Abstract
Defective viral genomes (DVGs) are generated during virus replication. DVGs bearing complementary ends are strong inducers of dendritic cell (DC) maturation and of the expression of antiviral and pro-inflammatory cytokines by triggering signaling of the RIG-I family of intracellular pattern recognition receptors. Our data show that DCs stimulated with virus containing DVGs have an enhanced ability to activate human T cells and can induce adaptive immunity in mice. In addition, we describe the generation of a short Sendai virus (SeV)-derived DVG RNA (DVG-324) that maintains strong immunostimulatory activity in vitro and in vivo. DVG-324 induced high levels of IFN-β expression when transfected into cells and triggered fast expression of pro-inflammatory cytokines and mobilization of dendritic cells when injected into the footpad of mice. Importantly, DVG-324 enhanced the production of antibodies to a prototypic vaccine after a single intramuscular immunization in mice. Notably, the proinflammatory cytokine profile induced by DVG-324 was different from that induced by poly I:C, the only viral RNA analogue currently used as an immunostimulant in vivo, suggesting a distinct mechanism of action. SeV-derived oligonucleotides represent novel alternatives to be harnessed as potent adjuvants for vaccination.
Keywords: Adjuvant, Sendai virus, defective genomes, dendritic cells, immunization
INTRODUCTION
New effective adjuvants to improve vaccine efficacy are needed. Advances in the understanding of molecular mechanisms that initiate the inflammatory response have revealed novel pathways that could be harnessed for adjuvant development. One of these pathways involves a family of intracellular helicases best represented by the retinoic acid-inducible gene 1 (RIG-I). RIG-I like receptors (RLRs) bind to virus-derived oligonucleotides leading to the expression of antiviral and pro-inflammatory molecules [1, 2]. RNA molecules that mimic natural viral-derived oligonucleotides can therefore be developed as novel RLR-targeted adjuvants.
Defective viral genomes (DVGs) are byproducts of viral replication that result from the loss of processivity of the viral polymerase during virus replication [3, 4]. DVGs are truncated viral genomes that lack essential genes for replication but that retain the molecular motifs necessary for stimulation of RLRs. DVGs derived from the mouse paramyxovirus Sendai (SeV) have potent immunostimulatory properties [5, 6]. Stimulatory SeV DVGs, known as copy-back genomes, contain complementary ends allowing the formation of double-stranded RNA structures. These structures trigger signaling by both RIG-I and the related helicase melanoma differentiation-associated protein 5 (MDA5) [5–7] promoting the expression of antiviral and proinflammatory cytokines in infected cells and inducing the complete maturation of mouse and human dendritic cells (DCs) [5, 6].
In this study, we tested the hypothesis that SeV DVGs can be harnessed as potent immunostimulants to be used during immunization. We specifically investigated whether SeV DVGs can provide immunostimulatory activity to human DCs, and whether they can be used as adjuvants in protocols using DCs as immunization vehicles. In addition, we set out to generate a shorter, optimized, synthetic DVG-derived molecule that retains the stimulatory properties of complete DVGs, but that is more amenable to be transitioned to vaccine development.
RESULTS
SeV containing DVGs enhance the ability of DCs to activate adaptive immune responses
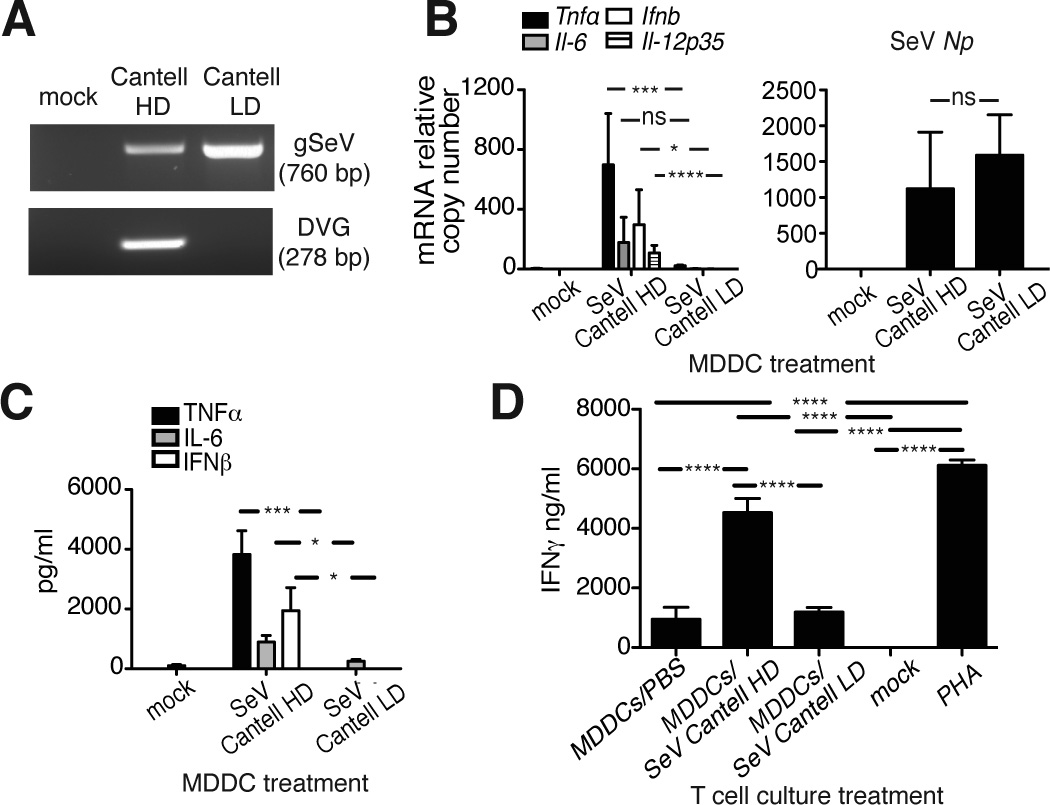
Stocks of SeV strain Cantell with a high content of copy-back DVGs (SeV Cantell HD) can efficiently induce the maturation of mouse and human DCs [6]. DVG content on infected cells can be visualized by PCR (Fig. 1A). To test if SeV Cantell HD enhances the ability of DCs to activate human T cells, we infected human monocyte-derived DCs (MDDCs) with SeV Cantell HD or SeV Cantell depleted of DVG-containing particles (LD) and co-cultured those infected MDDCs with allogeneic purified human CD4+ T cells. MDDCs infected with SeV Cantell HD expressed Ifnb, Il-6, Il-12p35, and Tnfα mRNA, but those infected with SeV Cantell LD did not express these genes despite normal expression of mRNA for the viral protein Np (Fig. 1B). Production of cytokines was confirmed from the culture supernatants using ELISA (Fig. 1C). Remarkably, IFNγ was produced at high levels in co-cultures containing MDDCs infected with SeV Cantell HD, but not in those containing cells infected with SeV Cantell LD (Fig. 1D). As controls, T cells were either not treated or treated with the unspecific activator phytohemagglutinin (PHA). This study demonstrates that viral particles containing DVGs can be used to enhance DC-mediated activation of human T cells.
Figure 1. Activation of human DCs upon SeV Cantell HD infection induces strong CD4+ T cell response.
(A) BMDCs were mock-infected or infected with a MOI=1.5 TCID50/cell of SeV Cantell HD or SeV Cantell LD. Infected cells were harvested 6 h post-infection and total RNA was analyzed by PCR to detect copy-back DVGs and standard viral genomic RNA (gSeV). Our DVG PCR is designed to detect most copy-back genomes generated in infected cells. SeV Cantell HD has one predominant copy-back genome that is seen as an amplicons of 278 nt. (B) Human MDDCs were infected with SeV Cantell HD or SeV Cantell LD (MOI=1.5 TCID50/cell). After 6 h, total RNA was extracted and analyzed by RT-qPCR for the expression of viral Np mRNA and cytokines. Data correspond to the average of five independent experiments. Each experiment was performed in triplicates. Bars correspond to SEM. p<0.0001 (Tnf), p=0.0340 (Il-6), p=0.0082 (Ifnb), p<0.0001 (Il-12p35) by one-way ANOVA. Significance after Bonferroni post-hoc test among different conditions is indicated in the graphs as *=p<0.05, ***= p<0.001, and ****= p<0.0001. Bonferroni denominator = 3. (C) Cytokine proteins were measured from the culture supernatants using ELISA. Data correspond to the average of two independent experiments. Each experiment was performed in triplicates. Bars correspond to SEM. p<0.0001 (Tnf), p=0.0027 (Il-6), p=0.0127 (Ifnb) by one-way ANOVA. Significance after Bonferroni post-hoc test among different conditions is indicated in the graphs as *=p<0.05, and ***= p<0.001. Bonferroni denominator = 3. (D) Activated MDDCs were co-cultured with human allogeneic CD4+ T cells (DCs: T cells ratio=1:5). After 5 days, the supernatant was collected and IFNγ was quantified by ELISA. Phytohaemagglutinin (PHA) was used for CD4+ T cell activation as a positive control. Data correspond to the average of three independent experiments. Each experiment was performed in triplicates. Bars correspond to SEM. p<0.0001 by one-way ANOVA. Significance after Bonferroni post-hoc test is indicated in the graphs as ****= p<0.001. Bonferroni denominator = 5. Data are expressed as copy numbers relative to the housekeeping genes Tuba1b and Rps11.
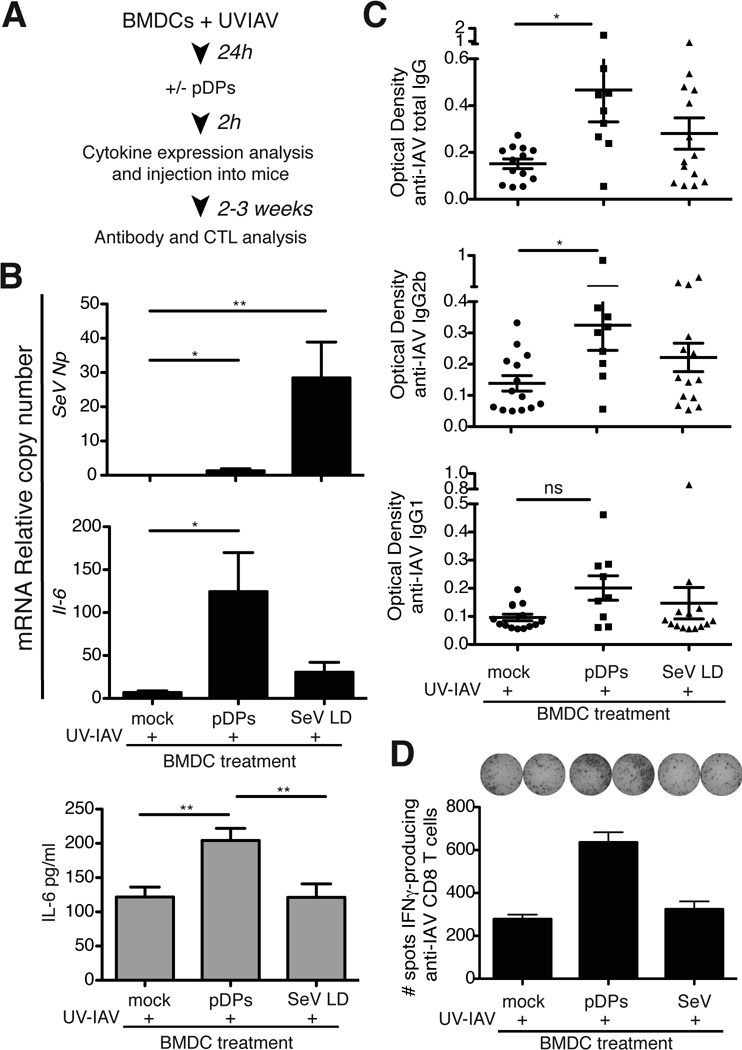
To evaluate whether DCs exposed to SeV DVGs show an enhanced ability to trigger adaptive immunity in vivo, we tested purified SeV defective particles containing DVGs (pDPs) as immunostimulants on a model of DC immunization in mice [8]. Bone marrow-derived DCs (BMDCs) were treated with UV-inactivated influenza virus (IAV) A/New Caledonia/20/99 (H1N1) as antigen for 24h followed by either treatment with pDPs or mock-treatment (Fig. 2A). Treatment with pDPs enhanced the expression of IL-6 by DCs compared to mock-treated cells (Fig. 2B) despite only marginal expression of SeV Np mRNA, as expected due to the inability of pDPs to replicate in the absence of helper virus [5]. In contrast, control infection with SeV Cantell LD showed high levels of SeV Np while cytokine expression was lower than in cells treated with pDPs. Remarkably, mice immunized with UV-IAV-BMDCs treated with pDPs showed enhanced production of total anti-IAV IgG as well as antibodies of the IgG2b and IgG1 isotypes compared with mice immunized with UV-IAV-BMDCs alone (Fig. 2C). Mice immunized with BMDCs treated with pDPs also showed higher frequency of anti-IAV specific heterosubtypic IFNγ-producing CD8+ T cells upon in vitro restimulation with splenocytes infected with IAV A/X-31 (H3N2) compared to controls (Fig. 2D). Overall these data demonstrate that SeV DVGs promote the ability of DCs to trigger specific adaptive immune responses in vivo.
Figure 2. SeV defective viral particles (DPs) enhance the ability of DCs to promote adaptive immune responses in vivo.
(A) Representation of the DC immunization protocol. BMDCs pretreated for 24 h with UV-inactivated IAV were incubated with 125 HA Units of pDPs or mock treated. (B) Expression of viral proteins (Np) mRNA and Il-12p40 was determined from a sample of the cells 2 h post-infection by RT-qPCR. Data correspond to the average of three independent experiments. Each experiment was performed in triplicates. Bars correspond to SEM. p=0.0039 (SeV Np), p=0.0130 (Il-6), p=0.0030 (IL-6 protein) by one-way ANOVA. Significance after Bonferroni post-hoc test among different conditions is indicated in the graphs as *=p<0.05, **= p<0.01. Bonferroni denominator = 3. Data are expressed as copy numbers relative to the housekeeping genes Tuba1b and Rps11. (C) Anti-influenza virus IgG, IgG2b, and IgG1 in sera of mice 14 days after immunization. The experiment was repeated independently three times. Data shown is a compilation of all experiments (total number of mice = 9–14/group). p=0.03 (total IgG), p=0.0367 (IgG2b) by Kruskal-Wallis test. Significance after Dunn’s post-hoc test for each treatment is indicated in the graphs as *=p<0.05. Dunn’s denominator = 3. The data was also reanalyzed after eliminating a maximum of one outlier from each group based on the Grubb’s test with a significance level of 0.05. Significance was maintained at p<0.05 for Total IgG. (D) Number of influenza virus specific IFNγ-producing CD8+ T cells in splenocytes from immunized mice. For these experiments splenocytes from 4 mice per each group were pooled for in vitro restimulation. The experiment was independently repeated twice. Bars correspond to SEM of the assay.
Recombinant SeV defective particles retain a strong immunostimulatory activity
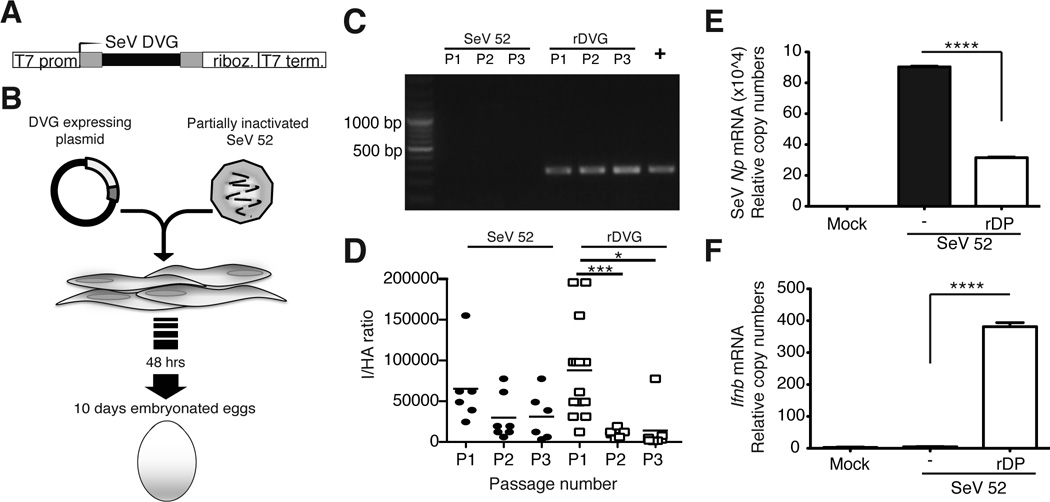
SeV can produce multiple different DVGs during its replication cycle. A copy-back DVG of 546 nucleotides (DVG-546) was identified to be strongly immunostimulatory and the predominant DVG in laboratory stocks of SeV Cantell [9]. We hypothesized that DVG-546 alone was sufficient to confer immunostimulatory activity to SeV. To test this hypothesis we established a reverse genetics system for rescue of SeV particles containing DVG-546. For this system, DVG-546 was cloned under the control of the T7 polymerase promoter and flanked at the 3’ end by the hepatitis delta ribozyme followed by the T7 terminator to create the precise 3’ end (Fig. 3A). This construct was transfected into cells expressing the T7 polymerase and infected with partially inactivated SeV 52 helper virus that provided the necessary proteins for virus replication and packaging. SeV 52 does not produce highly immunostimulatory copy-back DVGs [5], thus all stimulatory activity is provided by the recombinant DVG. Recombinant viral particles containing DVGs (rDPs) were amplified in embryonated hen eggs (Fig. 3B). DVG-546 was detectable by PCR (amplicons of 278 bp) in virus stocks obtained after one passage in eggs (Fig. 3C) and was enriched in subsequent passages as determined by the ratio between infectious (I) and total hemagglutinating viral particles (HA) in the allantoic fluid (I/HA) (Fig. 3D). rDP particles inhibited the replication of the helper virus shown by reduced expression of SeV Np in cells infected with rDP-containing allantoic fluid (Fig. 3E). In addition, rDP had strong ability to induce expression of type I IFNs in infected cells (Fig. 3F). These data confirmed that the immunostimulatory activity of SeV DPs could be reproduced with a recombinant virus containing DPs that carry DVG-546.
Figure 3. Recombinant SeV copy back DVG preserves strong stimulatory activity.
(A) Schematic of the construct for the expression of SeV DVG-546. (B) BHK-21 cells expressing the T7 polymerase (BSR-T7) were infected with partially inactivated SeV 52 and transfected with the plasmid encoding DVG-546. Cells and supernatant were collected 48 h later and inoculated into 10-day embryonated hen eggs. (C) LLC-MK2 cells were infected at a MOI of 5 TCID50/cell with virus obtained from three consecutive passages in eggs (PI-P3) of control SeV 52 alone or SeV 52 in the presence of rDPs. DVG-546 in the infected cells was detected 15 h after infection by PCR. DVG from SeV Cantell was used as a positive control (+). (D) Passages PI-P3 of allantoic fluid from eggs containing control SeV 52 alone or in the presence of rDPs were analyzed for their content of DI particles by determining the ratio of infectious particles over total hemagglutinating particles (HA). Data from each individual egg is shown (n=5–8). p=0.0001 (DVG) by Kruskal-Wallis test. Significance after Dunn’s post-hoc test among different conditions is indicated in the graphs as *=p<0.05 and ***= p<0.001. Dunn’s denominator = 3. (E) LLC-MK2 cells infected with SeV 52 alone or containing rDP (P3) were analyzed by RT-qPCR for the expression of the viral protein Np mRNA and (F) Ifnb mRNA. The experiment was independently repeated more than three times. Data shown correspond to the average of three experiments. p=0.0001 by one-way ANOVA with Bonferroni post-hoc test. Significance after Bonferroni post-hoc test among different conditions is indicated in the graphs as ****=p<0.0001. Bonferroni denominator = 3. Data are expressed as copy numbers relative to the housekeeping genes Tuba1b and Rps11
Naked SeV DVG RNA maintains potent immunostimulatory activity in vitro
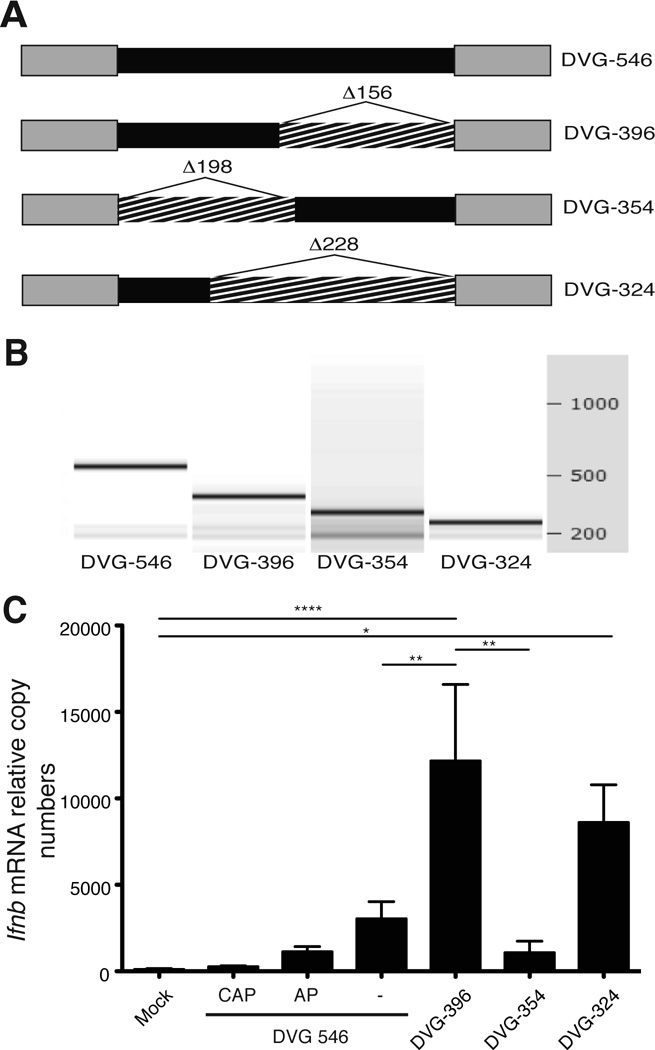
We next evaluated whether SeV DVG RNA induced the expression of Ifnb outside of the context of infection. To do this, we generated in vitro transcribed (ivt)RNA from the plasmid expressing DVG-546 (Fig. 4A). The integrity of the ivtRNA was confirmed by automated electrophoresis (Fig. 4B). ivtDVG-546 induced the expression of Ifnb mRNA in transfected cells while adding a 5’cap or treatment with alkaline phosphatase (AP) to remove 5’PPP significantly reduced the ability of ivtDVG-546 to trigger Ifnb expression (Fig. 4C). These data demonstrated that naked DVG-546 RNA retained its ability to trigger Ifnb expression through a mechanism that is largely dependent on the recognition of the 5’-triphosphate motif on the viral RNA [10–14].
Figure 4. Naked DVG-derived ivtRNA preserves immunostimulatory activity.
(A) Representation of deletion mutants of the DVG genome. Deletions (hatched) were performed in the internal sequence (black) without compromising the DVG complementary ends (grey). (B) The electrophoretic analysis for each ivtRNA was performed in an Agilent's 2100 Bioanalyzer. (C) LLC-MK2 cells were transfected with DVG-546, capped DVG-546 (CAP), DVG-546 treated with alkaline phosphatase (AP) or the different mutants and 6 h post-transfection cells were harvested and total cellular RNA was extracted to determine expression of Ifnb mRNA by RT-qPCR. The experiment was independently repeated three times. Each assay was performed in triplicates. Data corresponds to the average of all experiments (total n = 3–8/group). p=0.0001 by one-way ANOVA with Bonferroni post-hoc test. Significance after Bonferroni post-hoc test among different conditions is indicated in the graphs as ****=p<0.0001, **=p<0.01, and **=p<0.05. Bonferroni denominator = 3. Data are expressed as copy numbers relative to the housekeeping genes Actb and Tuba1b.
We next thought to generate shorter SeV DVG-derived oligonucleotides with similar or better immunostimulatory activity as more practical agents to be used as vaccine adjuvants. In these mutants we preserved the complementary ends of the DVG-546 molecule that provide a double-strand RNA motif that is presumed essential for the activation of RLRs [15–17]. Our strategy involved reducing the length of the internal non-complementary sequence (Fig. 4A). We first generated two DVG variants of similar length lacking different segments of the DVG internal sequence (DVG-396 and DVG-354, Fig. S1). These mutant DVGs were in vitro transcribed and their integrity was confirmed by automated electrophoresis (Fig. 4B). All mutants transfected into cells with similar efficiency (Fig. S2). Remarkably, cells transfected with ivtDVG-396 expressed higher levels of Ifnb mRNA than the parental ivtDVG-546 RNA, while ivtDVG-354 failed to induce high levels of Ifnb mRNA. We next reduced the length of the immunostimulatory DVG even further to generate DVG-324. This shorter DVG-derived RNA preserved a strong ability to induce the expression of Ifnb mRNA in infected cells (Fig. 4C). Mutants DVG-354 and DVG-324 have approximately the same size, suggesting that the immunostimulatory activity of DVGs depends mainly on their sequences and is not a direct function of the oligonucleotide length.
The cellular helicase RIG-I recognizes DVG-derived naked RNA
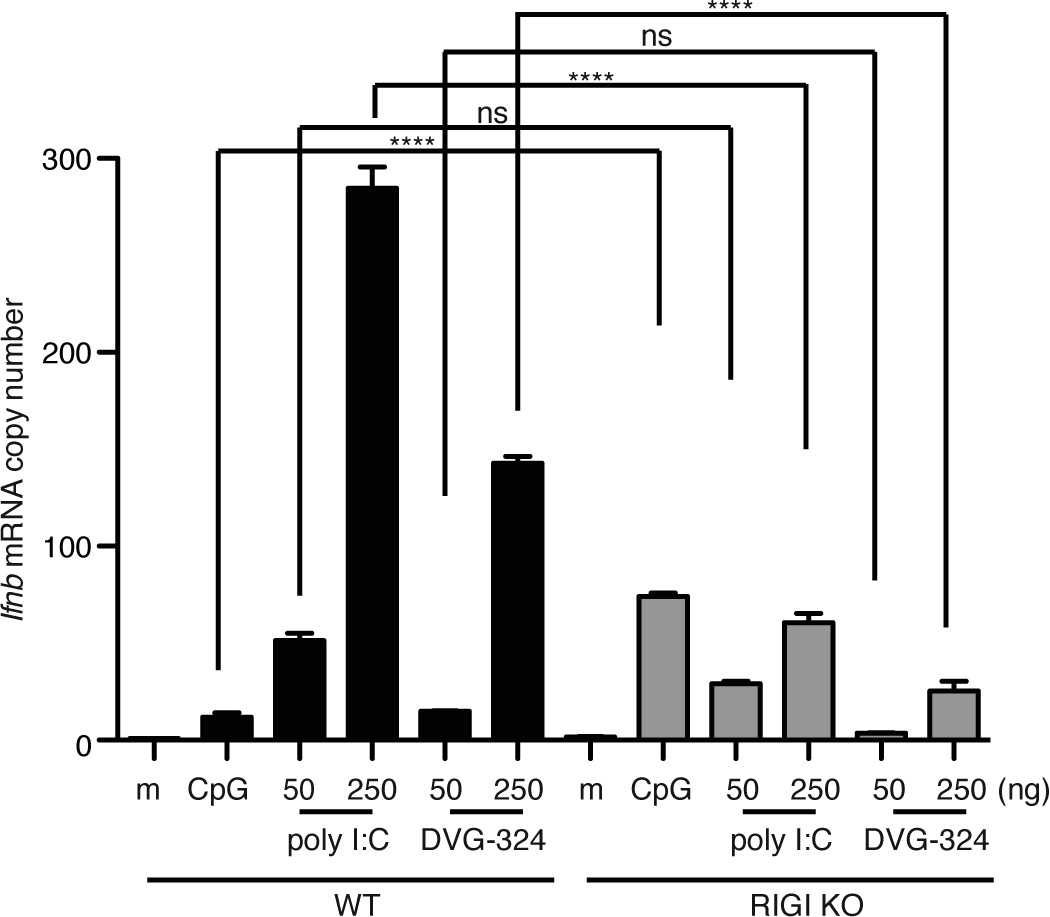
We and others have reported that SeV DVGs are recognized by RLRs [5–7, 18]. To determine whether our newly generated DVG-derived RNA was also recognized by RIG-I, we transfected RIG-IKO mouse embryo fibroblasts (MEFs) with increasing doses of poly I:C or ivtDVG-324. We treated the cells with the TLR ligand CpG as a control. Expression of Ifnb mRNA 6 h after transfection of ivtDVG-324 or poly I;C was largely reduced, although not completely eliminated, in cells lacking RIG-I, while expression of Ifnb in response to CpG was higher in RIG-I KO cells, suggesting that our RIG-I KO MEFs were functional and hyperresponsive to TLR ligands (Fig. 5). These data demonstrate that RIG-I is a primary sensor molecule for all our DVG-derived RNAs and agree with our previous reports of a role for additional cellular virus-sensing proteins in DVG recognition [6].
Figure 5. DVG-derived naked RNA trigger RIG-I signaling.
WT and RIG-I KO MEFs were transfected with 1 µg of CpG or with increasing doses of poly I:C or in vitro transcribed DVG-324 as indicated. Total RNA was extracted 8 h later and expression of Ifnb mRNA was determined by RT-qPCR. Data corresponds to the average of three experiments. p=0.0001 by one-way ANOVA with Bonferroni post-hoc test. Significance after Bonferroni post-hoc test among different conditions is indicated in the graphs as ****=p<0.0001. Bonferroni denominator = 12. Data are expressed as copy numbers relative to the housekeeping genes Actb and Tuba1b.
DVG-derived naked RNA induces localized immunostimulatory activity in mice
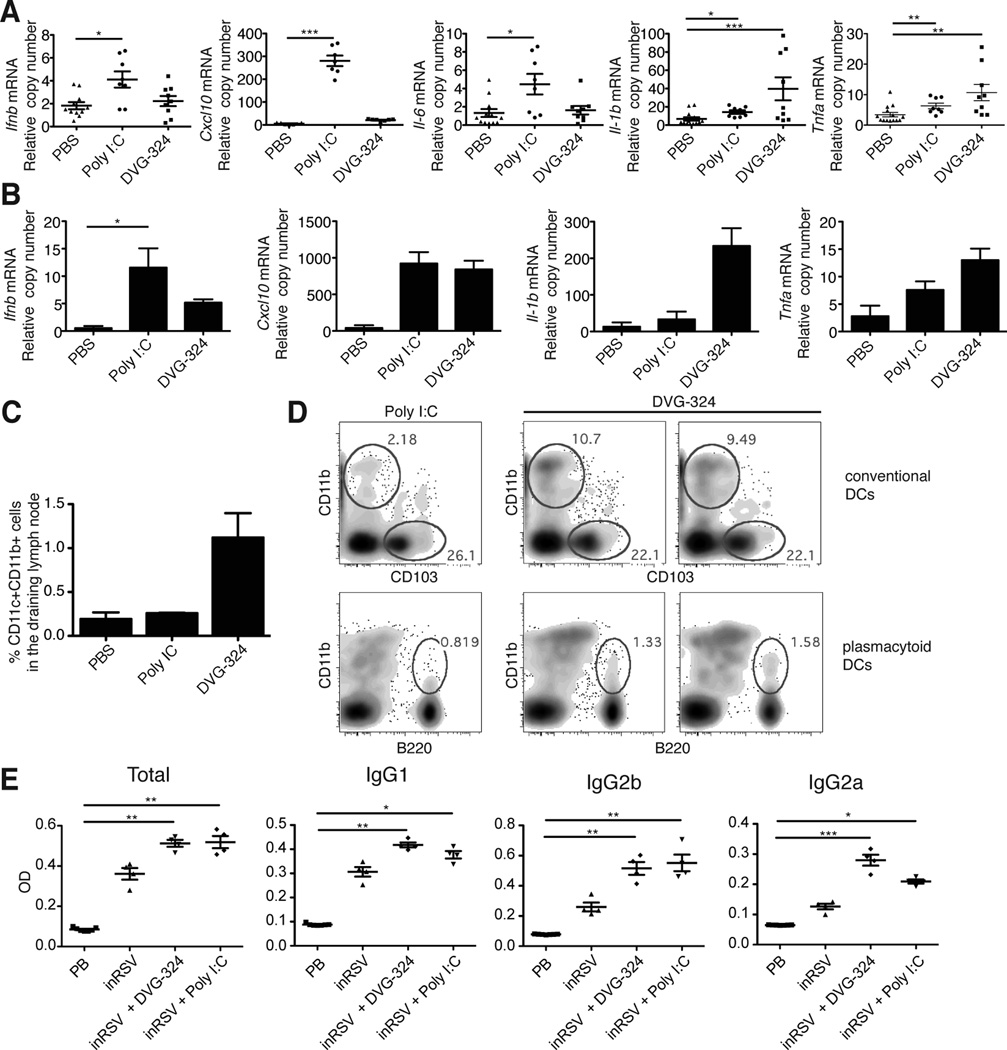
To test the immunostimulatory activity of DVG-derived RNA in vivo, we injected footpads of mice subcutaneously with ivtDVG-324 or, as a control, the synthetic viral analog poly I:C. Higher levels of Ifnb mRNA, as well as mRNA for the IFN-inducible gene Cxcl10 and for the cytokine il-6 were expressed at the infection site 36 h after poly I:C injection than after injection of ivtDVG RNA. However, the pro-inflammatory cytokines Il-1β and Tnf-α were only significantly expressed in tissue injected with DVG-324, but not upon poly I:C injection (Fig. 6A). A similar pattern of cytokine expression of Il-1β and Tnf-α was evident as early as 6 h after treatment with ivtDVG-324, while no differences in the expression of Ifnb and Cxcl10 were observed at this time point (Fig. 6B). Altogether, these data indicate that naked DVG-derived RNA preserves its high immunostimulatory activity in vivo and that it induces a different localized inflammatory environment than poly I:C.
Figure 6. DVG-derived naked RNA shows strong localized pro-inflammatory activity in mice.
Mice were injected subcutaneously in the footpad with 50 µg of DVG-324 or poly I:C (high molecular weight). (A–B) Footpad tissue was harvested after 36 h and RNA was extracted for the analysis of cytokine expression. The experiment was independently repeated three times. Each assay was performed in triplicates. Data shown is a compilation of all experiments. p=0.0192 (Ifnb), p=0.0201 (Il6), p=0.006 (Il1b), p=0.009 (Tnf) and p=0.0001 (Cxcl10) by Kruskal-Wallis test. Significance after Dunn’s post-hoc test among different conditions is indicated in the graphs as *=p<0.05, **=p<0.05 and ***= p<0.001. Dunn’s denominator = 3. To control for experimental variation, data was also analyzed by two-way ANOVA for the parameters treatment/experiment. Significance of variation among treatments are: p=0.0158 (Ifnb), p=0.0002 (Il6), p=<0.0001 (Il1b), p=<0.0001 (Tnf), and p<0.0001 (Cxcl10). (B) Analysis of cytokine expression in mice footpads collected at 6 h post infection. Data shown is an average of two independent experiments (n=4–5/group). Each assay was performed in triplicates. Data are expressed as copy numbers relative to the housekeeping gene rps11. p=0.0357 (Ifnb), p=0.0374 (Il1b), p=0.0490 (Tnf), and p=0.0979 (Cxcl10) by Kruskal-Wallis test. Significance after Dunn’s post-hoc test among different conditions is indicated in the graphs as *=p<0.05. Dunn’s denominator = 3. (C–D) Single cell suspensions from draining popliteal lymph nodes were analyzed by flow cytometry. (C) Percentage of CD11c+CD11b+ cells from the live cell gate is shown (n = 2 for PBS and 5 for poly I:C and DVG 324). p=0.387 by one-way ANOVA with Bonferroni post-hoc test. Bonferroni denominator = 3. (D) CD11c+CD11b+ cells were further gated for expression of CD103 and B220 to quantify CD11c+CD11b+CD103− DCs and CD11bloCD11cloB220+ plasmacytoid DCs. Data show a representative plot for poly I:C treatment and two representative plots for DVG-324 treatment. (E) Antibodies in the sera of Balb/c mice three weeks after immunization with a single i.m. dose of 180 µg of inactivated respiratory syncytial virus (inRSV) in the presence of 50 µg poly I:C, 50 µg DVG-324, or PBS. Sera pre-immunization (pre-bleed: PB) was also analyzed. n=4/group. p=0.0013 (total IgG), p=0.0011 (IgG1), p=0.0013 (IgG2b), and p=0.0008 (IgG2a) by Kruskal-Wallis test. Significance after Dunn’s post-hoc test among different conditions is indicated in the graphs as *=p<0.05, **p<0.01, and ***p<0.001. Dunn’s denominator = 4
In order to evaluate the capacity of DVG RNA to induce DC migration, we harvested the footpad draining popliteal lymph nodes and quantified the presence of different DCs populations by flow cytometry. Strikingly, a higher percentage of CD11b+CD11c+CD103− DCs, as well as a slightly larger percentage of plasmacytoid DCs CD11bloCD11cloB220+ DCs, were observed in the lymph node of mice injected with DVG-324 compared to poly I:C, suggesting that both DC populations participate in the response to DVG RNA (Fig. 6C–D). To determine whether DVG-324 has systemic effects that may compromise its safety, we determined the amount of type I IFN and IL-6 in the sera of mice injected in the footpad with DVG-324 or poly I:C. We were not able to detect significant levels of these proteins in the sera at 6 or 36 h post infection in any of the treatments by ELISA. To enhance the sensitivity of our analysis, we looked for the expression of type I IFN responsive genes in the spleen. Corresponding with the protein data, this bioassay did not show evidence of DVG-324 induced systemic type I IFN action (not shown) suggesting that DVG RNA acts locally without major systemic inflammatory effects. Overall, these data indicate that DVG-324 RNA can induce the expression of pro-inflammatory cytokines in the footpad tissue of mice and trigger the accumulation of DCs in the draining lymph node, representing a novel candidate adjuvant molecule.
DVG-derived naked RNA shows high pro-inflammatory activity in mice
To test the function of DVG-324 as a potential adjuvant, we evaluated the production of antibodies in mice after a single shot immunization with inactivated respiratory syncytial virus alone (inRSV) or in the presence of DVG-324 or, as a control, poly I:C. As shown in Fig. 6E, DVG-324 significantly enhanced the production of antibodies to the virus to levels comparable or higher than poly I:C. Remarkably, DVG-324 enhanced the breadth of antibodies produced with a particular improvement in the production of IgG1 and IgG2a when compared to poly I:C. These experiments serve as a proof of principle to demonstrate that the DVG-324 immunostimulatory activity can be pursued as an adjuvant for vaccine development.
DISCUSSION
SeV DVG-derived RNA oligonucleotides represent novel adjuvant candidates. Despite concerns related to RNA stability, several studies have demonstrated the successful use of naked mRNA as therapy in vivo [19–22]. The synthetic viral RNA analog poly I:C has entered clinical trials as an anti-cancer therapy [23, 24] and it has been shown to be safe in humans [20]. Here we show that SeV DVGs enhance the ability of mouse DCs to stimulate the development of adaptive immunity, and that naked DVG RNA maintains its immunostimulatory activity when injected subscutaneously in the footpad. In addition, we show that SeV DVGs enhance the ability of human DCs to activate T cells, supporting their role as potent novel immunostimulants.
To harness DVGs as adjuvants, we have generated an optimized SeV DVG-derived synthetic RNA that has high immunostimulatory capacity both in vitro and in vivo. Importantly, by comparing DVG mutants of similar lengths, we have identified a region in the SeV DVG-RNA that is essential for its potent immunostimulatory activity. Work is in progress to further characterize this region. Our data suggest that in addition to the 5’-trophosphate motif, the sequence composition is critical for effective RLR stimulation by natural viral agonists. This oligonucleotide has several advantages over poly I:C. First, poly I:C is a heterogeneous mixture of molecules of different sizes. In contrast, DVG RNA is a single molecule. Second, the molecular characteristics responsible for the immunostimulatory ability of DVG-derived RNA can be defined in detail allowing product optimization and modification, while it is impossible to do so for the variable mixture of poly I:C oligonucleotides. Third, DVG-derived RNA is shorter, and therefore more cost-effective to produce than poly I:C.
Interestingly, different inflammatory profiles were observed in vivo in the footpad tissue upon injection of poly I:C or DVG-derived RNA, suggesting that these molecules engage at least partially different signaling pathways in mice. Notably, DVG-324 induced significantly higher expression of mRNAs from pro-inflammatory genes than poly I:C, in particular, that of Il-1β. Production of IL-1 is a desired feature of an effective immunostimulant as it improves the immunogenicity of viral vaccines [25]. IL-1β plays an essential role in the effectiveness of the widely used aluminum salts adjuvants [26–30] and is required for induction of Th1 responses by the TLR7 agonist Resiquimod (R484) [31]. Higher levels of pro-inflammatory cytokines in response to DVG-324 also correlated with a higher percentage of CD11c+CD11b+CD103− DCs and CD11bloCD11cloB220+ plasmacytoid DCs in the draining lymph node of mice treated with DVG-324, suggesting that the pro-inflammatory environment triggered the migration of DCs from the skin to the lymph node [32, 33].
In our analysis of more than 10 independent preparations of DVGs and mutant constructs, we have determined that in vitro transcription of DVGs is a robust and highly consistent process in standard laboratory conditions. We routinely obtained around 100 µg per ivt reaction with a well-conserved level of purity. In all batches analyzed independently, in vitro transcribed DVGs showed strong immunostimulatory activity. Importantly, mRNA molecules of approximately 2 kb have been produced using a similar methodology for clinical applications, establishing the feasibility of large-scale production of RNA [34]. In addition, DVG RNA is stable for more than 6 months at −80°C and for up to one month at −20°C. Strategies such as incorporation into liposomes or cationic matrices may be used to further enhance RNA stability and efficiency.
SeV-DVG-546 was shown to induce protective immunity in mice when administered intranasally or intramuscularly [18]. Here we demonstrate that SeV-DVG RNA can also enhance the DC immunostimulatory capacity during a protocol of DC-based immunization. In addition, a shorter version of SeV-DVG RNA (DVG-324) can potently induce the expression of cytokines and the migration of DCs to the draining lymph node when injected subcutaneously. DVG-324 is a potent novel candidate adjuvant molecule to be tested in various vaccine platforms.
MATERIALS AND METHODS
Cell lines and mice
Rhesus monkey kidney epithelial cells (LLC-MK2) were obtained from ATCC, baby hamster kidney-21 (BHK-21) cells expressing the T7 RNA polymerase (BSR-T7) were provided by Dr. C. Basler, and RIG-I KO mouse embryo fibroblasts (MEFs) were provided by Dr. L. Gitlin. C57BL/6 and Balb/c mice (Taconic Farms) were housed in pathogen free conditions and used under institutionally approved protocols (specific approval for this project was obtained from the University of Pennsylvania Institutional Animal Care and Use Committees).
Viruses
SeV Cantell and 52, and IAV A/New Caledonia/20/99 and X-31 were grown in 10 days old hen embryonated eggs (B & E Eggs, Silver Springs, PA) for 40 h at 37°C [35]. SeV strain Cantell depleted of DVGs-containing particles (LD) was generated after 2 passages of SeV Cantell highly diluted as described elsewhere [5]. IAV was purified through a 40% sucrose cushion in sterile conditions. IAV was inactivated by UV light for 10 min. Inactivation was confirmed by establishing the virus inability to replicate. SeV 52 (LD) was partially inactivated by UV light for 45 sec. Defective particles were purified as previously described [5]. Respiratory syncytial virus strain A2 was obtained from ATCC and was grown in Hep-2 cells. Virus was inoculated at a MOI of 0.02 and harvested at day 4 post infection. Cells and supernatants were freeze-thawed three times and centrifuged 15 min at 1,200 rpm to eliminate debris. The clarified virus was filtered through a 0.22 µm filter and incubated with 1:4000 formalin dilution (37% formaldehyde ACS grade Fisher Scientific) as previously described [36] followed by ultracentrifugation for 2 h at 25,000 rpm (Beckman, SW32). Inactivation was confirmed by establishing the virus inability to replicate in Hep-2 cells. The pellet was suspended in RPMI media and protein concentration was determined by Bradford assay (Thermo Scientific).
Dendritic cell preparation
Human MDDCs were prepared as previously described [37]. Murine bone marrow-derived dendritic cells (BMDCs) were prepared from C57BL/6 mice according to a standard protocol [38].
Isolation of human naive CD4+ T cells and allogenic co-cultures
Naive CD4+ T cells were negative selected from PBMCs using a CD4+T cell isolation kit (Miltenyi Biotec). HLA-DR+ cells were removed using anti-HLA-DR microbeads (Miltenyi Biotec). Infected MDDCs were incubated with human naive CD4+ T cells in a 1:5 ratio for 5 days. Cytokines in the supernatant were measured by ELISA (eBiosciences).
Immunizations, antibody measurement, and ELISPOT
For immunizations using BMDCs as a vehicle, the cells were treated with 2.5 µg of UV-inactivated IAV A/New Caledonia/20/99 (H1N1) for 24 h prior to treatment with 125 HA Units of purified DPs. Control BMDCs were infected with SeV 52. Treated BMDCs were injected intraperitoneally into mice (5 × 105 cells/mouse). For immunization using inactivated RSV, mice were injected intramuscularly with 180 µg of formalin inactivated virus (1.5 × 108 pfu of virus) alone or in the presence of 50 µg of poly I:C or DVG-324. Sera were analyzed at day 14 for anti-influenza virus or at day 20 for anti-RSV total IgG, IgG1, IgG2b, and/or IgG2a antibodies by ELISA. IAV-immunized mice were sacrificed at day 21 and CD8+ T cells were isolated from splenocytes using positive selection (Miltenyi). T cells were co-cultured with irradiated splenocytes isolated from naive mice and infected with IAV strain X-31 (H3N2) in an ELISPOT plate coated with anti-IFN-γ at a 1:1 ratio. Plates were incubated for 24h before performing IFN-γ ELISA.
PCR and RT-qPCR
RNA was isolated using TRIzol and 2 µg of RNA was reversed transcribed using Transcriptor First Strand cDNA Synthesis Kit (Roche) and the primer 5’GGTGAGGAATCTATACGTTATAC3’. cDNA was amplified using Platinum Taq polymerase (Invitrogen) and the primers: for- 5’GGTGAGGAATCTATACGTTATAC3’ and rev- 5’ACCAGACAAGAGTTTAAGAGATATGTATT3’. For RT-qPCR 0.5–2.0 µg of RNA were reverse transcribed using High Capacity RNA-to-cDNA kit (Applied Biosystems). qPCR assay were performed using SYBR Green PCR Master Mix (Applied Biosystems) in a Viia7 Applied Biosystems Lightcycler. Primers used can be found in Table S1.
Plasmids and constructs
DVG-546 flanked at the 3’end by the SpeI-T7 promoter sequence and at the 5’ end by the hepatitis delta virus ribozyme, T7 polymerase terminator, and SapI sequences was synthetically prepared (DNA 2.0) and cloned into the pSL1180 vector (Amersham Pharmacia Biotech). In order to optimize the transcription of the DVG, 3 G residues were introduced downstream of the T7 promoter by site-directed mutagenesis (Stratagene, CA). To generate DVG mutants, restriction enzyme sites were introduced to the construct using the QuikChange II XL site-directed mutagenesis kit (Stratagene). Sequences and explanation of all the constructs can be found in Fig. S1.
Rescue of recombinant virus
BSR-T7 cells were infected with a MOI of approximately 60 of partially inactivated SeV 52 for one hour. Infected cells were transfected with 3µg of plasmid encoding DVG using XtremeGENE transfection reagent (Roche). Cells and supernatant were harvested after 48 h and 200 µL of the suspension was inoculated in the allantoic cavity of 10 days old embryonated hen eggs. After 40 h, allantoic fluid was harvested and egg inoculation was repeated for three consecutive passages.
In vitro transcription and transfections
DVG-expressing plasmids were linearized and in vitro transcribed using the MEGAscript T7 kit (Ambion) in the presence of RNase inhibitor (Fermentas). Capped RNA was synthetized using Cap Analog (m7G(5)ppp(5)G) (Ambion). RNA products were treated with DNase followed by LiCl precipitation. Integrity of ivtRNA was analyzed in an Agilent Bioanalyzer 2100. OD 260/280 (260/230) ratios for the ivtRNA were as follows (n > 4): DVG-546, 2.025 ± 0.04950 (2.730 ± 0.05657); DVG-396, 2.167 ± 0.06377 (2.662 ± 0.1455); DVG-354, 2.112 ± 0.04494 (2.488 ± 0.05975); DVG-324, 2.113 ± 0.04494 (2.483 ± 0.07234). RNA dephosphorylation was carried out using FastAP thermosensitive alkaline phosphatase (Fermentas). MEFs were transfected with 50 or 250 ng of ivtRNA or poly I:C, or with 1µg of CpG (Invivogen) using Lipofectamine 2000 (Invitrogen). LLC-MK2 cells transfections were performed using 250 ng of ivtRNA.
Mice footpad injection and flow cytometry
C57BL/6 mice were injected subcutaneously in the footpad with 50 µg of ivtRNA or PBS as control. Mice were euthanized at 6 or 36 h after infection for tissue collection. Cells were stained with antibodies against CD11c, CD11b, B220, Live Dead (eBioscience), and CD103 (BioLegend) for FACS analysis.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 5.00, GraphPad Software, San Diego California USA, www.graphpad.com.
Supplementary Material
Highlights.
Defective viral genomes enhance the maturation of human dendritic cells.
Their use as immunostimulants improve dendritic cell immunization efficacy.
Defective viral genome naked RNA has strong immunostimulatory activity.
A modified defective viral genome RNA induces strong innate immunity in mice.
This RNA improves the generation of antibodies to a prototypic vaccine in mice.
ACKNOWLEDGEMENTS
The authors wish to acknowledge Dillon Birdwell and Dr. Susan Weiss (UPenn) for RNA electrophoretic analysis, Dr. Christopher Basler (MSSM) for BSR-T7 cells, L. Gitlin for RIG-I KO MEFs, and Deepika Jain and Cristhian Cadena for technical support. In addition, the authors thank the valuable support of the Center for Clinical Epidemiology and Biostatistics at the University of Pennsylvania. The work was supported by NIH grant R01AI083284 and its associated Diversity Supplement.
Abbreviations
- DVG
defective virus genome
- DPs
defective particles
- IFN
interferon
- MDA5
melanoma differentiation-associated protein 5
- Poly I:C
polyinosinic:polycytidylic acid
- RIG-I
retinoic acid-inducible gene 1
- RLRs
RIG-I-like receptors
- RSV
respiratory syncytial virus
- inRSV
formalin-inactivated RSV
- SeV
Sendai virus
- SeV HD
SeV high content of defective particles
- SeV LD
SeV depleted of defective particles
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Xiao T. Innate immune recognition of nucleic acids. Immunol Res. 2009;43:98–108. doi: 10.1007/s12026-008-8053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 3.Huang AS, Baltimore D. Defective viral particles and viral disease processes. Nature. 1970;226:325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- 4.Lazzarini RA, Keene JD, Schubert M. The origins of defective interfering particles of the negative-strand RNA viruses. Cell. 1981;26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 5.Yount JS, Kraus TA, Horvath CM, et al. A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J Immunol. 2006;177:4503–4513. doi: 10.4049/jimmunol.177.7.4503. [DOI] [PubMed] [Google Scholar]
- 6.Yount JS, Gitlin L, Moran TM, Lopez CB. MDA5 Participates in the Detection of Paramyxovirus Infection and Is Essential for the Early Activation of Dendritic Cells in Response to Sendai Virus Defective Interfering Particles. J Immunol. 2008;180:4910–4918. doi: 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]
- 7.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez CB, Fernandez-Sesma A, Czelusniak SM, et al. A mouse model for immunization with ex vivo virus-infected dendritic cells. Cell Immunol. 2000;206:107–115. doi: 10.1006/cimm.2000.1736. [DOI] [PubMed] [Google Scholar]
- 9.Strahle L, Garcin D, Kolakofsky D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology. 2006;351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Takahasi K, Yoneyama M, Nishihori T, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 11.Lu C, Xu H, Ranjith-Kumar CT, et al. The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 14.Hornung V, Ellegast J, Kim S, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 15.Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Takeuchi O, Mikamo-Satoh E, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlee M, Hartmann G. The chase for the RIG-I ligand--recent advances. Mol Ther. 2010;18:1254–1262. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Gil L, Goff PH, Hai R, et al. A Sendai Virus-Derived RNA Agonist of RIG-I as a Virus Vaccine Adjuvant. J Virol. 2013;87:1290–1300. doi: 10.1128/JVI.02338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tewari K, Flynn BJ, Boscardin SB, et al. Poly(I:C) is an effective adjuvant for antibody and multi-functional CD4+ T cell responses to Plasmodium falciparum circumsporozoite protein (CSP) and alphaDEC-CSP in non human primates. Vaccine. 2010;28:7256–7266. doi: 10.1016/j.vaccine.2010.08.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caskey M, Lefebvre F, Filali-Mouhim A, et al. Synthetic double-stranded RNA induces innate immune responses similar to a live viral vaccine in humans. J Exp Med. 2011;208:2357–2366. doi: 10.1084/jem.20111171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stahl-Hennig C, Eisenblatter M, Jasny E, et al. Synthetic double-stranded RNAs are adjuvants for the induction of T helper 1 and humoral immune responses to human papillomavirus in rhesus macaques. PLoS Pathog. 2009;5:e1000373. doi: 10.1371/journal.ppat.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longhi MP, Trumpfheller C, Idoyaga J, et al. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J Exp Med. 2009;206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenfeld SJ, Young NS. Viruses and bone marrow failure. Blood Rev. 1991;5:71–77. doi: 10.1016/0268-960x(91)90037-d. [DOI] [PubMed] [Google Scholar]
- 24.Butowski N, Chang SM, Junck L, et al. A phase II clinical trial of poly-ICLC with radiation for adult patients with newly diagnosed supratentorial glioblastoma: a North American Brain Tumor Consortium (NABTC01-05) J Neurooncol. 2009;91:175–182. doi: 10.1007/s11060-008-9693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kayamuro H, Yoshioka Y, Abe Y, et al. Interleukin-1 family cytokines as mucosal vaccine adjuvants for induction of protective immunity against influenza virus. J Virol. 2010;84:12703–12712. doi: 10.1128/JVI.01182-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Nookala S, Re F. Aluminum hydroxide adjuvants activate caspase-1 and induce IL-1beta and IL-18 release. J Immunol. 2007;178:5271–5276. doi: 10.4049/jimmunol.178.8.5271. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum's adjuvant effect are mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kool M, Petrilli V, De Smedt T, et al. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J Immunol. 2008;181:3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- 29.Kool M, Soullie T, van Nimwegen M, et al. Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J Exp Med. 2008;205:869–882. doi: 10.1084/jem.20071087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosca F, Tritto E, Muzzi A, et al. Molecular and cellular signatures of human vaccine adjuvants. Proc Natl Acad Sci U S A. 2008;105:10501–10506. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madera RF, Wang JP, Libraty DH. The combination of early and rapid type I IFN, IL-1alpha, and IL-1beta production are essential mediators of RNA-like adjuvant driven CD4+ Th1 responses. PLoS One. 2011;6:e29412. doi: 10.1371/journal.pone.0029412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginhoux F, Merad M. Ontogeny and homeostasis of Langerhans cells. Immunol Cell Biol. 2010;88:387–392. doi: 10.1038/icb.2010.38. [DOI] [PubMed] [Google Scholar]
- 33.Merad M, Ginhoux F, Collin M. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat Rev Immunol. 2008;8:935–947. doi: 10.1038/nri2455. [DOI] [PubMed] [Google Scholar]
- 34.Pascolo S. Vaccination with messenger RNA (mRNA) Handb Exp Pharmacol. 2008:221–235. doi: 10.1007/978-3-540-72167-3_11. [DOI] [PubMed] [Google Scholar]
- 35.Lopez CB, Yount JS, Hermesh T, Moran TM. Sendai virus infection induces efficient adaptive immunity independently of type I interferons. J Virol. 2006;80:4538–4545. doi: 10.1128/JVI.80.9.4538-4545.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham BS, Henderson GS, Tang YW, et al. Priming immunization determines T helper cytokine mRNA expression patterns in lungs of mice challenged with respiratory syncytial virus. J Immunol. 1993;151:2032–2040. [PubMed] [Google Scholar]
- 37.Cotter CR, Nguyen ML, Yount JS, et al. The virion host shut-off (vhs) protein blocks a TLR-independent pathway of herpes simplex virus type 1 recognition in human and mouse dendritic cells. PLoS One. 2010;5:e8684. doi: 10.1371/journal.pone.0008684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez CB, Fernandez-Sesma A, Schulman JL, Moran TM. Myeloid dendritic cells stimulate both Th1 and Th2 immune responses depending on the nature of the antigen. J Interferon Cytokine Res. 2001;21:763–773. doi: 10.1089/107999001753124499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.