Abstract
Aims
Listeriosis is a frequent silage-associated disease in ruminants. The slugs Arion vulgaris are invaders in gardens, vegetable crops and meadows for silage production. Field and laboratory studies were conducted to clarify whether slugs could host Listeria monocytogenes and thereby constitute a threat to animal feed safety.
Methods and Results
Selective culture of L. monocytogenes from 79 pooled slug samples (710 slugs) resulted in 43% positive, 16% with mean L. monocytogenes values of 405 CFU g−1 slug tissues. Of 62 individual slugs cultured, 11% also tested positive from surface/mucus. Multilocus sequence typing analysis of 36 isolates from different slug pools identified 20 sequence types belonging to L. monocytogenes lineages I and II. Slugs fed ≅4·0 × 105 CFUL. monocytogenes, excreted viable L. monocytogenes in faeces for up to 22 days. Excretion of L. monocytogenes decreased with time, although there were indications of a short enrichment period during the first 24 h.
Conclusions
Arion vulgaris may act as a vector for L. monocytogenes.
Significance and Impact of the Study
Highly slug-contaminated grass silage may pose a potential threat to animal feed safety.
Keywords: Arion vulgaris, bacterial vectors, feed safety, Listeria monocytogenes, listeriosis, multilocus sequence typing, silage quality, slug invasion
Introduction
The invasive slug Arion vulgaris, in name confusions also referred to as Arion lusitanicus, is considered a major pest by garden owners and recently also by farmers in some European countries (Kozlowski 2007; Spörndly and Haaga 2010; Hatteland et al. 2013). Massive slug invasions have been reported involving cultured vegetables, berries and meadows for silage production. Slug contaminated silage has been suspected as the cause of animal health problems in Sweden during wet summers (Spörndly and Haaga 2010). Densities of more than 50 slugs per square meter have been reported from wildflower strips and meadows (Briner and Frank 1998b). As a consequence, high numbers of slugs might contaminate grass silage and cause a potential threat to the safety and quality of animal feed.
Listeriosis is one of the most frequent silage associated diseases in ruminants. Infection by Listeria monocytogenes can affect the central nervous system resulting in encephalitis, but may also cause septicemia, abortion, enteritis and mastitis (Blood 2000; Quinn et al. 2011). In Norway, 72–235 cases or outbreaks of listeriosis, mostly in sheep, have been reported annually (Anon. 2001; Hofshagen et al. 2002, 2003). However, as in Switzerland, where listeriosis was found to be surprisingly frequent in small ruminant fallen stock during a neuropathological survey (Oevermann et al. 2008) the incidence is probably higher, due to non-reported and undiagnosed cases. Listeria spp. are widely distributed in the environment although usually in low levels, and can be isolated from a variety of sources such as soil, plants, decaying vegetation and animal faeces (Fenlon 1988; Quinn et al. 1994). Feeding of contaminated and poorly preserved silage is a common cause of listeriosis in ruminants (Quinn et al. 2011). Such silage is also considered an important source of raw milk contamination which can lead to human listeriosis when unpasteurized milk is used for cheese production (Driehuis 2013).
Listeria spp. were detected in live land snails Helix pomatia, during a study of microbiological quality of snail meat (Temelli et al. 2006). However, whether A. vulgaris may carry Listeria monocytogenes is to, the best of our knowledge, not yet confirmed. The aim was to investigate whether naturally infected slugs could host L. monocytogenes, to quantify the bacterial load and assess the importance of slugs as vectors for transmission of the bacterium to silage. Additionally, a laboratory feeding experiment was conducted to determine whether L. monocytogenes could proliferate in living slugs.
Materials and methods
Field survey
Hundred sampling kits were distributed nationally to gardeners and farmers in September 2012. In addition, one kit was send to Denmark and one to Sweden. The kits included sampling procedures, two 0·5 l ventilated sterile plastic boxes, gloves for sampling and a short questionnaire relating to slug collection. The volunteers were instructed to pick 10 living adult slugs, five slugs in each box, on the same day or late in the evening prior to overnight transport to the laboratory by mail.
Slugs for the feeding experiment
Laboratory hatched slugs were used in the feeding experiment, to avoid natural contamination. Eggs from A. vulgaris were collected in September 2010 from South-East Norway (Rørestrand in Horten) from a location known to be pesticide free. The eggs hatched in October following incubation at 16 ± 2°C, and the slugs were kept on moistened paper at 2–3°C in ventilated plastic boxes and fed carrot and Chinese cabbage during the winter. The temperature was raised to 16 ± 2°C in May 2011, and for 6 weeks the slugs were fed on an intensive diet of carrot, white cabbage, cucumber, apple and commercial piglet feed for protein enrichment (Format Kvikk 160; Felleskjøpet Agri, Oslo, Norway). The slugs were not fed for 65 h prior to the start of the feeding experiment, to ensure rapid ingestion of the inoculated feed. Slugs used in the feeding experiment weighed on average 2·5 g (range 1·7–3·7 g) at the start of the experiment.
Preparation of inoculated feed
A suspension of L. monocytogenes (serotype1/2a lineage II, CCUG 15527, Culture Collection University of Göteborg, Sweden) was prepared by swabbing fresh colonies, grown on blood agar at 37 ± 1°C for 24 h, into sterile saline water (0·9% NaCl). A theoretical bacterial concentration of 1·5 × 108 cells ml−1 (0·5 McFarland) was obtained using VITEK DENSICHEK (bioMerieux, Marcy l'Etoile, France). To ensure a more accurate quantification of the suspension, 0·1 ml of ten-fold dilutions were plated onto blood agar (CM 0271; Oxoid, Basingstoke, UK) and incubated at 37 ± 1°C for 24 h prior to enumeration. Bacterial suspensions of L. monocytogenes were used in the experiment the same day they were made.
Pieces of cucumber (Cucumis sativus) were sliced and kept at room temperature overnight resulting in a drier texture capable of some absorption. Small squares were then cut from the outer cucumber layer, with the bottom of each piece consisting of peel to prevent penetration of fluid. A small well was made with a scalpel in each cucumber segment prior to application of the L. monocytogenes suspension. Each cucumber segment weighed 0·2–0·3 g and was inoculated with 0·02 ml of suspension corresponding to ≅4·0 × 105 CFU L. monocytogenes (blood agar enumeration).
The feeding experiment
Figure 1 summarizes the slug feeding experiment. Slugs were kept individually in ventilated boxes in order to monitor their appetite and to collect individual samples. The boxes were changed daily (except at weekends) to reduce the risk of recontamination from the slugs own faeces and mucus. Tissue paper (2 × 2 × 0·2 cm) moistened in distilled water was added to each box to prevent dehydration of the slugs. Feed intake was monitored after two, four and 24 h. From day two of the experiment, slugs were fed daily with white cabbage (Brassica oleracea convar. capitata). One of the slugs did not produce faeces on the last day of the experiment (day 17). Considering that this slug had been positive for L. monocytogenes during the entire experimental period, additional faecal samples were taken on day 19 and 22 from this slug only. The slugs were weighed individually both at the start of the experiment and on euthanization.
Figure 1.
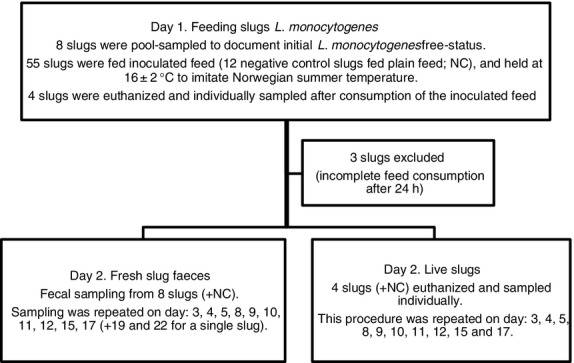
Overview of the 17 day feeding experiment. Slugs were fed inoculated feed containing Listeria monocytogenes on day one (or plain feed for negative controls), followed by sampling of fresh slug faeces and live slugs.
Detection and enumeration of Listeria monocytogenes
NMKL culture method No. 136 was followed with one modification (NMKL 2010). Slugs were finely chopped using a sterile scalpel prior to homogenization by hand for 2–4 min, instead of using a stomacher.
From each submitted field survey sample, 25 g of homogenized slugs and faeces were analysed, starting on the day of arrival at the laboratory. Since detection and enumeration were carried out simultaneously, the test portion was homogenized and diluted 1 : 10 in Half-Fraser broth (Oxoid) without selective agents. In addition to plating 0·1 ml of ten-fold dilutions onto the growth media of ALOA (Agar Listeria according to Ottaviani and Agosti; AES Chemunex, Bruze, France), 0·5 ml of the homogenized suspension was spread on each of two parallel ALOA plates to improve the enumeration limit to 1 log10 CFU g−1. Selective agents were then added to the Half-Fraser broth (Oxoid), followed by incubation at 30 ± 1°C for 24 ± 3 h. Subsequently, a second step of enrichment was performed by transferring 0·1 ml of the primary enrichment culture to 10 ml Fraser broth (Oxoid). Both primary and secondary enrichment cultures were plated in parallel on ALOA and RAPID'L.mono (Biorad, Hercules, CA) and incubated for 24 and 48 ± 3 h at 37 ± 1°C.
For confirmation, up to five typical colonies were spread onto blood agar and incubated at 37 ± 1°C for 24 ± 3 h. Typical haemolytic colonies were further confirmed as L. monocytogenes by testing for catalase production and rhamnose but not xylose fermentation. A selection of isolates were also checked for motility, CAMP reaction, Gram staining and verification using the commercial Listeria test, API Listeria (bioMerieux).
In the feeding experiment only quantification was performed. Consequently, homogenization and ten-fold serial dilution of individual slugs or faeces samples were done in saline peptone water (NMKL 2010). To prevent the pipette from clogging, BagFilter®′s (Interscience, St Nom la Bretêche, France) were used for the slug samples. The dilutions were plated on ALOA for enumeration.
Cultivation of Listeria monocytogenes from slug mucus
To examine for the presence of L. monocytogenes in external slug mucus, some slugs were individually placed on either RAPID'L.mono (field study) or ALOA plates (feeding experiment) and allowed to move freely for 2–4 min. The incubation of the growth media followed the same procedure as previously described (NMKL 2010). From the field study, single slugs from each of the first 62 Norwegian samples received were examined for external L. monocytogenes. The feeding experiment included 16 slugs, evenly sampled throughout the whole experimental period.
Multilocus sequence typing (MLST) of Listeria monocytogenes from slugs
One L. monocytogenes isolate from each positive pooled field study sample was analysed. DNA extraction was performed as described by Pospiech and Neumann (1995). PCR and MLST were performed according to the MLST scheme of Ragon et al. (2008). This MLST scheme is based on sequence analysis of the following seven housekeeping genes: acbZ (ABC transporter), bglA (beta-glucosidase), cat (catalase), dapE (Succinyl diaminopimelate desuccinylase), dat (D-amino acid aminotransferase), ldh (lactate deshydrogenase), and lhkA (histidine kinase). Profiles were submitted to the Listeria MLST database at the Pasteur Institute France (http://www.pasteur.fr/mlst). The neighbour-joining tree, based on concatenated allele sequences from the MLST analysis, was made using the online analysis tool (advanced mode) accessed date 10.09.2014: http://www.phylogeny.fr/ (Dereeper et al. 2008, 2010).
Animal welfare and euthanization
Slugs are not included in the European animal welfare legislation. Despite the lack of requirement for study permission, focus was maintained on careful management and rapid euthanization of slugs prior to analysis. The euthanization was performed by making a sagittal scalpel cut between the cephalic tentacles to cut the nerve ring in the head region of the slug.
Statistics
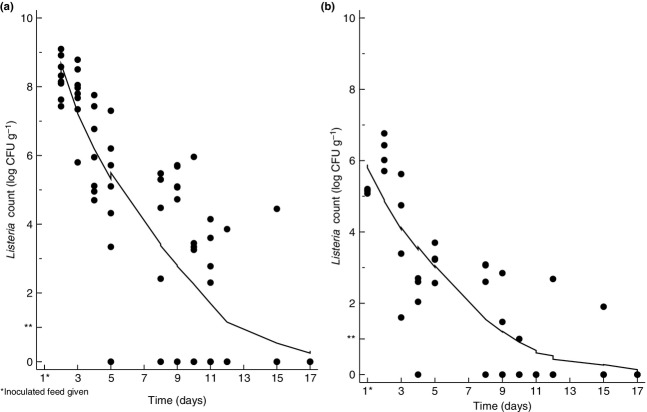
Statistical analysis was performed with the Stata ver. 12 software package (StataCorp LP, College Station, TX). Non-parametric locally weighted scatterplot smoothing (Lowess) functions were used to visualize the reduction in L. monocytogenes with time in the feeding experiment (Fig. 4). In addition, the two-sample Wilcoxon rank-sum (Mann–Whitney) test was used to prove an increase in L. monocytogenes within the slugs during the first 24 h of the feeding experiment.
Figure 4.
Listeria monocytogenes load in (a) slug faeces and (b) slugs following consumption of inoculated feed. **Detection limit of cultivation method.
Results
Field survey
In total, 710 slugs from different regions of Norway were analysed. Of 100 sampling kits distributed, 79 were returned containing a minimum 25 g of A. vulgaris. The majority (59 samples) contained 10 slugs as requested, but 25% (20 samples) contained on average six slugs (range 4–9 slugs). Fifty-nine samples (75%) represented garden environments, while 12 samples (15%) were collected from farm environments (defined as crops, pastureland or silage/hay meadows). In addition, five samples (6%) were mixtures of slugs collected from garden and farm environments, while the remaining three samples (4%) were collected from footpaths and woodlands. Slug faeces were also included in the pooled slug samples, but not all were analysed due to the dehydrated state of some faeces material. Of the pooled slug samples, 16% (13 of 79) tested positive, with a mean of 405 CFU L. monocytogenes g−1 slug (range 10–1205 CFU g−1). In addition, 21 further samples (27%) were positive after enrichment, resulting in a summarized prevalence of 43% in Norwegian samples. As shown in Fig. 2, the positive samples were geographically dispersed. The two samples from Denmark and Sweden were both positive for L. monocytogenes. The Danish sample, which experienced a longer delivery period, contained in excess of 30 000 CFU g−1 slug.
Figure 2.
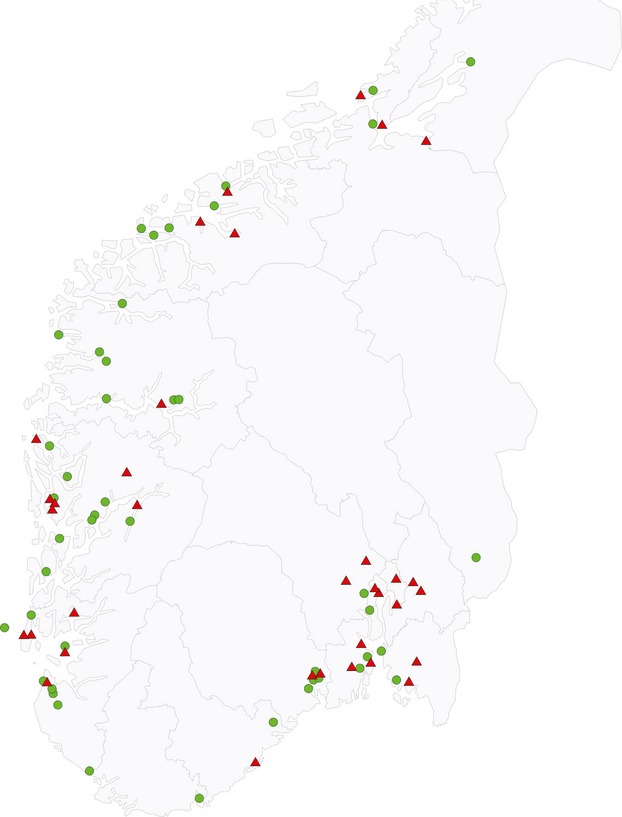
Geographic origins of the 79 Norwegian pooled slug samples. Triangles represent Listeria monocytogenes positive samples (43%) and circles non-detections.
Of 62 individual slugs (from different pools) tested, seven (11%) showed growth of L. monocytogenes from surfaces/mucus with a mean of 22 CFU per slug (range 1–57 CFU per slug).
Listeria ivanovii and Listeria innocua were also detected in some of the samples, but were not quantified or further identified in all samples.
Genetic diversity
MLST revealed a high diversity among slug derived L. monocytogenes isolates. The 36 isolates were differentiated into 20 different STs (Fig. 3). The majority of isolates belonged to lineage II (67%, 24/36) and lineage I 33% (12/36). Thirteen STs were represented by single isolates. The most common STs were ST1 (six isolates or 17%, including one isolate from Denmark), and ST91 (five isolates or 14%). ST7 and ST8, the latter including the Swedish strain, were each represented by three isolates (8%). The sequence types were dispersed over different geographical regions.
Figure 3.
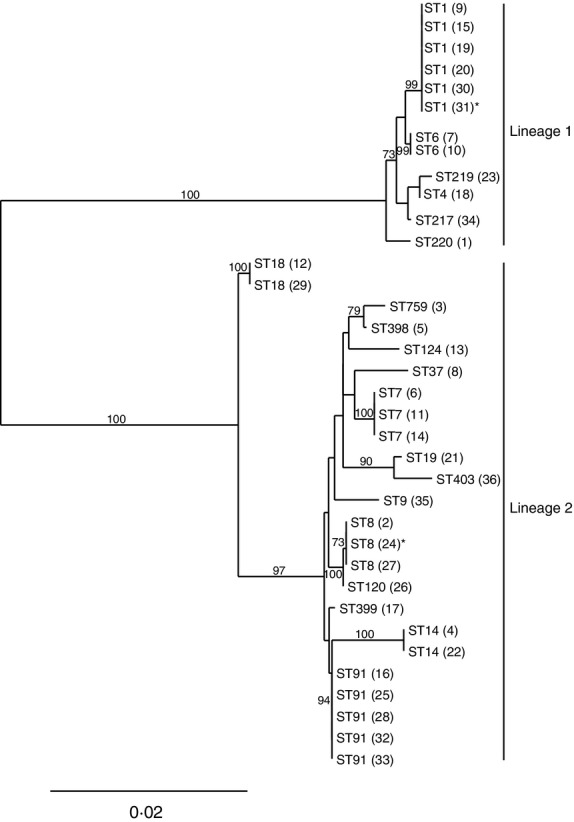
Neighbour-joining phylogenetic tree based on concatenated gene sequences from 36 Listeria monocytogenes isolates. The concatenated allele sequences representing each strain were used to generate the phylogenetic tree using MEGA. Bootstrap values >50% are shown based on 500 replicates. Labels represent ST numbers with strain designations in parentheses. Lineage I and II are indicated by a vertical line. The scale bar denotes 0·02 nucleotide substitutions per site. All isolates are from Norway except 31* (from Sweden) and 24*(from Denmark).
The feeding experiment
Viable L. monocytogenes was excreted in slug faeces for up to 22 days (range 5–22). The load of L. monocytogenes declined gradually (Fig. 4a). The single slug which excreted L. monocytogenes on day 15, did not give any faeces on day 17, but was positive on day 19 and 22 (results not shown).
As with slug faeces, the slug samples also showed declining L. monocytogenes counts, but L. monocytogenes was in general detected at lower levels and for shorter periods (see Fig. 4a,b). Slugs were positive for L. monocytogenes for up to 15 days (range 4–15). There was an increase in mean L. monocytogenes counts 24 h after receiving the inoculated feed, from 5·2 to 6·2 log10 CFU g−1 (P = 0·02).
Six of eight whole slugs (75%) positive for L. monocytogenes, also carried the bacterium on their surface up to day 12. Enumeration was not possible due to overgrowth of L. monocytogenes. Listeria monocytogenes was not detected in negative control slugs during the feeding experiment.
Discussion
Our study is the first to confirm that the invasive slug A. vulgaris can act as a vector for pathogenic Listeria spp. and that L. monocytogenes is commonly carried by slugs. Listeria monocytogenes is widely distributed in nature, and decomposing plant material is an important natural habitat (Welshimer and Donker-Voet 1971; Weis and Seeliger 1975; Brugere-Picoux 2008). Slugs live and feed in that sort of habitat, which can explain the high number of positive samples in our study. We have also shown that slug mucus and faeces, which are considerably less obvious than the slugs themselves, can contain viable L. monocytogenes. Results from the feeding experiment suggest that L. monocytogenes can be found in the surface/mucus of the majority of infected slugs. The lateral position of the anus behind the head region of A. vulgaris makes faecal contamination of the slug surface likely. Furthermore, there is a risk of pathogen transfer from slug faeces to crops, as defecation is stimulated by feeding (Shrewsbury and Barson 1947).
In the pooled Norwegian field samples, 16% of slugs carried an average of 405 CFU g−1 slug. Considering adult slug weights of 3–27 g in August (Briner and Frank 1998a), this represents 103–104 CFU per slug. Interestingly, the Danish sample contained more than 74 times this level with >30 000 CFU L. monocytogenes g−1 slug. Although no conclusions can be drawn from a single sample, this indicates that A. vulgaris (or their faeces) has the potential to contain a considerably higher number of L. monocytogenes than detected in Norwegian samples. Fenlon (1988) reported levels from 150 to ≥106 CFU of L. monocytogenes g−1 in silage fed to sheep with confirmed listeriosis. Given the levels of L. monocytogenes found in this study, direct consumption of a few slugs during grazing would probably not lead to disease in healthy ruminants. However, as contaminants in silage, slugs may increase initial L. monocytogenes levels. Under inadequate ensiling conditions, in particular poor sealing of big bale silage and pH >4·2, L. monocytogenes may proliferate to high levels and result in a greater risk of disease (Fenlon 1988; Driehuis and Oude Elferink 2000; Driehuis 2013).
The majority (75%) of field samples in this study were collected from gardens. Listeria monocytogenes is suggested to be more prevalent in farm environments, due to circles of contamination in which animal faeces are a well-known component (Nightingale et al. 2004; Oevermann et al. 2010; Santorum et al. 2012). Slugs find mammalian faeces attractive as food (Kozlowski 2007) and Arion ater is described as coprophagous (Shrewsbury and Barson 1947), a behaviour that can contribute to bacterial persistence. After experimental feeding with L. monocytogenes in this study, slugs carried and excreted L. monocytogenes in faeces for up to 22 days. Each slug consumed a quantity of bacteria (≅4·0 × 105 CFU) that probably represents an uncommonly high load under natural conditions. However, environmental load and the possibility of reinfection with L. monocytogenes may be factors affecting the length of carriage and excretion in natural surroundings. Our experiment showed large individual differences in the period of excretion (5–22 days). Similar L. monocytogenes loads on day one excludes insufficient consumption as a major source of error. Slugs showed an increase in L. monocytogenes level (P = 0·02) 24 h after the inoculated feed was given, suggesting a short period of bacterial enrichment. This initial enrichment can explain the relatively long excretion period, and potential differences in its efficiency could explain some of the individual variations in excretion-time. No such initial bacterial enrichment was reported after feeding Gray Field Slugs (Deroceras reticulatum) with Escherichia coli (Sproston et al. 2006) or A. vulgaris with Clostridium botulinum spores (Gismervik et al. 2014). Both studies reported short excretion time (up to 3–4 days), indicating direct bacterial passage through the digestive system.
Listeria monocytogenes consists of at least four evolutionary lineages (I, II, III, IV) with different but overlapping ecological niches (Wang et al. 2012). All L. monocytogenes isolates from A. vulgaris derived in the present field study belonged to the most common lineages, I and II (Wang et al. 2012).The majority of slug isolates were assigned to lineage II, which is widespread in natural and farm environments, is common in foods and commonly isolated from animal and human listeriosis cases (Wang et al. 2012). Lineage I is overrepresented in human listeriosis outbreaks, and has been reported to be more virulent in animal models, demonstrating an increased ability to invade and spread from cell to cell, compared to lineage II (Wang et al. 2012). ST1, belonging to lineage I, was predominant among the slug isolates and has also been shown to be prevalent from a wide range of sources and geographical regions worldwide (Chenal-Francisque et al. 2011; Wang et al. 2012; Haase et al. 2014). ST91, belonging to lineage II, was the second most common ST among the slug isolates. According to the Institute Pasteur MLST database, ST91 has been previously isolated from animals, food (cheese), compost and silage. ST7 and ST8, each represented by three slug isolates, have been previously identified from multiple sources, but ST7 is clearly dominated by animal isolates (Haase et al. 2014).
Our findings suggest that the invasive slug A. vulgaris may act as a vector for L. monocytogenes. Slug associated isolates clearly have the potential to cause disease in ruminants as well as humans. Dependent on ensiling conditions, highly slug-contaminated silage could pose a listeriosis risk.
Acknowledgments
The Research Council of Norway; Foundation for Research Levy on Agricultural Products and Agricultural Agreement Research Found and TINE is acknowledged for funding this project (199401) and the Ph.D. studentships of K.G. Thanks to Atilla Tarpai at the Norwegian Veterinary Institute for providing the map showing geographically dispersal of samples and Rune Pedersen and Hege Vinje for help during laboratory work. Thanks to Anja Kristoffersen and Malin Jonsson for statistical advices.
Conflict of Interest
The authors have no conflicts of interest to declare.
References
- Anon. Trends and Sources of Zoonotic Agents in Animals, Feedingstuffs, Food and Man in Norway 2000. Oslo, Norway: Norwegian Zoonosis Centre, Norwegian Veterinary Institute; 2001. [Google Scholar]
- Blood D. Pocket Companion to Veterinary Medicine. 9th edn. London: W.B.Saunders; 2000. [Google Scholar]
- Briner T, Frank T. Egg laying activity of the slug Arion lusitanicus Mabille in Switzerland. J Conchol. 1998a;36:9–15. [Google Scholar]
- Briner T, Frank T. The palatability of 78 wildflower strip plants to the slug Arion lusitanicus. Ann Appl Biol. 1998b;133:123–133. [Google Scholar]
- Brugere-Picoux J. Ovine listeriosis. Small Rumin Res. 2008;76:12–20. [Google Scholar]
- Chenal-Francisque V, Lopez J, Cantinelli T, Caro V, Tran C, Leclercq A, Lecuit M, Brisse S. Worldwide distribution of major clones of Listeria monocytogenes. Emerg Infect Dis. 2011;17:1110–1112. doi: 10.3201/eid1706.101778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driehuis F. Silage and the safety and quality of dairy foods: a review. Agric Food Sci. 2013;22:16–34. [Google Scholar]
- Driehuis F, Oude Elferink SJ. The impact of the quality of silage on animal health and food safety: a review. Vet Q. 2000;22:212–216. doi: 10.1080/01652176.2000.9695061. [DOI] [PubMed] [Google Scholar]
- Fenlon D. Listeriosis. In: Stark B, Wilkinson J, editors. Silage and Health. Marlow: Chalcombe Publications; 1988. pp. 7–18. [Google Scholar]
- Gismervik K, Bruheim T, Rorvik LM, Haukeland S, Skaar I. Invasive slug populations (Arion vulgaris) as potential vectors for Clostridium botulinum. Acta Vet Scand. 2014;56:65. doi: 10.1186/s13028-014-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase JK, Didelot X, Lecuit M, Korkeala H, Achtman M, Grp LMMS. The ubiquitous nature of Listeria monocytogenes clones: a large-scale Multilocus Sequence Typing study. Environ Microbiol. 2014;16:405–416. doi: 10.1111/1462-2920.12342. [DOI] [PubMed] [Google Scholar]
- Hatteland BA, Roth S, Andersen A, Kaasa K, Støa B, Solhøy T. Distribution and spread of the invasive slug Arion vulgaris Moquin-Tandon in Norway. Fauna Norv. 2013;32:13–26. [Google Scholar]
- Hofshagen M, Aavitsland P, Kruse H. Trends and Sources of Zoonotic Agents in Animals, Feedingstuffs, Food and Man in Norway 2001. Oslo, Norway: Norwegian Zoonosis Centre, Norwegian Veterinary Institute; 2002. [Google Scholar]
- Hofshagen M, Aavitsland P, Kruse H. Trends and Sources of Zoonotic Agents in Animals, Feedingstuffs, Food, and Man in Norway 2002. Oslo, Norway: Norwegian Zoonosis Centre, Norwegian Veterinary Institute; 2003. [Google Scholar]
- Kozlowski J. The distribution, biology, population dynamics and harmfulness of Arion lusitanicus Marbille, 1868 (Gastropoda: Pulmonata: Arionidae) in Poland. J Plant Prot Res. 2007;47:219–230. [Google Scholar]
- Nightingale KK, Schukken YH, Nightingale CR, Fortes ED, Ho AJ, Her Z, Grohn YT, McDonough PL, et al. Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl Environ Microbiol. 2004;70:4458–4467. doi: 10.1128/AEM.70.8.4458-4467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NMKL. Listeria monocytogenes. Detection in Foods and Feeding Stuffs and Enumeration in Foods. No 136. 5th edn. Oslo, Norway: Nordic Committee on Food Analysis; 2010. [Google Scholar]
- Oevermann A, Botteron C, Seuberlich T, Nicolier A, Friess M, Doherr MG, Heim D, Hilbe M, et al. Neuropathological survey of fallen stock: active surveillance reveals high prevalence of encephalitic listeriosis in small ruminants. Vet Microbiol. 2008;130:320–329. doi: 10.1016/j.vetmic.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Oevermann A, Zurbriggen A, Vandevelde M. Rhombencephalitis caused by Listeria monocytogenes in humans and ruminants: a zoonosis on the rise? Interdiscip Perspect Infect Dis. 2010;2010:632513. doi: 10.1155/2010/632513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- Quinn P, Carter M, Markey B, Carter G. Clinical veterinary microbiology. London: Mosby; 1994. [Google Scholar]
- Quinn PJ, Markey BK, Leonard FC, FitzPatrick ES, Fanning S, Hartigan PJ. Veterinary Microbiology and Microbial Disease. Chichester, West Sussex, UK: Wiley-Blackwell; 2011. [Google Scholar]
- Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008;4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santorum P, Garcia R, Lopez V, Martinez-Suarez JV. Review. Dairy farm management and production practices associated with the presence of Listeria monocytogenes in raw milk and beef. Span J Agric Res. 2012;10:360–371. [Google Scholar]
- Shrewsbury J, Barson GJ. 1947. pp. 70–76. A contribution to the study of bacterial flora of Arion ater. In Proc of the Society for Applied Bacteriology.
- Spörndly R, Haaga C. 2010. Silage quality when the crop is infected with Arion lusitanicus. In Nordic Feed Science Conference Uppsala, Sweden Swedish University of Agricultural Sciences. Department of Animal Nutrition and Management.
- Sproston EL, Macrae M, Ogden ID, Wilson MJ, Strachan NJC. Slugs: potential novel vectors of Escherichia coli O157. Appl Environ Microbiol. 2006;72:144–149. doi: 10.1128/AEM.72.1.144-149.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temelli S, Dokuzlu C, Sen MKC. Determination of microbiological contamination sources during frozen snail meat processing stages. Food Control. 2006;17:22–29. [Google Scholar]
- Wang Y, Zhao A, Zhu R, Lan R, Jin D, Cui Z, Wang Y, Li Z, et al. Genetic diversity and molecular typing of Listeria monocytogenes in China. BMC Microbiol. 2012;12:119. doi: 10.1186/1471-2180-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis J, Seeliger HP. Incidence of Listeria monocytogenes in nature. Appl Microbiol. 1975;30:29–32. doi: 10.1128/am.30.1.29-32.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshimer HJ, Donker-Voet J. Listeria monocytogenes in nature. Appl Microbiol. 1971;21:516–519. doi: 10.1128/am.21.3.516-519.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]