Abstract
Background & objectives:
Of the three major genotypes of Mycobacterium avium subspecies paratuberculosis (MAP), ‘Bison type’ is most prevalent genotype in the domestic livestock species of the country, and has also been recovered from patients suffering from Crohn's disease. Recently, a new assay based on IS1311 locus 2 PCR- restriction endonuclease analysis (REA) was designed to distinguish between ‘Indian Bison type’ and non-Indian genotypes. The present study investigated discriminatory potential of this new assay while screening of a panel of MAP isolates of diverse genotypes and from different geographical regions.
Methods:
A total of 53 mycobacterial isolates (41 MAP and 12 Mycobacterium other than MAP), three MAP genomic DNA and 36 MAP positive faecal DNA samples from different livestock species (cattle, buffaloes, goat, sheep and bison) and geographical regions (India, Canada, USA, Spain and Portugal) were included in the study. The extracted DNA samples (n=92) were analyzed for the presence of MAP specific sequences (IS900, ISMav 2 and HspX) using PCR. DNA samples were further subjected to genotype differentiation using IS1311 PCR-REA and IS1311 L2 PCR-REA methods.
Results:
All the DNA samples (except DNA from non-MAP mycobacterial isolates) were positive for all the three MAP specific sequences based PCRs. IS1311 PCR-REA showed that MAP DNA samples of Indian origin belonged to ‘Bison type’. Whereas, of the total 19 non-Indian MAP DNA samples, 2, 15 and 2 were genotyped as ‘Bison type’, ‘Cattle type’ and ‘Sheep type’, respectively. IS1311 L2 PCR-REA method showed different restriction profiles of ‘Bison type’ genotype as compared to non-Indian DNA samples.
Interpretation & conclusions:
IS1311 L2 PCR-REA method successfully discriminated ‘Indian Bison type’ from other non-Indian genotypes and showed potential to be future epidemiological tool and for genotyping of MAP isolates.
Keywords: Genotypes, Indian bison type, IS1311 L2 PCR-REA, Mycobacterium avium subspecies paratuberculosis, paratuberculosis
Mycobacterium avium subspecies paratuberculosis (MAP) is a cause of chronic granulomatous enteritis and lymphadinits (paratuberculosis or Johne's disease) in different species of animals1 including primates2. Johne's disease (JD) is responsible for inflicting huge economic losses to the livestock industry worldwide due to reduced milk production, increased premature culling and mortality and reduced fertility3. MAP-infected animals (sub-clinical and clinical) shed live bacilli in faeces and milk, thereby increasing the risk of transmission to newborn and other susceptible animals. Recently MAP has also been shown to be associated with Crohn's disease4,5 in human beings. MAP escapes standard pasteurization temperatures, and, therefore, presence of live MAP bacilli in the milk supplies is of great concern and poses potential risk to human population6,7.
Control of MAP infection is the priority of developed countries to secure animal productivity and to reduce chances of human exposure. Inspite of the availability of sensitive tests8,9, ‘Test and cull’ method is not effective for the control of disease in livestock herds/flocks. Vaccination is considered as the method of choice for the control of JD in animals10,11. However, efficacy of vaccines depends on the genotype of candidate strain used11,12. Therefore, knowledge of the genotypes infecting domestic livestock species is critical for designing disease control strategies.
Based on IS1311 polymerase chain reaction-restriction endonuclease analysis (PCR-REA) method, MAP isolates have been grouped into three genetically distinct genotypes (’Cattle type’, ‘Sheep type’ and ‘Bison type)13,14. Studies reported host preferences of these MAP geno-groups, however, host adaptation is not absolute and inter-species sharing of MAP genotypes has been reported15. ‘Bison type’ genotype was first reported from wild bison of Montana, USA13, and later similar genotypes have been reported as major genotypes infecting domestic livestock, wild ruminants and human population in India16. Recently, ‘Bison type’ genotype has also been reported from other regions of Asia (Korea)17 and Africa (Uganda)18. We have earlier identified genomic variations in terms of genetic rearrangements, in-del polymorphisms and locus polymorphisms in ‘Bison type’ genotype of Indian origin when compared with genotypes reported from other parts of the world19,20. Therefore, new nomenclature ‘Indian Bison type’ was assigned to ‘native Bison type’ genotype20. Another molecular signature (sequence variations of native genotype) was deletion of two base pairs (TG) at 64th and 65th positions of IS1311 element particularly at locus 2 as compared to non-Indian isolates19,20. Taking the advantage of this molecular signature, a new IS1311 locus 2 specific PCR-REA (IS1311 L2 PCR-REA) assay was optimized for discrimination of ‘Indian Bison type’ from other isolates21. However, validation of this newly optimized assay was not done on a large panel of MAP isolates from different geographical regions of the world. The present study was carried out to investigate applicability of IS1311 L2 PCR-REA assay on samples of diverse genotypes (Cattle type, Sheep type and Bison type) and different geographical regions (India, Canada, USA, Spain and Portugal).
Material & Methods
A total of 53 mycobacterial isolates (41 MAP and 12 mycobacteria other than MAP), three MAP genomic DNA and 36 MAP faecal DNA samples from different hosts and geographical regions were processed for the investigation of genomic variations using IS1311 PCR-REA and IS1311 L2 PCR-REA. The study was performed between August 2010 and July 2013 at the Department of Microbiology and Molecular Biology, National JALMA Institute for Leprosy and Other Mycobacterial Diseases (NJIL&OMD) located in Agra, Uttar Pradesh, India. Of the 41 MAP isolates, 25, 3, 2 and 11 were from India, Canada, Spain and USA, respectively (Table). Three MAP genomic DNA samples of Portugal origin (provided by Dr Maria Gazouli, School of Animal Science, Agricultural University, Athens, Greece) were also included in this study.
Table.
Genotyping of MAP isolates/ DNA using IS1311 PCR-REA and locus 2 specific IS1311 PCR-REA method
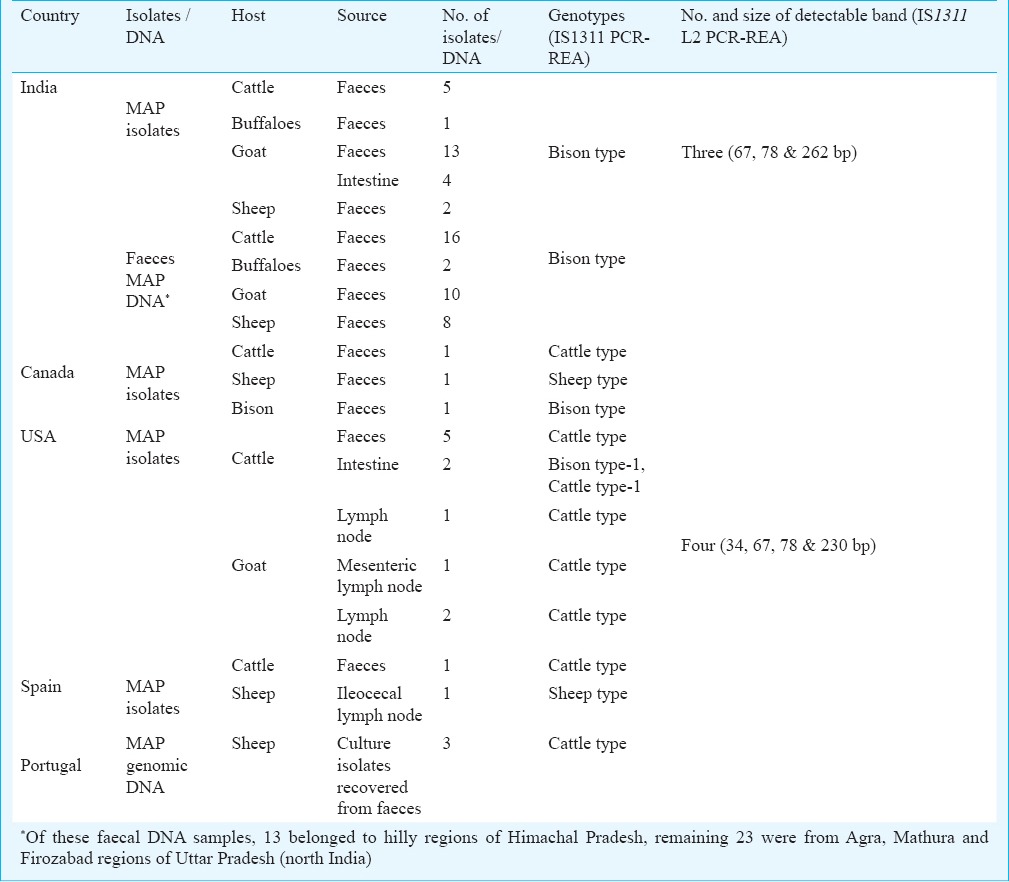
MAP isolates (n=25) and faecal DNA samples (n=36) of Indian origin were collected from epidemiological studies conducted in Agra, Mathura, Jhansi and hilly regions of northern India (data not shown). To check the specificity of IS1311 L2 PCR-REA, a total of 12 mycobacterial isolates other than MAP [M. smegmatis, M. vaccae, M. marinum, M. chelonae, M. flavescens, M.fortuitum, M. kansasii, M. bovis, M. bovis (BCG), M. avium, M.gastri, M. indicus pranii] were obtained from mycobacterial repository, NJIL&OMD, Agra. All the mycobacterial isolates (n=53) were subjected to isolation of genomic DNA as per the method of van Soolingen et al22. DNA samples recovered from isolates and faecal samples were subjected to IS90023, ISMav224 and HspX25 specific PCRs for the molecular identification of MAP.
IS1311 PCR-REA (Genotyping of isolates/DNA): Genotyping of each MAP DNA obtained from mycobacterial isolates or faecal samples was carried out by IS1311 PCR-REA method14. Briefly, reaction was carried out in 30 µl volume, containing 20 µl positive IS1311 PCR product, 3 µl 10X buffer (Fermentas, USA), 2 units of each endonuclease HinfI and MseI (Fermentas). Reaction mixture was incubated at 37⁰C for 1.5 h. Band patterns were visualized after electrophoresis on 4 per cent high resolution agarose gel stained with ethidium bromide and genotype profile interpretation was done as described by Sevilla et al14.
IS1311 locus 2 PCR-REA (Sub-genotyping of MAP isolates): IS1311 L2 PCR-REA optimized by Sohal et al21 was used to characterize all MAP isolates/faecal DNA included in this study. Firstly, locus 2 of IS1311 was amplified using the specific primers (P1: CACCAACCATGCAGAGGTAA; P2: GGAATCCGCAACTCCAAAT) and then amplicons were subjected to restriction digestion using BsaJ1. PCR reaction mix contained primers (10 pmoles), Taq polymerase (1 unit), MgCl2 (1.5 mM), dNTPs (0.2 mM), buffer 10X (2.5 µl) and template DNA (5 ng) in a final volume of 25.0 µl at thermocycler conditions: denaturation at 95 °C for 5 min followed by 40 cycles of de-naturation at 95 °C for 30 sec, annealing at 55 °C at 30 sec, extension at 72 °C for 1 min followed final extension at 72 °C for 7 min. PCR products were visualized on 1.5 per cent agarose gel. Amplification products (~425 bp) were digested with BsaJI enzyme for 2 h at 37°C and band pattern was observed on 4 per cent agarose gel.
The study protocol was approved by the institute's ethical committee.
Results
MAP specific IS900, ISMav 2 and HspX PCRs showed presence of MAP DNA in all samples (Figs. 1–3) except those extracted from isolates of mycobacteria other than MAP.
Fig. 1.
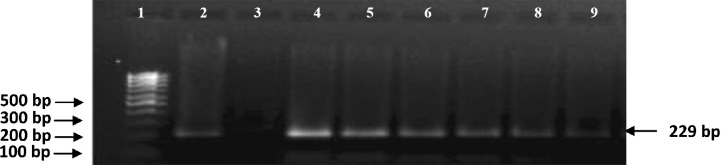
Molecular identification of MAP isolates by using PCR targeting IS900 sequence (specific product size 229bp). Lane 1: 100bp marker; Lane 2: Positive control (Indian Bison type MAP S5), Lane 3: Negative control (deionized distilled water); Lane 4: isolates no. T7 (MAP positive); Lane 5: isolates no. 22G (MAP positive); Lane 6: isolates no. R27 (MAP positive); Lane 7: isolates no. MAP 16 (MAP positive); Lane 8: isolates no. MAP 18 (MAP positive), Lane 9: isolates no.W213 (MAP positive).
Fig. 3.
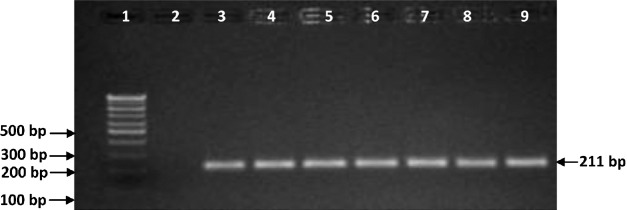
Molecular identification of MAP isolates using PCR targeting HspX sequence (Specific product size 211bp). Lane 1: 100bp marker; Lane 2: Negative control (deionized distilled water); Lane 3: Positive control (Indian Bison type MAP S5); Lane 4: isolates no.T7 (MAP positive); Lane 5: isolates no.22G (MAP positive); Lane 6: isolates no. R27 (MAP positive); Lane 7: isolates no. MAP 16 (MAP positive); Lane 8: isolates no. MAP 18 (MAP positive), Lane 9: isolates no.W213 (MAP positive).
Fig. 2.
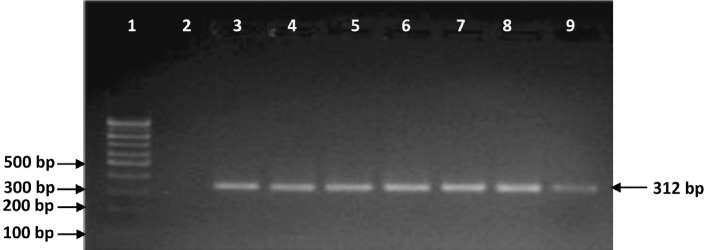
Molecular identification of MAP isolates by using PCR targeting ISMav02 sequence (specific product size 312 bp). Lane 1: 100bp marker; Lane 2: Negative control (deionized distilled water); Lane 3: Positive control (Indian Bison type MAP S5); Lane 4: isolates no.T7 (MAP positive); Lane 5: isolates no.22G (MAP positive); Lane 6: isolates no.R27 (MAP positive); Lane 7: isolates no.MAP 16 (MAP posi-tive); Lane 8: isolates no. MAP 18 (MAP positive), Lane 9: isolates no.W213 (MAP positive).
IS1311 PCR-REA (Genotyping): Of the 80 MAP-DNA samples, 63, 15 and 2 showed the pattern of ‘Bison type’, ‘Cattle type’ and ‘Sheep type’ genotypes, respectively (Table, Fig. 4). All MAP DNA samples of Indian origin belonged to ‘Bison type’. Of the three isolates of Canada, one each was genotyped as ‘Cattle type’, ‘Sheep type’ and ‘Bison type’. Of the 11 isolates of USA origin, one and 10 were genotyped as ‘Bison type’ and Cattle type, respectively. Of the two isolates from Spain, one was ‘Cattle type’ and another was identified as ‘Sheep type’, whereas, all three isolates of Portugal origin were identified as ‘Cattle type’ (Table).
Fig. 4.
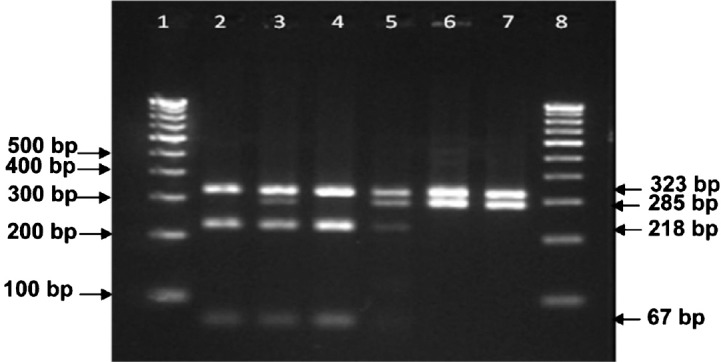
Genotyping of MAP isolates using IS1311 polymerase chain reaction-restriction endonuclease analysis. Lane 1: 100bp ladder; Lane 2: isolates no. MAP16 (Bison type of Indian origin); Lane 3: isolates no.429 (Cattle type of Spain origin); Lane 4: isolates no. B1 (Bison type of Canada origin); Lane 5: isolates no.W213 (Cattle type of USA origin); Lane 6: isolates no.22G (Sheep type of Spain origin), Lane 7: isolates no.MAPS1 (Sheep type of Canada origin), Lane 8: 100 bp DNA ladder.
IS1311 L2 PCR-REA (Sub-genotyping): Restriction profile and band pattern for marker IS1311 L2 PCR-REA distinguished ‘Bison type’ DNA of Indian origin from all non-Indian MAP samples (Table, Fig. 5). After restriction digestion of ~425 bp product (belonging to IS1311 element at locus 2) with BsaJI enzyme, four digestion products were visualized in the 4 per cent high resolution agarose gel (34, 67, 78 & 230 bp) for non-Indian MAP. However, digestion of IS1311 locus 2 amplicon of MAP DNA of Indian origin resulted into only three detectable bands in the gel (67, 78 & 262 bp).
Fig. 5.
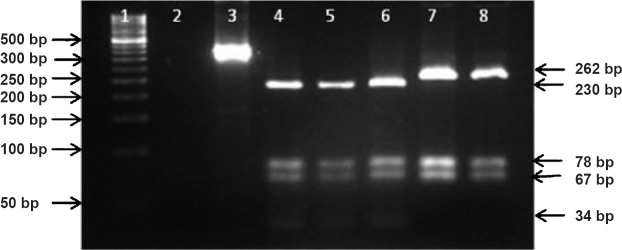
Discriminatory restriction pattern of representative MAP isolates of Indian and non-Indian origin using IS1311 PCR locus 2-REA method. Lane 1 Marker (50bp); Lane 2: Negative control, Lane 3: Undigested PCR product (~425bp); Lane 4: isolates no. W213 (Cattle type of USA origin); Lane 5: isolates no. 22G (Sheep type of Spain origin); Lane 6: isolates no. MAP B1 (Bison type of Canada origin); Lane 7: isolates no. MAP 16 (Bison type of Indian origin); Lane 8: isolates no. MAP 18 (Bison type of Indian origin).
On evaluating the specificity of IS1311 L2 PCR-REA on 12 non-MAP mycobacterial DNA, none were amplified except DNA of M. avium. However, the restriction profile of M. avium was different to that of MAP.
Discussion
Molecular epidemiology of MAP has been poorly understood due to slow growth of the pathogen in artificial medium. Rapid strain differentiation methods are pre-requisite to understand the origin of infection, disease transmission and to design disease control strategies. In the present study, IS1311 PCR-REA method was applied on MAP isolates/DNA from India, Canada, USA, Spain and Portugal, and MAP positive faecal DNA samples of domestic ruminants of north India for determining the genotype. Further, potential of recently described IS1311 PCR L2 PCR-REA method was evaluated for genomic marker based differentiation between Indian and non-Indian strains.
In the present study, genomic and faecal DNA samples obtained from MAP infected animals of north India were genotyped as Bison type. These results were similar to that reported in previous epidemiological investigations conducted in north India16, and ‘Bison type’ genotype was identified as the dominant genotype. Contrary to the present study, ‘Cattle type’ genotype of MAP was found as predominant genotype infecting domestic livestock, wild ruminant and non-ruminant species in other countries17,26. ‘Sheep type’ strains are rarely associated with paratuberculosis in species other than sheep15,27. In present study, we could not detect any ‘Cattle type’ and ‘Sheep type’ genotype of MAP in animals from north India. Previously ‘Cattle type’ strains have been reported from cattle and human population of northern India16.
The results showed that new IS1311 L2 PCR-REA assay successfully discriminated ‘Bison type’ genotype of Indian origin (’Indian Bison type’) from MAP isolates of other genotypes (Cattle type, Sheep type and Bison type) of non-Indian origin. The test was found to be very specific as all mycobacterial isolates (except M. avium) other than MAP could not be amplified by the MAP specific primers. Due to high genetic similarity between M. avium and MAP, IS1311 L2 PCR amplified the DNA of both. However, the restriction profiles of IS1311 locus 2 by BsaJ1 restriction enzyme were different between both species. The present finding confirmed that ‘TG’ gap deletion at 64th and 65th position of IS1311 element at locus 2 was a stable marker and could be used in future strain typing as ‘Molecular signature’ and in epidemiological investigations. This newly optimized tool successfully worked on the clinical DNA samples obtained from faecal samples of MAP infected animals. Further, this assay can give results much faster (1 day) than culture based typing methods (i.e. RFLP or PFGE) and is particularly suitable in conditions where we may have culture negative results.
In conclusion, our study demonstrates that IS1311 L2 PCR-REA assay is a rapid, and easy to perform method for the differentiation of ‘Bison type’ MAP isolates of Indian origin from non-Indian MAP isolates of different genotypes. MAP is an important livestock pathogen world over which besides inviting trade restrictions, is a potential human pathogen.
Acknowledgment
Authors acknowledge the Indian Council of Agricultural Research, New Delhi, for financial support and Indian Council of Medical Research, New Delhi for Post Doctoral Fellowship to the first and the fourth authors (AVS and PKS). Authors thank Dr R.A. Juste, NEIKER, Spain; Dr Maria Gazouli, School of Animal Science, Agricultural University, Athens, Greece and Dr M.T. Collins, School of Veterinary Medicine, University of Wisconsin, Madison, for providing M. avium subsp. paratuberculosis isolates/DNA samples.
References
- 1.Harris NB, Barletta RG. Mycobacterium avium subsp. paratuberculosis in Veterinary Medicine. Clin Microbiol Rev. 2001;14:489–512. doi: 10.1128/CMR.14.3.489-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClure HM, Chiodini RJ, Anderson DC, Swenson RB, Thayer WR, Coutu JA. Mycobacterium paratuberculosis infection in a colony of stumptail macaques (Macaca arctoides) J Infect Dis. 1987;155:1011–9. doi: 10.1093/infdis/155.5.1011. [DOI] [PubMed] [Google Scholar]
- 3.Hasonova L, Pavlik I. Economic impact of paratuberculosis in dairy cattle herds: a review. Vet Med Czech. 2006;51:193–211. [Google Scholar]
- 4.Pierce ES. Where are all the Mycobacterium avium subspecies paratuberculosis in patients with Crohn's disease? PLoS Pathog. 2009;5:e1000234. doi: 10.1371/journal.ppat.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wynne JW, Bull TJ, Seemann T, Bulach DM, Wagner J, Kirkwood CD, et al. Exploring the zoonotic potential of Mycobacterium avium subspecies paratuberculosis through comparative genomics. PLoS One. 2011;6:e22171. doi: 10.1371/journal.pone.0022171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant IR, Ball HJ, Rowe MT. Incidence of Mycobacterium paratuberculosis in bulk raw and commercially pasteurized cows’ milk from approved dairy processing establishments in the United Kingdom. Appl Environ Microbiol. 2002;68:2428–35. doi: 10.1128/AEM.68.5.2428-2435.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shankar H, Singh SV, Singh PK, Singh AV, Sohal JS, Greenstein RJ. Presence characterization and genotype profiles of Mycobacterium avium subspecies paratuberculosis from unpasteurized individual and pooled milk, commercial pasteurized milk and milk products in India by culture, ELISA, PCR and PCR-REA methods. Int J Infect Dis. 2010;14:121–6. doi: 10.1016/j.ijid.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 8.Wadhwa A, Bannantine JP, Byrem TM, Stein TL, Saxton AM, Speer CA, et al. Optimization of serum EVELISA for milk testing of Johne's disease. Foodborne Patho Dis. 2012;9:749–54. doi: 10.1089/fpd.2011.1115. [DOI] [PubMed] [Google Scholar]
- 9.Singh SV, Singh PK, Kumar H, Sohal JS, Singh AV. Evaluation of blood PCR with blood culture, ELISA and microscopic examination for the diagnosis of Mycobacterium avium subspecies paratuberculosis infection in goat kids. Indian J Small Rumin. 2010;16:67–73. [Google Scholar]
- 10.Perez V, Garcia Marin JF, Bru R, Moreno B, Badiola JJ. Results of a paratuberculosis vaccination trial in adult sheep. Med Vet. 1995;12:196–201. [Google Scholar]
- 11.Singh SV, Singh PK, Singh AV, Sohal JS, Gupta VK. Comparative efficacy of an indigenous ‘Inactivated vaccine’ using highly pathogenic field strain of Mycobacterium avium subspecies paratuberculosis ‘Bison type’ with a commercial vaccine for the control of Capri - Paratuberculosis in India. Vaccine. 2007;25:7102–10. doi: 10.1016/j.vaccine.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 12.Uzonna JE, Chilton P, Whitlock RH, Habecker PL, Scott P, Sweeney RW. Efficacy of commercial and field-strain Mycobacterium paratuberculosis vaccinations with recombinant IL-12 in a bovine experimental infection model. Vaccine. 2003;21:3101–9. doi: 10.1016/s0264-410x(03)00261-5. [DOI] [PubMed] [Google Scholar]
- 13.Whittington RJ, Marsh IB, Whitlock RH. Typing of IS1311 polymorphisms confirms that bison (Bison bison) with paratuberculosis in Montana are infected with a strain of Mycobacterium avium subsp. paratuberculosis distinct from that occurring in cattle and other domestic livestock. Mol Cell Probes. 2001;15:139–45. doi: 10.1006/mcpr.2001.0346. [DOI] [PubMed] [Google Scholar]
- 14.Sevilla I, Singh SV, Garrido JM, Aduriz G, Rodriguez S, Geijo MV, et al. Molecular typing of Mycobacterium avium subspecies paratuberculosis strains from different hosts and regions. Rev Sci Tech Rev. 2005;24:1061–6. [PubMed] [Google Scholar]
- 15.Stevenson K, Alvarez J, Bakker D, Biet F, de Juan L, Denham S, et al. Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants. BMC Microbiol. 2009;9:212. doi: 10.1186/1471-2180-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh SV, Sohal JS, Singh PK, Singh AV. Genotype profiles of Mycobacterium avium subspecies paratuberculosis isolates recovered from animals, commercial milk and human beings in North India. Int J Infect Dis. 2009;13:221–7. doi: 10.1016/j.ijid.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 17.Kim JM, Ku BK, Lee HN, Jang YB, Her M, Kim JW, et al. In: International Association for Paratubersulosis (Ed.), Proceedings of the 11th International Colloquium on Paratuberculosis. Sydney, Australia: 2012. Mycobacterium avium subsp. paratuberculosis in wild boards from Korea; p. 175. [Google Scholar]
- 18.Okuni JB, Dovas CI, Loukopoulos P, Bouzalas IG, Kateete DP, Joloba ML, et al. Isolation of Mycobacterium avium subspecies paratuberculosis from Ugandan cattle and strain differentiation using optimised DNA typing techniques. BMC Vet Res. 2012;8:99. doi: 10.1186/1746-6148-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohal JS, Sheoran N, Narayanasamy K, Brahmachari V, Singh S, Subodh S. Genomic analysis of local isolate of Mycobacterium avium subspecies paratuberculosis. Vet Microbiol. 2009;134:375–82. doi: 10.1016/j.vetmic.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 20.Sohal JS, Singh SV, Singh PK, Singh AV. On the evolution of ‘Indian Bison type’ Mycobacterium avium subspecies paratuberculosis. Microbiol Res. 2010;165:163–71. doi: 10.1016/j.micres.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Sohal JS, Singh SV, Singh PK, Singh AV, Kumar N. A new marker IS 1311 L2 PCR-REA for identification of ‘Indian Bison Type’ Mycobacterium avium subspecies paratuberculosis. Indian J Biotech. 2013;12:204–7. [Google Scholar]
- 22.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–86. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vary PH, Andersen PR, Green E, Hermon-Taylor J, McFadden JJ. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J Clin Microbiol. 1990;28:933–7. doi: 10.1128/jcm.28.5.933-937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mobius P, Hotzel H, Rassbach A, Kohler H. Comparison of 13 single-round and nested PCR assays targeting IS900, ISMav2, f57 and locus 255 for detection of Mycobacterium avium subsp. paratuberculosis. Vet Microbiol. 2008;126:324–33. doi: 10.1016/j.vetmic.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Ellingson JLE, Stabel JR, Radcliff RP, Whitlock RH, Miller JM. Detection of Mycobacterium avium subspecies paratuberculosis in free-ranging bison (Bison bison) by PCR. Mol Cell Probes. 2005;19:219–25. doi: 10.1016/j.mcp.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Motiwala AS, Li L, Kapur V, Sreevatsan S. Current understanding of the genetic diversity of Mycobacterium avium subsp. paratuberculosis. Microbes Infect. 2006;8:1406–18. doi: 10.1016/j.micinf.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Sevilla I, Garrido J, Geijo M, Juste R. Pulsed-field gel electrophoresis profile homogeneity of Mycobacterium avium subsp. paratuberculosisisolates m from cattle and heterogeneity of those from sheep and goats. BMC Microbiol. 2007;7:12. doi: 10.1186/1471-2180-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]


