Abstract
Background & objectives:
There is a worldwide emergence of fluoroquinolone resistance in Shigella species. To understand the molecular mechanisms associated with fluoroquinolone resistance, naturally occurring fluoroquinolone-resistant strains and laboratory-induced spontaneous mutants of Shigella spp. were used and the relative contributions of acrAB-tolC efflux pumps, gyrase and topoisomerase target gene mutations towards fluoroquinolone resistance were determined.
Methods:
Eight Shigella flexneri and six S. dysenteriae clinical isolates were studied. Three consecutive mutants resistant to ciprofloxacin for S. flexneri SFM1 (≥0.25 µg/ml), SFM2 (≥4 µg/ml) and SFM3 (.32 µg/ml) were selected in 15 steps from susceptible isolates by serial exposure to increasing concentrations of nalidixic acid and ciprofloxacin. Similarly, two mutants for S. dysenteriae SDM1 (≥0.25 µg/ml) and SDM2 (≥4 µg/ml) were selected in eight steps. After PCR amplification sequence analyses of gyrase and topoisomerase target genes were performed. Expression of efflux genes acrA, acrB, acrR and tolC was measured using real-time PCR.
Results:
Mutations were observed in gyrA Ser83 →Leu, Asp87 →Asn/Gly, Val196 →Ala and in parC Phe93 →Val, Ser80 →Ile, Asp101 →Glu and Asp110 →Glu. Overall, acrA and acrB overexpression was associated with fluoroquinolone resistance (P<0.05); while tolC and acrR expression levels did not.
Interpretation & conclusions:
Fluoroquinolone resistance in Shigella spp. is the end product of either a single or a combination of mutations in QRDRs and/ or efflux activity. Novel polymorphisms were observed at Val196 →Ala in gyrA in clinical isolates and Phe93 →Val, Asp101 →Glu, Asp110 →Glu and in parC in majority of laboratory-grown mutants.
Keywords: Efflux pumps, fluoroquinolone resistance, quinolone resistance determining regions (QRDRs), Shigella
Resistance to fluoroquinolones is usually the result of a synergistic action of activation of acrAB-tolC efflux pumps and/or mutations in bacterial quinolone resistance determining regions (QRDRs), and/or plasmid-encoded mechanisms1,2,3,4,5. The mutations in bacterial targets gyrA and gyrB encoding for DNA gyrase and topoisomerase IV cause a change either in the target structure or its binding strength, resulting in less susceptibility6. The plasmid-mediated fluroquinolone resistance is either due to protection of DNA gyrase by a pentapeptide repeat family protein called qnr or ciprofloxacin-modification by an enzyme (aminoglycoside acetyltransferase) encoded by gene aac(6΄)-Ib-cr5,7. Further, the overexpression of efflux pumps results in overall decreased accumulation of antibiotic inside the bacterial cells further resulting in decreased susceptibility8,9. In Gram-negative bacteria, these drug efflux pumps contribute to intrinsic antibiotic resistance and mainly include three superfamilies namely, resistance-nodulation-cell division (RND) family, major facilitator superfamliy (MFS), and the ATP-binding cassette (ABC)4,8. In particular, the proton/drug antiporters belonging to RND family play an important role in clinically relevant resistance in Gram-negative bacteria, and the archetypal bacterial RND pump – acrAB-tolC has been studied extensively in Escherichia coli10,11. This consists of a tripartite system containing a periplasmic membrane fusion protein (MFP) encoded by acrA, an outer membrane protein (OMP) encoded by tolC, and a cytoplasmic membrane transporter (RND) encoded by acrB. This complex forms a channel spanning the entire membrane, governing the transportation of antibiotics from the periplasmic space and cytoplasm to the extracellular environment10,11.
Since several mechanisms are known by which shigellae become resistant to fluroquinolones, we undertook this study to determine the relative contributions of acrAB-tolC efflux pumps, gyrase and topoisomerase target gene mutations towards ciprofloxacin (CIP)/fluoroquinolone (FQ) resistance in Shigella. We used a set of laboratory-induced mutants and naturally occurring fluoroquinolone-resistant isolates of S. flexneri and S. dysenteriae serotype 1 to study the step-wise evolution of quinolone resistance, occurrence of mutations in the QRDR region and changes in the expression of efflux pump encoding genes.
Material & Methods
This study was conducted at the Enteric Laboratory, Department of Medical Microbiology, Postgraduate Institute of Medical Education and Research, Chandigarh, India. The study protocol was approved by the institutional ethics committee.
Eight S. flexneri (618, 497, 44, 488, 804, 710, 592, 993) and six S. dysenteriae (1169, 677, 553, 115, 622, 53) isolates collected from stool specimens of patients suffering from dysentery between 2001-2007 were included in the present study. These isolates were confirmed by biochemical tests and serotyping (Denka-Seiken, Japan). The antibiotic susceptibility for nalidixic acid (NA, 30 µg) and ciprofloxacin (5 µg) (Oxoid Limited, Hampshire, UK) was determined and minimum inhibitory concentrations (MICs) by an E-test (bioMérieux SA, Marcy l’Etoile, France), according to the Clinical and Laboratory Standards Institute (CLSI) guidelines, 201012.
Selection of nalidixic acid and ciprofloxacin-resistant mutants in vitro: Wild-type spontaneous mutants of Shigella exhibiting increased resistance to nalidixic acid and ciprofloxacin were selected from nalidixic acid and ciprofloxacin-susceptible S. dysenteriae 1169 and S. flexneri 497 isolates as described previously for E. coli13. The mutants were selected by in vitro step-wise exposure to increasing concentrations of nalidixic acid (Sigma-Aldrich, St. Louis, MO, USA) and subsequently to ciprofloxacin (Sigma-Aldrich). Briefly, nalidixic acid and ciprofloxacin-susceptible S. flexneri 497 and S. dysenteriae 1169 were grown in increasing concentration of nalidixic acid (0.5,1,2,4,16,32,64,128 µg/ml) on subsequent days in brain heart infusion broth (BHI; BD Difco™; Becton, Dickinson and Co., Detroit, MI, USA) at 37°C overnight. A 0.5 McFarland of overnight culture prepared in normal saline was spread on Mueller-Hinton agar (MHA) (BD Difco™) supplemented with similar concentrations of nalidixic acid (0.5,1,2,4,16,32,64, 128 µg/ml) and was incubated for 24 to 48h at 37°C. Single colonies grown at the respective concentrations were selected and subcultured into BHI broth supplemented with the next higher concentration of antibiotic used in MHA plate. The overnight culture was spread again onto MHA plates supplemented with the next higher concentration of nalidixic acid. S. flexneri (SFM) and S. dysenteriae (SDM) mutants selected from plates with 128 µg/ml of nalidixic acid were subsequently exposed to ciprofloxacin at concentrations ranging from 0.0325 to 44 µg/ml. In vitro selection procedures for ciprofloxacin were similar to those described for the nalidixic acid resistance selections. The mutants were stocked in stock media (BHI with 20% glycerol) supplemented with desired concentration of antibiotic.
PCR analyses for quinolone resistance genes: The gyrA14, gyrB15, parC16, qnrA17 and aac(6’)Ib-cr5 were amplified using primers and conditions as described previously. The nucleotide sequencing was performed for representative amplicons and submitted to GenBank (NCBI, Bethesda, MD, USA).
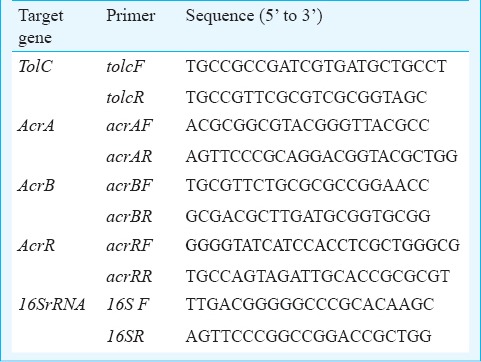
Real time PCR assay for expression of efflux pump genes: The bacterial total RNA was extracted18 and cDNA was synthesized from 1.0µg of RNA using RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas Life Sciences, USA) using random hexamer primers and stored at -80°C. Expression for acrA, acrB, tolC and acrR genes for spontaneous mutants and the clinical isolates was studied. The gene specific primers were designed using Primer 3 software and oligonucleotide sequences are listed in Table I. The cDNA levels of target genes were determined by relative quantitative real time PCR in 10.0 µl reaction mixture containing: 1.0 µl synthesized cDNA solution, 7.0 µl 1X SYBR master mix (Fermentas Life Sciences, USA), 100 nmol/µl of each forward and reverse primer. The amplification was performed under the following reaction conditions: enzyme activation -95°C for 10 min, followed by 35 cycles of denaturation 95°C for 15 sec, annealing 68°C for 20 sec, extension 72°C for 30 sec using Light Cycler 480 (Roche Diagnostics, USA). Threshold cycle (CT) values were calculated from the amplification plots, and the gene expression levels were compared by normalization with endogenous control 16SrRNA gene. The cDNA was diluted 10-5 time for 16SrRNA amplification. The expression for target genes acrA, acrB, tolC, and acrR was measured by 2- ΔΔCT method, where ΔΔCT = (CTtarget – CTreference)S - (CTtarget -CTreference) T19, where S was sensitive strain and T was resistant test strain.
Table I.
Oligonucleotide sequences used in the study
Synergy test: Synergy test was performed for ciprofloxacin using CIP agar method. The efflux pump inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to MHA plate at a concentration of 100 µM2,20. Susceptibility testing for ciprofloxacin by E-test was performed, both in presence and absence of CCCP.
Ciprofloxacin accumulation assay: Accumulation of ciprofloxacin was studied as described by Charvalos et al.21 with some modifications. Cells were grown up to an OD600 of 0.4, harvested and washed with 50 mM phosphate buffer, PH 7.2. Cells were resuspended in the same buffer maintaining an OD600 of 0.20 and energized with 0.2 per cent glucose for 20 min at 30°C. Ciprofloxacin was added at a concentration of 10 µg/ml. Aliquots of 0.5 ml were withdrawn at different time intervals (0, 10 and 20 min) and dispensed into 1 ml glycine hydrochloride, incubated for 60 min at 37°C and fluorescence was measured in a spectrofluorimeter (ciprofloxacin: excitation wavelength 275 nm, emission wavelength 440 nm). CCCP was added (100 µM/ml) to each aliquot at 20 minutes and fluorescence was measured again as described above.
Results
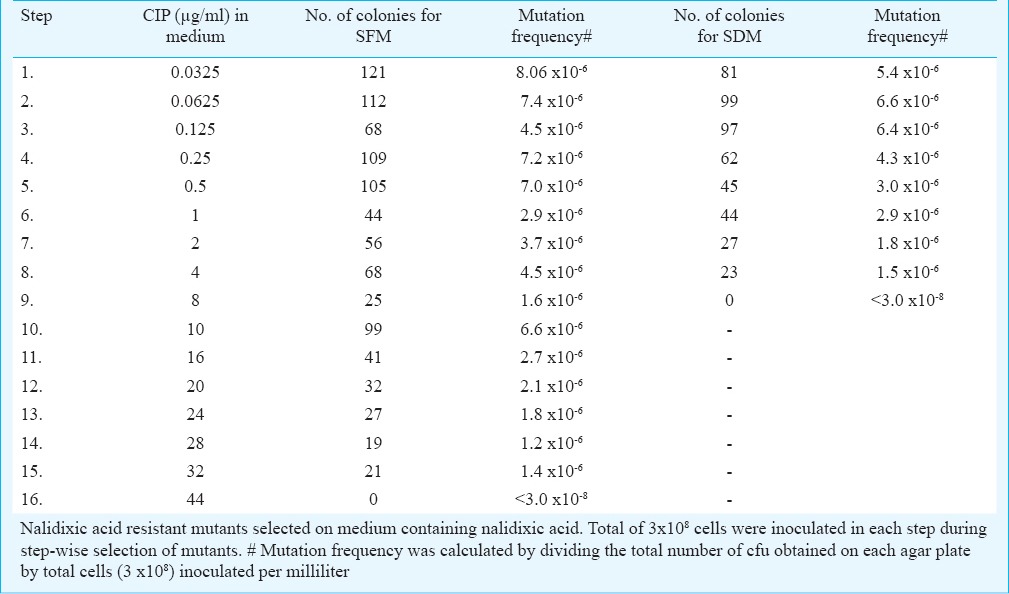
In vitro selection of Shigella mutants resistant to ciprofloxacin: Three consecutive mutants resistant to ciprofloxacin for S. flexneri SFM1 (≥0.25 µg/ml), SFM2 (≥4 µg/ml) and SFM3 (≥32 µg/ml) were selected in 15 steps. Similarly, two mutants for S. dysenteriae SDM1 (≥0.25 µg/ml) and SDM2 (≥4 µg/ml) were selected in eight steps. Table II shows the mutation frequencies for different steps varying between 8.06-1.2 x 10-6.
Table II.
Step-wise selection of ciprofloxacin (CIP) resistant spontaneous mutants of S. flexneri (SFM) and S. dysenteriae (SDM)
Antibiotic susceptibility for clinical isolates: Three resistance patterns were identified in S. flexneri: group I consisted of two susceptible isolates (618, 497) to nalidixic acid (NAS) and ciprofloxacin (CIPS); group II consisted of two isolates (44 and 488) resistant to NAR and susceptible to CIPS; and group III consisted of four isolates (710, 592, 804 and 993) resistant to both NAR and CIPR. For S. dysenteriae: group I consisted of NAS and CIPS (1169, 677) and group II NAR, CIPMS (moderately sensitive to CIP) consisted of 533 and 622, while group III NAR and CIPR consisted of isolates 52 and 115 (Table III).
Table III.
Antibiotic susceptibilities, mutation analyses and synergy testing of study isolates
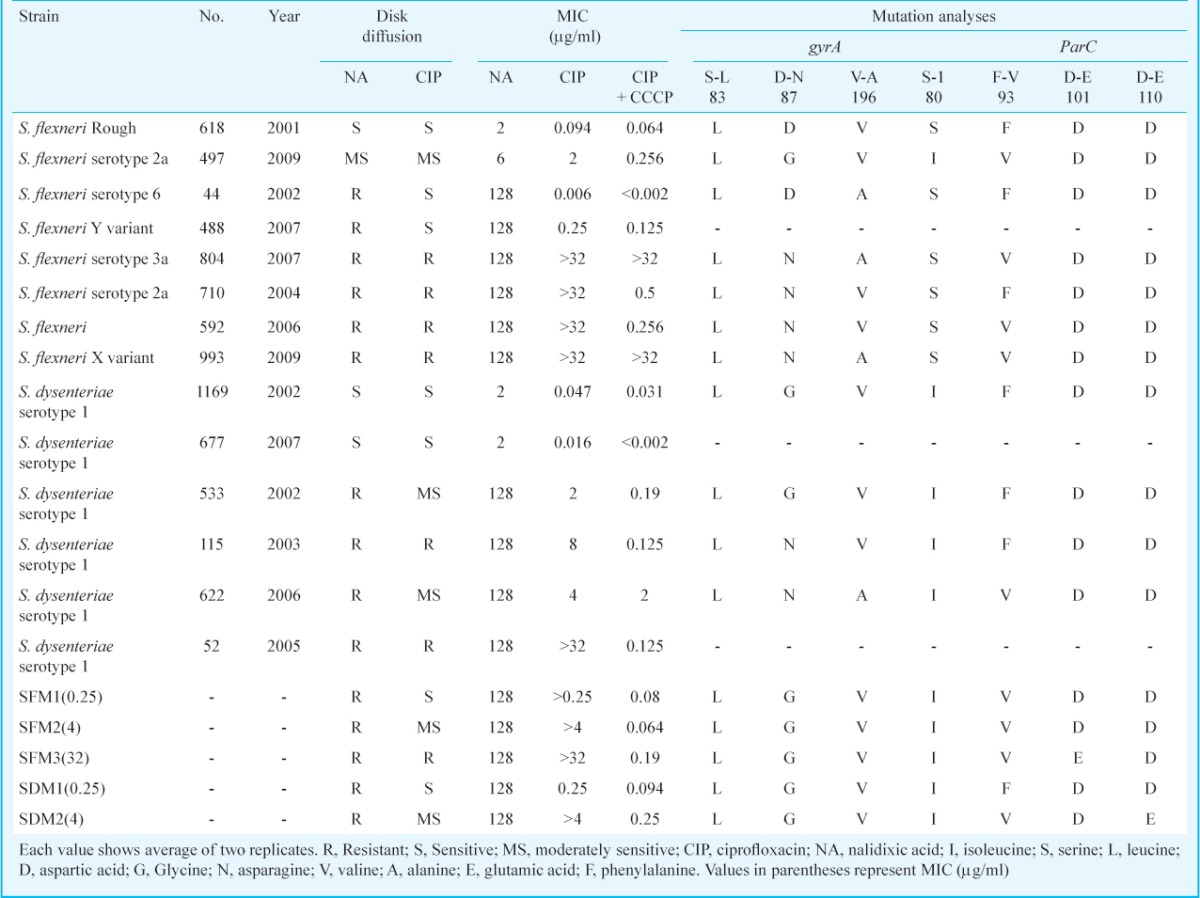
Identification of mutations in clinical isolates and mutants in gyrA, gyrB and parC: On the sequence analyses, mutations were detected in the sequences of gyrA and parC genes. Mutations observed in gyrA were Ser83 →Leu (618, 497, 44, 804, 710, 592, 993, 1169, 533, 622 and 115), Asp87 →Asn/Gly (497, 804, 710, 592, 993, 1169, 533, 622 and 115) and Val196 →Ala (44, 804, 993 and 622). In parC mutations detected were Phe93 →Val (497, 804, 592, 622 and 993) and Ser80 →Ile (497, 1169, 533, 115 and 622). S. flexneri mutants SFM1, SFM2, SFM3 exhibited same genetic profile as wild type 497 strain for gyrA, while substitution in parC, Asp101 →Glu was observed in SFM3. S. dysenteriae mutants SDM1 and SDM2 exhibited same genetic profiles for gyrA to wild type 1169 strain, while SDM2 showed substitutions in parC, Phe93 →Val and Asp110 →Glu (Table III). No mutation was observed in gyrB either in isolates or mutants. The qnrA and aac(6’)Ib-cr genes, which are plasmid mediated quinolone resistance gene, were found absent in all isolates.
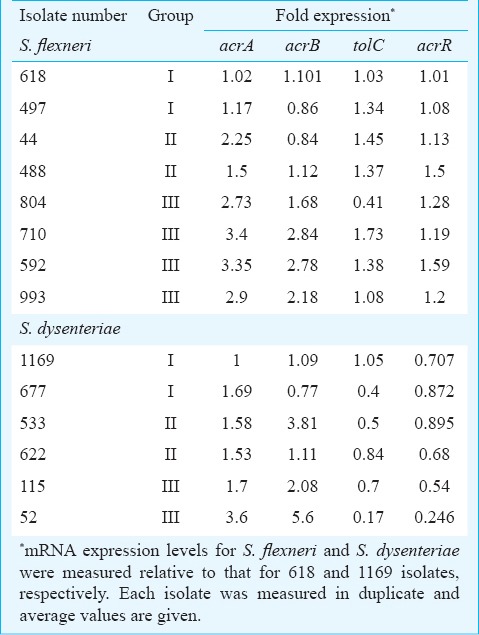
Expression of efflux pump genes: Relative quantification of acrA, acrB, acrR and tolC genes showed highly variable expression in clinical isolates (Table IV). Up- and downregulation of these genes was investigated after culture under the same conditions for all isolates. The normalization of the expression values was done against the sensitive isolates viz., 497 for S. flexneri and 1169 for S. dysenteriae using 16SrRNA as an endogenous control gene. The expression for S. flexneri isolates varied between 1.02-3.4 for acrA, 0.84-2.84 for acrB, 0.41-1.73 for tolC, 1.01-1.59 for acrR (Table IV). Overall a higher expression level of acrA and acrB mRNA in the ciprofloxacin resistant isolates (group III) was seen as compared with susceptible isolates (group I). In case of tolC and acrR, there was no difference in expression among the three groups. The mRNA expression for S. dysenteriae isolates varied between 1.0-3.6 for acrA, 0.77-5.6 for acrB, 0.17-1.05 for tolC and 0.24-0.895 for acrR.
Table IV.
mRNA expression levels for acrA, acrB, tolC and acrR genes for S. flexneri and S. dysenteriae clinical isolates
The spontaneous bacterial mutant colonies were passed in BHI broth in absence and presence of designated concentration of ciprofloxacin and after overnight growth the mRNA expression levels for acrA, acrB, tolC and acrR were measured (Table V). It was observed that the mRNA expression levels for various efflux pump genes in mutants in absence of ciprofloxacin were comparable to the expression observed in resistant clinical isolates. An up-regulation was observed in the expression for acrA, acrB and tolC for mutants in presence of ciprofloxacin while expression for acrR was comparable either in absence or presence of ciprofloxacin.
Table V.
Expression of acrA, acrB, acrR and tolC in spontaneous mutants of S. flexneri 497 and S. dysenteriae 1169

Synergy testing and ciprofloxacin accumulation: To confirm the role of efflux pump towards ciprofloxacin resistance, the susceptibilities of various clinical isolates and spontaneous mutants were checked against ciprofloxacin in absence and presence of CCCP. Overall, a change in susceptibility was observed with 1.5 to 256-fold difference in MIC values on addition of CCCP when compared to those obtained in CCCP-free medium (Table III). The clinical isolates and mutants susceptible to ciprofloxacin (618, 44, 488, 1169, 677, SFM1 and SDM1) showed almost comparable MIC values (only 2.5-7.5-fold difference), showing that there was not much difference in efflux pump activity in these susceptible isolates. Some resistant clinical isolates (804 and 993) showed similar MIC values (>32) both in presence and absence of CCCP showing that resistance towards ciprofloxacin in these clinical isolates seemed to be independent of efflux pump activity. A few resistant clinical isolates (710, 592, 115 and 52) and spontaneous mutants (SFM2, SFM3 and SDM2) showed 64 to 256-fold change in the MIC values in presence and absence of CCCP. In these isolates, after addition of CCCP, the MICs reverted to levels <1 µg/ml in the susceptible range, thus, showing efflux pump activity to be the main mechanism of ciprofloxacin resistance in these isolates.
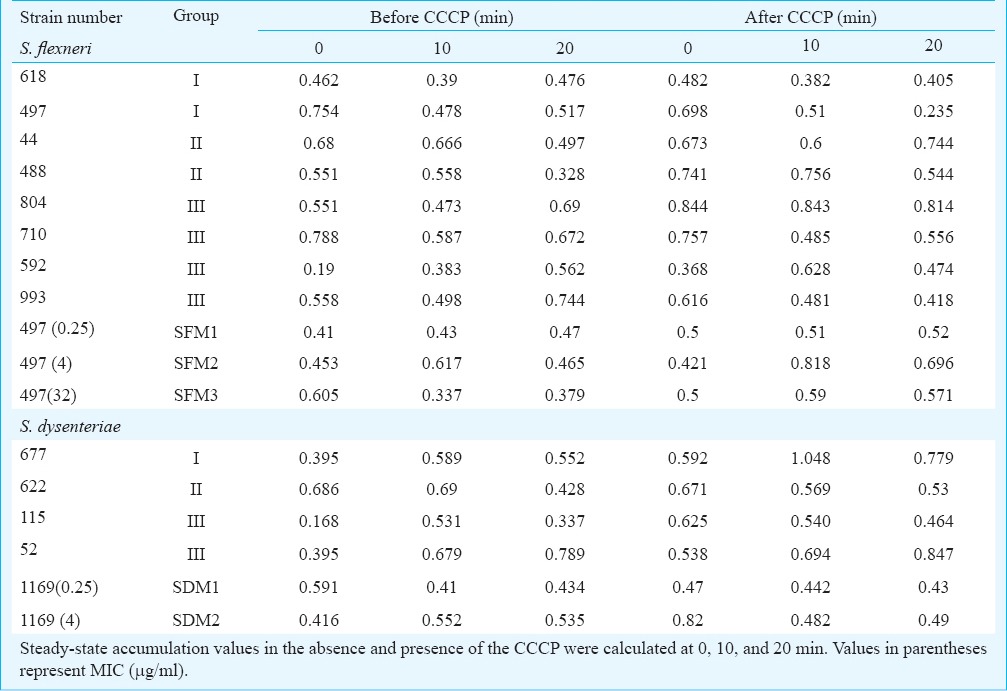
Ciprofloxacin accumulation was further carried out to validate efflux activity (Table VI). The efflux pump activity could be demonstrated for strain numbers 44,592,488,52,677, SFM2 and SFM3 whereas no increase in ciprofloxacin accumulation after CCCP uncoupling could be demonstrated for strain numbers 618,497,804,710,993,622,115, SDM1 and SDM2. Overall, without CCCP, the pump was active and ciprofloxacin was extruded from cells as measured by decreased accumulation inside the cells. Generally, the pump reached its peak activity at 10 min. After inhibition by CCCP, ciprofloxacin started accumulating. An initial increase in ciprofloxacin level reaching a peak between 10 to 20 min was observed (Table VI).
Table VI.
Ciprofloxacin (CIP) accumulation before and after carbonyl cyanide chlorophenyl hydrazone (CCCP) treatment
Discussion
The fluoroquinolone resistance in Shigella spp. is on rise in our region22,23,24. To understand the molecular mechanisms associated with rising fluoroquinolone resistance in Shigella spp., laboratory-induced spontaneous mutants and clinical isolates of S. flexneri and S. dysenteriae exhibiting varied susceptibilities against fluoroquinolones were used to determine the relative contributions of acrAB-tolC efflux pumps and mutations in QRDRs towards ciprofloxacin resistance. The partial sequences of gyrA, gyrB and parC in clinical isolates and mutants were analysed. It is known that atleast two mutations in gyrA are required to reach CLSI breakpoint. Substitution of the highly conserved residue Serine-83 (to Leucine) in gyrA is the commonest mutation. The alteration of the residue Aspartic acid-87 (to Asparagine) further increases the effect of Serine-83 mutation for resistance increase13. All isolates in our study exhibited mutation in gyrA at position 83 (Ser83 →Leu) and a few isolates at position 87 (Asp87 →Asn/Gly) also. These mutations had already been reported from India5, Bangladesh and Nepal25.
Additionally, a novel alteration in gyrA at position 196 (Val196 →Ala) was observed in a few isolates. A mutation in parC at position 80 (Ser80 →Ile) was also detected as previously reported from Kolkata26. A new mutation was observed in parC at position 93 (Phe93 →Val). Two other novel substitutions were observed Asp101 →Glu in SFM3 and Asp110 →Glu in SDM2. Further studies are needed to establish association of these polymorphisms with ciprofloxacin resistance. But it is evident that due to the selection pressure exerted by the overuse of fluoroquinolones, new mutations are expected.
Using real-time PCR based relative quantification, expression of acrA, acrB, tolC and acrR genes in Shigella clinical isolates and spontaneous mutants was studied; acrA and acrB overexpression was found to be associated with fluoroquinolone resistance. Therefore, acrAB overexpression could be an indicator of fluoroquinolone resistance. The acrAB overexpression has already been reported from fluoroquinolone resistant clinical isolates of S. flexneri27. Overall, the level of acrA and acrB mRNA expression in the ciprofloxacin-resistant isolates was significantly higher than that in the susceptible isolates, which supported that overexpression was associated with resistance in certain clinical isolates of S. flexneri and S. dysenteriae. Expression of RND efflux pump target genes in laboratory-grown mutants in absence of ciprofloxacin was comparable to ciprofloxacin-resistant clinical isolates (group III). However, when mutants were grown in the presence of ciprofloxacin, tolC was also upregulated significantly in addition to acrA and acrB. This observation suggested that acrAB-tolC efflux pumps were further overexpressed in presence of ciprofloxacin, leading to decreased accumulation of antibiotic, and reduced susceptibility.
It is known that acrAB-tolC overproduction results in decreased accumulation of antibiotic inside the cells and increased MICs8. These acrAB-tolC pumps are proton/drug antiporters and utilize the energy of proton-motive force to transport antibiotics out of cell28. Therefore, to determine the direct role of efflux pumps overexpression in ciprofloxacin resistance uncoupler CCCP (which inhibits efflux activity) was used. After the disruption of the efflux pump, the MICs for certain ciprofloxacin resistant isolates were almost comparable to ciprofloxacin susceptible isolates. Expectedly, in the ciprofloxacin accumulation assay, a significant decrease in CIP accumulation upon addition of efflux pump inhibitor (CCCP) was observed in a few ciprofloxacin resistant isolates. As evident, efflux pump when operative leads to extrusion of antibiotic and after addition of CCCP antibiotic gets accumulated in the cytosol. This suggested that efflux pump could be one of the factors responsible for the development of resistance in isolates in our region. The role of active efflux using CCCP has also been elucidated previously in Shigella spp.5 and enterotoxigenic E. coli29 from India.
Some of the resistant isolates did not show any decrease in MIC after disruption of efflux activity and exhibited high MIC values. Similar results were obtained with CIP accumulation assay, suggesting strain variations in efflux pump activity. This indicates that resistance in such isolates was due to mutations in QRDRs and independent of efflux activity. In addition to mutations in QRDRs, role of other plasmid-mediated quinolone resistant (qnr) determinants5 and porin proteins30 needs to be studied. Ciprofloxacin is a hydrophilic quinolone that enters the Gram-negative bacteria by outer membrane porins13. Though qnr gene was absent in the present isolates there might be other differences due to porins as described for E.coli30.
The present findings suggest that acrAB overexpression may be used as an indicator of fluoroquinolone resistance in S. flexneri and S. dysenteriae. The expression of these efflux pump genes was found to vary in different clinical isolates with varying susceptibilites. This can be due to the presence of single/double or multiple mutations in QRDRs. Therefore, to remove the bias contributed by these mutations, we induced spontaneous mutants resistant to fluoroquinolones. These spontaneous mutants showed that the resistance was contributed mainly by enhanced expression of acrA and acrB genes, validating the expression in clinical isolates. Further, the use of CCCP abolished efflux activity and some resistant isolates showed comparable MICs to susceptible isolates proving the role of efflux activity towards fluoroquinolone resistance. Some resistant isolates showed no decrease in the MIC values, indicating that resistance was due to a mechanism other than efflux activity. It appears that the mechanism of fluoroquinolone resistance is unique to each isolate and is due to either a single or combination of mutations in QRDRs and/ or efflux activity.
Acknowledgment
This work was financially supported by the Indian Council of Medical Research (ICMR), New Delhi, India.
References
- 1.Chu C, Su LH, Chu CH, Baucheron S, Cloeckaert A, Chiu CH. Resistance to fluoroquinolones linked to gyrA and parC mutations and overexpression of acr AB efflux pump in Salmonella enterica serotype Choleraesuis. Microb Drug Resist. 2005;11:248–53. doi: 10.1089/mdr.2005.11.248. [DOI] [PubMed] [Google Scholar]
- 2.Kim JY, Kim SH, Jeon SM, Park MS, Rhie HG, Lee BK. Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clin Microbiol Infect. 2008;14:760–5. doi: 10.1111/j.1469-0691.2008.02033.x. [DOI] [PubMed] [Google Scholar]
- 3.Oethinger M, Kern WV, Jellen-Ritter AS, McMurry LM, Levy SB. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the acrAB efflux pump. Antimicrob Agents Chemother. 2000;44:10–3. doi: 10.1128/aac.44.1.10-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin Microbiol Infect. 2004;10:12–26. doi: 10.1111/j.1469-0691.2004.00763.x. [DOI] [PubMed] [Google Scholar]
- 5.Pazhani GP, Niyogi SK, Singh AK, Sen B, Taneja N, Kundu M, et al. Molecular chararcterization of multidrug resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J Med Microbiol. 2008;57:856–63. doi: 10.1099/jmm.0.2008/000521-0. [DOI] [PubMed] [Google Scholar]
- 6.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–92. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pu XY, Pan JC, Wang HQ, Zhang W, Huang ZC, Gu YM. Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother. 2009;63:917–20. doi: 10.1093/jac/dkp087. [DOI] [PubMed] [Google Scholar]
- 8.Saier MH, Jr, Paulsen IT, Sliwinski MK, Pao SS, Skurray RA, Nikaido H. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 1998;12:265–74. doi: 10.1096/fasebj.12.3.265. [DOI] [PubMed] [Google Scholar]
- 9.Van Bambeke F, Balzi E, Tulkens PM. Antibiotic efflux pumps. Biochem Pharmacol. 2000;60:457–70. doi: 10.1016/s0006-2952(00)00291-4. [DOI] [PubMed] [Google Scholar]
- 10.Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69:1555–623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zgurskaya HI. Multicomponent drug efflux complexes: architecture and mechanism of assembly. Future Microbiol. 2009;4:919–32. doi: 10.2217/fmb.09.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wayne, PA, USA: CLSI; 2010. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing, 20 th informational supplement M100-S20. [Google Scholar]
- 13.Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994;38:1284–91. doi: 10.1128/aac.38.6.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman M, Mauff G, Levy J, Couturier M, Pulverer G, Glasdorff N, et al. Detection of 4-quinolone resistance mutation in gyrA gene of Shigella dysenteriae type 1 by PCR. Antimicrob Agents Chemother. 1994;38:2488–91. doi: 10.1128/aac.38.10.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gharib AA, Norhan K Abd El-Aziz. Molecular an analysis of quinolone resistance-determining regions in avian pathogenic Escherichia coli. Int J Adv Res. 2013;1:145–57. [Google Scholar]
- 16.Vila J, Ruiz J, Goni P, De Anta MT. Detection of mutations in parC in quinolone resistant clinical isolates of E. coli. Antimicrobial Agents Chemother. 1996;40:491–3. doi: 10.1128/aac.40.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Sahm DF, Jacoby GA, Hooper DC. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob Agents Chemother. 2004;48:1295–9. doi: 10.1128/AAC.48.4.1295-1299.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chomczynski P, Mackey K. Modification of the Trizol reagent procedure for isolation of RNA from polysaccharide and proteoglycan-rich sources. Biotechniques. 1995;19:942–5. [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2- ÄÄCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Pournaras S, Maniati M, Spanakis N, Ikonomidis A, Tassios PT, Tsakris A, et al. Spread of efflux pump-overexpressing, non-metallo-beta-lactamase-producing, meropenem-resistant but ceftazidime-susceptible Pseudomonas aeruginosa in a region with bla VIM endemicity. J Antimicrob Chemother. 2005;56:761–4. doi: 10.1093/jac/dki296. [DOI] [PubMed] [Google Scholar]
- 21.Charvalos E, Tselentis Y, Hamzehpour MM, Köhler T, Pechere JC. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob Agents Chemother. 1995;39:2019–22. doi: 10.1128/aac.39.9.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taneja N, Mewara A, Kumar A, Verma G, Sharma M. Cephalosporin-resistant Shigella flexneri over 9 years (2001-09) in India. J Antimicrob Chemother. 2012;67:134–53. doi: 10.1093/jac/dks061. [DOI] [PubMed] [Google Scholar]
- 23.Taneja N. Changing epidemiology of Shigellosis and emergence of ciprofloxacin-resistant Shigellae in India. J Clin Micobiol. 2007;45:678–9. doi: 10.1128/JCM.02247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taneja N, Lyngdoh V, Vermani A, Mohan B, Rao P, Singh M, et al. Re-emergence of multi-drug resistant Shigella dysenteriae with added resistance to ciprofloxacin in north India and their plasmid profiles. Indian J Med Res. 2005;122:348–54. [PubMed] [Google Scholar]
- 25.Talukder KA, Khajanchi BK, Islam MA, Dutta DK, Islam Z, Safa A, et al. Genetic relatedness of ciprofloxacin resistant Shigella dysenteriae type 1 strains isolated in South Asia. J Antimicrob Chemother. 2004;54:730–4. doi: 10.1093/jac/dkh425. [DOI] [PubMed] [Google Scholar]
- 26.Dutta S, Kawamura Y, Ezaki T, Nair GB, Iida K, Yoshida S. Alteration in the gyrA subunit of DNA gyrase and the parC subunit of topoisomerase IV in quinolone-resistant Shigella dysenteriae serotype 1 clinical isolates from Kolkata, India. Antimicrob Agents Chemother. 2005;49:1660–1. doi: 10.1128/AAC.49.4.1660-1661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H, Duan G, Zhu J, Lv R, Xi Y, Zhang W, et al. The AcrAB-TolC pump is involved in multidrug resistance in clinical Shigella flexneri isolates. Microb Drug Resist. 2008;14:245–9. doi: 10.1089/mdr.2008.0847. [DOI] [PubMed] [Google Scholar]
- 28.Paulsen IT, Brown MH, Skurray RA. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pazhani GP, Chakraborty S, Fujihara K, Yamasaki S, Ghosh A, Nair GB, et al. QRDR mutations, efflux system and antimicrobial resistance genes in enterotoxigenic Escherichia coli isolated from an outbreak of diarrhoea in Ahmedabad, India. Indian J Med Res. 2011;134:214–23. [PMC free article] [PubMed] [Google Scholar]
- 30.Lou H, Chen M, Black SS, Bushell SR, Ceccarelli M, Mach T, et al. Altered antibiotic transport in OmpC mutants isolated from a series of clinical strains of multi-drug resistant E. coli. PLoS One. 2011;6:e25825. doi: 10.1371/journal.pone.0025825. [DOI] [PMC free article] [PubMed] [Google Scholar]


