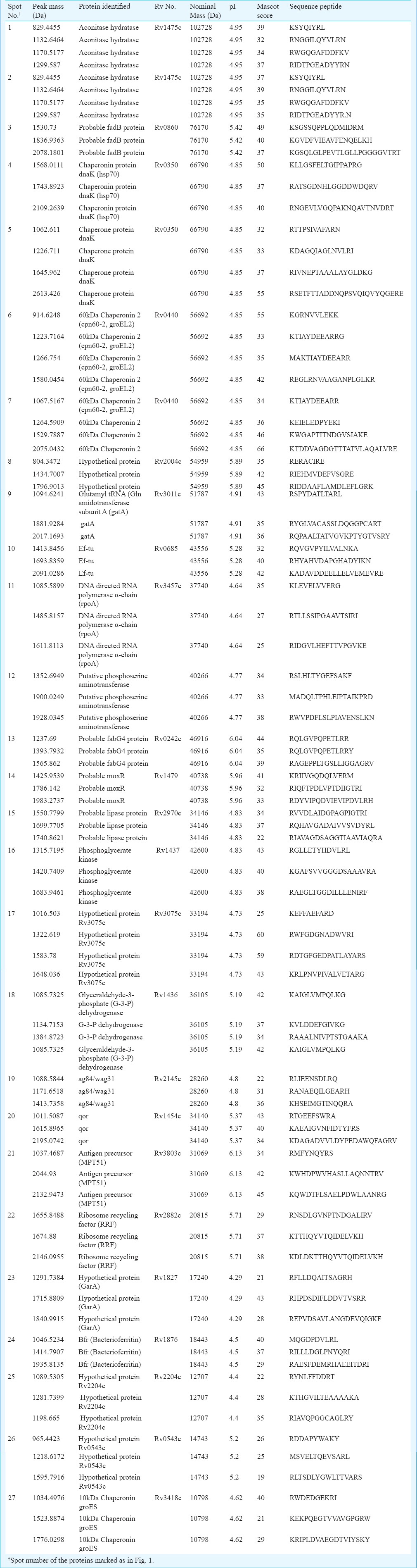
Table III.
Isolates specific proteins overexpression in MDR-TB isolates
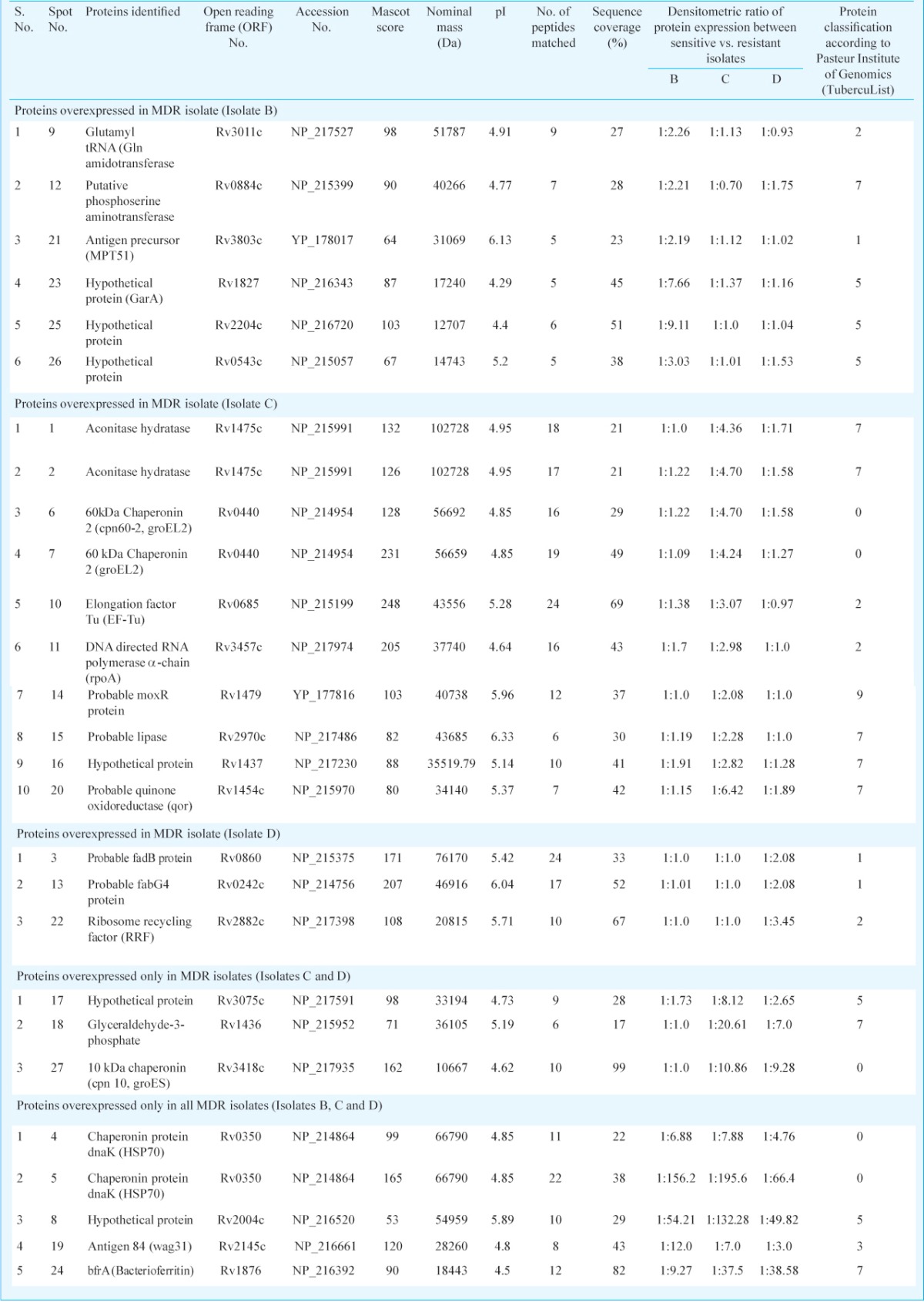
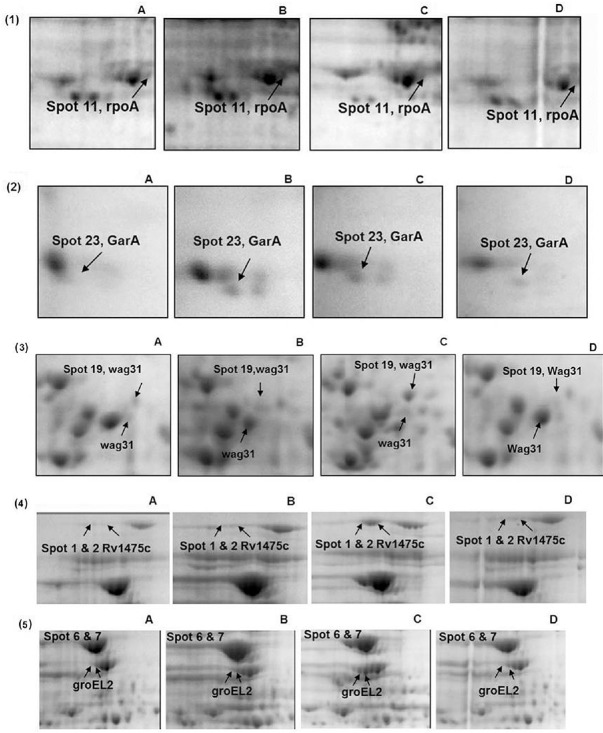
Fig. 3.
Magnified region of overexpressed proteins present in MDR isolates. Zoomed in regions of 2D-gel images showing the overexpressed proteins in drug resistant isolates: (1) rpoA (2) GarA (3) wag31 (4) Rv1475c (5) groEL2.
Among the identified proteins, we were more interested in studying the possible role of Rv1827 (GarA) and wag31 in drug resistance, since these proteins have been identified as physiological substrates for protein kinases G (pknG). Our result revealed that GarA and wag31 were upregulated in the drug resistant isolates. We analyzed the mRNA expression of Rv1827 and its cognate protein kinases, pknG, pknB, pknA and wag31.
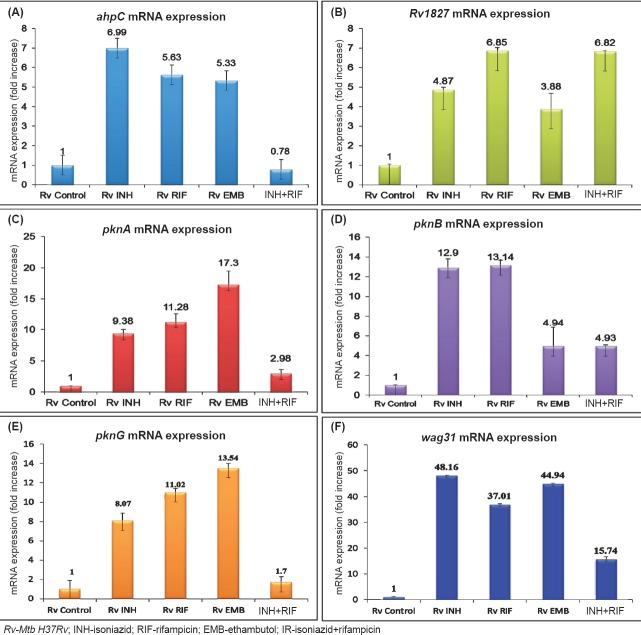
Drug induced changes in mRNA expression of protein kinases: To verify our protein expression observations, we studied the mRNA expression profile to see the effect of the four drugs on the standard strain of Mtb (H37Rv) which was sensitive to all anti-TB drugs. For this, the mRNA from H37Rv strain was isolated before and after exposing it to INH (0.1µg/ml), EMB (5.0 µg/ml), RIF (1.0 µg/ml) and INH+RIF (0.1 + 1.0µg/ml) for 6 h. Consistent with the proteomic data, seen in clinical isolates, Rv1827 expression was upregulated in all the tested conditions. As expected, the upregulation was 6.82 fold when the Mtb standard strain (H37Rv) was exposed to INH and RIF together, but other tested genes had relatively diminished expression. While combining the EMB, the expression of wag31 was higher and pknA and pknG expressions were highest (Fig. 4).
Fig. 4.
Detection of mRNA expression in drug treated Mtb isolates. Isolates treated with isoniazid (INH, 0.1μg/ml), rifampicin (RIF, 1.0μg/ml), ethambutol (EMB, 5μg/ml) and INH (0.1μg/ml) +RIF (1.0μg/ml). The expression is represented in fold increase. The 16sRNA was used as internal control for mRNA expression analysis. mRNA expression of (A) ahpC, (B) Rv1827, (C) pknA, (D) pknB, (E) pknG and (F) wag31. Calculation and statistical analysis carried out by using the 2-∆∆CT method and results presented in fold increase.
Glycogen storage: GarA, which is a glycogen regulatory protein, was found upregulated in our MDR isolates. It was found that as compared to sensitive isolates the glycogen accumulation in MDR isolates was higher. Consistent with GarA protein levels, the glycogen accumulation measured after seven days was 1.8, 2.0 and 2.1 folds higher in isolates B, C and D, respectively, as compared to sensitive isolate A. Interestingly, after 15 days the glycogen storage remained almost unchanged (Fig. 5).
Fig. 5.
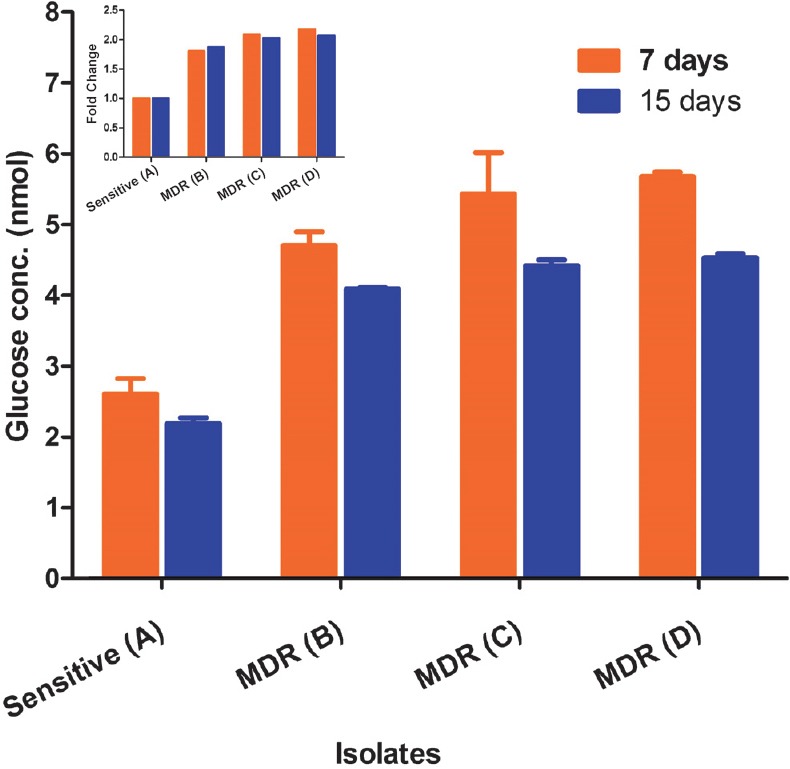
Glycogen content of sensitive and MDR isolates of Mtb. glycogen was extracted after 7 and 15 days and glycogen storage was determined by the estimation of glucose. The bars indicate glucose concentration of Mtb cells after 7 and 15 days, respectively. The inset shows the glycogen content in terms of fold increase after normalization in comparison to sensitive vs. MDR TB clinical isolates. Values are mean ± SD (n=3).
Discussion
Emergence of drug resistance in Mtb has become a major concern for TB control programme managers and treating physicians. Though advances in genome sequencing methods have provided better opportunities to our understanding about functional genomics and proteomics of the Mtb, the knowledge about mechanism of drug resistance still remains limited only to the association of genetic polymorphism. Most often, the data from proteomic studies are used to understand host-pathogen interaction, virulence, drug resistance and drug tolerance8,9,19,20,21,22,23. Such studies have provided a comprehensive list of Mtb proteins that are found differentially regulated in laboratory maintained standard H37Rv strain exposed to drug pressure. However, in such studies protein (s) that could be modulated in vivo during or after acquiring in vivo drug resistance are missed out22. The ever-increasing evidences suggest that the expression of genes22 and/or proteins23 in clinical isolates is markedly different from the laboratory maintained H37Rv strain; suggesting that majority of these observations may not have direct impact in real life scenario.
In the present study, several proteins were identified that were upregulated in drug resistant isolates. Of the 27 upregulated proteins, five were upregulated in all sequential resistant isolates (B, C and D). Approximately half of these overexpressed proteins are reported as essential for in vitro growth of Mtb21,22,23,24. Although their functional role in drug resistance is elusive, validation of these proteins as a biomarker of drug resistance will provide a scope for finding an effective candidate drug for MDR-TB. It also needs to be emphasized that identifying a protein as “upregulated” does not necessarily imply that it is a true determinant of drug resistance, because it is possible that some or all of these proteins are associated with adaptability of the Mtb to survive longer in the host system.
Among the upregulated proteins, rpoA (Rv3457c) was found 2.9 folds upregulated in the drug resistant isolates. In a similar study compensatory mutations in rpoA and rpoC (Rv0668) were identified particularly in more than 30 per cent of RIF-resistant strains and the authors proposed that mutation in these alleles could also be associated with MDR24. Though we have not screened our resistant isolate for these mutations, this study supports our proteomic approach and identification of an upregulated rpoA protein in drug resistant isolates. Dussurget et al25 reported that in M. smegmatis, IdeR negatively controls iron-uptake and expression of BfrA and BfrB. In our study, BfrA was upregulated in drug resistant isolates, suggesting a role of this protein in inducing resistance to INH. However, such conclusions could not be validated by deletion mutant of bfrA and bfrB in Mtb22.
We found that the identified proteins spots (Rv1475c, dnaK, groEL2, groES and wag31) had different electrophoretic mobilities in resistant and sensitive isolates. Similar observations have been reported by Mattow et al21 when the protein profile of intra-phagosomal Mtb H37Rv was analyzed. It has also been suggested that this shift is determined by the protein modifications rendered during sample preparation and growth conditions adopted by various laboratories8. However, this may be the unlikely factor in our study, as we strictly followed the same protocol throughout the study. Further, our analysis was stringent and we considered a particular protein to be “upregulated” only if the observation was consistent in three independent experiments. Even then, we were cautious not to conclude whether the changes in protein mobility necessarily reflect protein modification. Further studies are warranted to establish the role of post-translation modifications in these proteins during drug resistance.
Antigen 84 (Wag31/ Rv2145c) has been demonstrated to be involved in the regulation of cell morphology and pknG mediates the survival of Mtb inside the host macrophages, and genome-wide transcriptional analysis reveals that the pknG is upregulated during the exposure of INH26. Further, pknG is associated with the intrinsic resistance of Mtb to various anti-TB drugs26. Wag31 protein has been found to be overexpressed and involved in regulation of cell morphology. It also plays important role in survival of mycobacteria under oxidative stress27 and provides optimal substrate for pknA and pknB. In addition, mRNA expression of wag31 was increased by 15.7 folds during the INH and RIF exposure. Earlier also, wag31 has been reported to be over expressed in the MDR isolates8, supporting our hypothesis that wag31 plays an important role in drug resistance.
Of the five hypothetical proteins (Rv2004c, Rv1437, Rv3075c, Rv2204c and Rv1827) overexpressed in MDR isolates, three (Rv2004c, Rv2204c and Rv1437) could not be assigned to any function in survival or pathogenesis of the bacteria, though Rv3075c has been reported to be overexpressed in streptomycin resistant isolates11. In our study, Rv1827 (GarA) was found 7.6 folds upregulated in drug resistant isolates as compared to that in the susceptible isolate A. It has been identified as an optimal substrate for PknB and PknG. The protein is also reported to act as a phosphorylation-dependent molecular switch in mycobacterial signalling process mediated by protein kinases28,29,30. Further, GarA has been found to be predominantly expressed under the exponential growth phase and has been suggested as a regulatory model for glycogen degradation and glutamate metabolism28,29,30. To infer whether the protein overexpression of Rv1827 in drug resistant isolates facilitated increased glycogen accumulation, we quantified the glycogen content of drug resistant and sensitive isolates. The findings were consistent with this hypothesis and the glycogen content was relatively more in resistant isolates (B, C and D) than the sensitive isolate A. However, the difference in accumulation was significant up to 7th day, but not after 15th day of growth, suggesting that the expression of Rv1827 might be an important marker of dormancy. While the precise role of glycogen storage in mycobacteria is not known, glycogen stores may serve as a reservoir of carbon and energy that can be mobilized by mycobacteria for survival during periods of carbon starvation. Such observations have been made in other bacteria such as Vibrio cholerae during transition stage between host and aquatic environments31.
In our study, the identified proteins cannot be categorically classified as drug resistance-specific alterations, as this can also happen due to host immune response or stresses encountered by the individual strain in vivo. Most of these proteins are essential for the survival of mycobacteria in phagosome or as virulence factor (unpublished observation), but their role in drug resistance cannot be ruled out completely.
The 2D gel electrophoresis followed by MS-based proteomic analysis on sequential isolates showed approximately 500 proteins per gel. This resolution is much less than expected, as there are 4000 predicted genes in Mtb. The poor sensitivity observed in the present study could be attributed to various reasons such as the extraction protocol; low resolution power of the coomassie brilliant blue stain used or due to the IEF-strips (PH 4-7). These strips resolve only the proteins having isoelectric point in the range of 4-7.
In conclusion, our study highlights the intricacies associated with sequential clinical isolates of Mtb, a rare opportunity for any laboratory, which is a natural phenomenon and cannot be generated artificially in the laboratory. The sequential isolation of four isolates from the same patient during the treatment period showed a phenomenon where a sensitive isolate turned to a multidrug resistant isolate. It is possible that some of the upregulated proteins identified from MDR clinical isolates of Mtb in the present study may prove as potential biomarkers of drug resistance in future.
Acknowledgment
This work was supported by grant from Department of Biotechnology, Government of India to the last author (SS) (BT/ PR7880/ MED/ 14/1161/2006). The first author (AS) received financial assistance in the form of senior research fellowship from the Indian Council of Medical Research, Government of India. The authors thank Shri Sandeep Kadian for his help in spoligotyping data analysis and Dr V.M. Katoch, Director-General of Indian Council of Medical Research, New Delhi, for allowing us to use his laboratory facilities for MALDI-TOF/TOF MS at JALMA, Agra.
Patent Applied: Sarman Singh, Gopinath K., Amit Singh and Niti Singh. Novel protein markers of drug resistance in Mycobacterium tuberculosis (1752/DEL/2008).
References
- 1.World Health Organization. Global Tuberculosis Report 2014. [accessed on November 3, 2014]. Available from: http://apps.who.int/iris/bitstream/10665/137094/1/9789241564809_eng.pdf?ua=1 .
- 2.Rattan A, Kalia A, Ahmad N. Multidrug-resistant Mycobacterium tuberculosis: molecular perspectives. Emerg Infect Dis. 1998;4:195–209. doi: 10.3201/eid0402.980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramaswamy SV, Reich R, Dou SJ, Jasperse L, Pan X, Wanger A, et al. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47:1241–50. doi: 10.1128/AAC.47.4.1241-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gryadunov D, Mikhailovich V, Lapa S, Roudinskii N, Donnikov M, Panikov S, et al. Evaluation of hybridization on oligonucleotide microarrays for analysis of drug-resistant Mycobacterium tuberculosis. Clin Microbiol Infect. 2005;11:531–9. doi: 10.1111/j.1469-0691.2005.01183.x. [DOI] [PubMed] [Google Scholar]
- 5.Louw GE, Warren RM, Gey van Pittius NC, McEvoy CR, Van Helden PD, Victor TC. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob Agents Chemother. 2009;53:3181–9. doi: 10.1128/AAC.01577-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson M, DeRisi J, Kristensen HH, Imboden P, Rane S, Brown PO, et al. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–8. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R, Marcotte EM. The proteomic response of Mycobacterium smegmatis to anti-tuberculosis drugs suggests targeted pathways. J Proteome Res. 2008;7:855–65. doi: 10.1021/pr0703066. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Zhang W, Gao F, Huang Y, Lu C, Wang H. Comparison of the proteome of isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis. Microb Drug Resist. 2006;12:231–8. doi: 10.1089/mdr.2006.12.231. [DOI] [PubMed] [Google Scholar]
- 9.Sharma P, Kumar B, Singhal N, Katoch VM, Venkatesan K, Chauhan DS, et al. Streptomycin induced protein expression analysis in Mycobacterium tuberculosis by two-dimensional gel electrophoresis & mass spectrometry. Indian J Med Res. 2010;132:400–8. [PubMed] [Google Scholar]
- 10.Jungblut PR, Schaible UE, Mollenkopf HJ, Zimny-Arndt U, Raupach B, Mattow J, et al. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol Microbiol. 1999;33:1103–17. doi: 10.1046/j.1365-2958.1999.01549.x. [DOI] [PubMed] [Google Scholar]
- 11.Sharma P, Kumar B, Gupta Y, Singhal N, Katoch VM, Venkatesan K, et al. Proteomic analysis of streptomycin resistant and sensitive clinical isolates of Mycobacterium tuberculosis. Proteome Sci. 2010;8:59. doi: 10.1186/1477-5956-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopinath K, Singh S. Multiplex PCR assay for simultaneous detection and differentiation of Mycobacterium tuberculosis, Mycobacterium avium complexes and other Mycobacterial species directly from clinical specimens. J Appl Microbiol. 2009;107:425–35. doi: 10.1111/j.1365-2672.2009.04218.x. [DOI] [PubMed] [Google Scholar]
- 13.Singh A, Gopinath K, Singh N, Singh S. Deciphering the sequential events during in vivo acquisition of drug resistance in Mycobacterium tuberculosis. Int J Mycobacteriol. 2014;3:36–40. doi: 10.1016/j.ijmyco.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Patricia TK, Cubica GP. Atlanta, Ga: U.S Department of Health and Human Services, Public Health Service, Centers for Disease Control; 1995. Public health mycobacteriology: a guide for the level III laboratory. [Google Scholar]
- 15.Demay C, Liens B, Burguière T, Hill V, Couvin D, Millet J, et al. SITVITWEB - a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12:755–66. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 17.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 18.Schulze U, Larsen ME, Villadsen J. Determination of intracellular trehalose and glycogen in Saccharomyces cerevisiae. Anal Biochem. 1995;228:143–9. doi: 10.1006/abio.1995.1325. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ÄÄCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Reddy PV, Puri RV, Khera A, Tyagi AK. Iron storage proteins are essential for the survival and pathogenesis of Mycobacterium tuberculosis in THP-1 macrophages and the guinea pig model of infection. J Bacteriol. 2012;194:567–75. doi: 10.1128/JB.05553-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattow J, Siejak F, Hagens K, Becher D, Albrecht D, Krah A, et al. Proteins unique to intraphagosomally grown Mycobacterium tuberculosis. Proteomics. 2006;6:2485–94. doi: 10.1002/pmic.200500547. [DOI] [PubMed] [Google Scholar]
- 22.Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010;6:e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pheiffer C, Betts JC, Flynn HR, Lukey PT, van Helden P. Protein expression by a Beijing strain differs from that of another clinical isolate and Mycobacterium tuberculosis H37Rv. Microbiology. 2005;151:1139–50. doi: 10.1099/mic.0.27518-0. [DOI] [PubMed] [Google Scholar]
- 24.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2011;44:106–10. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dussurget O, Rodriguez M, Smith I. Protective role of the Mycobacterium smegmatis IdeR against reactive oxygen species and isoniazid toxicity. Tuber Lung Dis. 1998;79:99–106. doi: 10.1054/tuld.1998.0011. [DOI] [PubMed] [Google Scholar]
- 26.Wolff KA, Nguyen HT, Cartabuke RH, Singh A, Ogwang S, Nguyen L, et al. Protein kinase G is required for intrinsic antibiotic resistance in mycobacteria. Antimicrob Agents Chemother. 2009;53:3515–9. doi: 10.1128/AAC.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukherjee P, Sureka K, Datta P, Hossain T, Barik S, Das KP, et al. Novel role of Wag31 in protection of mycobacteria under oxidative stress. Mol Microbiol. 2009;73:103–19. doi: 10.1111/j.1365-2958.2009.06750.x. [DOI] [PubMed] [Google Scholar]
- 28.O’Hare HM, Durán R, Cerveñansky C, Bellinzoni M, Wehenkel AM, Pritsch O, et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70:1408–23. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 29.Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.England P, Wehenkel A, Martins S, Hoos S, André-Leroux G, Villarino A, et al. The FHA-containing protein GarA acts as a phosphorylation-dependent molecular switch in mycobacterial signaling. FEBS Lett. 2009;583:301–7. doi: 10.1016/j.febslet.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 31.Bourassa L, Camilli A. Glycogen contributes to the environmental persistence and transmission of Vibrio cholerae. Mol Microbiol. 2009;72:124–38. doi: 10.1111/j.1365-2958.2009.06629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]