Abstract
DNA arrays are useful tools for simultaneously studying the expressions of a large number of genes. Herein, we describe the construction and the optimization of conditions for a low-density DNA macroarray specific for the porcine immune system. This specific DNA macroarray contains 63 gene products, including 20 cytokines, 11 chemokines, and 12 immunologically relevant receptors. It was constructed by designing gene-specific oligonucleotide primers from porcine sequences available in the EMBL or TIGR expressed sequence tag data bank and using primers from conserved regions of aligned sequences from other species for sequences unavailable for swine. Amplicons produced by reverse transcription-PCR were cloned, sequenced, and spotted onto nylon filters. A trial DNA array was first produced to optimize the intensity, specificity, and variability of signals from amplicons amplified with either gene-specific or universal primers. The DNA macroarray was then validated by comparing the gene expression profile of nonstimulated peripheral blood mononuclear cells (PBMCs) to that of phorbol 12-myristate 13-acetate and ionomycin (PMA-Iono)-stimulated PBMCs from three different animals over a 48-h time period. As already described for more conventional techniques, we showed that certain genes, such as those for CD40, gamma interferon, interleukin 2 (IL-2), the IL-2 receptor, and tumor necrosis factor alpha, were upregulated in PMA-Iono-stimulated PBMCs. A detailed analysis also indicated a downregulation of several genes which are expressed mainly by macrophages (IL-1, IL-8, AMCF-1, natural-resistance-associated macrophage protein, neutrophil chemotactic protein, DAP-12, and monocyte chemoattractant protein) in samples stimulated for 24 h with PMA-Iono compared to their levels of expression in control samples. These results indicate that the DNA macroarray that we constructed can be a useful tool for simultaneously monitoring the mRNA expression of immunologically relevant genes in different porcine samples.
The recent availability of a number of complete genomic sequences, such as those of bacteria (6), plants (1), animals (25), and humans (19, 33), has opened up new avenues in the study of the biology of these organisms with DNA arrays. Indeed, DNA arrays offer the advantage of providing simultaneous quantitative measurements of the expression levels of hundreds to thousands of genes.
Different DNA arrays have been used to analyze the host response to infection (31, 37). For example, microarrays constructed from the mouse genome have been used to analyze the in vitro response of mice macrophages to Salmonella enterica serovar Typhimurium (30) or Legionella pneumophila (26) infection as well as the in vivo response to Schistosoma mansoni infection (18). Microarrays have also been used to study the infection of human monocytes with Staphylococcus aureus (34), human foreskin fibroblasts with Toxoplasma gondii (5), and human cell lines with Listeria monocytogenes (8) or Bordetella pertussis (3). Analysis of the gene expression of human peripheral blood mononuclear cells (PBMCs) in response to bacteria, bacterial products, and pharmacological stimulants has also been performed with a DNA array (7).
In contrast, very few studies of the host response to infection have been carried out using specific immunological DNA arrays for domesticated animals. For example, a chicken DNA array was prepared from nonredundant cDNAs and used to study the reaction of chicken fibroblasts infected with Marek's disease virus (24). Also, a bovine-specific cDNA array system containing leukocyte expressed sequence tags (ESTs) and amplicons representing known genes was used to study cattle PBMCs infected by Mycobacterium avium subsp. paratuberculosis (9). Some workers have also carried out cross-species microarray hybridization experiments. Indeed, calf γδ-T-cell-subset cDNAs were hybridized to human arrays, showing that they have markedly different tissue-specific functions (17).
Although the pig genome is currently being sequenced and a number of ESTs are available (13), no results of a study using a DNA array specific for the porcine immune system have been published. One of the main interests of our laboratory is to analyze the effect of microbial pathogens and toxins on the immune system, using the pig as a target and model species. In the present paper, we describe a pilot macroarray that can be used as a tool to analyze the changes in gene expression of the pig immune response.
MATERIALS AND METHODS
Isolation and culture of porcine PBMCs.
Blood from healthy conventionally reared boars from a slaughterhouse facility at Pamiers, France, was used to isolate PBMCs as previously described (10). PBMCs isolated from each animal were adjusted to 5 × 106 cells/ml in Dulbecco's minimal essential medium (Eurobio, Les Ulis, France) containing 2 mM l-glutamine, 10% fetal bovine serum (HyClone; Perbio, Brebières, France), 100 U of penicillin/ml, and 0.1 mg of streptomycin (Eurobio) per ml. Cells were distributed in 24-well plates (Greiner bio-one, Poitiers, France) and stimulated or not stimulated with 50 ng of phorbol 12-myristate 13-acetate per ml and 1 μg of ionomycin per ml (PMA-Iono; Sigma, St. Quentin Fallavier, France). After 3 to 48 h of incubation, cells were harvested for RNA isolation. RNA samples were also obtained from PBMCs collected immediately after isolation and used as time zero samples.
Extraction of RNA.
Total RNA was extracted from 10 × 106 PBMCs conserved in Trizol (Invitrogen, Cergy Pontoise, France) as recommended by the manufacturer. Poly(A)+ mRNA was isolated from 30 × 106 PBMCs with the Quickprep mRNA purification kit (Amersham Biosciences, Orsay, France). All RNAs [total RNA and poly(A)+ mRNA] were resuspended in ultrapure water containing 0.02% (wt/vol) diethyl pyrocarbonate (Sigma). RNAs were quantified by measuring the optical density at 260 nm (OD260). Purity was assessed by determining the OD260/OD280 ratio, which was between 1.8 and 2.
Design of primers and production of amplicon.
A global list of immunologically relevant porcine genes, especially those for cytokines and chemokines, was prepared by searching the scientific literature and publicly available databases (PubMed, GenBank, and TIGR) and adding our own isolated sequences (7a). Oligonucleotide primer pairs for the porcine sequences, selected for size (18 to 27 bp), annealing temperature (55 to 68°C), GC% (20 to 80%), and amplicon length (200 to 650 bp), were designed by using the software Primer3 (http://www.broad.mit.edu/cgi-bin/primer/primer3_www.cgi Whitehead Institute, Massachusetts Institute of Technology, Boston, Mass.). For the 5′ extension of the RANTES protein sequence, primers were designed from the conserved regions of human-, mouse-, and rat-aligned sequences. The sizes of the amplicons and the accession or EST numbers of the genes which were spotted on the macroarray are listed in Table 1. The amplicons were produced by reverse transcription (RT)-PCR using 40 cycles of PCR as previously described (10) and analyzed by electrophoresis. The correct band was purified with a QIAquick gel extraction kit (QIAGEN, Courtaboeuf, France) and cloned into the PCR 2.1 TOPO vector (Invitrogen). Plasmid DNA for each amplicon was isolated with the GenElute plasmid miniprep kit (Sigma) and checked by sequencing (Genome Express, Grenoble, France).
TABLE 1.
List of the amplicons spotted onto the DNA macroarray
| Amplicon name | Gene group | Size (bp) | Accession no. or EST | Gene product description |
|---|---|---|---|---|
| ICAM | Adhesion molecule | 204 | AF156712 | Intercellular adhesion molecule |
| VCAM | Adhesion molecule | 675 | L43124 | Vascular cell adhesion molecule |
| CD1 | CD antigen | 433 | AF059492 | CD1 antigen |
| CD40 | CD antigen | 606 | AF326598 | CD40 antigen |
| CD59 | CD antigen | 210 | AF020302 | CD59 antigen |
| CD80 | CD antigen | 222 | AF203442 | CD80 antigen |
| CD95 | CD antigen | 397 | AB027297 | CD95 antigen/Fas ligand |
| IL-8 | Chemokine | 269 | X61151 | IL-8 |
| AMCF-1 | Chemokine | 487 | M99367 | Alveolar macrophage chemotactic factor 1 (homologue of IL-8) |
| AMCF-2 | Chemokine | 284 | M99368 | Alveolar macrophage-derived chemotactic factor 2 |
| BRAK | Chemokine | 237 | AY308800 | Breast and kidney derived |
| MCP-1 | Chemokine | 320 | Z48479 | Monocyte chemoattractant protein 1 |
| MCP-2 | Chemokine | 558 | Z48480 | Monocyte chemoattractant protein 2 |
| MIP-3-β | Chemokine | 157 | AY312065 | Macrophage inflammatory protein 3 beta |
| RANTES | Chemokine | 290 | F14636 | Regulated on activation of normal T cell expressed and secreted |
| SDF-1β | Chemokine | 446 | AY312066 | Stromal-cell-derived factor 1 |
| SLC | Chemokine | 419 | AY312067 | Secondary lymphoid tissue chemokine |
| TECK | Chemokine | 284 | AY312064 | Thymic expressed chemokine |
| NCP-1 | Chemotactic factor | 283 | X77935 | Neutrophil chemotactic protein 1 |
| COX1 | Cyclooxygenase | 471 | AF207823 | Cyclooxygenase 1 |
| GM-CSF | Cytokine | 407 | U61139 | Granulocyte-macrophage colony-stimulating factor |
| IFN-α | Cytokine | 403 | M28623 | IFN-α |
| IFN-γ | Cytokine | 379 | S63967 | IFN-γ |
| IL-1α | Cytokine | 337 | X52731 | IL-1 alpha |
| IL-1β | Cytokine | 286 | X86725 | IL-1 beta |
| IL-2 | Cytokine | 338 | X58428 | IL-2 |
| IL-4 | Cytokine | 324 | X68330 | IL-4 |
| IL-5 | Cytokine | 236 | AJ133452 | IL-5 |
| IL-6 | Cytokine | 493 | M86722 | IL-6 |
| IL-7 | Cytokine | 362 | AB035380 | IL-7 |
| IL-10 | Cytokine | 446 | L20001 | IL-10 |
| IL-12p35 | Cytokine | 255 | L35765 | IL-12 (p35 subunit) |
| IL-12p40 | Cytokine | 375 | U08317 | IL-12 (p40 subunit) |
| IL-13 | Cytokine | 346 | AF385626 | IL-13 |
| IL-15 | Cytokine | 286 | U58142 | IL-15 |
| IL-16 | Cytokine | 315 | AB091290 | IL-16 |
| IL-18 | Cytokine | 259 | AF176949 | IL-18 |
| TGF-β1 | Cytokine | 337 | M23703 | TGF-β1 |
| TGF-β2 | Cytokine | 268 | L08375 | TGF-β2 |
| TNF-α | Cytokine | 351 | X57321 | TNF-α |
| GST-A3 | Glutathione S-transferase | 435 | Z69585 | Glutathione S-transferase alpha3 |
| GST-M1 | Glutathione S-transferase | 405 | BE232478 (EST) | Glutathione S-transferase mu1 |
| GST-O1 | Glutathione S-transferase | 434 | AF188838 | Glutathione S-transferase omega1 |
| GST-P1 | Glutathione S-transferase | 428 | BF440652 (EST) | Glutathione S-transferase pi1 |
| GST-T2 | Glutathione S-transferase | 461 | BI344427 (EST) | Glutathione S-transferase theta2 |
| β-Actin | Housekeeping | 233 | U07786 | Beta actin |
| Cyclo | Housekeeping | 368 | AY008846 | Cyclophilin |
| GAPDH | Housekeeping | 603 | AF017079 | Glyceraldehyde-3-phosphate dehydrogenase |
| HPRT | Housekeeping | 248 | AF143818 | Hypoxanthine phosphoribosyltransferase |
| ICE | IL-1β-converting enzyme | 398 | AB027296 | IL-1β-converting enzyme |
| DAP-12 | Immunoreceptor | 259 | AF152021 | Associated adapter protein DAP-12 |
| NKG2D | Immunoreceptor | 405 | AF285448 | Natural killer immunoreceptor NKG2D |
| IL-2Rα | Cytokine receptor | 427 | U78317 | IL-2R alpha chain |
| IL-6Rα | Cytokine receptor | 220 | AF147881 | IL-6R alpha chain |
| IL-8R1 | Cytokine receptor | 300 | AF296552 | IL-8R1 |
| TGF-β 3R | Cytokine receptor | 329 | L07595 | TGF-β type III receptor |
| TNF-R | Cytokine receptor | 520 | U19994 | p55 TNF receptor |
| MHC class II | MHC class II | 471 | AB010577 | Major histocompatibility complex class II SLA-DQa beta chain |
| 16S | Mitochondrial gene | 300 | AF107224 | Mitochondrial 16S RNA |
| NRAMP | NRAMP | 307 | AF132037 | Natural-resistance-associated macrophage protein |
| CTLA4 | T-cell activation marker | 261 | AF220248 | Cytotoxic-T-lymphocyte-associated protein 4 |
| iNOS | Toxicity response gene | 593 | U59390 | Inducible nitric oxide synthase |
SLA-DQ, swine leukocyte antigen DQ.
Macroarray production.
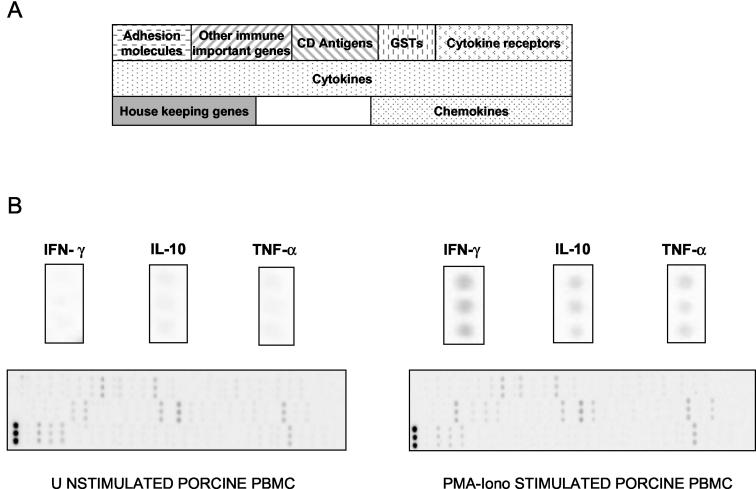
To prepare all the amplicon templates for spotting on the nylon membrane, PCR amplifications were performed on the purified diluted plasmid with either specific primers for each amplicon or primers designed close to the cloning site of the vector (universal TOPO primers; sense, TAGTAACGGCCGCCAGTGTGCT; antisense, CCGCCAGTGTGATGGATATCTGCA). PCR amplicons were precipitated, washed, and resuspended in distilled water. The amplicon concentration was quantified by measuring the OD260 and adjusted to 0.2 mg/ml. As seen, the amplicons were spotted in triplicate onto Immobilon-NY+ membranes (Millipore, Guyancourt, France) (100 nl of the PCR template per spot) at the Toulouse Génopôle (http://genopole.toulouse.inra.fr) by using a Eurogridder robot (Eurogentec, Seraing, Belgium), and the preliminary testing of arrays revealed good spot homogeneity and good reproducibility (Fig. 1).
FIG. 1.
Macroarray specific for the pig immune system. (A) Gene layout on the DNA macroarray; (B) hybridization of DNA macroarray with 33P-labeled cDNA probes from unstimulated and PMA-Iono-stimulated porcine PBMCs cultured for 3 h. GSTs, glutathione S-transferases.
Synthesis of labeled cDNA and DNA macroarray hybridization.
Radioactively labeled cDNA probes were prepared according to an already published protocol (21) by using 5 μg of total RNA or 1 μg of poly(A)+ RNA and 9 μg of a random hexamer primer or 1 μg of an oligo(dT)12-18 primer (Invitrogen). cDNA was purified with ProbeQuant G-50 micro columns (Amersham Biosciences), and similar levels of 33P incorporation were verified.
The macroarrays were prehybridized at 65°C for 4 h in 2 ml of Church's buffer (0.5 M sodium phosphate buffer, pH 7.2, 1% sodium dodecyl sulfate, 1 mM EDTA) before being hybridized for 48 h at 65°C. They were then washed extensively (with 40 mM sodium phosphate buffer, pH 7.2, and 0.1% sodium dodecyl sulfate), dried, and exposed to a Kodak-Molecular Dynamics low-energy screen (Amersham Biosciences) for 3 to 5 days. The hybridization signals were visualized by scanning the screen with a Molecular Dynamics Storm 840 PhosphorImager (Amersham Biosciences) at a resolution of 50 μm.
DNA array data analysis.
After image acquisition, the scanned images were imported into the Image Master array software (Amersham Biosciences) to quantify the intensities for each spot. The background hybridization intensity was corrected for by subtracting the mean intensity value of water samples spotted randomly over the membrane. All data were analyzed with the software programs Bioplot and Bioclust (https://bio71.gba.insa-tlse.fr/) or imported into a Microsoft Excel worksheet for graphical analysis.
RESULTS
Optimization of the DNA array.
A trial DNA array containing 14 immunologically relevant genes spotted in triplicate was produced to optimize different factors. We first investigated the intensity, specificity, and variability of signals from amplicons amplified with either gene-specific or universal primers. Analysis of the DNA arrays did not reveal any difference in the signal intensities of the amplicons amplified with specific or universal primers (data not shown). Due to the convenience of producing the amplicons with this method, the next macroarray generation was created by using universal TOPO primers. Several RNA purification and labeling techniques were also compared. Trial membranes were hybridized with cDNA probes produced with either a random hexamer primer or an oligo(dT) primer. At the same time, a comparison between total and poly(A)+ mRNA template samples was carried out. Although there were no differences in intensity between total and poly(A)+ samples, those transcribed with the random primer always had stronger intensities than those transcribed with the oligo(dT) primer (data not shown). Therefore, the next generation of cDNA probes was produced from total RNA with random hexamer primers.
The trial array was checked for the within-sample variability of the spotted amplicons. The coefficient of variation for this parameter was between 2 and 18% depending on which gene was studied and was higher for genes with low expression levels, such as those for transforming growth factor β1 (TGF-β1) (10%), interleukin 15 (IL-15) (15%), and IL-16 (18%). This DNA macroarray was also tested by comparing the levels of gene expression of porcine PBMCs cultured for 3 h in the presence and in the absence of PMA-Iono. Figure 1 reveals good spot homogeneity and demonstrates that mitogenic stimulation induced the overexpression of several immunological genes, such as those for gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), and IL-10.
Selection of a housekeeping gene.
As a differential expression of housekeeping genes has been described recently (14, 22, 35), the expression levels of four putative housekeeping genes included in the array were determined. Compared to those of immunological genes, the levels of expression of three out of these four housekeeping genes were high: median arbitrary units were 58 × 104, 23 × 104, and 13 × 104 for the β-actin, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and cyclophilin genes, respectively, and 2.1 × 104 for the immunological genes. In contrast, the housekeeping gene for hypoxanthine phosphoribosyltransferase (HPRT) had a level of expression similar to that of the immunological genes. The median value of HPRT expression through the different time points was more reproducible than that of the other housekeeping genes, and HPRT expression was very reproducible (median arbitrary units, 2.6 × 104). Therefore, in all later procedures the HPRT gene was used to normalize the results.
Kinetic analysis of porcine gene expression.
The global change in gene expression was investigated kinetically in both unstimulated and PMA-Iono-stimulated porcine PBMCs. Gene expression in unstimulated PBMCs was first analyzed. The immunity-relevant genes were expressed at low levels in all animals, with expression varying less than twofold between the different animals studied. A global analysis also revealed that in vitro culture for 3 h modulated the expression of several immunity-relevant genes in both unstimulated and PMA-Iono-stimulated PBMCs. Indeed, in vitro culture induced the expression of 10 and 16 genes and suppressed the expression of 2 and 6 genes in unstimulated and PMA-Iono-stimulated cultures, respectively (Tables 2 and 3).
TABLE 2.
Expression profile of unstimulated porcine PBMCs cultivated in vitro for 3, 12, 24, and 48 h
| Gene productb | Relative expression (AU) ata:
|
|||
|---|---|---|---|---|
| 3 h | 12 h | 24 h | 48 h | |
| ICAM | 81.9 ± 10.2 | 28.0 ± 1.3 | 5.3 ± 1.6 | 5.7 ± 1.2 |
| MCP-1 | 2.5 ± 0.5 | 10.4 ± 2.0 | 11.9 ± 2.6 | 27.8 ± 2.9 |
| NCP-1 | 3.0 ± 0.2 | 4.7 ± 0.2 | 3.3 ± 1.0 | 2.9 ± 0.8 |
| IL-6Rα | 2.9 ± 1.1 | 2.4 ± 1.2 | 2.3 ± 2.2 | 1.0 ± 0.1 |
| IL-2Rα | 2.3 ± 0.9 | 1.4 ± 0.2 | 1.2 ± 0.2 | 1.1 ± 0.1 |
| IL-4 | 2.1 ± 0.5 | 1.4 ± 0.1 | 1.3 ± 0.1 | 1.2 ± 0.2 |
| IL-8 | 2.6 ± 0.8 | 1.1 ± 0.3 | 1.3 ± 0.6 | 0.9 ± 0.4 |
| GST-O1 | 2.3 ± 0.7 | 1.9 ± 0.2 | 1.8 ± 0.5 | 1.7 ± 0.0 |
| MCP-2 | 1.3 ± 0.0 | 3.2 ± 0.5 | 5.8 ± 3.8 | 7.6 ± 0.3 |
| NRAMP | 1.5 ± 0.0 | 2.3 ± 0.1 | 2.9 ± 0.1 | 2.1 ± 0.2 |
| VCAM | 1.8 ± 0.4 | 1.7 ± 0.0 | 1.4 ± 0.1 | 1.9 ± 0.3 |
| CD1 | 1.7 ± 0.1 | 1.5 ± 0.1 | 1.3 ± 0.2 | 1.2 ± 0.1 |
| CD59 | 1.8 ± 0.0 | 1.5 ± 0.3 | 1.3 ± 0.2 | 1.0 ± 0.3 |
| CD80 | 1.4 ± 0.0 | 1.3 ± 0.1 | 1.1 ± 0.0 | 1.0 ± 0.1 |
| CD95 | 1.0 ± 0.0 | 1.1 ± 0.0 | 1.1 ± 0.2 | 0.9 ± 0.1 |
| AMCF-1 | 1.5 ± 0.1 | 1.1 ± 0.3 | 1.2 ± 0.6 | 0.5 ± 0.4 |
| BRAK | 1.4 ± 0.2 | 1.1 ± 0.4 | 1.0 ± 0.1 | 0.9 ± 0.1 |
| MIP-3β | 1.5 ± 0.1 | 1.3 ± 0.2 | 1.0 ± 0.2 | 0.9 ± 0.2 |
| RANTES | 1.1 ± 0.1 | 1.4 ± 0.2 | 1.4 ± 0.1 | 0.6 ± 0.1 |
| SDF-1β | 2.0 ± 0.3 | 1.4 ± 0.1 | 1.1 ± 0.1 | 1.1 ± 0.0 |
| SLC | 1.7 ± 0.2 | 1.5 ± 0.4 | 1.3 ± 0.0 | 1.1 ± 0.2 |
| TECK | 1.8 ± 0.1 | 1.6 ± 0.3 | 1.1 ± 0.2 | 1.1 ± 0.2 |
| COX1 | 1.3 ± 0.1 | 1.0 ± 0.3 | 0.9 ± 0.2 | 0.8 ± 0.1 |
| GM-CSF | 1.8 ± 0.6 | 1.7 ± 0.6 | 1.5 ± 0.7 | 0.9 ± 0.2 |
| IFN-α | 1.3 ± 0.2 | 1.2 ± 0.1 | 0.9 ± 0.1 | 1.2 ± 0.1 |
| IFN-γ | 1.9 ± 0.1 | 1.3 ± 0.1 | 1.2 ± 0.2 | 1.0 ± 0.0 |
| IL-2 | 1.5 ± 0.1 | 1.3 ± 0.3 | 1.0 ± 0.2 | 0.9 ± 0.1 |
| IL-5 | 1.7 ± 0.1 | 1.5 ± 0.1 | 1.3 ± 0.1 | 1.2 ± 0.3 |
| IL-6 | 2.0 ± 0.2 | 1.5 ± 0.3 | 1.1 ± 0.3 | 1.1 ± 0.2 |
| IL-7 | 1.4 ± 0.1 | 1.1 ± 0.2 | 1.0 ± 0.2 | 0.9 ± 0.2 |
| IL-10 | 1.7 ± 0.8 | 1.2 ± 0.3 | 0.8 ± 0.2 | 0.7 ± 0.2 |
| IL-12p35 | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.1 ± 0.1 | 1.0 ± 0.2 |
| IL-12p40 | 1.5 ± 0.2 | 1.3 ± 0.0 | 1.1 ± 0.1 | 1.1 ± 0.1 |
| IL-13 | 1.7 ± 0.2 | 1.5 ± 0.1 | 1.2 ± 0.0 | 1.4 ± 0.1 |
| IL-15 | 1.3 ± 0.1 | 1.2 ± 0.3 | 1.0 ± 0.3 | 1.0 ± 0.3 |
| IL-16 | 1.5 ± 0.6 | 1.3 ± 0.1 | 1.2 ± 0.1 | 1.2 ± 0.2 |
| IL-18 | 1.8 ± 0.3 | 1.5 ± 0.2 | 1.2 ± 0.3 | 1.0 ± 0.3 |
| TGF-β2 | 1.8 ± 0.6 | 1.2 ± 0.2 | 1.1 ± 0.0 | 1.2 ± 0.2 |
| TNF-α | 1.0 ± 0.1 | 0.8 ± 0.2 | 0.7 ± 0.2 | 0.7 ± 0.1 |
| GST-A3 | 1.4 ± 0.2 | 1.3 ± 0.3 | 0.9 ± 0.3 | 0.9 ± 0.2 |
| GST-M1 | 1.8 ± 0.6 | 1.6 ± 0.3 | 1.5 ± 0.1 | 1.5 ± 0.3 |
| GST-P1 | 0.8 ± 0.1 | 1.5 ± 0.2 | 1.4 ± 0.1 | 1.8 ± 0.2 |
| GST-T2 | 1.5 ± 0.2 | 1.3 ± 0.4 | 1.1 ± 0.3 | 0.9 ± 0.3 |
| IL-8R1 | 1.4 ± 0.2 | 1.1 ± 0.2 | 0.8 ± 0.2 | 0.7 ± 0.1 |
| TGF-β 3R | 1.9 ± 0.6 | 1.6 ± 0.1 | 1.3 ± 0.0 | 1.2 ± 0.1 |
| TNF-R | 1.3 ± 0.0 | 1.6 ± 0.1 | 1.4 ± 0.1 | 1.4 ± 0.1 |
| NKG2D | 1.5 ± 0.2 | 1.1 ± 0.3 | 0.9 ± 0.2 | 0.6 ± 0.1 |
| DAP-12 | 1.3 ± 0.2 | 1.7 ± 0.1 | 1.6 ± 0.1 | 1.6 ± 0.2 |
| CTLA4 | 2.0 ± 0.2 | 1.5 ± 0.2 | 1.1 ± 0.1 | 1.0 ± 0.2 |
| ICE | 1.9 ± 0.2 | 1.5 ± 0.3 | 1.1 ± 0.1 | 1.1 ± 0.3 |
| iNOS | 1.4 ± 0.1 | 1.1 ± 0.2 | 0.9 ± 0.2 | 0.9 ± 0.1 |
| IL-1α | 1.1 ± 0.1 | 0.9 ± 0.3 | 0.7 ± 0.3 | 0.4 ± 0.2 |
| IL-1β | 1.3 ± 0.1 | 0.9 ± 0.5 | 0.9 ± 0.4 | 0.3 ± 0.2 |
| CD40 | 0.7 ± 0.1 | 0.5 ± 0.1 | 0.4 ± 0.1 | 0.4 ± 0.1 |
| MHC class II | 0.6 ± 0.1 | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.2 ± 0.0 |
| TGF-β1 | 0.6 ± 0.1 | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.3 ± 0.1 |
Data are expressed as the means ± standard errors of the means of values obtained from three animals relative to those at time zero. Boldface values (mean, >2.0) indicate gene overexpression, and underlined values (mean, <0.5) indicate gene underexpression. AU, arbitrary units.
Abbreviation explanations can be found in the gene product description column of Table 1.
TABLE 3.
Expression profile of PMA-Iono-stimulated porcine PBMCs cultivated in vitro for 3, 12, 24, and 48 h
| Gene productb | Relative expression (AU) ata:
|
|||
|---|---|---|---|---|
| 3 h | 12 h | 24 h | 48 h | |
| ICAM | 9.0 ± 2.9 | 16.8 ± 6.7 | 16.2 ± 8.9 | 10.0 ± 4.5 |
| MCP-1 | 0.9 ± 0.2 | 1.0 ± 0.3 | 1.5 ± 0.7 | 1.4 ± 0.8 |
| NCP-1 | 0.8 ± 0.3 | 0.8 ± 0.1 | 0.8 ± 0.3 | 1.6 ± 0.5 |
| IL-6Rα | 2.5 ± 1.8 | 1.5 ± 1.10 | 0.4 ± 0.1 | 0.3 ± 0.2 |
| IL-2Rα | 3.7 ± 0.5 | 2.8 ± 0.7 | 3.2 ± 0.6 | 3.7 ± 1.0 |
| IL-4 | 2.2 ± 0.6 | 1.3 ± 0.5 | 1.1 ± 0.5 | 1.1 ± 0.2 |
| IL-8 | 2.9 ± 1.5 | 1.4 ± 1.1 | 0.8 ± 0.7 | 0.8 ± 0.4 |
| GST-O1 | 1.0 ± 0.1 | 1.2 ± 0.2 | 2.3 ± 0.4 | 1.8 ± 0.0 |
| MCP-2 | 0.4 ± 0.1 | 0.4 ± 0.1 | 0.4 ± 0.0 | 0.6 ± 0.1 |
| NRAMP | 1.3 ± 0.2 | 1.1 ± 0.4 | 0.8 ± 0.2 | 0.6 ± 0.4 |
| VCAM | 1.9 ± 0.1 | 1.8 ± 0.4 | 1.1 ± 0.1 | 1.0 ± 0.2 |
| CD1 | 1.4 ± 0.4 | 1.0 ± 0.2 | 0.7 ± 0.3 | 1.0 ± 0.1 |
| CD59 | 1.5 ± 0.2 | 1.2 ± 0.1 | 0.7 ± 0.0 | 1.3 ± 0.1 |
| CD80 | 1.9 ± 0.2 | 1.1 ± 0.0 | 0.7 ± 0.0 | 1.0 ± 0.1 |
| CD95 | 1.2 ± 0.2 | 1.1 ± 0.2 | 1.1 ± 0.2 | 1.1 ± 0.1 |
| AMCF-1 | 1.4 ± 0.0 | 0.5 ± 0.2 | 0.2 ± 0.1 | 0.2 ± 0.1 |
| BRAK | 1.1 ± 0.1 | 0.8 ± 0.1 | 0.6 ± 0.1 | 0.6 ± 0.1 |
| MIP-3β | 1.3 ± 0.3 | 1.0 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 |
| RANTES | 3.3 ± 1.3 | 0.9 ± 0.0 | 0.9 ± 0.1 | 0.8 ± 0.3 |
| SDF-1β | 1.3 ± 0.5 | 1.0 ± 0.4 | 1.0 ± 0.2 | 1.1 ± 0.3 |
| SLC | 0.9 ± 0.1 | 1.0 ± 0.1 | 0.9 ± 0.0 | 1.1 ± 0.0 |
| TECK | 1.6 ± 0.3 | 1.3 ± 0.2 | 1.0 ± 0.2 | 1.2 ± 0.1 |
| COX1 | 1.4 ± 0.0 | 1.0 ± 0.1 | 0.7 ± 0.2 | 1.1 ± 0.1 |
| GM-CSF | 1.9 ± 0.3 | 2.0 ± 0.3 | 2.0 ± 0.4 | 1.9 ± 0.3 |
| IFN-α | 1.0 ± 0.5 | 0.7 ± 0.1 | 0.7 ± 0.1 | 0.8 ± 0.3 |
| IFN-γ | 35.8 ± 15.3 | 10.4 ± 1.9 | 9.4 ± 0.1 | 7.8 ± 1.2 |
| IL-2 | 15.5 ± 10.0 | 11.1 ± 1.7 | 12.8 ± 5.8 | 1.4 ± 0.0 |
| IL-5 | 1.5 ± 0.2 | 1.2 ± 0.1 | 0.7 ± 0.1 | 0.9 ± 0.1 |
| IL-6 | 2.6 ± 0.5 | 3.2 ± 0.8 | 1.3 ± 0.4 | 1.0 ± 0.3 |
| IL-7 | 1.2 ± 0.2 | 1.0 ± 0.1 | 0.7 ± 0.1 | 0.8 ± 0.0 |
| IL-10 | 9.8 ± 2.0 | 1.0 ± 0.5 | 0.6 ± 0.3 | 0.5 ± 0.0 |
| IL-12p35 | 1.1 ± 0.2 | 0.9 ± 0.1 | 0.7 ± 0.1 | 0.9 ± 0.0 |
| IL-12p40 | 1.1 ± 0.0 | 1.5 ± 0.4 | 1.5 ± 0.6 | 1.0 ± 0.1 |
| IL-13 | 2.1 ± 0.2 | 1.2 ± 0.3 | 1.1 ± 0.4 | 1.1 ± 0.4 |
| IL-15 | 1.0 ± 0.1 | 0.8 ± 0.1 | 0.7 ± 0.0 | 0.7 ± 0.0 |
| IL-16 | 1.1 ± 0.6 | 1.1 ± 0.6 | 1.0 ± 0.4 | 0.9 ± 0.2 |
| IL-18 | 1.2 ± 0.2 | 1.1 ± 0.2 | 0.9 ± 0.2 | 0.9 ± 0.2 |
| TGF-β2 | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.3 | 1.3 ± 0.2 |
| TNF-α | 9.7 ± 0.9 | 3.6 ± 0.5 | 3.4 ± 0.6 | 1.1 ± 0.2 |
| GST-A3 | 1.1 ± 0.1 | 1.0 ± 0.2 | 0.7 ± 0.1 | 1.0 ± 0.3 |
| GST-M1 | 2.3 ± 0.9 | 1.7 ± 0.4 | 1.4 ± 0.5 | 1.4 ± 0.3 |
| GST-P1 | 0.5 ± 0.0 | 0.8 ± 0.1 | 0.8 ± 0.1 | 1.1 ± 0.2 |
| GST-T2 | 1.3 ± 0.2 | 1.0 ± 0.1 | 0.7 ± 0.0 | 0.7 ± 0.0 |
| IL-8R1 | 1.0 ± 0.1 | 0.7 ± 0.0 | 0.6 ± 0.0 | 0.6 ± 0.0 |
| TGF-β 3R | 2.0 ± 0.6 | 1.2 ± 0.5 | 1.0 ± 0.4 | 1.1 ± 0.3 |
| TNF-R | 0.8 ± 0.1 | 0.7 ± 0.0 | 0.6 ± 0.0 | 0.6 ± 0.1 |
| NKG2D | 1.3 ± 0.1 | 0.6 ± 0.2 | 0.4 ± 0.1 | 0.5 ± 0.1 |
| DAP-12 | 0.8 ± 0.1 | 0.6 ± 0.1 | 0.4 ± 0.0 | 0.4 ± 0.0 |
| CTLA4 | 2.1 ± 0.2 | 1.3 ± 0.1 | 1.1 ± 0.1 | 1.5 ± 0.0 |
| ICE | 0.9 ± 0.0 | 0.9 ± 0.2 | 0.8 ± 0.1 | 1.1 ± 0.1 |
| iNOS | 1.0 ± 0.1 | 0.7 ± 0.1 | 0.6 ± 0.1 | 0.8 ± 0.0 |
| IL-1α | 0.7 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.0 | 0.1 ± 0.0 |
| IL-1β | 0.9 ± 0.2 | 0.2 ± 0.1 | 0.2 ± 0.0 | 0.1 ± 0.0 |
| CD40 | 2.7 ± 0.6 | 1.4 ± 0.1 | 0.7 ± 0.1 | 0.7 ± 0.1 |
| MHC class II | 0.9 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| TGF-β1 | 2.0 ± 0.3 | 0.7 ± 0.3 | 0.5 ± 0.2 | 0.4 ± 0.1 |
Data are expressed as the means ± standard errors of the means of values obtained from three animals relative to those at time zero. Boldface values (mean, >2.0) indicate gene overexpression, and underlined values (mean, <0.5) indicate gene underexpression. AU, arbitrary units.
Abbreviation explanations can be found in the gene product description column of Table 1.
Stimulation with PMA-Iono induced an early upregulation of the expression of several genes; however, most of the genes returned to their baseline expression level by 48 h (Table 3). This stimulation also downregulated, in a time-dependent manner, the expression of the immunity-relevant genes, with a minimum expression observed 24 h poststimulation. Certain genes, such as the IL-10, CD40, and RANTES genes, were upregulated at the beginning of the stimulation by PMA-Iono and downregulated thereafter (Table 3). Of note is that other genes, such as those for NCP-1 and MCP-1, were modulated only in unstimulated PBMCs (Table 2) and not affected in PMA-Iono-stimulated cultures.
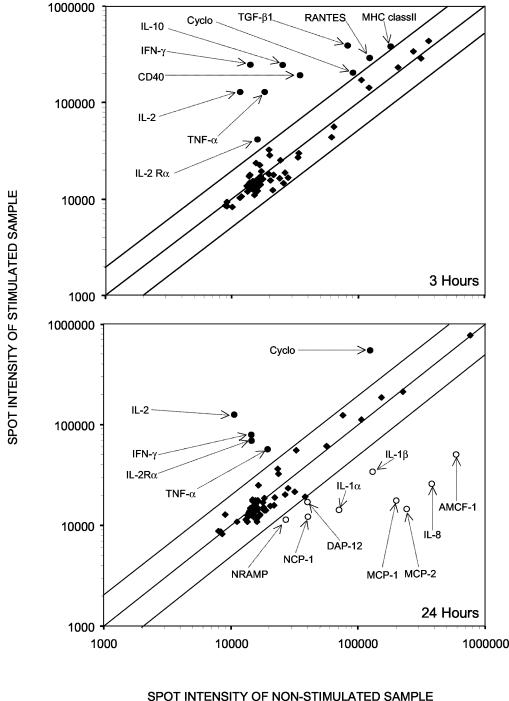
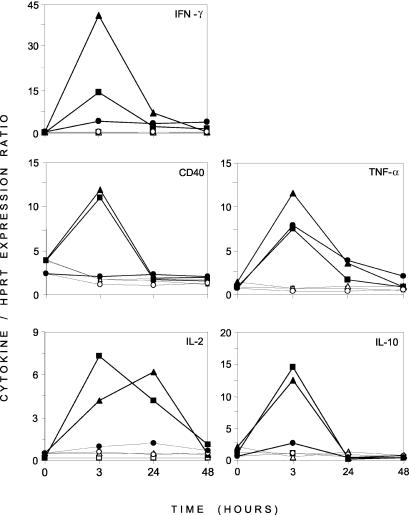
A differential analysis between unstimulated and PMA-Iono-stimulated PBMCs cultivated in vitro for 3 and 24 h was carried out (Fig. 2). It indicated that at the early time point (3 h), 10 genes expressed mainly by lymphocytes were overexpressed upon PMA-Iono-stimulation. At the later time point (24 h), a few genes remained overexpressed in stimulated samples, and several other genes (the MCP-1, MCP-2, IL-1α, IL-1β, IL-8, AMCF-1, DAP-12, natural-resistance-associated macrophage protein [NRAMP], and NCP-1 genes) expressed mainly by macrophages appeared to be downregulated by the mitogenic stimulation compared with levels of expression in unstimulated samples. We further analyzed the expression of five genes in control and PMA-Iono-stimulated porcine PBMCs. Figure 3 clearly illustrates the induction of the expression of these genes by PMA-Iono stimulation but also reveals the variation between the responses of animals. Indeed, one of the three animals responded poorly to PMA-Iono stimulation for the expression of genes coding for CD40, IL-2, and IL-10 but had a response similar to that of the other animals for the expression of TNF-α.
FIG. 2.
Scatterplot showing the differential levels of expression of the genes between unstimulated and PMA-Iono-stimulated porcine PBMCs cultured in vitro for 3 and 24 h. Labeled cDNA probes were prepared from lymphocytes of the same animal stimulated or not stimulated with PMA-Iono for 3 or 24 h. Genes that were overexpressed are indicated by filled circles, those that were expressed normally are indicated by filled diamonds, and those that were underexpressed are indicated by open circles. Abbreviations can be found in the gene product description column of Table 1.
FIG. 3.
Kinetics of mRNA expression of five immune genes (the CD40, IFN-γ, IL-2, TNF-α, and IL-10 genes) in PBMCs isolated from three different pigs. RNAs obtained from unstimulated and PMA-Iono-stimulated PBMCs which had been cultured for different times (0, 3, 24, and 48 h) were reverse transcribed, labeled, and hybridized to the macroarray. Relative mRNA levels for each gene plotted against time are presented as the mean levels of intensity of expression standardized to HPRT gene expression. In each panel, unstimulated and PMA-Iono-stimulated PBMCs from the same pig are represented by the same symbol. Open symbols represent unstimulated PBMCs. Filled symbols represent PMA-Iono-stimulated PBMCs.
DISCUSSION
We have developed a DNA macroarray containing immunologically relevant porcine genes. This array has been validated on pig PBMCs cultured in vitro. RT conditions to label the cDNA were optimized to obtain the best reproducibility and yield. We showed that using total RNA and a random hexamer primer gave the strongest reproducible signals (Fig. 1).
When DNA arrays are used, there are several sources of systematic variation which can be corrected by normalization. Normalization can be done in a number of ways: by using either all genes on the array, housekeeping genes, or spiked controls (36). Our macroarray contains fewer than 100 genes, and all of them have the potential to be differentially expressed. In contrast, housekeeping genes are considered to be expressed constantly. However, several reports have indicated that the expression levels of these genes may change in different situations and tissues (11, 14, 15, 22, 32, 35). In this paper, we demonstrated that the gene coding for HPRT was expressed uniformly among the tested samples and at the same level as the other immunological genes. Therefore, we used this method to normalize our results, and we are currently working on the development of spiked controls.
As was already observed in other studies (10, 28), variations in the individual responses to gene expression upon mitogenic stimulation were observed (Fig. 3). As the animals were randomly chosen from a commercial farm, even if they were from the same breed, the differences in their genetic backgrounds could have contributed to the observed variations.
Treatment with a combination of PMA and ionomycin activates lymphocytes, as well as other leukocytes, by mimicking the intracellular signals that occur during natural responses to antigens (20, 29). A number of studies have investigated the effect of PMA-Iono on the proliferation and expression of cytokines in human and animal PBMCs. As observed in the present study, PMA-Iono stimulation increased the expression of the gene coding for IFN-γ in human PBMCs (4) but also in PMA-differentiated THP-1 cells (2) and in a porcine trophectoderm cell line (23). Similarly, PMA-Iono stimulation has been reported to increase the expression of both IL-2 and the IL-2 receptor (IL-2R) in murine mixed leukocytes (20), human T cells (16), and porcine PBMCs (Table 3). Increased expression of TNF-α and CD40 upon PMA-Iono stimulation was also observed in CD3-positive human T cells and B cells, respectively (12, 27). Using a DNA macroarray specific to the porcine immune system, we confirm the induction of the expression of IFN-γ, IL-2, the IL-2R, TNF-α, and CD40 in pig PBMCs stimulated with PMA-Iono. Of note is that the kinetic expression of these genes resembles previous results that were obtained with concanavalin A-stimulated PBMCs monitored by RT-PCR (10).
PBMCs display both innate and adaptive immune functions which have well-established roles in surveillance for infectious threats, both directly through contact with infectious agents and indirectly through interactions with infected cells and tissues by means of secreted molecules acting as messages. DNA arrays can monitor the expression of genes involved in the immune response as well as that of the secreted messages. Using a DNA macroarray specific to the porcine immune system, we were able to measure and quantify the changes in gene expression in PBMCs during in vitro culture in the absence or presence of PMA-Iono. This array is currently used to compare the gene expression levels of tissues obtained from pig-ligated loops inoculated with various strains of Escherichia coli and demonstrating different levels of pathogenicity (T. N. Ledger, I. Taranu, and I. P. Oswald, unpublished results).
Acknowledgments
We thank Veronique Le Berre and Adilia Dagkesamanskaya for the spotting of the PCR amplicons onto the membranes and Thierry Pineau for the use of the TOPO universal primers. We appreciate the help of Segei Sokol for his aid with the computer analysis of data obtained and that of Pascal Martin for helpful discussion.
We also thank the Toulouse Génopôle, INRA Transversalité “Immunité Muqueuse,” the AGENAE program, and the European Union (project AEEC infection, QLK2-CT-2000-006000) for financial support.
We are solely responsible for the work described in this article, and our opinions are not necessarily those of the European Union.
REFERENCES
- 1.Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796-815. [DOI] [PubMed] [Google Scholar]
- 2.Asseffa, A., L. A. Dickson, S. Mohla, and T. A. Bremner. 1993. Phorbol myristate acetate-differentiated THP-1 cells display increased levels of MHC class I and class II mRNA and interferon-gamma inducible tumoricidal activity. Oncol. Res. 5:11-18. [PubMed] [Google Scholar]
- 3.Belcher, C. E., J. Drenkow, B. Kehoe, T. R. Gingeras, N. McNamara, H. Lemjabbar, C. Basbaum, and D. A. Relman. 2000. The transcriptional responses of respiratory epithelial cells to Bordetella pertussis reveal host defensive and pathogen counter-defensive strategies. Proc. Natl. Acad. Sci. USA 97:13847-13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biller, H., B. Bade, H. Matthys, W. Luttmann, and J. C. Virchow. 2001. Interferon-γ secretion of peripheral blood CD8+ T lymphocytes in patients with bronchial asthma: in vitro stimulus determines cytokine production. Clin. Exp. Immunol. 126:199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blader, I. J., I. D. Manger, and J. C. Boothroyd. 2001. Microarray analysis reveals previously unknown changes in Toxoplasma gondii-infected human cells. J. Biol. Chem. 276:24223-24231. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Boldrick, J. C., A. A. Alizadeh, M. Diehn, S. Dudoit, C. L. Liu, C. E. Belcher, D. Botstein, L. M. Staudt, P. O. Brown, and D. A. Relman. 2002. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc. Natl. Acad. Sci. USA 99:972-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Bourges, D., C. H. Wang, C. Chavaleyre, and H. Salmon. T and sIgA B lymphocytes of the pharyngeal and palatine tonsils: differential expression of adhesion molecules and chemokines. Scand. J. Immunol., in press. [DOI] [PubMed]
- 8.Cohen, P., M. Bouaboula, M. Bellis, V. Baron, O. Jbilo, C. Poinot-Chazel, S. Galiegue, E. H. Hadibi, and P. Casellas. 2000. Monitoring cellular responses to Listeria monocytogenes with oligonucleotide arrays. J. Biol. Chem. 275:11181-11190. [DOI] [PubMed] [Google Scholar]
- 9.Coussens, P. M., C. J. Colvin, G. J. M. Rosa, J. P. Laspiur, and M. D. Elftman. 2003. Evidence for a novel gene expression program in peripheral blood mononuclear cells from Mycobacterium avium subsp. paratuberculosis-infected cattle. Infect. Immun. 71:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dozois, C. M., E. Oswald, N. Gautier, J. P. Serthelon, J. M. Fairbrother, and I. P. Oswald. 1997. A reverse transcription-polymerase chain reaction method to analyze porcine cytokine gene expression. Vet. Immunol. Immunopathol. 58:287-300. [DOI] [PubMed] [Google Scholar]
- 11.Eickhoff, B., B. Korn, M. Schick, A. Poustka, and J. Van der Bosch. 1999. Normalization of array hybridization experiments in differential gene expression analysis. Nucleic Acids Res. 27:e33. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, S. R., D. M. Roberton, H. Zola, and P. J. Macardle. 2000. Expression of the costimulator molecules, CD40 and CD154, lymphocytes from neonates and young children. Hum. Immunol. 61:378-388. [DOI] [PubMed] [Google Scholar]
- 13.Fahrenkrug, S. C., T. P. L. Smith, B. A. Freking, J. Cho, J. White, J. Vallet, T. Wise, G. Rohrer, G. Pertea, R. Sultana, J. Quackenbush, and J. W. Keele. 2002. Porcine gene discovery by normalized cDNA-library sequencing and EST cluster assembly. Mamm. Genome 13:475-478. [DOI] [PubMed] [Google Scholar]
- 14.Foss, D. L., M. J. Baarsch, and M. P. Murtaugh. 1998. Regulation of hypoxanthine phosphoribosyltransferase, glyceraldehyde-3-phosphate dehydrogenase and β-actin mRNA expression in porcine immune cells and tissues. Anim. Biotechnol. 9:67-78. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs, P. J., C. Cameron, L. C. Tan, S. A. Sadek, and W. M. Howell. 2003. House keeping genes and gene expression analysis in transplant recipients: a note of caution. Transplant Immunol. 12:89-97. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto, S., Y. Takahashi, Y. Tomita, T. Hayama, S. Sawada, T. Horie, C. C. McCombs, and J. P. Michalski. 1991. Mechanism of calcium ionophore and phorbol ester-induced T-cell activation. Scand. J. Immunol. 33:393-403. [DOI] [PubMed] [Google Scholar]
- 17.Hedges, J. F., D. Cockrell, L. Jackiw, N. Meissner, and M. Jutila. 2003. Differential mRNA expression in circulating γδ T lymphocyte subsets defines unique tissue-specific functions. J. Leukoc. Biol. 73:306-314. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann, K. F., T. C. McCarty, D. H. Segal, M. Charamonte, M. Hesse, E. M. Davis, A. W. Cheever, P. S. Meltzer, H. C. Morse III, and T. A. Wynn. 2001. Disease fingerprinting with cDNA microarrays reveals distinct gene expression profiles in lethal type 1 and type 2 cytokine-mediated inflammatory reactions. FASEB J. 15:2545-2547. [DOI] [PubMed] [Google Scholar]
- 19.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. (Erratum, 412:565.) [DOI] [PubMed] [Google Scholar]
- 20.Isakov, N., and A. Altman. 1985. Tumor promoters in conjunction with calcium ionophores mimic antigenic stimulation by reactivation of alloantigen-primed murine T lymphocytes. J. Immunol. 135:3674-3680. [PubMed] [Google Scholar]
- 21.Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken, J. Arroyo, J. D. Hoheisel, and J. Francois. 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278:20345-20357. [DOI] [PubMed] [Google Scholar]
- 22.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 23.Marcelo, P., M. Bernoin, and F. Lefèvre. 2003. Atypical mechanisms regulate the PMA-induced expression of IFN-γ in a porcine trophectoderm cell line. Vet. Immunol. Immunopathol. 92:163-172. [DOI] [PubMed] [Google Scholar]
- 24.Morgan, R. W., L. Sofer, A. S. Anderson, E. L. Bernberg, J. Cui, and J. Burnside. 2001. Induction of host gene expression following infection of chicken embryo fibroblasts with oncogenic Marek's disease virus. J. Virol. 75:533-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 26.Nakachi, N., K. Matsunaga, T. W. Klein, H. Friedman, and Y. Yamamoto. 2000. Differential effects of virulent versus avirulent Legionella pneumophila on chemokine gene expression in murine alveolar macrophages determined by cDNA expression array technique. Infect. Immun. 68:6069-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen, L. T., M. Ramanathan, B. Weinstock-Guttman, M. Baier, C. Brownscheidle, and L. D. Jacobs. 2003. Sex differences in in vitro pro-inflammatory cytokine production from peripheral blood of multiple sclerosis patients. J. Neurol. Sci. 209:93-99. [DOI] [PubMed] [Google Scholar]
- 28.Oswald, I. P., C. M. Dozois, R. Barlagne, S. Fournout, M. V. Johansen, and H. O. Bøgh. 2001. Cytokine mRNA expression in pigs infected with Schistosoma japonicum. Parasitology 122:299-307. [DOI] [PubMed] [Google Scholar]
- 29.Pinzon-Charry, A., J. P. Vernot, R. Rodriguez, and M. E. Patarroyo. 2003. Proliferative response of peripheral blood lymphocytes to mitogens in the owl monkey Aotus nancymae. J. Med. Primatol. 32:31-38. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberger, C. M., M. G. Scott, M. R. Gold, R. E. Hancock, and B. B. Finlay. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 164:5894-5904. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberger, C. M., A. J. Pollard, and B. B. Finlay. 2001. Gene array technology to determine host responses to Salmonella. Microbes Infect. 3:1353-1360. [DOI] [PubMed] [Google Scholar]
- 32.Savonet, V., C. Maenhaut, F. Miot, and I. Pirson. 1997. Pitfalls in the use of several “housekeeping” genes as standards for quantitation of mRNA: the example of thyroid cells. Anal. Biochem. 247:165-167. [DOI] [PubMed] [Google Scholar]
- 33.Venter, J. C., M. D. Adams, E. W. Myers, P. M. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, et al. 2001. The sequence of the human genome. Science 291:1304-1351. (Erratum, 292:1838.) [DOI] [PubMed] [Google Scholar]
- 34.Wang, Z. M., C. Liu, and R. Dziarski. 2000. Chemokines are the main proinflammatory mediators in human monocytes activated by Staphylococcus aureus, peptidoglycan, and endotoxin. J. Biol. Chem. 275:20260-20267. [DOI] [PubMed] [Google Scholar]
- 35.Wu, X., K. Englund, B. Lindblom, and A. Blanck. 2003. mRNA-expression of often used house-keeping genes and the relation between RNA and DNA are sex steroid-dependent parameters in human myometrium and fibroids. Gynecol. Obstet. Investig. 55:225-230. [DOI] [PubMed] [Google Scholar]
- 36.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. http://nar.oupjournals.org/cgi/reprint/30/4/e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yowe, D., W. J. Cook, and J. C. Gutierrez-Ramos. 2001. Microarrays for studying the host transcriptional response to microbial infection and for the identification of host drug targets. Microbes Infect. 3:813-821. [DOI] [PubMed] [Google Scholar]