Abstract
By using a recombinant severe acute respiratory syndrome coronavirus (SARS-CoV) nucleocapsid protein-based enzyme-linked immunosorbent assay (ELISA) and serum specimens serially collected (from day 0 to day 240 after symptom onset) from patients with pneumonia due to SARS-CoV, we analyzed the longitudinal profiles of immunoglobulin G (IgG), IgM, and IgA antibodies against the SARS-CoV nucleocapsid protein in patients with pneumonia due to SARS-CoV. For IgG, the median optical density at 450 nm (OD450) turned positive at day 17 and a biphasic response was observed. At day 240, all patients were still positive for anti-nucleocapsid protein IgG antibody. For IgM, the median OD450 turned positive at day 20.5, peaked at about day 80, and fell to below the baseline level at about day 180. At day 240, 36% of the patients were still positive for anti-nucleocapsid protein IgM antibody. For IgA, the median OD450 turned positive at day 17, peaked at about day 50, and fell to below the baseline level at about day 180. At day 240, 36% of the patients were still positive for anti-nucleocapsid protein IgA antibody. The time of seroconversion detected by the recombinant SARS-CoV nucleocapsid protein-based ELISA and that detected by indirect immunofluorescence assay were similar. The median times of seroconversion for IgG, IgM, and IgA detected by the indirect immunofluorescence assay were 17 days (17 days by ELISA), 16.5 days (20.5 days by ELISA), and 17.5 days (17 days by ELISA), respectively, after disease onset. One, four, and one of the six patients who died did not produce any IgG, IgM, and IgA antibodies against the nucleocapsid protein of SARS-CoV, respectively, although these antibodies were detected in all six patients by the indirect immunofluorescence assay. Further studies should be performed to see whether SARS-CoV nucleocapsid protein antibody positivity has any prognostic significance.
Severe acute respiratory syndrome (SARS) has affected 30 countries in five continents, with more than 8,000 cases and 750 deaths. A novel virus, the SARS coronavirus (SARS-CoV), has been confirmed to be the etiological agent, and its genome has been completely sequenced (4, 6-8). Recently, SARS-CoV-like viruses have been isolated from Himalayan palm civets found in a live animal market in Guangdong Province of China (3). This finding implies that animals could be the reservoir for the ancestor of SARS-CoV. As for the detection of antibodies against SARS-CoV, at the moment, the most widely used methods are antibody detection in acute- and convalescent-phase sera by indirect immunofluorescence assay and enzyme-linked immunosorbent assay (ELISA) with cell culture extracts (4, 7). However, antibody detection by indirect immunofluorescence assay and ELISA with cell culture extracts may be less reproducible, more difficult to standardize, and more labor-intensive than ELISA-based antibody detection tests with recombinant antigens. Furthermore, production of the infected cell lines used to coat the ELISA plates and the slides for indirect immunofluorescence requires cultivation of the SARS-CoV, for which biosafety level 3 laboratory facilities are required. Such facilities are not available in most clinical microbiology laboratories.
ELISA-based antibody detection tests with recombinant antigens are well known to offer higher reproducibilities, are easier to standardize, and are less labor-intensive than antibody detection by indirect immunofluorescence assay and ELISA with cell culture extracts and do not require cultivation of SARS-CoV (1, 2, 9, 12). Recently, investigators have reported on the use of recombinant SARS-CoV nucleocapsid protein ELISA-based antibody tests for serodiagnosis of SARS-CoV pneumonia and study of the seroprevalence of nonpneumonic SARS-CoV infections (10, 11). In the study described in this article, using serially collected serum specimens from patients with SARS-CoV pneumonia, we analyzed the longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the SARS-CoV nucleocapsid protein in patients with pneumonia due to SARS-CoV. The time of seroconversion detected by the recombinant SARS-CoV nucleocapsid protein-based ELISA and that detected by indirect immunofluorescence assay were also compared.
MATERIALS AND METHODS
ELISA for detection of IgG, IgM, and IgA antibodies against nucleocapsid protein of SARS-CoV.
The methods for the cloning and purification of His6-tagged recombinant nucleocapsid protein and optimization of the ELISA for detection of IgG, IgM, and IgA against SARS-CoV were reported previously (10, 11).
ELISA was performed as described previously (10, 11). Briefly, each well of an immunoplate (Nunc, Roskilde, Denmark) was coated with purified His6-tagged recombinant nucleocapsid protein (20 ng for IgG detection, 80 ng for IgM detection, and 30 ng for IgA detection) for 12 h and then blocked in phosphate-buffered saline (PBS) with 2% bovine serum albumin. Diluted human sera (1:40 for IgG detection, 1:10 for IgM detection, and 1:10 for IgA detection) were added to the wells of the His6-tagged recombinant nucleocapsid protein-coated plates in a total volume of 100 μl, and the plates were incubated at 37°C for 2 h. After the plates were washed with washing buffer five times, 100 μl of diluted horseradish peroxidase-conjugated goat anti-human IgG (1:4,000), mouse anti-human IgM (1:500), and mouse anti-human IgA (1:1,000) antibodies (Zymed Laboratories Inc., South San Francisco, Calif.) were added to the wells and the plates were incubated at 37°C for 1 h. After the plates were washed with washing buffer five times, 100 μl of diluted 3,3′,5,5′-tetramethylbenzidine (Zymed Laboratories Inc.) was added to each well and the plates were incubated at room temperature for 15 min. One hundred microliters of 0.3 M H2SO4 was added, and the absorbance at 450 nm of each well was measured. Each sample was tested in duplicate, and the mean absorbance for each serum sample was calculated. The cutoff values of the optical density at 450 nm (OD450) for IgG, IgM, and IgA detection were 0.263, 0.488, and 0.217, respectively.
Longitudinal profile of IgG, IgM, and IgA antibodies against SARS-CoV nucleocapsid protein in patients with pneumonia due to SARS-CoV.
Serial serum samples (obtained from day 0 to day 240 after symptom onset) were collected from 20 patients with pneumonia due to SARS-CoV who were positive for antibodies against SARS-CoV, as detected by an indirect immunofluorescence assay (7). The serum samples were subject to IgG, IgM, and IgA detection by the recombinant nucleocapsid protein ELISA for IgG, IgM, and IgA detection. The median OD450 values for the serum specimens from all patients were calculated at each time point.
Comparison of times of seroconversion detected by recombinant nucleocapsid protein ELISA and indirect immunofluorescence assay for IgG, IgM, and IgA detection.
The serum samples from the 20 patients mentioned above were collected in the first 40 days after the onset of the illness and were subjected to IgG, IgM, and IgA detection by indirect immunofluorescence assay, as described below. The times of seroconversion detected by the two antibody tests were compared. The day of seroconversion was defined as the day when the OD450 of the serum sample reached a value above the cutoff for the ELISA and the day when a serum sample at ≥1:10 dilution gave a positive signal for the indirect immunofluorescence assay.
Indirect immunofluorescence assay.
The indirect immunofluorescence assay was performed as described previously (7). Briefly, microscopic slides were coated with SARS-CoV-infected fetal rhesus kidney cells that were fixed in acetone. The proportion of infected cells was adjusted to 60 to 70%, as judged with a positive control serum sample obtained from a patient with pneumonia due to SARS-CoV. Ten microliters of serum sample (diluted at 1:10) was added to the slides, and the slides were incubated at 37°C for 30 min. After the slides were washed with PBS three times, 10 μl of fluorescein isothiocyanate-conjugated goat anti-human IgG, IgM, or IgA antibody (INOVA Diagnostics Inc., San Diego, Calif.) was added to the slides and the slides were incubated at 37°C for 30 min. After the slides were washed with PBS three times they were examined under UV light illumination with an immunofluorescent microscope. Serum samples positive at the screening dilution of 1:10 were retested by using twofold serial dilutions.
RESULTS
Longitudinal profile of IgG, IgM, and IgA antibodies against SARS-CoV nucleocapsid protein in patients with pneumonia due to SARS-CoV.
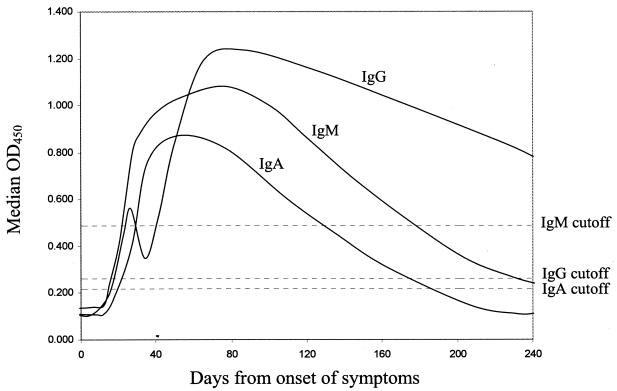
Six of the 20 patients with pneumonia due to SARS-CoV died before day 240 (on days 24, 25, 26, 33, 34, and 53, respectively, after the onset of illness). In addition, 3 of the remaining 14 patients who survived refused further venipuncture after day 40. The median OD450 results for the available serum specimens from all patients at each time point were calculated and were plotted against the number of days from the time of onset of symptoms (Fig. 1). For IgG, the median OD450 rose above the baseline level at day 17. After an initial surge, the IgG antibody level dropped transiently at about days 30 to 40 and then rose again. At day 240, all 11 patients from whom serum samples were available were still positive for anti-nucleocapsid protein IgG antibody. For IgM, the median OD450 rose above the baseline level at day 20.5, peaked at about day 80, and fell to below the baseline level at about day 180. At day 240, 4 of the 11 patients from whom serum samples were available were still positive for anti-nucleocapsid protein IgM antibody. For IgA, the median OD450 rose above the baseline level at day 17, peaked at about day 50, and fell to below the baseline level at about day 180. At day 240, 4 of the 11 patients from whom serum samples were available were still positive for anti-nucleocapsid protein IgA antibody.
FIG. 1.
Longitudinal profile of IgG, IgM, and IgA antibodies to SARS-CoV nucleocapsid protein in patients with pneumonia due to SARS-CoV.
Comparison of times of seroconversion detected by recombinant nucleocapsid protein ELISA and indirect immunofluorescence assay for IgG, IgM, and IgA detection.
The times of seroconversion for the 20 patients from whom serial serum samples were collected and who were confirmed to have pneumonia due to SARS-CoV by the indirect immunofluorescence assay are shown in Table 1. For IgG detection, the median time of seroconversion detected by the indirect immunofluorescence assay was 17 days (range, 13 to 39 days) after disease onset, whereas that detected by the recombinant nucleocapsid protein ELISA was 17 days (range, 2 to 39 days) after disease onset. In 4 of the 20 patients, IgG was detected by the ELISA earlier than by the indirect immunofluorescence assay, whereas in another 5 of the 20 patients, IgG was detected by the indirect immunofluorescence assay earlier than by the ELISA. For IgM detection, the median time of seroconversion detected by the indirect immunofluorescence assay was 16.5 days (range, 13 to 22 days) after disease onset, whereas that detected by the recombinant nucleocapsid protein ELISA was 20.5 days (range, 12 to more than 33 days) after disease onset. In 4 of the 20 patients, IgM was detected by the ELISA earlier than by the indirect immunofluorescence assay, whereas in another 13 of the 20 patients, IgM was detected by the indirect immunofluorescence assay earlier than by the ELISA. For IgA detection, the median time of seroconversion detected by the indirect immunofluorescence assay was 17.5 days (range, 11 to 27 days) after disease onset, whereas that detected by the recombinant nucleocapsid protein ELISA was 17 days (range, 4 to more than 32 days) after disease onset. In 10 of the 20 patients, IgA was detected by the ELISA earlier than by the indirect immunofluorescence assay, whereas in another 6 of the 20 patients, IgA was detected by the indirect immunofluorescence assay earlier than by the ELISA.
TABLE 1.
Comparison of times of IgG, IgM, and IgA seroconversion detected by indirect immunofluorescence assay and recombinant nucleocapsid protein ELISA in 20 patients
| Patient no. | Day of death | Day of seroconversion
|
|||||
|---|---|---|---|---|---|---|---|
| IgG
|
IgM
|
IgA
|
|||||
| Indirect immunofluorescence assay | Recombinant nucleocapsid protein ELISA | Indirect immunofluorescence assay | Recombinant nucleocapsid protein ELISA | Indirect immunofluorescence assay | Recombinant nucleocapsid protein ELISA | ||
| 1 | 25 | 16 | 19 | 14 | 25a | 16 | 14 |
| 2 | 17 | 17 | 16 | 31 | 16 | 17 | |
| 3 | 17 | 12 | 16 | 20 | 13 | 4 | |
| 4 | 53 | 19 | 19 | 18 | 18 | 18 | 19 |
| 5 | 33 | 13 | 32a | 16 | 32a | 13 | 32a |
| 6 | 27 | 17 | 22 | 27 | 27 | 27 | |
| 7 | 17 | 17 | 16 | 24 | 17 | 16 | |
| 8 | 24 | 17 | 17 | 17 | 23a | 20 | 20 |
| 9 | 18 | 18 | 18 | 12 | 22 | 28 | |
| 10 | 18 | 22 | 19 | 19 | 22 | 19 | |
| 11 | 34 | 14 | 14 | 17 | 33a | 17 | 14 |
| 12 | 26 | 13 | 21 | 13 | 21 | 13 | 21 |
| 13 | 28 | 28 | 16 | 13 | 19 | 16 | |
| 14 | 17 | 14 | 14 | 17 | 17 | 10 | |
| 15 | 39 | 39 | 21 | 31 | 21 | 21 | |
| 16 | 26 | 2 | 17 | 26 | 26 | 17 | |
| 17 | 13 | 13 | 16 | 13 | 20 | 16 | |
| 18 | 16 | 16 | 14 | 16 | 19 | 16 | |
| 19 | 13 | 17 | 17 | 17 | 11 | 17 | |
| 20 | 17 | 17 | 20 | 17 | 17 | 17 | |
The patient results were still negative by the indicated day.
DISCUSSION
The present study is the first in the literature to describe the longitudinal IgG, IgM, and IgA antibody profiles in patients with pneumonia due to SARS-CoV with serum samples serially collected up to day 240 after the onset of illness. In a previous study (5) that described the profile of IgG and IgM antibodies against SARS-CoV in patients with pneumonia due to SARS-CoV by an ELISA (the type of antigen used was not mentioned), both IgG and IgM antibody levels increased to detectable levels at the second week of illness. In the present study, the levels of all three antibodies (IgG, IgM, and IgA) increased to detectable levels at the third week of illness. We speculate that this difference could be due to the use of corticosteroids in our patients. Most of the patients with pneumonia due to SARS-CoV in our locality were given corticosteroids in the first 21 to 28 days of illness. In fact, some patients received a cumulative methylprednisolone dose of up to 4.5 g throughout the treatment period. This high dose of corticosteroid could have had a suppressive effect on the patients' antibody responses. In both studies, IgM antibodies became detectable later than IgG antibodies, which may be due to the earlier development of IgG antibodies than IgM antibodies or the differential sensitivities of the class-specific ELISAs. This is in contrast to the phenomena described for most other pathogens, against which IgM antibodies often appear a few days earlier than the IgG antibodies. In the previous study (5), the mean IgM antibody titers of the patients dropped to below a detectable level at about day 90. On the other hand, the median IgM antibody levels of our patients were still positive at day 90. In fact, all 11 patients from whom serum samples were available still had detectable IgM and IgA antibodies at day 90.
The biphasic IgG response in our patients could have been due to the use of corticosteroids. In the previous study (5), the mean IgG antibody levels of the patients rose steadily to a maximum at about day 30 after the onset of illness. On the other hand, the median IgG antibody level in our patients dropped slightly from about days 30 to 40 after the onset of illness (Fig. 1). This slight drop in the IgG antibody level was followed by a further increase in the median IgG antibody level to a peak at about day 80. In fact, this phenomenon of a biphasic IgG antibody response was seen in 6 of the 11 patients, and in 1 other patient, the IgG level plateaued transiently at about days 30 to 40 before rising to the peak level. Similar to the delayed appearance of antibodies, we speculate that this was also due to the use of corticosteroids. As described above, most of our patients received high-dose corticosteroids in the first 21 to 28 days of their illness. At about days 21 to 28, withdrawal of the corticosteroid treatment from the patients may have led to a secondary rise in IgG levels, giving rise to the apparent biphasic IgG antibody response seen in our patients.
The present recombinant nucleocapsid protein-based IgG ELISA had a sensitivity similar to that of the indirect immunofluorescence assay for the serodiagnosis of pneumonia due to SARS-CoV. In a previous study (10), it was shown that 6 of 106 (5.7%) serum samples that tested positive by the indirect immunofluorescence assay for IgG detection tested negative by the recombinant nucleocapsid protein IgG ELISA. However, of five patients with pneumonia due to SARS-CoV who were seronegative by the indirect immunofluorescence assay for IgG detection but reverse transcription-PCR positive for SARS-CoV, two had IgG antibodies against the nucleocapsid protein of SARS-CoV, as detected by the recombinant nucleocapsid protein IgG ELISA (unpublished data). In the present study, as shown in Table 1, in 4, 4, and 10 of the 20 patients, IgG, IgM, and IgA antibodies, respectively, were detected by the ELISA earlier than by the indirect immunofluorescence assay. On the other hand, in another 5, 12, and 6 of the 20 patients, the respective antibodies were detected by the indirect immunofluorescence assay earlier than by the ELISA. These findings show that the present recombinant nucleocapsid protein-based ELISA has sensitivity similar to that of the indirect immunofluorescence assay for the serodiagnosis of pneumonia due to SARS-CoV. As ELISA is much less labor-intensive and the interpretation of the results is more objective and the ELISA does not require cultivation of SARS-CoV, the present data further confirm that the ELISA could be an alternative to the indirect immunofluorescence assay as a screening test for antibodies against SARS-CoV in clinical microbiology laboratories. Interestingly, one, four, and one of the six patients who died did not produce any detectable IgG, IgM, and IgA antibodies against the nucleocapsid protein of SARS-CoV, respectively, even though these antibodies were detected in all of them by the indirect immunofluorescence assay. Further studies should be performed to see whether SARS-CoV nucleocapsid protein antibody positivity has any prognostic significance.
Acknowledgments
This study was supported by the Research Grant Council Grant (HKU 7532/03 M); the Vice-Chancellor SARS Research Fund, The University of Hong Kong; Research Fund for the Control of Infectious Diseases; William Benter Infectious Disease Fund; Kai Cheong Tong SARS Research Fund; Suen Chi Sun Charitable Foundation SARS Research Fund; and HKU DBS Bank SARS Research Fund.
REFERENCES
- 1.Briese, T., C. G. Hatalski, S. Kliche, Y. S. Park, and W. I. Lipkin. 1995. Enzyme-linked immunosorbent assay for detecting antibodies to Borna disease virus-specific proteins. J. Clin. Microbiol. 33:348-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, C. M., P. C. Y. Woo, A. S. P. Leung, S. K. P. Lau, X. Y. Che, L. Cao, and K. Y. Yuen. 2002. Detection of specific antibodies to an antigenic cell wall galactomannoprotein for serodiagnosis of Aspergillus fumigatus aspergillosis. J. Clin. Microbiol. 40:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 4.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and The SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 5.Li, G., X. Chen, and A. Xu. 2003. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 349:508-509. [DOI] [PubMed] [Google Scholar]
- 6.Peiris, J. S. M., C. M. Chu, V. C. C. Cheng, K. S. Chan, I. F. N. Hung, L. L. M. Poon, K. I. Law, B. S. F. Tang, T. Y. W. Hon, C. S. Chan, K. H. Chan, J. S. C. Ng, B. J. Zheng, W. L. Ng, R. W. M. Lai, Y. Guan, K. Y. Yuen, and Members of the HKU/UCH, SARS Study Group. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia—a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, K. Y. Yuen, and Members of the SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peiris, J. S. M., K. Y. Yuen, A. D. Osterhaus, and K. Stohr. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431-2441. [DOI] [PubMed] [Google Scholar]
- 9.Woo, P. C. Y., K. T. K. Chong, A. S. P. Leung, S. S. Y. Wong, S. K. P. Lau, and K. Y. Yuen. 2003. AFLMP1 encodes an antigenic cell wall protein in Aspergillus flavus. J. Clin. Microbiol. 41:845-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo, P. C. Y., S. K. P. Lau, H. W. Tsoi, K. H. Chan, B. H. L. Wong, X. Y. Che, V. K. P. Tam, S. C. F. Tam, V. C. C. Cheng, I. F. N. Hung, S. S. Y. Wong, B. J. Zheng, Y. Guan, and K. Y. Yuen. 2004. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet 363:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo, P. C. Y., S. K. P. Lau, B. H. L. Wong, H. W. Tsoi, A. M. Y. Fung, K. H. Chan, V. K. P. Tam, J. S. M. Peiris, and K. Y. Yuen. 2004. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 42:2306-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuen, K. Y., C. M. Chan, K. M. Chan, P. C. Y. Woo, X. Y. Che, A. S. P. Leung, and L. Cao. 2001. Characterization of AFMP1: a novel target for serodiagnosis of aspergillosis. J. Clin. Microbiol. 39:3830-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]