Abstract
Though cytoplasmic male sterility (CMS) in peppers is associated with the orf507 gene, definitive and direct evidence that it directly causes male sterility is still lacking. In this study, differences in histochemical localization of anther cytochrome c oxidase between the pepper CMS line and maintainer line were observed mainly in the tapetal cells and tapetal membrane. Inducible and specific expression of the orf507 gene in the pepper maintainer line found that transformants were morphologically similar to untransformed and transformed control plants, but had shrunken anthers that showed little dehiscence and fewer pollen grains with lower germination rate and higher naturally damaged rate. These characters were different from those of CMS line which does not produce any pollen grains. Meanwhile a pollination test using transformants as the male parent set few fruit and there were few seeds in the limited number of fruits. At the tetrad stage, ablation of the tapetal cell induced by premature programmed cell death (PCD) occurred in the transformants and the microspores were distorted and degraded at the mononuclear stage. Stable transmission of induced semi-male sterility was confirmed by a test cross. In addition, expression of orf507 in the maintainer lines seemed to inhibit expression of atp6-2 to a certain extent, and lead to the increase of the activity of cytochrome c oxidase and the ATP hydrolysis of the mitochondrial F1Fo-ATP synthase. These results introduce the premature PCD caused by orf507 gene in tapetal cells and semi-male sterility, but not complete male sterility.
Keywords: Capsicum annuum L., orf507, cytochrome c oxidase, tapetum, transgenic semi-male sterility
Introduction
Cytoplasmic male sterility (CMS) is a maternally inherited trait that prevents a plant from producing functional pollen grains. CMS has been used widely to produce F1hybrids in order to increase fruit yield and decrease the energetic (Budar and Pelletier, 2001). The economically viable production of hybrid seeds requires a good pollination control system to avoid self-pollination of the female line. The CMS lines, that can-not produce functional pollen grains, are often used as female lines in hybrid seed production, to increase the purity of seeds (Schnable and Wise, 1998). However, the several bottlenecks faced during the development of CMS lines through conventional breeding require biotechnological intervention. The plant mitochondrial genome cannot be manipulated directly, so investigations of mitochondrial contributions to male sterility must be made by engineering nuclear genes encoding mitochondria-targeted proteins, including the expression of unedited forms of mitochondrial genes in the maintainer line (Hernould and Suharsono, 1993), and the expression of the CMS-associated mitochondrial orfs (Yamamoto et al., 2008). In most cases, CMS was related with premature degradation of the tapetal cell, a sporogenous tissue that nurtures the pollen mother cells (Hernould et al., 1998). In sunflowers, the CMS-associated open reading frame (ORF), orfH522, induced male sterility after being introduced into tobacco plants and expressed under the tapetum-specific promoter TA29. Bright-field microscopy demonstrated ablation of the tapetal cell layer in transgenic sterile plants that contain the orfH522 gene. Premature DNA fragmentation and programmed cell death (PCD) were observed at meiosis stage in the anthers of sterile plants (Nizampatnam et al., 2009).
It is generally believed that CMS results from the rearrangement of mitochondrial genomes, which is attributed to the generation of novel orfs (Schnable and Wise, 1998; Budar and Pelletier, 2001; Hanson and Bentolila, 2004; Linke and Börner, 2005; Yang and Zhang, 2007). Some experimental evidence has confirmed the correlation between CMS-associated orfs and the occurrence of CMS (Hanson and Bentolila, 2004). In Brassica juncea, transgenic plants that contained the CMS-associated orf220 gene and a mitochondrial-targeting sequence of the β subunit of F1-ATPase were male-sterile. In addition, transgenic stem mustard plants had aberrant floral development, identical to what has been observed in the CMS stem mustard phenotype (Yang et al., 2010). As described above, targeted expression in mitochondria of novel orfs has been shown to lead to male sterility or semi-sterility in some cases (He et al., 1996; Wang et al., 2006; Kim et al., 2007; Yamamoto et al., 2008), while in other cases, there was no effect (Chaumont et al., 1995; Wintz et al., 1995; Duroc et al., 2006). Most of the orfs in CMS lines are related to the mitochondrial energy metabolism complex. For example, orf522 in sunflower PET1-CMS encodes a protein sharing 18 amino acids with ORFB which is homologous to ATP8 in Reclinomonas (Balk and Leaver, 2001), and orf79 in rice is co-transcribed with the B-atp6 gene and encodes a cytotoxic peptide (Wang et al., 2006). All novel orfs have been confirmed to be associated with CMS. In addition, mutation of genes encoding subunits of mitochondrial enzymes also induces male sterility. For instance, mutation in the MGP1 gene, encoding the FAd subunit of F1Fo-ATP synthase in Arabidopsis thaliana, led to the destruction of mitochondria in pollen grains and subsequently the death of the pollen grains (Li et al., 2010).
In pepper (Capsicum annuum L.), CMS was first isolated from an Indian C. annuum accession (PI164835) (Peterson, 1958). Molecular investigations revealed that male sterility in many CMS lines is associated with the expression of orf456, which is located downstream and co-transcribed with the mitochondrial coxII gene (Kim et al., 2007), and the pseudogene ψatp6-2, 3′-truncated form of atp6-2 in maintainer line (Kim and Kim, 2006). Expression of the mitochondrion-targeted orf456 gene under the tapetum-specific promoter TA29 in transgenic Arabidopsis has shown that 45% of the T1 transgenic population is male-sterile and had no seed set, and the pollen grains of semi-sterile T1 plants have exine layer defects and vacuolated pollen phenotypes (Kim et al., 2007). However, an altered transcript, orf507, was found in a sequencing error at the 3′ end of orf456 (Gulyas et al., 2010). Expression of the orf507 gene in the flowers and leaves encodes a toxic protein that inhibits cell growth, and specifically interacts with a subunit of ATP synthase 6 (Li et al., 2013). In CMS plants, ATP synthase activity and content was reduced by more than half, compared normal plants. Based on these results, the researchers speculated that ORF507 might cause MtATP6 to be nonfunctional (Li et al., 2013). However, empirical proof that ORF507 is sufficient to induce male sterility has not been provided. In a previous study, we found that mitochondrial cytochrome c oxidase and F1Fo-ATPase were dysfunctional in the CMS line HW203A (Ji et al., 2013), but it remains unclear whether expression of orf507 is responsible for the dysfunction of the mitochondrial enzyme.
Therefore, we hypothesized that if orf507 is expressed under the tapetum-specific promoter TA29 from tobacco, and the protein is targeted into mitochondria using the coxIV pre-sequence as the transit peptide, ORF507 protein introduced into maintainer lines might result in male sterility and dysfunction of the mitochondrial enzyme in pepper plants. The coxIV pre-sequence have been proved to target the in-frame proteins into mitochondria in heterologous systems without any error into the chloroplast (Schmitz and Lonsdale, 1989). Kim et al. (2007) demonstrated that the coxIV pre-sequence was successfully used for targeting ORF456 from pepper into mitochondria in a heterologous plant. Simultaneously, we analyzed the histochemical localization of cytochrome c oxidase in anthers of different developmental stages in the pepper CMS line and maintainer line to evaluate the relationship between cytochrome c oxidase and male sterility.
Materials and methods
Plant materials and growth conditions
The pepper (Capsicum annuum L.) CMS line HW203A and its near-isogenic maintainer line HW203B (WT) were provided by the Asian Vegetable Research and Development Center (AVRDC). The CMS line was used as non-recurrent parent and maintainer line (WT), as recurrent parent. Eight generations of back-crosses were developed to obtain a CMS line similar to a maintainer line with CMS transferred from non-recurrent parents (CMS line). After eight backcrosses, the developed line (CMS line) is an isogenic line with it's maintainer line (WT), but the cytoplasmic background was different in the CMS and WT lines.
Capsicum annuum used in this study were grown in plastic trays containing steam-sterilized growing medium at 25°C with a 16 h light/8 h dark cycle.
Histochemical localization of cytochrome c oxidase in anthers at different developmental stages
Anthers were collected from the CMS line (HW203A) and the maintainer line (HW203B) at the microspore mother cell stage (MMC), tetrad stage and mononuclear microspore (MNM) stage at 4°C to avoid the inactivity of the enzyme. At each stage of anther development, five anthers were randomly selected from five pepper plants. The anthers were soaked in a fixation solution (2.5% glutaraldehyde, 1%TritonX-100) for 20 min at 4°C, washed with 0.1 M PBS (pH 7.4), and then incubated in a PBS buffer solution containing 0.1 mg/mL catalase for 10 min. The anthers were washed again in PBS three times, and then incubated in a 0.1 M PBS buffer solution containing 1, 0.1, and 1 mg/mL cyt c for 1 h. 1 mM KCN was added in the control. Sample fixation and observation was conducted following the protocols outlined in Ji et al. (2014a).
Vector construction
To construct the vector, a tapetum-specific promoter (TA29) from tobacco and a mitochondrial transit sequence (the first 25 amino acids of COXIV) from yeast were cloned. The orf507 gene was taken from the HW203A CMS line of C. annuum. Primers used in this study (Table 1) were designed for amplification of the component sequences from their respective sources, and restriction enzyme sites were introduced at the 5′ end of the primers. The PCR amplified component sequences were first cloned in the pMD19-T vector, sequenced, and excised with specific enzymes that were introduced into the primers. The TA29 promoter, coxIV pre-sequence and orf507 were cloned sequentially into the pCAMBIA 2300 vector. The resultant clone pTCON (pCAMBIA 2300: TA29-coxIV pre-sequence—orf507—nos terminator) was used for plant transformation. The control was constructed without the objective gene orf507, pTCN (pCAMBIA 2300: TA29-coxIV pre-sequence—nos terminator). The pSCON vector (pCAMBIA 2300: CaMV35S-coxIV pre-sequence—orf507—nos terminator) was also constructed for the comparison with the pTCON transgenic plants. The clones were confirmed by restriction analysis, PCR and sequencing. The confirmed clones were then transformed into Agrobacterium tumefaciens strain GV3101, following the standard freeze thaw method of transformation (Ji et al., 2014a).
Table 1.
List of primers used in this study.
| Primer code | Sequence (5′–3′) | Purpose of primers |
|---|---|---|
| TA29F | 5′-ACACAAATAGCCGGCTATAT-3′ | Amplification of TA29 promoter |
| TA29R | 5′-GCTTCGAATAGCTAATTTCTTTAAGTAAAAAC-3′ | |
| BamH I coxIVF | 5′-CGCGGATCCATGCTTTCACTACGTCAATCTATAAG-3′ | Amplification of the transit peptide |
| Kpn I coxIVR | 5′-CGGGGTACCACCCTCTTTAGCACCAGGACC-3′ | |
| Kpn I orf507F | 5′-CGGGGTACCATGCCCAAAAGTCCCATGT-3′ | Amplification of orf507 gene |
| Sac I orf507R | 5′-CGAGCTCTTAAAAAGCGCTAAACAAAT-3′ | |
| caubi F | 5′-TGTCCATCTGCTCTCTGTTG-3′ | qRT-PCR |
| caubi R | 5′-CCCAAGCACAATAAGAC-3′ | qRT-PCR |
| qcoxII F | 5′-ATCCAGCCATTACTATCAAA-3′ | qRT-PCR |
| qcoxII R | 5′-GTACAGCCCAACTATCAGG-3′ | qRT-PCR |
| q507F | 5′-ATGCCCAAAAGTCCCATGT-3′ | qRT-PCR |
| q507R | 5′-AAAAGCGCTAAACAAATTGC-3′ | qRT-PCR |
| r atp6-2F | 5′-GCTTCGCCTTCGTATAGTAGTTC-3′ | qRT-PCR |
| r atp6-2R | 5′-AGTCATAGTGCTCACCCTGTTTG-3′ | qRT-PCR |
The restriction enzyme sites added at 5′ end of primers are underlined.
Plant transformation
The constructed TCON was used to transform the maintainer line of pepper (HW203B), via leaf disk transformation procedure. Sterile explants were infected with the GV3101 strain (OD600 = 0.5~0.8) containing TCON for 10 min. The infected explants were co-cultured in a solid MB medium for 3 d until the single colony had formed, and then transferred into another medium (MB + 50 mg/L Cef). When adventitious buds began to grow, the explants were cultured in a medium (MB + 50 mg/L Kan + 50 mg/L Cef). Induction, differentiation and elongation of the adventitious buds were executed according to procedures outlined by Wang et al. (Wang, 2007). Finally, the elongated adventitious buds were cultured in a solid medium containing 1/2 MS + 50 mg/L Kan + 50 mg/L Cef (pH 5.8) for root production. The plants were acclimatized initially in a tissue culture room and later transferred to an illumination incubator. The transformed control plants were infected with the Agrobacterium strain containing TCN. The T0 male sterile plants obtained were pollinated with the pollen from transformed control pepper plants to obtain BC1T0 generation seeds.
Selection and detection of the transformed pepper
Genomic DNA was isolated from the leaves of transformed and transformed control plants using the CTAB method to check for transgenicity (Ji et al., 2014b). PCR analysis using BamHI coxIVF and SacI orf507R primers was performed to confirm the presence of the gene cassette in the transgenic plants. Plants that were positive for gene-specific PCR were subjected to RT-PCR (reverse transcription PCR) and qRT-PCR (quantitative real time PCR) analysis using cDNA from flower buds at the tetrad stage, leaves as the template, and the ubiquitin-conjugating protein gene (caubi F, caubi R) as the reference gene. The cDNA of transformed control plants was used as the negative control, with the coxII gene as the reference gene. RNA isolation of the flower buds and leaves was conducted according to Ji et al. (2013). The two sets of primers used for RT-PCR and qRT-PCR analysis were Kpn I orf507F, SacI orf507R and q507F, q507R, respectively (sequences listed in Table 1). The TCON vector was used as the positive control.
Pollen analysis
After flowering, transgenic and control plants were scored for pollen abundance, pollen characteristics and germination. Non-dehisced anthers from 10 flowers selected randomly from different plants were collected into 2 mL tubes containing 400 μL 1% sodium metaphosphate and placed in a dryer to release the pollen grains. Natural damage rate of the pollen grains was then observed. Fresh pollen grains from five flowers collected from different plants were mixed randomly to assess the germination rate according to the protocol described by Ji et al. (2014a). Pollen viability was tested with the acetocarmine staining method (Malayeri et al., 2012). The nucleus of the viable pollen grains can be stained deep red, while that of the non-viable pollen grains cannot be stained. The CMS lines and the transformed control plants were used as the controls. All the experiments were replicated three times.
Based on the rate of pollen damage and germination results, plants were classified as sterile, semi-sterile, and fertile. If no pollen grains were released and the anthers were wizened and browning, such plants were classified as completely male-sterile. If the anthers were wizened, more than 30% of pollen grains were damaged and less than 5% of pollen grains germinated, then such plants were classified as semi-sterile, while plants were designated as fertile if greater than 40% pollen grains were able to germinate.
Light microscopy
At the mother microspore cell stage, tetrad stage and MNM stage, anthers were fixed in glutaraldehyde, dehydrated in an ethanol series, and embedded in Epon812 resin (Ji et al., 2014a). Using an automatic microtome (ULTRACUT, Germany) sections of 1 μm were then cut. A bright-field photograph of each anther cross-section was taken using a microscope.
Pollination test
The untransformed pepper plants (female parent) were crossed with transgenic plants (male parent). Fifteen flowers were pollinated in each combination. Then the fruit-set and seeds in each fruit were counted. The transformed control pepper plant was used as male parent for the control.
Progeny analysis
Transgenic plants (female parent) were crossed with untransformed plants (male parent) to obtain BC1T0 plants. The seeds obtained were subjected to a germination test on filter paper containing 150 mg/L Kanamcin. Seeds were scored as positively germinating if the radicles could grow normally and root hairs were present, and they were scored as negatively germinating if the radicles browned and died. The transgenic line with 1:1 ratio of positive:negative seeds was chosen as the material for molecular analysis of the test cross progeny.
Seeds of the selected transgenic line were cultured in plastic trays containing steam-sterilized growing medium at 25°C on a cycle of 16 h light/8 h dark. Gene-specific PCR analysis of the leaves was performed with BamHI coxIVF and SacI orf507R primers. RT-PCR detection of the BC1T0 progeny was similar to that for the T0 generation plants, using the primers Kpn I orf507F and SacI orf507R. The 80 randomly selected progeny were used in the PCR.
Expression pattern of the orf507 and atp6-2 gene in BC1T0 progeny
To assess interactions between the orf507 and atp6-2 genes, expression patterns of the two genes in BC1T0, CMS line and maintainer line were analyzed with qRT-PCR. RNA was isolated from anthers at the microspore mother cell (MMC) stage, tetrad stage and MNM stage, and transcribed to the cDNA template (Ji et al., 2013). Two sets of primers (q507F, q507R and r atp6-2F, r atp6-2R) were used to analyze the expression patterns of the orf507 and atp6-2 genes, respectively. In BC1T0, ubiquitin-conjugating protein gene was used as the reference gene for the analysis of the orf507 gene and coxII gene was used as the reference gene for atp6-2, while for the analysis of the orf507 and atp6-2 genes in the CMS line and maintainer line, mitochondrial coxII was used as the reference gene. All of the sequences of the primers were listed in Table 1. All experiments were replicated three times.
Activity of mitochondrial cytochrome c oxidase and ATP hydrolysis of F1Fo-ATP synthase
At the tetrad stage in the BC1T0 progeny, mitochondria were isolated from anthers to measure the activity of mitochondrial cytochrome c oxidase and the ATP hydrolysis of F1Fo-ATP synthase following established protocols (Ji et al., 2014a), using untransformed plants and the CMS line as controls. All the experiments were replicated three times.
Results
Histochemical localization of cytochrome c oxidase in anthers
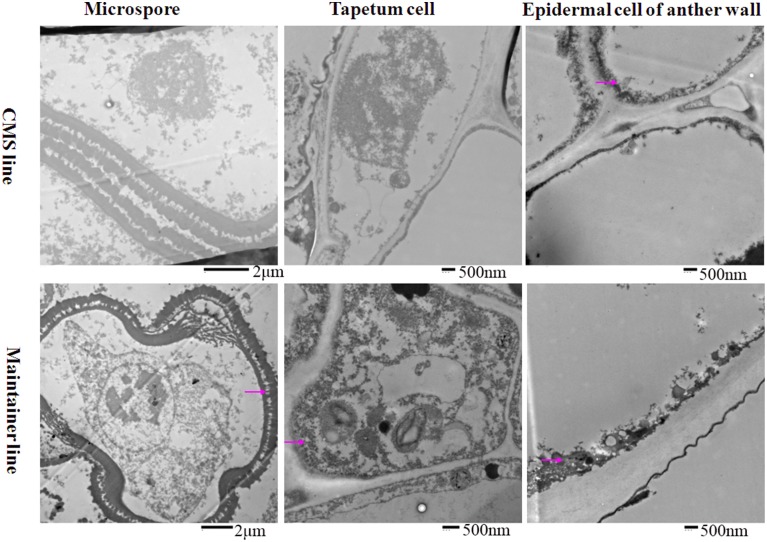
To analyze the relationship between cytochrome c (cyt c) oxidase and CMS, we performed histochemical localization of the enzyme in anthers of the CMS line (HW203A) and the maintainer line (HW203B). Reduction of oxidized cyt c by DAB leads to accumulation of DAB oxide that can react with the OsO4 to form osmium black, which can be viewed as black particles under TEM (Noda et al., 1994). The results showed that osmium black was localized on the membrane of the anther wall at the MMC stage in the CMS line, while hardly any osmium black particles were found in the microspore mother cell and the tapetum cell (Figure 1). All of these findings indicated that in the CMS line, cytochrome c oxidase was less active at the MMC stage than in the maintainer line. However, in the maintainer line osmium black particles were observed in the cytoplasm and nucleus of the microspore mother cell, the tapetum cell, and the membrane of the anther wall (Figure 1).
Figure 1.
Localization of cytochrome c oxidase in anthers at the MMC stage. Purple arrow indicated black particles of cyt c oxidase reaction.
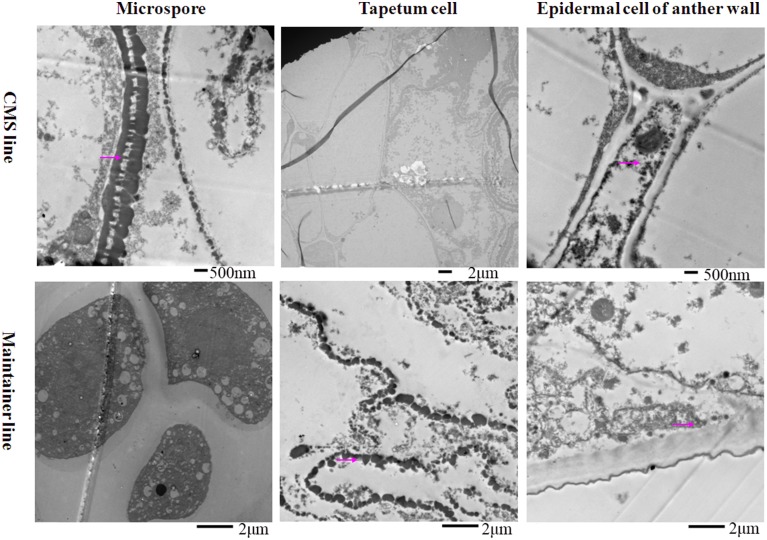
At the tetrad stage, in the CMS line in which the tapetal cells started to degrade, the activity of the enzyme was concentrated mainly in the membrane and intercellular space of the anther wall, the cytoplasm and the space between the exine and intine of the tetrad microspores (Figure 2). In contrast, osmium black particles were uniformly distributed in the cells of the anther wall in the maintainer line, and numerous osmium black particles were concentrated in the tapetal cell and tapetal membrane (Figure 2). Few osmium black particles were observed in the tetrad microspores (Figure 2).
Figure 2.
Localization of cytochrome c oxidase in anthers at the tetrad stage. Purple arrow indicated black particles of cyt c oxidase reaction.
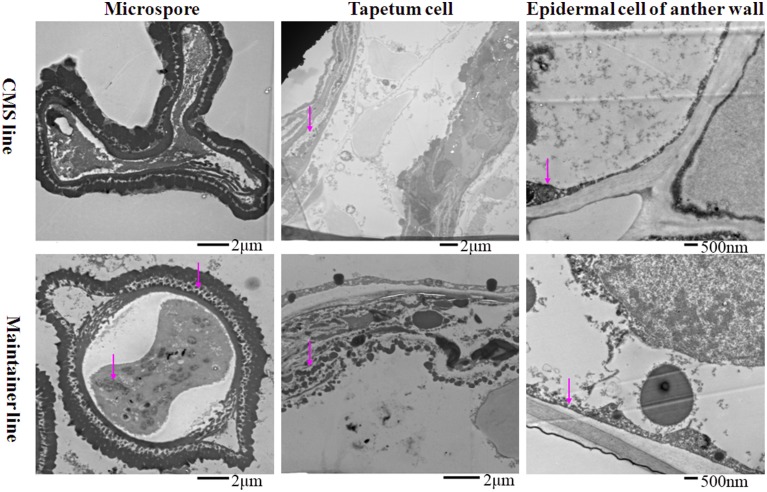
At the MNM stage in the CMS line, concentrated osmium particles were found in the membrane of the anther wall (Figure 3). The tapetal cells of the CMS line were degraded, and few osmium black particles were observed in the residuum (Figure 3). Meanwhile, the microspores were distorted (Figure 3). In the maintainer line, a strong enzymatic reaction occurred in the MNMs, the tapetal membrane and the residuum of the tapetum, especially in the cytoplasm and the space between the extine and intine of the MNMs (Figure 3). In addition, few osmium black particles were observed in the membrane and the nucleus of the anther wall (Figure 3).
Figure 3.
Localization of cytochrome c oxidase in anthers at the MNM stage. Purple arrow indicated black particles of cyt c oxidase reaction.
All of these findings indicated that throughout the development of anthers, the reaction in the anthers of the enzyme cytochrome c oxidase showed significant differences between the CMS line and the maintainer line, especially in the tapetum after the tetrad stage. This indirect evidence might indicate the dysfunction of cytochrome c oxidase in the CMS line, combing with a former study that demonstrated that the changing trend of cytochrome c oxidase at different pollen development stages in the CMS line was contrary to that in the maintainer line (Ji et al., 2013).
Transformation of pepper plants
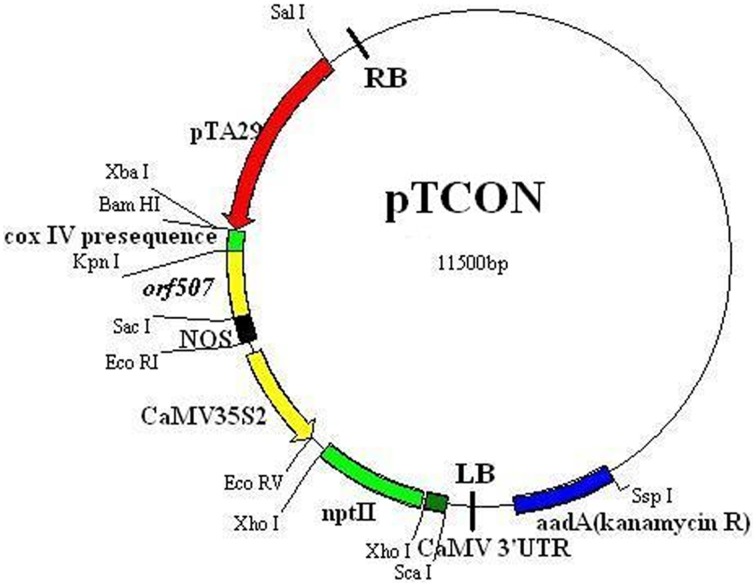
Analysis of the potential role of the orf507 gene in causing the dysfunction of cytochrome c oxidase and C. annuum CMS was performed via the construction of the TCON vector using a pCAMBIA2300 vector. Cytological observation of anther development showed premature PCD in the tapetum of the CMS line (HW203A), and significant differences in histochemical localization of cytochrome c oxidase also occurred in the anther tapetum. In this study, orf507 was expressed under a tapetum-specific TA29 promoter, and then the expression products were transferred into the mitochondria via the coxIV pre-sequence that consisted of the first 25 amino acids of the coxIV gene isolated from yeast. The schematic illustration of the gene construct was shown in Figure 4.
Figure 4.
Schematic illustration of pTCON vector construct.
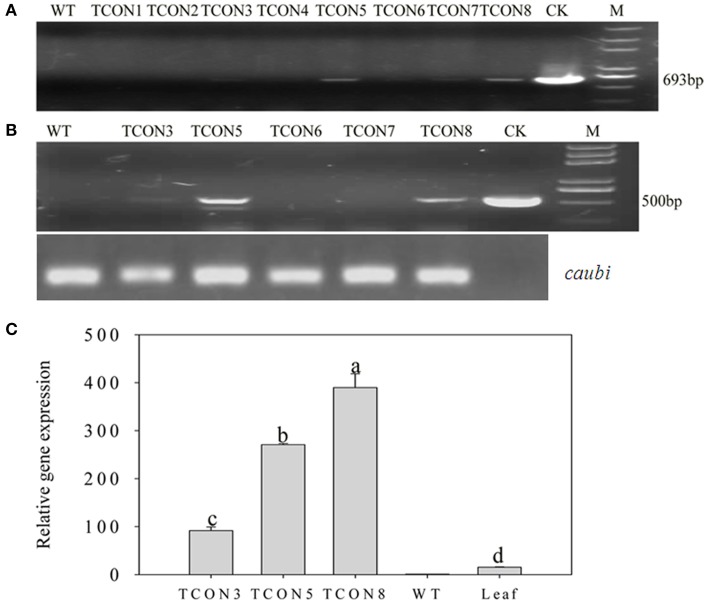
Using an improved genetic transformation system (Wang, 2007), transformed plants were obtained throughout the shoot differentiation, elongation, and rooting stages (Supplementary Figure 1). There were no phenotypic abnormalities between the transformants and the controls that had been infected with the TCN vector without the orf507 gene. The transformants were checked for the transgenicity using gene-specific PCR primers with the pTCON plasmid as the positive control and the non-transgenic plant as the negative control. Among the eight transformants obtained from explantation, five lines (TCON3, TCON5, TCON6, TCON7, and TCON8) showed the presence of the gene casette, while no PCR products were observed in the non-transgenic plants (WT) (Figure 5A).
Figure 5.
Molecular analysis of T0 transgenic plants. (A) PCR confirmation of TCON transgenic pepper plants using BamHI coxIVF and SacI orf507R primers that would amplify 693 bp fragment of the cassette in DNA level.(B) RT-PCR analysis of buds from TCON transgenic plants with Kpn I orf507F and SacI orf507R primers that would amplify 507 bp fragment with caubi gene as reference gene. (C) qRT-PCR analysis of buds from TCON transgenic plants. TCON1~8, transgenic TCON plants; WT, transformed control plant; CK, positive control (amplified from plasmid DNA); Leaf, leaves collected from the three transformants; M, Trans2K plus marker. Statistically significant differences between the means were determined using Fisher's LSD test (p < 0.05).
Inspection for the presence of orf507 transcripts in the anthers of PCR positive progeny plants revealed that only three transformants (TCON3, TCON5, and TCON8) were able to amplify the objective fragment. No fragments were amplified in the WT (Figure 5B). Further, qRT-PCR analysis showed that expression of the orf507 gene was restricted to the buds and was basically not detected in the leaves of transgenic plants (Figure 5C), which indicated the anther-specific expression of orf507. Meanwhile the gene expression in TCON8 was significantly higher than that in TCON3 and TCON5, with no expression detected in the WT (Figure 5C).
Morphological and cytological analysis of pollen grains
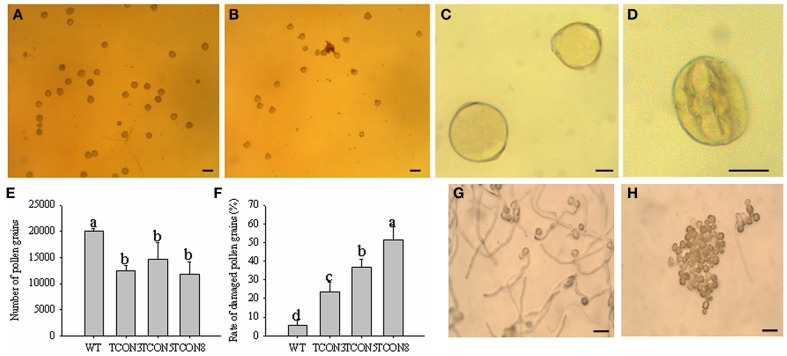
Anthers of transgenic plants had few pollen grains that were not plump and either did not show dehiscence or did so only slightly (Figure 6A) compared to non-transgenic plants (Figure 6B). This indicated that the transgenic plants were semi-sterile, different from the CMS line (HW203A) whose shriveled anthers did not produce any pollen grains (Figure 6C).
Figure 6.
The morphology of the anthers from the T0 transgenic plant (TCON8). (A) WT. (B) T0 transgenic plant (TCON8). (C) CMS lines HW203A. Bars = 1 mm.
To analyze the pollination ability, the transformants were used as male parent to pollinate the untransformed plants. The results showed that most of the pollinated flowers fell and nearly no fruit was set. Even though some small fruits were obtained, no seeds or very few seeds were observed (Table 2). These indicated that the pollen grains of the transformants were unable to pollinate.
Table 2.
Pollination test of T0 plants with non-transformed plants as female parent.
| T0 plant | No. of pollinated flowers (WT♀ × T0♂) | Fruit-setting | No. of seeds (means ± SE) |
|---|---|---|---|
| TCON3 | 15 | 2 | 1.50 ± 0.71 |
| TCON5 | 15 | 0 | 0.00 ± 0.00 |
| TCON8 | 15 | 0 | 0.00 ± 0.00 |
| WT | 15 | 8 | 58.42 ± 8.18 |
The quantity of pollen grains in transgenic plants (T0) was significantly lower than in WT, but higher than in the CMS line (Figures 7A,B,E), which may have resulted from the slight dehiscence of the anthers in transgenic plants. Moreover, the rate of naturally damaged pollen grains in T0 (51.4%) (Figures 7D,F) was significantly higher than in WT (5.6%) (Figures 7C,F). Few germination of pollen grains was observed in T0 (Figure 7H), while nearly all pollen grains of WT produced normal pollen tubes (Figure 7G).
Figure 7.
Cytological analysis of the pollen grains in T0 transgenic plants. (A,C) Pollen grains of WT. (B,D) Pollen grains of T0 transgenic plants. (E) Number of pollen grains in each anther. (F) Rate of damaged pollen grains. (G) Pollen germination of WT. (H) Pollen germination of T0 transgenic plants. (A,B) Bars = 100 μm; (C,D) Bars = 25 μm; (G,H) Bars = 200 μm.
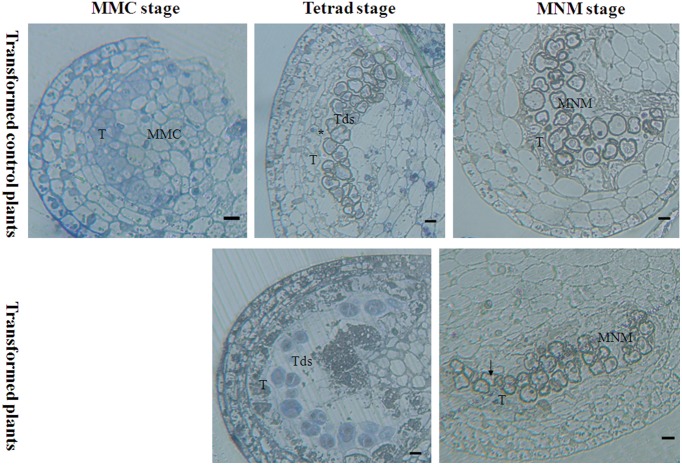
Light microscopy that was used to examine the effect of orf507 gene expression on the anther at different developmental stages revealed that at the mother microspore cell stage, the tapetal cell in the transformed plants showed no abnormalities compared to the transformed control plants, in which the tapetal cells were arranged closely with a dense cytoplasm (Figure 8). At the tetrad stage, tapetal cells of the transformed plants degraded, and no nucleus was observed, which is different from the CMS line in which the tapetum started to degrade but the cellular morph still existed until the mononuclear stage (Ji et al., 2013). In contrast, the tapetal cell of the transformed control plants developed normally and had many nuclei (Figure 8). Notably, at the mononuclear stage, the tapetum of the transformed control plants also degraded, but the anther chamber of the transformed plants became narrow, and the microspores lacked an integrated nucleus and were squeezed and distorted (Figure 8), compared to those of the transformed control plants (Figure 8). The results corroborated the analysis that showed the rate of natural damage to the pollen grains in transformed plants was significantly higher than in transformed control plants. All of these results demonstrated that expression of orf507 driven by TA29 promoter led to premature PCD and then tapetal degeneration in anthers of transgenic plants, even earlier than in the CMS line (HW203A).
Figure 8.
Bright-field photographs of sections of anthers at different stages of development from TCON male sterile transformant and transformed control pepper plants. At the MMC stage, anther development showed no differences in the transformant and control. T, tapetum; MMC, microspore mother cell; Tds, tetrad; MNM, mononuclear microspore; black arrow indicated the degraded mononuclear microspore; * nucleus of the tapetum cell. Bars = 25 μm.
Analysis of the BC1T0 generation of transgenic plants
The three transformants obtained were pollinated with untransformed plants. Germination tests of the BC1T0 seeds on filter paper containing kanamycin showed a 1:1 segregation ratio, which might indicated the presence of a single copy insert of the orf507, while no seeds of the untransformed plants germinated (Table 3).
Table 3.
Germination test of the BC1T0 seeds under selection with Kanamycin.

Left of the slash: No. of seeds germinated under selection.
Right of the slash: No. of seeds used in the test.
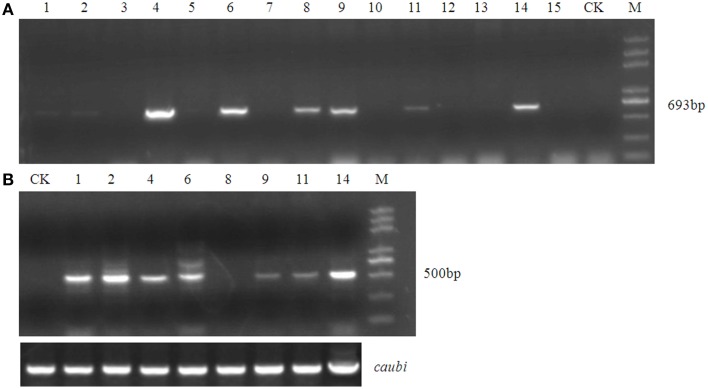
Fifteen plants were randomly selected from among the BC1T0 plants of TCON 8 and were grown without selection for PCR analysis. The results showed a 1:1 segregation for the transgene (Figure 9A), which might demonstrate the single copy insert of the transgene. However, this should be confirmed further by Southern blotting. RT-PCR for the presence of orf507 transcripts in the anthers of PCR positive progeny plants confirmed the results of PCR analysis (Figure 9B). Because the obtained transgenic plants were semi-male sterile, and the degree of sterility differed significantly among individual plants, it was difficult to clearly evaluate co-segregation of the male sterility trait and the transgene, although about 80% of PCR positive plants in the progeny of TCON 8 were partially sterile. The morphology of the BC1T0 flowers with the transgene was similar to T0 plants (Figure 10A), while the anthers of the BC1T0flowers without the transgene were plump and produced plenty of pollen grains, which was similar to the WT plants. Above 95% of pollen grains of all progeny plants that were positive in the PCR analysis could not be stained, while the negative plants stained dark red (Figure 10B). No pollen grains were found in the CMS line. This indicated a certain co-segregation of the sterility trait with the transgene, albeit with incomplete penetrance in the BC1T0 progeny.
Figure 9.
Molecular analysis of test cross progeny of transgenic male sterile TCON 8 plant. (A) PCR analysis using BamHI coxIVF and SacI orf507R primers. (B) RT-PCR analysis with Kpn I orf507F and SacI orf507R primers. CK, transformed control plant; M, Trans2K plus marker; Lane1~15, progeny plant numbers; caubi, reference gene.
Figure 10.
Morphology of BC1T0 anthers (A) and pollen viability of BC1T0 plants (B). (A) Bars = 1 cm (B) Bars = 100 μm.
Gene expression pattern in BC1T0 progeny
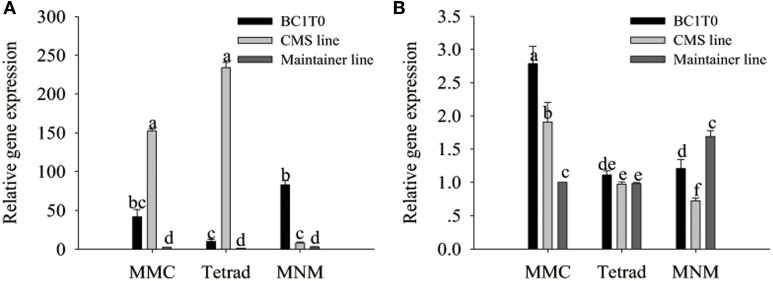
Previously, Li et al. (2013) confirmed the interaction of MtATP6 with ORF507 using the yeast two-hybrid analysis. Our investigation of the reaction of the orf507 gene with the atp6-2 gene using qRT-PCR showed that throughout all stages of development, the expression of the orf507 gene in BC1T0 increased, while in the CMS line its expression gradually decreased, showing the changing trend with BC1T0, with no gene expression observed in the maintainer line. This might be due to the expression driven by different promoters in the CMS line and BC1T0 progeny (Figure 11A).
Figure 11.
qRT-PCR analysis of orf507 (A) in buds of BC1T0, CMS line, Maintainer line, and atp6-2(B) in buds of BC1T0, Maintainer line (WT) and ψatp6-2 (B) in CMS line. Statistically significant differences between the means were determined using Fisher's LSD test (p < 0.05).
At the MMC and the MNM stages, we observed increased orf507 gene expression and decreased atp6-2 gene expression in BC1T0, which might indicate that expression of orf507 inhibited expression of atp6-2 to a certain degree (Figure 11B). Meanwhile, it might be confirmed by the up-regulated expression of the atp6-2 gene in the maintainer line and the down-regulated expression in BC1T0. However, at the MMC stage, expression of atp6-2 in BC1T0 was significantly higher than it was in the maintainer line, which might result from the regulation of other factors driven by the expression of orf507 in BC1T0. The contradiction might be due to the regulation of other CMS-related genes on the ψatp6-2 gene in the CMS line, or the mutation of the ψatp6-2 gene affected the interaction of ORF507 with ATP6-2.
Activity of mitochondrial enzymes in BC1T0 progeny
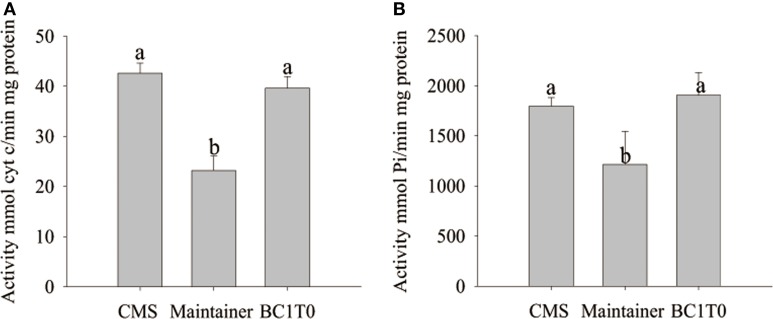
In the previous study (Ji et al., 2013), we have observed the dysfunction of the mitochondrial enzymes cytochrome c oxidase and the F1Fo-ATP synthase in the CMS line of pepper plants. Meanwhile, histochemical localization of cytochrome c oxidase in the anther tapetum after the tetrad stage showed the opposite trend between the CMS line and the maintainer line. In addition, premature PCD of the tapetum in the transformants of TCON occurred at the tetrad stage. However, to date, we still do not understand whether the dysfunction of cytochrome c oxidase and the F1Fo-ATP synthase in the CMS line was induced by expression of the orf507 gene. To investigate whether the dysfunction was induced by expression of orf507, we analyzed the activity of the two enzymes in the anthers of BC1T0 progeny at the tetrad stage.
At the tetrad stage of development, activity of cytochrome c oxidase and the ATP hydrolysis of the F1Fo-ATP synthase in BC1T0 progeny and the CMS line were not significantly different, and the levels in both types of plants were significantly higher than in the untransformants (maintainer line) (Figure 12). This indicated that expression of orf507 in BC1T0 might lead to the increased activity of cytochrome c oxidase and ATP hydrolysis of the F1Fo-ATP synthase at the tetrad stage, which was consistent with previous findings that suggested the enzyme activity in the CMS line was significantly higher than in the maintainer line at the tetrad stage (Ji et al., 2013). However, whether changes in ATP hydrolysis of the F1Fo-ATP synthase were induced by the direct interaction of ORF507 on F1Fo-ATP synthase or by a change in the activity of cytochrome c oxidase still needs to be confirmed by further studies.
Figure 12.
Analysis of cytochrome c oxidase activity (A) and ATP hydrolysis activity of mitochondrial F1Fo-ATP synthase (B) in buds of BC1T0, CMS line and Maintainer line (WT). Statistically significant differences between the means were determined using Fisher's LSD test (p < 0.05).
Discussion
CMS is known to be associated with novel orfs resulting from rearrangement of the mitochondrial genome (Schnable and Wise, 1998). In pepper, CMS is associated with the expression of a novel mitochondrial gene, orf507, which is expressed as a mitochondrial polypeptide in all the tissues of male-sterile plants (Kim et al., 2007; Gulyas et al., 2010; Ji et al., 2013). The CMS related orfs always regulate sterility by affecting the expression of the co-transcription gene, or by interacting with the protein and changing its biological function, such as the interaction of orfH79 with the atp6 gene in rice (Zhang et al., 2007). In pepper, orf507 is co-transcribed with the coxII gene, and we also observed the dysfunction of mitochondrial cytochrome c oxidase in the CMS line. However, it remains unclear which tissue of the anthers undergoes the mutation that leads to the dysfunction of cytochrome c oxidase. In this study, by histochemial localization of cytochrome c oxidase, we found that differences between the CMS line and the maintainer line occurred mainly in the tapetum, through all the developmental stages of anthers, especially at the tetrad stage, which was similar to our previous study (Ji et al., 2013). Dysfunction of the mitochondrial enzyme is known to be associated with CMS (Zhang et al., 2007). As the marker enzyme, the aberrant cytochrome c oxidase will directly cause a change in the function of the mitochondria (Rich and Bonner, 1978). The tapetum provides nutrition and energy for the development of pollen grains (Pacini, 1997), so enzyme dysfunction in the tapetal cell might lead to a reduction of energy, which might in turn induce degradation of the tapetal cells. Then, premature PCD of the tapetum causes nutrient deficiency of the microspores, leading ultimately to male sterility.
Although the dysfunction of cytochrome c oxidase in the CMS line has been confirmed, we still do not understand the relationship of the CMS-specific orf507 gene with cytochrome c oxidase and male sterility.
Cytochrome c oxidase is an enzyme complex that is being composed of 13 subunits and several metal prosthetic sites in mammals (Caar and Winge, 2003). Some CMS-related genes encode important subunits for or interact with the subunits of cytochrome c oxidase to affect its function. For example, in CMS-WA rice, the protein encoded by the CMS-specific WA352 gene interacts with COX11, which is encoded by a nuclear gene. In CMS-WA lines, WA352 accumulates in the anther tapetum, thereby inhibiting COX11 function in peroxide metabolism, triggering premature tapetum PCD, and consequent pollen abortion (Luo et al., 2013). For the co-transcription of the orf507 gene with the coxII gene, we postulated that the ORF507 interacts with COXII which is firmly bound to subunit I and is the only subunit to possess a polar domain containing the binuclear CuA center. The CuA binuclear center is reduced by cytochrome c and then passed by an electron to cytochrome a, which in turn passes an electron to the cytochrome a3-CuB binuclear center and, finally, to the dioxygen molecule (Ostermeier and Lwata, 1996). The interference of COXII might cause the dysfunction of cytochrome c oxidase. The transgenic plants were generated using cotyledon of the maintainer line HW203B as explants. In addition, orf507 was expressed under a constitutive expression promoter, CaMV35S, in the SCON vector. However, the elongated shoots infected with the SCON vector became etiolated and died gradually. We speculated that the toxic effect of ORF507 protein gradually accumulated on the shoot (Li et al., 2013), which inhibited development of the elongated shoots. Similarly, in CMS-HL rice, expression of orfH79 leads to the excessive accumulation of ROS, a decrease in ATP synthesis, damage of the mitochondrial membrane potential and a decline in the rate of respiration, which finally inhibits root growth (Peng et al., 2010). The ORF507 protein might play a similar role, but empirical proof, such as the content of ROS, would be needed to provide evidence to support this hypothesis.
In the detection of the transformants using PCR and RT-PCR analysis, three transformants were confirmed to be positive for the presence of the orf507 gene. Compared with the plump anthers of the maintainer line, the anthers of the transgenic plants were shriveled, barely dehisced, and could produce only few pollen grains; The quantity and germination rate of pollen was significantly lower in transformed plants than untransformed plants, while the natural damaged rate of pollen grains was higher in transgenic plants. Overall, this suggested that the transgenic plants were partially male-sterile. However, expression of the orf456 gene in Arabidopsis induces complete male sterility (Kim et al., 2007). Male sterility in Arabidopsis might result from sequence alteration of orf456, or regulation of other CMS-related genes. In addition, expression of orf507 was negatively correlated with the germination rate of pollen grains. Similar observations have been made in tobacco plants that were positive in the PCR analysis that were fertile and showed no expression or lower levels of expression of orfH522 transcripts (Nizampatnam et al., 2009). It appears that expression of the CMS-related genes needed to exceed a threshold level to impair mitochondrial activity, and thereby induce male sterility. However, the toxic effect of ORF507 driven by the TA29 promoter was restricted to the tapetal cells, although the number of transgenic plants is insufficient to make the conclusion, which might prevent the accumulation of ORF507 from reaching the threshold level needed to trigger ablation of the tapetal cells (Smart et al., 1994). In the CMS line, the orf507 gene is expressed in all tissues under the coxII promoter (Gulyas et al., 2010). The reason for the partial sterility of TCON transformants might include: (1) the CMS-related protein may not be localized in the appropriate compartments of mitochondria; (2) co-factors essential for the action of the CMS-related protein might not be found in heterologous systems; and (3) the protein might not be able to fold properly (Flavell, 1974; Duroc et al., 2006; Stockmeyer et al., 2007).
Expression of the orf507 gene in HW203B affected only the development of the tapetum, and not other tissues of the anthers or their maturation. Ablation of the tapetum occurred in a pattern that is typical of the reported TA29 expression profile (Koltunow et al., 1990). At the tetrad stage the tapetal cells in the transgenic plants almost completely degraded. At the MNM stage, though transgenic plants could produce microspores, the microspores were shrunken, distorted and damaged. After flowering, pollen grains collected from the anthers were also damaged.
As the anthers developed, the trend for the expression of the orf507 gene in BC1T0 was contrary to what was found in the CMS line, which might be due to the expression driven by different promoters in the CMS line and BC1T0 progeny. The interaction of ORF507 with MtATP6 reduced the activity of the mitochondrial F1Fo-ATP synthase and the ATP content (Li et al., 2013). In this study, expression patterns of orf507 and atp6-2 in BC1T0 progeny were opposite. Moreover, the opposing trends for the two genes in BC1T0 were different from what was seen in the CMS line. These findings might suggest that there were other CMS-related genes in the CMS line that interacted with the ATP6-2 subunit, or that the mutation of ψatp6-2 altered the interaction with ORF507.
We still do not understand whether dysfunction of cytochrome c oxidase and the F1Fo-ATP synthase in the CMS line was induced by the expression of orf507. Cytochrome c oxidase (EC 1.9.3.1) plays a crucial role in aerobic life due to its specific capability to accept an electron from cytochrome c and then transfer it to a dioxygen molecule to form water, while pumping protons across the mitochondrial membrane to produce 2.5 ATP (Wikström, 2000). The abnormal increase in the activity of cytochrome c oxidase induced by orf507 expression, causing an increased ability to transfer electrons, would disorder the transfer of electrons, and thus destroy the membrane potential (Pecina et al., 2004). Altering the pH of the cell and disrupting the electrochemical gradient may cause ATP synthase to hydrolyze ATP rather than synthesis ATP (Cabezón et al., 2000; García-Trejo and Morales-Ríos, 2008). With ATP hydrolysis, protons are transported from the matrix to the inter-membrane space to rectify the membrane potential (Crompton, 1999). Expression of orf507 increased the hydrolysis of ATP of F1Fo-ATP synthase, which resulted in premature ATP hydrolysis. Concurrently, the imbalance between ATP synthesis and consumption would aggravate the sustainable accumulation of ROS and lead to oxidative stress beyond the regulation of the antioxygen system in mitochondria. This would subsequently destroy the structure and the active site of the mitochondrial enzyme, thus disrupting the enzyme activity. ATP is an important determinant of the integrity of mitochondria. Lack of ATP will limit vegetative growth, therefore the CMS plants were ATP deficient (Teixeira et al., 2005). The imbalance between energy expenditure and production exacerbates the defects of the proteins in the respiratory chain complexes, and further deteriorates the state of mitochondrial disorder (Wan et al., 2007). These changes could lead to the premature PCD and ablation of plant cells that we observed.
Mitochondrion is a semi-self-organelle in plant cells. Unlike nuclear DNA, which has no protective histones combined with mtDNA, thus it is easily impaired by excessive ROS (Singh, 2004). Therefore, a test of mtDNA fragmentation would further ascertain the relationship between the orf507 gene and excessive ROS accumulation.
Conclusions
This report provides direct evidence that dysfunction of cytochrome c oxidase in the tapetum might be responsible for male sterility in pepper and the orf507 gene might be one of the causative factors of male sterility in pepper CMS line, building on the indirect evidences accumulated by several earlier reports. We have also established that orf507 alone is insufficient to induce complete male sterility in a heterologous system of pepper, which was different from orf456 inducing complete male sterility when transformed into A. thaliana. In future, discovery of more CMS-related genes and elucidation of their functions will help to unravel the mechanism of complete male sterility in pepper.
Author contributions
JJ, ZL, and ZG conceived the research. JJ, WH, YY, and ZL performed the research. JJ, WH, YL, DL performed interpretation of data. JJ, ZL, and DL took the photos. JJ, YY, and WC performed statistical analyses. JJ and ZG wrote the paper. JJ, DL, and YY revised the paper. ZG and WC provided the materials and resources for the research. JJ, WH, WC, and ZG performed the integrity of the work. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No. 31272163, No. 30771467), the Shaanxi Provincial Science and Technology Coordinating Innovative Engineering Project (2012KTCL02-09), Scientific Research Fund for the Doctoral Young Scholars, Northwest A&F University (2013BSJJ110), the Northwest A&F University Cyrus Tang Seed Development Fund and the Scientific Research Fund for the Doctoral Young Scholars, Northwest A&F University (2013BSJJ110). We gratefully acknowledge Dr. Rui Cai (Shanghai Jiaotong University, China) for providing pCAMBIA2300 vector. We thank Mogoedit Company (Xi'an, China) for providing language help. Seeds of pepper HW203A and HW203B were obtained from the Asian Vegetable Research and Development Center (AVRDC, Taiwan, China).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/journal/10.3389/fpls.2015.00272/abstract
Generation of the transformed plants by means of leaf disc method. (A) Bud differentiation. (B) Shoot elongation. (C,D) Rooting stage, Rooting plant. (E) Transplanted plant.
References
- Balk J., Leaver C. J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. 10.1105/tpc.13.8.1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budar F., Pelletier G. (2001). Male sterility in plant: occurrence, determinism, significance and use. Life Sci. 324, 543–550. 10.1016/S0764-4469(01)01324-5 [DOI] [PubMed] [Google Scholar]
- Caar H. S., Winge D. R. (2003). Assembly of cytochrome c oxidase within the mitochondria. Acc. Chem. Res. 36, 309–316 10.1021/ar0200807 [DOI] [PubMed] [Google Scholar]
- Cabezón E., Butler P. J., Runswick M. J., Walker J. E. (2000). Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J. Biol. Chem. 275, 25460–25464. 10.1074/jbc.M003859200 [DOI] [PubMed] [Google Scholar]
- Chaumont F., Bernier B., Buxant R., Williams M., Levings C. S., Boutry M. (1995). Targeting the maize T-urf13 product into tobacco mitochondria confers methomyl sensitivity to mitochondrial respiration. Proc. Natl. Acad. Sci. U.S.A. 92, 1167–1171. 10.1073/pnas.92.4.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton M. (1999). The mitochondrial permeability transition pore and its role in cell death. Biochem. J. 341, 233–249 10.1042/0264-6021:3410233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y., Gaillard C., Hiard S., Tinchant C., Berthomé R., Pelletier G., et al. (2006). Nuclear expression of a cytoplasmic male sterility gene modifies mitochondrial morphology in yeast and plant cell. Plant Sci. 170, 755–767 10.1016/j.plantsci.2005.11.008 [DOI] [Google Scholar]
- Flavell R. (1974). A model for the mechanism of cytoplasmic male sterility in plants, with special reference to maize. Plant Sci. Lett. 3, 259–263 10.1016/0304-4211(74)90096-0 [DOI] [Google Scholar]
- García-Trejo J. J., Morales-Ríos E. (2008). Regulation of the F1Fo-ATP synthase rotary nanomotor in its monomeric-bacterial and dimeric-mitochondrial forms. J. Biol. Physics 34, 197–212. 10.1007/s10867-008-9114-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyas G., Shin Y., Kim H., Lee J. S., Hirata Y. (2010). Altered transcript reveals an orf507 sterility-related gene in chili pepper (Capsicum annuum L.). Plant Mol. Biol. Rep. 28, 605–612 10.1007/s11105-010-0182-4 [DOI] [Google Scholar]
- Hanson M. R., Bentolila S. (2004). Interaction of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16, 154–169. 10.1105/tpc.015966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Abad A. R., Gelvin S. B., Mackenzie S. A. (1996). A cytoplasmic male sterility associated mitochondrial protein causes pollen disruption in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 93, 11763–11768. 10.1073/pnas.93.21.11763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernould M., Suharsono S. (1993). Male-sterility induction in transgenic tobacco plants with an unedited atp9 mitochondrial gene from wheat. Proc. Natl. Acad. Sci. U.S.A. 90, 2370–2374. 10.1073/pnas.90.6.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernould M., Suharsono S., Zabaleta E., Carde J. P., Litvak S., Araya A., et al. (1998). Impairment of tapetum and mitochondria in engineered male-sterile tobacco plants. Plant Mol. Biol. 36, 499–508. 10.1023/A:1005946104983 [DOI] [PubMed] [Google Scholar]
- Ji J. J., Huang W., Li D. W., Yin Y. X., Gong Z. H. (2014a). A CMS-related gene, ψatp6-2, causes increased ATP hydrolysis activity of the mitochondrial F1Fo-ATP synthase and induces male sterility in pepper (Capsicum annuum L.). Plant Mol. Biol. Rep. 32, 888–899 10.1007/s11105-014-0702-8 [DOI] [Google Scholar]
- Ji J. J., Huang W., Yin C. C., Gong Z. H. (2013). Mitochondrial cytochrome c oxidase and F1Fo-ATPase dysfunction in peppers (Capsicum annuum L.) with cytoplasmic male sterility and its association with orf507 and ψatp6-2 genes. Int. J. Mol. Sci. 14, 1050–1068. 10.3390/ijms14011050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J. J., Huang W., Yin Y. X., Li Z., Gong Z. H. (2014b). Development of a SCAR marker for early identification of S- cytoplasm based on the mitochondrial SRAP analysis in pepper (Capsicum annuum L.). Mol. Breed. 33, 679–690 10.1007/s11032-013-9984-z [DOI] [Google Scholar]
- Kim D. H., Kang J. G., Kim B. D. (2007). Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.). Plant Mol. Biol. 63, 519–532. 10.1007/s11103-006-9106-y [DOI] [PubMed] [Google Scholar]
- Kim D. H., Kim B. D. (2006). The organization of mitochondrial atp6 gene region in male fertile and CMS lines of pepper (Capsicum annuum L.). Curr. Genet. 49, 59–67. 10.1007/s00294-005-0032-3 [DOI] [PubMed] [Google Scholar]
- Koltunow M., Jessie T., Cox K. H., Wallroth M., Goldberg R. B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2, 1201–1224. 10.1105/tpc.2.12.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J., Pandeya D., Jo Y. D., Liu W. Y., Kang B. C. (2013). Reduced activity of ATP synthase in mitochondria causes cytoplasmic male sterility in chili pepper. Planta 237, 1097–1109. 10.1007/s00425-012-1824-6 [DOI] [PubMed] [Google Scholar]
- Li W. Q., Zhang X. Q., Xia C. (2010). Male gametophyte defective 1, encoding the FAd subunit of mitochondrial F1Fo-ATP synthase, is essential for pollen formation in Arabidopsis thaliana. Plant Cell Physiol. 51, 923–935. 10.1093/pcp/pcq066 [DOI] [PubMed] [Google Scholar]
- Linke B., Börner T. (2005). Mitochondrial effects on flower and pollen development. Mitochondrion 5, 389–402. 10.1016/j.mito.2005.10.001 [DOI] [PubMed] [Google Scholar]
- Luo D. P., Xu H., Liu Z. N., Guo J. X., Li H. Y., Chen L. T., et al. (2013). A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nature Genet. 45, 573–577. 10.1038/ng.2570 [DOI] [PubMed] [Google Scholar]
- Malayeri B. E., Noori M., Jafari M. (2012). Using the pollen viability and morphology for fluoride pollution biomonitoring. Biol. Trace Elem. Res. 147, 315–319. 10.1007/s12011-011-9290-8 [DOI] [PubMed] [Google Scholar]
- Nizampatnam N. R., Doodhi H., Narasimhan Y. K. (2009). Expression of sunflower cytoplasmic male sterility-associated open reading frame, orfH522 induces male sterility in transgenic tobacco plants. Planta 229, 987–1001. 10.1007/s00425-009-0888-4 [DOI] [PubMed] [Google Scholar]
- Noda K., Tani N., Nakamura Y., Kuwahara Y. (1994). Cytochrome c oxidase activity in osteoclasts appeared in an early stage of parathyroid hormone treatment. J. Electron. Microsc. 43, 168–172. [PubMed] [Google Scholar]
- Ostermeier C., Lwata S. (1996). Cytochrome c oxidase. Curr. Opin. Struc. Biol. 6, 460–466 10.1016/S0959-440X(96)80110-2 [DOI] [PubMed] [Google Scholar]
- Pacini E. (1997). Tapetum character states: analytical keys for tapetum types and activities. Can. J. Bot. 75, 1448–1459 10.1139/b97-859 [DOI] [Google Scholar]
- Pecina P., Houstkova H., Hansikova H., Zeman J., Houstek J. (2004). Genetic defects of cytocrhome c oxidase assembly. Physiol. Res. 53, S213–S223. Available online at: http://www.biomed.cas.cz/physiolres/pdf/53%20Suppl%201/53_S213.pdf [PubMed] [Google Scholar]
- Peng X. J., Wang K., Hu C. F., Zhu Y. L., Wang T., Yang J., et al. (2010). The mitochondrial gene orfH79 plays a critical role in impairing both male gametophyte development and root growth in CMS-Honglian rice. BMC Plant Biol. 10:125. 10.1186/1471-2229-10-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson P. A. (1958). Cytoplasmically inherited male sterility in Capsicum. Am. Nat. 92, 111–119 10.1086/282017 [DOI] [Google Scholar]
- Rich P. R., Bonner W. D. (1978). The sites of superoxide anion generation in higher plant mitochondria. Arch. Biochem. Biophys. 188, 206–213. 10.1016/0003-9861(78)90373-9 [DOI] [PubMed] [Google Scholar]
- Schmitz U. K., Lonsdale D. M. (1989). A yeast mitochondrial presequence functions as a signal for targeting to plant mitochondria in vivo. Plant Cell 8, 783–791. 10.1105/tpc.1.8.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Wise R. P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–180 10.1016/S1360-1385(98)01235-7 [DOI] [Google Scholar]
- Singh K. K. (2004). Mitochondria damage checkpoint in apoptosis and genome stability. FEMS Yeast Res. 5, 127–132. 10.1016/j.femsyr.2004.04.008 [DOI] [PubMed] [Google Scholar]
- Smart C. J., Moneger F., Leaver C. J. (1994). Cell-specific regulation of gene expression in mitochondria during anther development in sunflower. Plant Cell 6, 811–825. 10.1105/tpc.6.6.811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockmeyer K., Pring R. D., Kempken F. (2007). Heterologous expression of the cytoplasmic male sterility associated orf107 from Sorghum bicolor in Arabidopsis thaliana. Endocytobiosis Cell Res. 18, 11–23 Available online at: http://zs.thulb.uni-jena.de/servlets/MCRFileNodeServlet/jportal_derivate_00103271/ECR_18_2007_11-23_Stockmeyer.pdf [Google Scholar]
- Teixeira R. T., Knorpp C., Glimelius K. (2005). Modified sucrose, starch, and ATP levels in two alloplasmic male-sterile lines of B. napus. J. Exp. Bot. 56, 1245–1253. 10.1093/jxb/eri120 [DOI] [PubMed] [Google Scholar]
- Wan C. X., Li S. Q., Wen L., Kong J., Wang K., Zhu Y. G. (2007). Damage of oxidative stress on mitochondria during microspores development in Honglian CMS line of rice. Plant Cell Rep. 26, 373–382. 10.1007/s00299-006-0234-2 [DOI] [PubMed] [Google Scholar]
- Wang X. E. (2007). Transformation of Pepper with Cold-Induced CBF4 Gene and Analysis of Cold Hardness-Related Indexes. Dissertation, Northwest A&F University. [Google Scholar]
- Wang Z. H., Zou Y. J., Li X. Y., Zhang Q. Y., Chen L., Wu H., et al. (2006). Cytoplasmic male sterility of rice with Boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18, 676–687. 10.1105/tpc.105.038240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström M. (2000). Proton Translocation by cytochrome c oxidase: a rejoinder to recent criticism. Biochemistry 39, 3515–3519. 10.1021/bi9925322 [DOI] [PubMed] [Google Scholar]
- Wintz H., Chen H. C., Sutton C. A., Conley C. A., Cobb A., Ruth D., et al. (1995). Expression of the CMS-associated urfs sequence in transgenic petunia and tobacco. Plant Mol. Biol. 28, 83–92. 10.1007/BF00042040 [DOI] [PubMed] [Google Scholar]
- Yamamoto M. P., Shinada H., Onodera Y., Komaki C., Mikami T., Kubo T. (2008). A male sterility-associated mitochondrial protein in wild beets causes pollen disruption in transgenic plants. Plant J. 54, 1027–1036. 10.1111/j.1365-313X.2008.03473.x [DOI] [PubMed] [Google Scholar]
- Yang J. H., Liu X. Y., Yang X. D., Zhang M. F. (2010). Mitochondrially-targeted expression of a cytoplasmic male sterility-associated orf220 gene causes male sterility in Brassica juncea. BMC Plant Biol. 10:231. 10.1186/1471-2229-10-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. H., Zhang M. F. (2007). Molecular genetics of mitochondrial respiratory/ATP related genes in relation to cytoplasmic male-sterility of higher plants. Gene Genomes Genomics 1, 21–26. [Google Scholar]
- Zhang H., Li S. Q., Yi P., Wan C. X. (2007). A honglian CMS line of rice displays aberrant Fo of F1Fo-ATPase. Plant Cell Rep. 26, 1065–1071. 10.1007/s00299-006-0293-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of the transformed plants by means of leaf disc method. (A) Bud differentiation. (B) Shoot elongation. (C,D) Rooting stage, Rooting plant. (E) Transplanted plant.