Abstract
A newly developed severe acute respiratory syndrome (SARS)-specific enzyme-linked immunosorbent assay (ELISA) was further validated to confirm cutoff values and evaluate its diagnostic performance with clinical samples. In parallel, an immunochromatographic test was also evaluated. A total of 227 clinical serum specimens collected from SARS patients were used in the study, together with 385 samples from healthy donors. By use of an immunofluorescent (IF) test as the “gold standard, ” both the ELISA and the immunochromatographic test were able to detect immunoglobulin G antibodies to SARS not only from late-convalescent-stage samples (>21 days from the onset of clinical symptoms), as previously established, but also from early-acute-phase samples (1 to 10 days from onset). The ELISA, using an optical density (OD) of 0.25 as its cutoff value, produced the best sensitivity while maintaining high specificity. It detected SARS-specific antibodies in 58, 70, 75, and 95%, respectively, of the four groups of samples collected from patients 1 to 10 days, 11 to 20 days, 21 to 30 days, and more than 30 days after the onset of clinical symptoms. Similarly, the immunochromatographic test detected SARS-specific antibodies in 55, 68, 81, and 79% of the four groups, respectively. The overall specificities for the ELISA and the rapid test were 99.5 and 97.7%, respectively. Although the positive correlation observed between the ELISA OD values and the IF titers was moderate (r = 0.6915; P < 0.001), the detection rates of both the ELISA and the rapid test were found well in agreement with the IF titers.
The newly emerged severe acute respiratory syndrome (SARS) is a serious respiratory illness of global significance. Its highly contagious nature, combined with its high mortality rate, has proven disruptive and costly in an increasingly globalized world. As new cases appear to be reemerging in January 2004, the urgency for prompt identification and isolation of infected patients remains. Although the identification of the novel coronavirus SARS coronavirus (SARS-CoV) as the etiological agent for SARS (3, 4, 7, 9) made it possible for various tests to be developed, providing tools for laboratory diagnosis remains a priority, as suggested by the World Health Organization (WHO). To date, there is still no standardized test for diagnosing SARS regardless of whether tests are antigen based or antibody based (http://www.who.int/csr/sars/conference/june_2003/materials/presentations/en/laboratorydiagnosis.pdf). In this regard, various immunofluorescent (IF) tests were essentially used as the “gold standards” for serodiagnosis of SARS during the 2003 outbreak. Although these IF protocols were found to be well correlated with PCR and clinical outcomes (9), they differed from laboratory to laboratory and were technically very demanding. In addition, these protocols utilized virus-infected materials and thus required laboratories with biosafety-level-3 facilities under current regulations. Hence, alternative tests with standardized approaches addressing these shortcomings are very much needed. However, only a thorough evaluation with standardized panels or by a trial using large numbers of clinical specimens will help to firmly establish the usefulness of new tests.
In this study, two newly developed serological tests were evaluated and validated with large numbers of samples. The two tests, an enzyme-linked immunosorbent assay (ELISA) and an immunochromatographic test, are both based on recombinant proteins, and both demonstrated excellent performance when tested with convalescent-phase samples from SARS patients in a previous study (5). The present expansion to include earlier specimens from acute-phase patients as well as large numbers of relevant controls provided a more realistic assessment of the performances of the two tests for serological diagnosis of SARS in clinical settings.
MATERIALS AND METHODS
Serum specimens.
Serum specimens were collected from patients who presented with suspected clinical SARS according to the WHO definition (13) and who were admitted to one of three acute-care regional hospitals in Hong Kong between 18 March and 24 May 2003. A total of 227 serum samples from these patients were tested by using an IF test (14) and were confirmed to have IF titers of >1:10 to 1:2,560. In the meantime, 385 serum samples collected locally from healthy donors in Hong Kong were used as controls. These samples were also tested by the same IF test and confirmed to have IF titers of <1:10. In addition, 1,066 serum samples from healthy donors purchased from BioClinical Partners Inc. (Franklin, Mass.) were included in the study as additional healthy controls. For the disease controls, serum samples from various previous studies in the Genelabs Diagnostics (Singapore) archive were used; these included 50 samples each from patients who had non-SARS-related fever (confirmed as dengue fever) or suffered from non-SARS-related respiratory illness (confirmed as tuberculosis).
IF test.
The IF test was prepared and carried out as described by Yam et al. (14). Briefly, smears of SARS-CoV-infected Vero cells were prepared, fixed in acetone for 10 min, and stored at −80°C before use. Batches of smears with 60 to 70% SARS-CoV-infected cells were high-titer verified by using a positive-control serum sample before use. Patient samples prepared in serial twofold dilutions starting with 1:10 were added to the smears and incubated for 30 min at 37°C. The smears were washed twice in phosphate-buffered saline (PBS) before a further incubation for 30 min at 37°C with a goat anti-human immunoglobulin G (IgG) labeled with fluorescein isothiocyanate. A sample was scored as positive if the fluorescent intensity was equal to or higher than that of a weakly positive control included in the study.
ELISA.
The ELISA was produced by Genelabs Diagnostics Pte Ltd. in Singapore by utilizing two recombinant proteins (Gst-N and Gst-U274) as previously described (5). The assays were carried out by strictly following the instructions provided. Briefly, 10-μl quantities of serum specimens each in 200 μl of a Tris-based diluent containing bovine serum albumin and skim milk powder were added to the appropriate wells of the ELISA plates and incubated for 30 min at 37°C, followed by six washes with PBS-Tween. A horseradish peroxidase-conjugated goat anti-human IgG (dilution, 1:500) was added at 100 μl per well and incubated for a further 30 min at 37°C. The plates were then washed six times in PBS-Tween, and color development was carried out by the addition of 100 μl of the enzyme substrate tetramethylbenzidine (TMB) per well. After a 15-min incubation in the dark at 37°C, the reaction was stopped by adding 100 μl of 1 N HCl per well. Optical densities (OD) were measured at 450 nm with a 620-nm reference filter.
Rapid immunochromatographic test.
The membrane-based test device was also produced at Genelabs Diagnostics Pte Ltd. in Singapore by following the procedure previously described (5). The device consisted of a chromatography strip, a separator, and an absorbent pad, all housed in a cassette as described previously (5).
Two SARS-specific recombinant proteins, Gst-N and Gst-U274, were deposited separately on the chromatography strip according to the detailed descriptions of Guan et al. (5, 6). If a test sample contained an antibody to SARS-CoV, the antibody would bind to either or both of the immobilized recombinants, and the immunocomplex formed could be detected by the immobilized colloidal gold labeled with anti-human IgG when the latter was released by a reagent-releasing and washing buffer (5).
These assays also were carried out by strictly following the instructions provided by the manufacturer (5). Briefly, 25 μl of serum sample was added to the sample well and allowed to migrate laterally to cover a portion of the membrane in the result-viewing window. Three drops of the reagent-releasing and washing buffer were added to the second well when the serum sample wetting front reached the blue indicator line in the viewing window. The separator tab was pulled until resistance was felt, and an additional drop of the reagent-releasing and washing buffer was then added to the sample well.
The results can typically be read through the viewing window in 2 to 15 min, but they were all recorded at 15 min. A sample was scored as negative when only the control line appeared but as positive when the control line and either or both of the test lines were seen in the viewing window (5).
Statistical analysis.
Delta values, defined as the distance of the mean OD/cutoff ratio of the sample population from the cutoff, measured in standard deviations (SD) (2), were calculated for validation of the cutoff values. The kappa statistic was used to measure the strength of an agreement between the results by the new rapid test and the new ELISA. A kappa statistic of >0.75, 0.40 to 0.75, or <0.40 represents excellent, good to fair, or poor agreement, respectively (10).
RESULTS
Evaluation and validation of the ELISA.
As presented in Table 1, three cutoff values at OD of 0.45, 0.30, and 0.25 were used to evaluate the performance of the ELISA. With the cutoff value set at an OD of 0.45, the ELISA produced the best specificity, 100% (385 of 385), but a low overall sensitivity of 60% (136 of 227) (Table 1). However, improved performance was obtained when the cutoff value was adjusted lower, to an OD of 0.3 or 0.25. The ELISA detected almost 12% more SARS-associated samples overall with an OD of 0.25 as the cutoff value than with the OD 0.45 setting (Table 1). An improved delta value of 0.53 for positive results was also obtained. In a supplementary test, a further 1,066 samples from healthy controls were tested, and a mean OD of 0.0432 ± 0.0745 was obtained. An OD of 0.25 was found to be equivalent to the mean OD ± 3 SD, whereas an OD of 0.45 was found to be equivalent to the mean OD ± 5 SD. The two cutoff values of 0.25 and 0.45 produced similarly high specificities of 98 versus 99.2%, 100 versus 100%, and 92 versus 98% for the supplemental healthy-control group, the non-SARS controls with respiratory illness, and the non-SARS controls with fever, respectively (Table 1).
TABLE 1.
Different cutoff values selected for the ELISA with their corresponding sensitivities, specificities, PPVs, NPVs, and delta values
| Cutoff (OD) | Sensitivitya at:
|
Specificityb
|
PPVc,d | NPVd,e | Delta
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-10 Days | 11-20 Days | 21-30 Days | >30 Days | Total | HD (HK) | HD (USA) | Non-SARS (respiratory) | Non-SARS (fever) | Positive | Negative | |||
| 0.25 | 18/31 (58) | 84/120 (70) | 43/57 (75) | 18/19 (95) | 163/227 (71.8) | 383/385 (99.5) | 1,045/1,066 (98.0) | 50/50 (100) | 46/50 (92) | 163/165 (98.8) | 383/447 (85.7) | 0.53 | 2.42 |
| 0.30 | 16/31 (52) | 78/120 (65) | 42/57 (74) | 18/19 (95) | 155/227 (68.3) | 384/385 (99.7) | 1,050/1,066 (98.5) | 50/50 (100) | 48/50 (96) | 155/156 (99.4) | 384/456 (84.2) | 0.40 | 2.61 |
| 0.45 | 15/31 (48) | 65/120 (54) | 40/57 (70) | 16/19 (84) | 136/227 (59.9) | 385/385 (100) | 1,058/1,066 (99.2) | 50/50 (100) | 49/50 (98) | 136/136 (100) | 385/476 (80.8) | 0.10 | 3.03 |
Given as number of positive samples/number of samples tested (percent).
Given as number of negative samples/number of samples tested (percent). HD (HK), 385 serum samples from healthy donors in Hong Kong; HD (USA), 1,066 serum samples from healthy donors purchased from BioClinical Partners Inc. IF titers were not determined for the healthy controls from BioClinical Partners or for the non-SARS disease controls.
Given as number of true positives detected/total number of samples testing positive (percent).
PPV and NPV were calculated by using the results for healthy donors from Hong Kong [HD (HK)] and those for SARS patients (total) obtained at the same testing site in Hong Kong.
NPV, number of true negatives detected/total number of samples testing negative (percent).
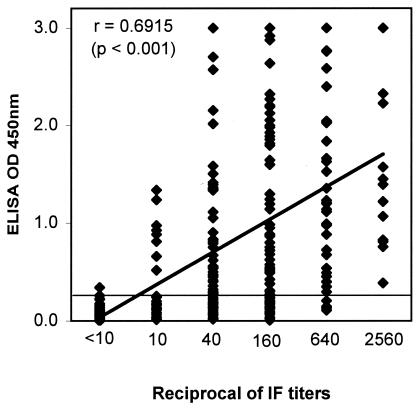
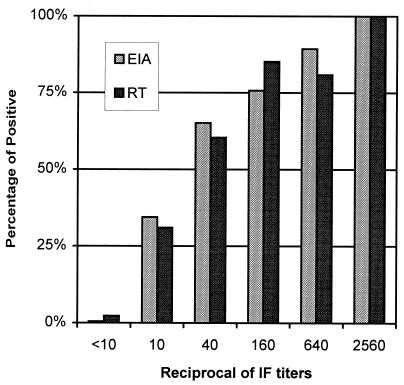
With the cutoff set at an OD of 0.25, the ELISA detected IgG antibodies to SARS-CoV not only in late-convalescent-stage samples (with a high sensitivity of 95%) but also in acute-phase specimens. It detected IgG antibodies to SARS-CoV in 58, 70, and 75% of samples collected from SARS patients 1 to 10 days, 11 to 20 days, and 21 to 30 days after the onset of clinical symptoms, respectively. The specificity obtained with the cutoff OD of 0.25 remained high at 99.5% (383 of 385) for the healthy-control group tested at the same testing site (Table 1). The test thus provided an overall positive predictive value (PPV) of 98.8% and an overall negative predictive value (NPV) of 85.7%. A moderate but positive correlation was also observed between the ELISA OD values and the IF titers (r = 0.6915; P < 0.001 [Fig. 1 ]). However, the correlation held firm, with the ELISA detecting antibodies to SARS-CoV in progressively more patient samples as IF titers increased (Fig. 2).
FIG. 1.
Correlation of OD obtained by ELISA and IF titers. For the group with IF titers of <10, the samples tested were from healthy donors (n = 385). For all other groups, samples tested were from SARS patients (total of 227 samples).
FIG. 2.
Distribution of detection rates (given as percentages) of the ELISA (EIA) and the rapid test (RT) in relation to IF titers. For the group with IF titers of <10, the samples tested were from healthy donors (n = 385). For all other groups, samples tested were from SARS patients (total of 227 samples).
Evaluation of the rapid immunochromatographic test.
When the same set of clinical specimens was tested by the rapid immunochromatographic test, a performance similar to that of the ELISA was obtained. The overall rate of detection of SARS-associated specimens by the rapid test was 70.5% (160 of 227), and its specificity was 97.7% (376 of 385) (Table 2). Detection rates were 55, 68, 81, and 79%, respectively, for the four groups of samples collected from SARS patients at 1 to 10 days, 11 to 20 days, 21 to 30 days, and more than 30 days after the onset of clinical symptoms (Table 2). Because this test utilized two recombinant proteins separately, represented by two resultant bands, the performances of the two proteins were also analyzed. The Gst-N protein alone detected antibodies to SARS-CoV in 157 of the 160 samples found to be positive by the two proteins combined (Table 2). Furthermore, the Gst-N protein presented less cross-reactivity, reacting to only 1 of the 385 specimens from the healthy-control group. In contrast, Gst-U274 detected only 8% (18 of 227) overall of the SARS-associated samples but contributed to most of the nonspecific reactivity with samples from healthy controls (8 of 385). Interestingly, the three samples that were detected by Gst-U274 only, but not by the Gst-N protein, were those collected from patients at an earlier stage, 1 to 20 days after the onset of clinical symptoms (Table 2). The test was therefore shown to have a PPV and an NPV of 94.7 and 84.9%, respectively, with the populations tested (Table 2). Again, in a supplementary test, the rapid test was further evaluated with disease control groups consisting of non-SARS patients who suffered from respiratory illness or fever and was found to cross-react to only 3 of the 50 or 4 of the 50 samples tested in the respective groups (Table 2). When the results were compared, the rapid test and the ELISA gave an excellent overall agreement of 92.5%, with a kappa statistic of 0.81 (Table 3). In addition, the rapid test was also found to perform well in concordance with the IF titers, detecting progressively more patient samples as the titers increased (Fig. 2).
TABLE 2.
Performance of the rapid immunochromatographic test in detecting IgG antibodies to SARS in samples from patient groups and healthy controls
| Recombinant protein | Sensitivitya at:
|
Specificityb
|
PPVc,d | NPVd,e | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1-10 Days | 11-20 Days | 21-30 Days | >30 Days | Total | HD (HK) | Non-SARS (respiratory) | Non-SARS (fever) | |||
| Combined | 17/31 (55) | 82/120 (68) | 46/57 (81) | 15/19 (79) | 160/227 (70.5) | 376/385 (97.7) | 47/50 (94) | 46/50 (92) | 160/169 (94.7) | 376/443 (84.9) |
| Gst-N | 16/31 (52) | 80/120 (67) | 46/57 (81) | 15/19 (79) | 157/227 (69.2) | 384/385 (99.7) | 48/50 (96) | 47/50 (94) | 157/158 (99.4) | 384/454 (84.6) |
| Gst-U274 | 1/31 (3) | 8/120 (7) | 5/57 (9) | 4/19 (21) | 18/227 (8) | 377/385 (97.9) | 49/50 (98) | 49/50 (98) | 18/26 (69.2) | 377/586 (64.3) |
Given as number of positive samples/number of samples tested (percent).
Given as number of negative samples/number of samples tested (percent). HD (HK), 385 serum samples from healthy donors in Hong Kong. IF titers were not determined for the non-SARS disease controls.
Given as number of true positives detected/total number of samples testing positive (percent).
PPV and NPV were calculated by using the results for healthy donors from Hong Kong [HD] (HK)] and those for SARS patients (total) obtained at the same testing site in Hong Kong.
NPV, number of true negatives detected/total number of samples testing negative (percent).
TABLE 3.
Agreement between the new rapid test and the new ELISA with sera from SARS patients and healthy controls
| Result with rapid test | No. of ELISA results that were:
|
Agreement (%)a | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 144 | 25 | 85.2 |
| Negative | 21 | 422 | 95.3 |
The kappa statistic obtained was 0.81. A kappa statistic of ≥0.75 represents excellent agreement, 0.40 to 0.75 represents good to fair agreement, and <0.40 represents poor agreement (11).
DISCUSSION
SARS is new to humans, and thus many aspects of the disease and its etiological agent, SARS-CoV, have yet to be fully understood. For example, the apparently contradictory phenomenon of a high infection rate for health care workers but a low infection rate for family members has yet to be fully explained. It appears that viral loads are low both during early infection and after about 3 weeks of infection. This situation renders PCR techniques relatively insensitive during the early stage of infection. Serological approaches such as IF tests, as demonstrated, have been welcome complementary tools for combating SARS. In fact, IF tests were practically used as the gold standard for serodiagnosis of SARS in hospitals during the outbreak despite their limitations. Nevertheless, new approaches are needed, and the IF test should be a useful reference for our evaluations.
In a previous study, the newly developed ELISA and the rapid immunochromatographic test demonstrated excellent performances, with PPVs and NPVs higher than 95%, suggesting great potential at least for exclusion of SARS or as study tools for epidemiologists (5). However, the study was limited to convalescent-phase samples from SARS patients, and the usefulness of the new tests for clinical diagnosis of SARS thus remained to be fully verified. In the present study, the two new tests were further evaluated for their performances with 227 patient samples and 385 samples from healthy controls. Furthermore, the study included a validation of the cutoff values using an additional 1,066 samples from healthy donors and 100 samples from disease controls. The study thus allowed us to fine-tune the cutoff value for the ELISA. The data showed that the adjusted cutoff OD of 0.25 produced the best sensitivity for the ELISA without compromising its specificity much (Table 1).
WHO is aware that the risk of false-positive results from SARS-CoV testing will be very high in the postoutbreak period and thus recommends a testing algorithm to minimize false- positive results in low-risk areas but to enhance surveillance in higher-risk areas (http://www.who.int/csr/sars/guidelines/en/SARSLabmeeting.pdf). In view of this recommendation, it may be appropriate to set a higher cutoff value of 0.45, with a PPV of 100%, for the ELISA when it is in use in low-risk areas in order to minimize the possibility of a false-positive result. Similarly, a lower cutoff value of 0.25, with an increased sensitivity and a higher NPV, may be desirable and appropriate when the ELISA is used in higher-risk areas. In this case, Western blot testing to further verify and confirm the results of samples repeatedly testing positive by the ELISA, especially in the case of borderline positives, will be helpful in resolving false-positives. Thus, further studies are warranted.
With the detection rate of 95% for samples obtained more than 30 days after the onset of clinical symptoms (Table 1), the present data confirmed previous findings that the ELISA was excellent at detecting antibodies to SARS in convalescent-phase samples (5). Although the ELISA yielded an overall sensitivity of about 72%, most of its shortcomings are attributable to its performance with samples obtained earlier, 1 to10 days after the onset of clinical symptoms (Table 1). This new test detected only 58% of this group (Table 1). However, it is noteworthy that this detection rate is a significant improvement over that of a capture ELISA, reported very recently, that detected IgG antibodies in only 46% (30 of 65) of an equivalent group (12). It is perhaps also noteworthy that IgG antibody to SARS CoV is believed to reach its peak value around 60 days after the onset of obvious symptoms (8). The Centers for Disease Control and Prevention has recommended including, as part of its case definition for exclusion of SARS, the absence of antibodies to the virus in convalescent-phase serum samples obtained more than 21 days after onset of the illness (1).
Similarly, the rapid test was found to have an overall detection rate comparable to that of the ELISA, at about 70%, with most of its shortcomings, again, due to results for the early group (Table 2). Although the rapid test utilized two recombinant proteins, Gst-N and Gst-U274, the latter appeared to contribute little to the sensitivity of the kit but more to the nonspecific reactivity with healthy controls. Gst-U274 detected only 18 of the 227 patient samples but cross-reacted with 8 of the 385 healthy-control samples (Table 2). As discussed previously, the biological function of Gst-U274 has yet to be unraveled, and the utility of this protein as a diagnostic marker thus has yet to be fully understood (5). The present study appears to suggest a limitation of this protein. However, it is interesting that the only three samples that were detected by Gst-U274 but not Gst-N were those collected from patients in the earlier group, 1 to 20 days from the onset of clinical symptoms.
The positive correlation observed between the ELISA ODs and the IF titers was moderate, with a correlation coefficient (r) of 0.6915 (P < 0.001 [Fig. 1]). This finding suggested that the ELISA OD and the IF titers were only moderately associated; the difference between the two approaches could be due to the fact that the new test utilized recombinant proteins rather than the virus-infected cells used in the IF test. However, it is noteworthy that the overall performances of both the ELISA and the rapid test were well in concordance with the IF titers. Both new tests detected progressively more patient samples as IF titers increased (Fig. 2). This finding, thus, suggested that the shortcomings of the new tests were mostly limited to samples with IF titers lower than 1:40 (Fig. 2). Nevertheless, the two new tests had similar overall sensitivities of 70.5 to 71.8% and specificities of 97.7 to 99.5% relative to the IF test, which was used as the gold standard (Tables 1 and 2). When the two tests were compared with each other, they gave an overall agreement of 92.4% with a kappa statistic of 0.81 (Table 3). This confirmed our previous findings and suggested that although the immunochromatographic test is a simple and rapid test that needs no special training to use, its performance was fully compatible with that of the ELISA. The immunochromatographic test should be a valuable addition to current options for combating SARS, especially in areas where laboratory facilities are not available.
Acknowledgments
This project was partially supported by a grant from the EDB (Economic Development Board of Singapore) under its Innovation Development Scheme.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2003. Updated interim surveillance case definition for severe acute respiratory syndrome (SARS)—United States, April 29, 2003. Morb. Mortal. Wkly. Rep. Dispatch 52:1-3. [PubMed] [Google Scholar]
- 2.Crofts, N., W. Maskill, and D. Gust. 1988. Evaluation of enzyme-linked immunosorbent assays: a method of data analysis. J. Virol. Methods 22:51-59. [DOI] [PubMed] [Google Scholar]
- 3.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brot, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 384:1967-1976. [DOI] [PubMed] [Google Scholar]
- 4.Fouchier, R. A. M., T. Kuiken, M. Schutten, G. van Amerongen, G. J. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. M. E. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan, M., H. Y. Chen, S. Y. Foo, Y. J. Tan, P. Y. Goh, and S. H. Wee. 2004. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 11:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan, M., H. Y. Chen, T. P. Chow, A. R. Pereira, and P. K. Mun. November 2001. Assay devices and methods of analyte detection. U.S. patent 6,316,205.
- 7.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and the SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 8.Nie, Q. H., X. D. Luo, and W. L. Hui. 2003. Advances in clinical diagnosis and treatment of severe acute respiratory syndrome. World J. Gastroenterol. 9:1139-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, K. Y. Yuen, and members of the SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pottumarthy, S., A. J. Morris, A. C. Harrison, and V. C. Wells. 1999. Evaluation of the tuberculin gamma interferon assay: potential to replace the Mantoux skin test. J. Clin. Microbiol. 37:3229-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 12.Shi, Y. L., Y. P. Yi, P. Li, T. J. Kuang, L. H. Li, M. Dong, Q. J. Ma, and C. Cao. 2003. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 41:5781-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization. 2003. Severe acute respiratory syndrome (SARS). Wkly. Epidemiol. Rec. 78:86-87. [Google Scholar]
- 14.Yam, W. C., K. H. Chan, L. L. M. Poon, Y. Guan, K. Yuen, W. H. Seto, and J. S. M. Peiris. 2003. Evaluation of reverse transcription-PCR assay for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 41:4521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]