Abstract
Microorganisms can be engineered for the production of chemicals utilized in the polymer industry. However many such target compounds inhibit microbial growth and might correspondingly limit production levels. Here, we focus on compounds that are precursors to bioplastics, specifically styrene and representative alpha-olefins; 1-hexene, 1-octene, and 1-nonene. We evaluated the role of the Escherichia coli efflux pump, AcrAB-TolC, in enhancing tolerance towards these olefin compounds. AcrAB-TolC is involved in the tolerance towards all four compounds in E. coli. Both styrene and 1-hexene are highly toxic to E. coli. Styrene is a model plastics precursor with an established route for production in E. coli (McKenna and Nielsen, 2011). Though our data indicates that AcrAB-TolC is important for its optimal production, we observed a strong negative selection against the production of styrene in E. coli. Thus we used 1-hexene as a model compound to implement a directed evolution strategy to further improve the tolerance phenotype towards this alpha-olefin. We focused on optimization of AcrB, the inner membrane domain known to be responsible for substrate binding, and found several mutations (A279T, Q584R, F617L, L822P, F927S, and F1033Y) that resulted in improved tolerance. Several of these mutations could also be combined in a synergistic manner. Our study shows efflux pumps to be an important mechanism in host engineering for olefins, and one that can be further improved using strategies such as directed evolution, to increase tolerance and potentially production. Biotechnol. Bioeng. 2015;112: 879–888. © 2015 The Authors. Biotechnology and Bioengineering Published by John Wiley & Periodicals, Inc.
Keywords: directed evolution, host engineering, olefin production, solvent tolerance
Introduction
Microbes can be engineered to produce a large variety of chemicals. Many of these chemicals impose varying degrees of toxicity on the microbial strain being engineered for their production. Host engineering to improve strain tolerance towards a target compound is essential to optimize the production levels. Different compounds may also require different host engineering approaches, as the nature of toxicity is compound dependent. Hydrophobic chemicals are hypothesized to impact membrane permeability and fluidity, diminishing energy transduction, interfering with membrane protein function, and affecting a range of essential cellular processes (Jarboe et al., 2010; Sikkema et al., 1995). Tolerance and defense mechanisms range from induction of chaperones, modification of membrane composition and cellular morphology, to induction of efflux pumps that export the compounds out of the cell membrane and the cell. Of these, export pumps have emerged as an important mechanism for use in host engineering (Chen et al., 2013; Doshi et al., 2013; Dunlop et al., 2011) and also good targets for directed evolution (Fisher et al., 2013; Foo and Leong, 2013).
In this study we focus on the role of an Escherichia coli transporter in improving tolerance and production of olefin compounds that are valuable precursors in the plastic and polymer industry (e.g., bioacrylic, biostyrene) (McKenna and Nielsen, 2011; Schubert et al., 1988). Transporters provide a general mechanism for the export of toxic compounds (Nikaido, 2009; Takatsuka et al., 2010) thus also improving tolerance. For chemicals produced in the microbial cytoplasm, transport out of the cell may also improve production levels. In gram negative bacteria, tolerance towards solvent-like compounds is conferred by the hydrophobe/amphiphile efflux family of resistance-nodulation-division (RND) pumps (Nikaido and Takatsuka, 2009; Ramos et al., 2002; Tseng et al., 1999). RND efflux pumps are composed of three subunits. An inner membrane unit binds the substrate and with a proton motive force, exports it through the outer membrane subunit. The periplasmic subunit connects and stabilizes the inner and outer membrane units (Fig. 1A).
Figure 1.
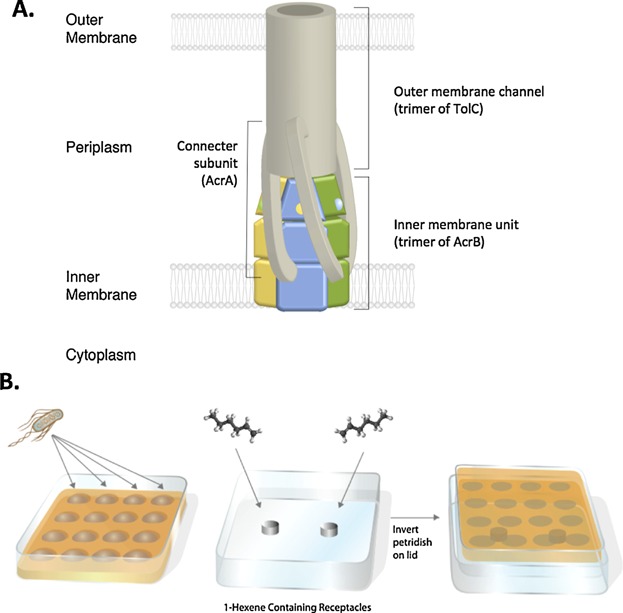
Schematic representation of the AcrAB-TolC efflux pump (A), and assay developed to determine the impact of 1-hexene on cell survival (B). A: Organization of the three proteins forming the tripartite efflux pump complex and their localization in the E. coli membrane is represented. The trimer of the AcrB subunit is shown in color. The newly discovered AcrZ domain (Hobbs et al., 2012) is not depicted in this diagram. B: Assay with a saturated atmosphere using the volatile compound to impose toxicity. The arrows represent the inoculation cell culture on the agar surface; the open lid of the petri-dish is used to place small receptacles containing a volatile compound. The chamber is assembled by inverting the petri-dish on the lid, cells facing the receptacles and sealed using parafilm (See methods). This method was used for 1-hexene survival assays as well as to screen AcrB mutants. The saturated atmosphere assay was also used with other alpha-olefins like 1-octene and 1-nonene.
In E. coli, the AcrAB-TolC efflux pump (Fig. 1A) composed of AcrB (inner membrane protein), TolC (outer membrane protein), and AcrA (periplasmic) has been extensively studied (Du et al., 2014; Murakami et al., 2002; Tikhonova et al., 2011) and has broad substrate specificity, ranging from detergents to antibiotics and solvents. This pump is reported to play a major role in the secretion of various alkanes such as hexane, heptane, octane, and nonane (Takatsuka et al., 2010) and also in imparting tolerance to various terpene based biofuel compounds (Dunlop et al., 2011). Several reports focus on the mechanism of this pump, especially that of AcrB, describing its rotational conformation changes (Seeger et al., 2006; Seeger et al., 2008; Sennhauser et al., 2007; Takatsuka and Nikaido, 2009, 2010), its binding pockets and the amino acids involved in substrate recognition (Eicher et al., 2012; Husain and Nikaido, 2010; Vargiu and Nikaido, 2012). In this study we test the role of this RND efflux pump in providing tolerance towards various olefins and evaluate its impact on the production of styrene. We use a random mutagenesis based strategy to successfully identify variants of the AcrB protein for improved 1-hexene tolerance. We discuss the potential modes of action of the AcrB mutations that lead to the improved function.
Materials and Methods
Media, Chemicals and Strains
Chemicals: styrene (99%), 1-hexene (99%), 1-octene (98%), nonene (96%), isopropyl b-D-1-thiogalactopyranoside (IPTG), dodecane, arabinose (ara), bile salts, chloramphenicol (Cm), kanamycin (Km), ampicillin (Amp), lysozyme, and antibodies (ab), were purchased from Sigma–Aldrich (St. Louis, MO), dimethylformamide (DMF) from Fisher Scientific (Hampton, NH), and glucose from VWR (Westchester, PA). All strains, plasmids and oligonucleotides are listed in supplemental data Table S1. All plasmids and strains listed in Table S1 are available through the JBEI registry (http://public-registry.jbei.org), (Ham et al., 2012). Strains were cultured in Luria-Bertani (LB) broth supplemented with appropriate antibiotics, or inducers as necessary, and with 15 g/L agar (BD, Sparks, MD) for plate culture and assays. Oligonucleotides synthesis was conducted at Integrated DNA Technologies (Coralville, IA) and sequencing by Quintara (Albany, CA). BLAST (NCBI) was used to identify the unexpected insertion sequences. E. coli DH10B was used for sub-cloning, E. coli K12 BW25113 (Baba et al., 2006) (Table S1) was used for toxicity assay and E. coli NST74 (ATCC 31884) was used for styrene production. Arabidopsis thaliana (ecotype Col-0) cDNA and Saccharomyces cerevisiae S288C were used to amplify genes required for styrene production: PAL2 and FDC1 as previously described (McKenna and Nielsen, 2011).
Construction of Gene Deletion Mutants
The E. coli K12 BW25113 ΔacrAB and E. coli NST74 ΔacrAB strains were constructed using the one-step inactivation of chromosomal genes using published methods (Datsenko et al., 2000). In order to generate a ΔacrAB strain, the E. coli K12 BW25113 ΔacrA was used. The genomically encoded Km marker in the ΔacrA strain was removed using the pCP20, and the plasmid pKD46 was introduced into the resulting strain. The PCR product used to mediate gene replacement for acrB was amplified from the E. coli K12 BW25113 ΔacrB strain using the primers DelB F and DelB R and introduced in the Km marker-free E. coli K12 BW25113 ΔacrA strain (pKD46). KmR colonies were tested for the loss of acrB. After curing the pKD46, the KmR gene was removed generating the strain E. coli K12 ΔacrAB. To generate E. coli NST74 ΔacrAB, the PCR used to mediate the replacement of acrA and acrB in E. coli NST74 was amplified from E. coli K12 BW25113 ΔacrAB KmR using the primers DelAB F and DelAB R, introduced into E. coli NST74 (pKD46) and KmR colonies tested for the loss of operon acrAB. Here again, after curing the pKD46, the KmR gene was removed generating the final strain E. coli NST74 ΔacrAB.
Construction of Plasmids
High fidelity phusion polymerase (Thermo Scientific, Waltham, MA) was used for all gene amplifications, except for generating the acrB variant library. All restriction enzymes were fast digest enzymes (Thermo Scientific, Waltham, MA). tolC was amplified using the primers TolC F and TolC R and cloned in the pBbS5a-RFP (p0a) (Lee et al., 2011) with the restriction sites NdeI and XhoI generating the plasmid pTolCa. acrA was amplified using the primers AcrA F and AcrA R and cloned into the pBbA8c-RFP (p0c) (Lee et al., 2011) using the restriction sites BglII and NdeI generating the plasmid pAc. acrB was amplified using the primers AcrB F and AcrB R and cloned in pAc using the restriction site NdeI and XhoI generating the plasmid pABc.
Genes encoding PAL2 and FDC1, were amplified from A. thaliana and S. cerevisiae S288C cDNA, respectively. FDC1 was amplified using the primers Fdc1 F and Fdc1 R. PAL2 was amplified using the primers Pal2 F and Pal2 R. The two overlapping PCR products were extracted and used to generate a long fragment containing the two genes. Overlap PCR included 5 PCR cycles without primers and 25 cycles with the external primers Fdc1 F and Pal2 R. The resulting PCR was digested with BglII and XhoI, and cloned in pBbE5a-RFP (plac0) or pBbE1k-RFP (ptrc0) between the restriction site BglII and XhoI generating the plasmids placSty and ptrcSty, respectively. To generate the acrB variant library by error prone PCR, the Dream Taq polymerase (Thermo Scientific, Waltham, MA) was used. PCR products were obtained by amplifying acrB from the pABc (0.5 ng/μL), using the primers Cam F and Cam R (0.1 μM), the kit buffer (containing 2 mM MgCl2), 0.2 mM dNTP mix (Thermo Scientific, Waltham, MA), 1% DMSO, and 0.1 unit/μL polymerase. Twenty cycles were performed resulting in a pool of mutated acrB sequences. The resulting library contained, on average, 2.2 amino acid mutations per protein, as determined by sequencing randomly selected acrB variants. PCR fragments were gel extracted, cloned via Gibson cloning (Gibson et al., 2009) into pAc digested with NdeI and XhoI, and then introduced in the E. coli K12 ΔacrAB strain.
The acrB variants with desired mutations were generated using overlap PCR with the high fidelity phusion polymerase. For each single mutation variant, two PCR products were obtained by amplification from the pABc vector as listed in Table S1. PCR 1 and 2 were extracted for every variant and used to generate the full length genes. The resulting products were introduced using Gibson assembly (Gibson et al., 2009) into pAc digested with NdeI and XhoI, and transformed into E. coli DH10B. Plasmids with confirmed sequences were then introduced into the E. coli K12 ΔacrAB strain. The variants with multiple mutations were obtained by introducing additional mutation(s) in a given variant, using the overlap PCR workflow described above. PyMOL (http://www.pymol.org/) was used to visualize the structure of AcrB and the localization of the individual mutations.
Growth Impact of acrAB Overexpression
Colonies of E. coli K12 ΔacrAB containing the plasmid pABc were used to inoculate a 6 mL LB-Cm pre-culture tube. Hundred microliter of cells at OD600 = 1 were used to inoculate 10 mL LB-Cm supplemented with glucose (2 g/L) and various concentrations of arabinose (0, 5, 10, 20, 30, 40, and 50 mM). Cell growth at 37 °C was followed by OD600 measurements using a spectrophotometer (Beckman Coulter DU800, Brea, CA).
Impact of Alpha-Olefins, Styrene, and Bile Salts on Growth
Cells were pre-cultured in 6 mL LB-Cm with 2 g/L glucose and 5 mM arabinose at 37 °C. At OD600 = 1, 100 μL culture was transferred to 15 mL glass culture containing 10 mL of LB, supplemented with the appropriate antibiotic and inducer. Cultures tubes fitted with a septum lid (VWR, Westchester, PA) were used for the volatile chemicals. DMF was used as co-solvent to prepare stock solutions for all three olefin compounds (styrene at 10, 12, 14, and 16 g/L, 1-hexene at 5, 6, 7, and 8 g/L, and 1-octene at 10, 20, 50, and 100 g/L). Bile salts solution at 105 g/L was prepared in LB, and sterilized by filtration. At an OD600 of 0.2, 100 μL of the olefin stock solutions or 500 μL of bile salts solution was added to the 10 mL cultures. After 2 or 2.5 h at 37 °C for the olefins, and 1.5 h for the bile salts, OD600 was measured. All cultures were done in duplicate or triplicate, as indicated, starting from separate single colonies.
For agar plate based assays (Fig. 1B), cells were first streaked overnight and three colonies were used to inoculate a preculture (6 mL LB-Cm with 2 g/L glucose, 5 mM arabinose). At OD600 = 1, the precultures were used to prepare a dilution series. Dilution 0 (D0) corresponds to the OD600 = 1, and contains ∼109 cells/mL. Dilution 1 (D1) is a 10-fold dilution of D0, dilution 2 (D2) a 10-fold dilution of D1 up to dilution 5 (D5) which contains ∼104 cells/mL. Ten microliter of each dilution was spotted in triplicate on LB-agar plates (single well plates, Thermo Scientific, Waltham, MA) containing 30 mL of LB-agar (with 30 mg/L Cm, 2g/L glucose, 5 mM arabinose). Small receptacles (microfuge tube caps) were placed on the open lid of the agar plate, and filled with 400–600 μL of 1-hexene, 1-octene or 75 μL of 1-nonene. The agar plates were then inverted over the filled caps, cells facing the olefin compound, sealed with parafilm, placed in a plastic bag (the original packaging for the petri dishes) and incubated for 2 days at room temperature (Fig. 1B). For 1-octene, 2 mL of compound was overlaid on top of the agar, submerging the cells. For assays relating to TolC the LB media and agar were also supplemented with ampicillin (100 mg/L) and IPTG (100 μM).
For the high throughput screening of AcrB variants, two methods were used. For the first screen, individual clones were collected and inoculated in 96 deep well plates (USA Scientific, Ocala, FL) using 800 μL LB (with 30 mg/L Cm, 2 g/L glucose, 5 mM arabinose) and cultured overnight at 37 °C with agitation. Cells were then diluted 100-fold and 5 μL of the dilution was spotted on agar plates containing 25 mL of LB-agar-Cm and inducer, using a liquid handler robot (Biomek FX, Beckman Coulter, Brea, CA). Six hundred microliter of 1-hexene was used per plate to create a saturated atmosphere. Plates were sealed and incubated for 2 days as described earlier. For the second screen, after transformation of the acrB variant library in E. coli K12 ΔacrAB, cells were plated on LB-agar-Cm plates directly, with or without a 1-hexene selection pressure, using 300 μL to saturate the atmosphere (Fig. 1B). For each screen we counted the number of cells able to grow on agar plates after exposure to the olefin compound. We assessed that the strain(s) expressing a variant had better survival than the wild type when the corresponding liquid culture, spotted on agar plates, resulted in at least three times more colonies relative to strain expressing the wild type.
Measuring the Toxic Effect of Styrene
E. coli NST74 ΔacrAB containing pABc or p0c and either placSty, ptrcSty, plac0, or ptrc0, were inoculated into 6 mL LB (with 15 g/L glucose, 10 mM arabinose and appropriate antibiotics). At OD600 = 1, 5 μL cells were inoculated in 95 μL of LB-antibiotic (with 15 g/L glucose, 10 mM arabinose), biolog redox dye (MixA) solution (Biolog, Hayward, CA) and various concentration of IPTG (0, 10, 20, 50 100, 200, and 300 μM). The experiment was performed in 96 well plates (Biolog, Hayward, CA), at 37 °C and growth was monitored using the Omnilog system (Biolog, Hayward, CA). In these conditions, strains containing p0c did not grow. For the strains with ptrcSty, the impact of styrene production on growth was also tested on LB-agar plate (with 25 mg/L Km, 15 g/L glucose, 10 mM arabinose, and 100 μM IPTG). For the glass tube based growth of E. coli NST74 ΔacrAB (with pABc or p0c and either placSty or plac0), colonies were inoculated in 10 mL of LB (with 100 mg/L ampicillin, 15 g/L glucose, 10 mM arabinose) and grown overnight at 37 °C. From this culture, cells were inoculated in glass tubes (starting OD600 = 0.02) containing 10 mL of LB (with 100 mg/L amp, 15 g/L glucose, 10 mM arabinose, and 100 μM IPTG). Growth was monitored using a spectrophotometer (Beckman Coulter DU800, Brea, CA).
Styrene Production and GC-MS Analysis
E. coli NST74 ΔacrAB, placSty and pABc or p0c strains were inoculated in 10 mL LB (with 100 mg/L amp, 15 g/L glucose, 10 mM arabinose) and cultured at 30 °C. At OD600 = 0.6, 100 μM IPTG was added and cultured at 25 °C with agitation. The culture supernatant was extracted with 1 mL dodecane for 1 h with agitation at room temperature. One hundred microliter of the dodecane layer was analyzed by gas-chromatography mass-spectrometry (GC–MS, GC model 6890, Hewlett Packard, Palo Alto, CA; MS model 5973, Agilent Technologies, Santa Clara, CA) using a DB-5MS (30 m × 0.25 mm × 0.25 μm) fused-silica capillary column and helium as the carrier gas. The GC analytical method was as follows: hold at 40 °C for 2 min after injection, increase to 100 °C at 30 °C/min, then to 300 °C at 60 °C/min hold at 300 °C for 2 min. An m/z range from 40 to 200 was used for styrene detection.
Western Blot Assays
The pellet of a 2 mL culture at OD600 = 1 (LB-Cm, 2 g/L glucose, 5 mM arabinose, 37 °C) was resuspended and incubated for 15 min in 30 μL B-Per buffer (Thermo Scientific, Waltham, MA) supplemented with 1 mg/mL lysozyme. Each sample was sonicated (SP Scientific, Gardiner, NY) for 5 s at the lowest power setting at 4 °C, and clarified via centrifugation at 4 °C for 30 min at 15 rpm. Total protein present in the clarified fraction was quantified using the DC Lowry Reagent as per manufacturers instructions (Bio-Rad, Hercules, CA). Twenty microliter of normalized protein samples were mixed with SDS loading dye buffer (Life technologies, Carlsbad, CA) and DTT (Life technologies, Carlsbad, CA) and incubated at 98 °C for 20 min. Ten microliter of samples were run on 4–12% Bis-Tris gel at 165 V with SDS MES Buffer (Life technologies, Carlsbad, CA) and transferred to a PVDF membrane using the iBlot transfer system (Life technologies, Carlsbad, CA). The membrane was washed in PBS buffer (20 mM NaPO4, 50 mM NaCl, pH = 7.4) and blocked overnight at 4 °C with 25 mL of 3% of BSA in PBS-Tween-20 (EMD Millipore, Billerica, MA). The primary ab; a monoclonal mouse anti-His was diluted 1:5,000, the secondary ab; a goat anti-mouse HRP conjugated was diluted 1:10,000, in 1% BSA-PBS-Tween-20 and used to incubate the membrane for 1 h each at room temperature. Three washes in PBS-Tween20 were performed after antibody incubation. The membrane was then incubated in 2 mL of HRP detection solution (Bio-Rad, Hercules, CA) for 10 min.
Results
Toxicity of Olefins, and AcrB Involvement
We found that the toxicity of olefin compounds varies broadly as a function of side groups and chain length. We defined a toxic dose as the concentration of compound that decreased the growth by half, measured after 2 h of exposure. For E. coli K12, the toxic dose is 110 mg/L for styrene, 65 mg/L for 1-hexene, while 1-octene did not affect the growth in liquid culture (Fig. 2A). On agar plates, we observed that E. coli K12 survival under an overlay of 1-octene, is equivalent to the control (Fig. 2B), suggesting that cells are minimally affected by its presence. To examine the involvement of AcrAB-TolC in styrene or alpha-olefin tolerance, we constructed acrA and acrB mutants (E. coli K12 ΔacrAB), and transformed the strain with a plasmid borne acrAB operon under the control of an arabinose inducible promoter (pABc) or a control plasmid (p0c). For styrene, we used liquid cultures with exogenously added styrene (Fig. 3A). For 1-hexene, 1-octene, and 1-nonene we developed an agar plate survival assay (see methods, Fig. 1B), in an atmosphere saturated with the volatile compound. For all tested chemicals, the pump provided an advantage in the growth (Fig. 3A) and survival (Fig. 3B) of the strain.
Figure 2.
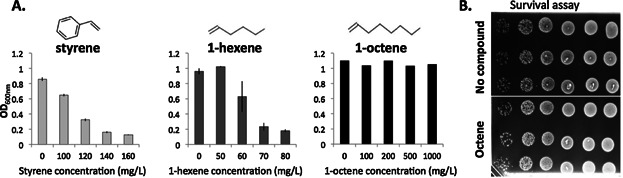
Impact of styrene, 1-hexene, and 1-octene on cell growth. A: The indicated concentrations of olefin compounds were added to an E. coli K12 culture at an OD600 = 0.2. OD was monitored after 2 h at 37 °C. The bars on the two first panels represent standard deviation between duplicates. B: A survival test in presence of 1-octene was performed. Six different dilutions of a culture (10 cells to 107 cells, left to right) of E. coli K12 were spotted in triplicate on an agar plate. An overlay of 1-octene was added over the cells (control had no overlay). Plates were incubated at room temperature for 2 days, and the lid of the petri-dishes were removed for recording images.
Figure 3.
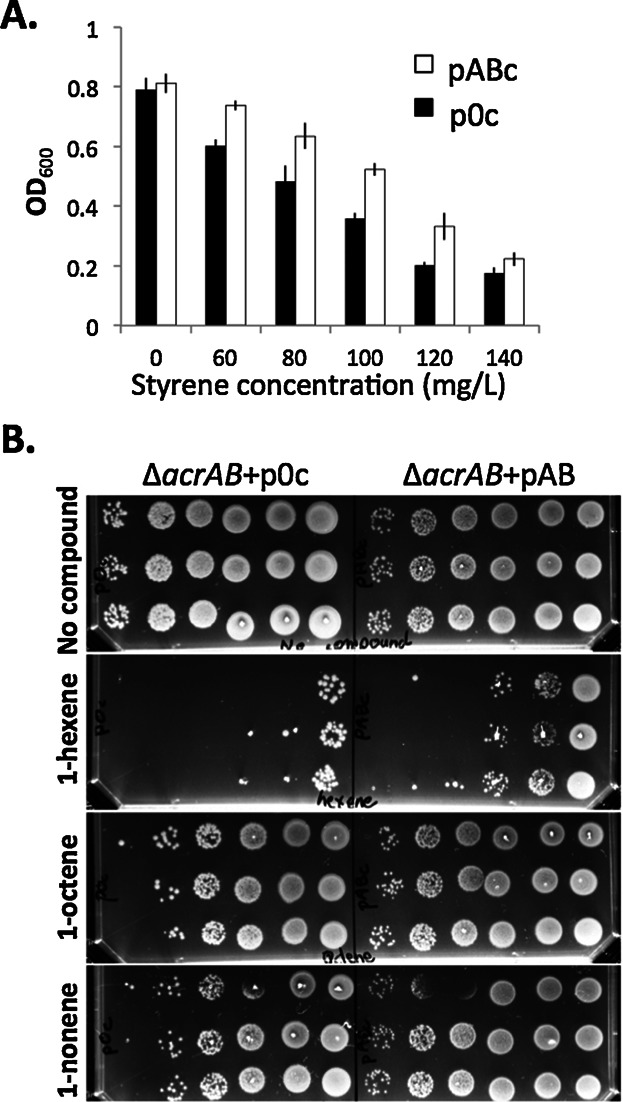
Impact of AcrAB production on growth in the presence of styrene, 1-hexene, 1-octene, and 1-nonene. A: Different concentrations of styrene were added to an OD600 = 0.2 culture of E. coli K12 ΔacrAB containing the pABc (white) or the control plasmid p0c (black). OD was monitored after 2 h of growth at 37 °C. The bars represent standard deviation between duplicates. B: Survival tests in presence of 1-hexene, 1-octene, or 1-nonene saturated atmosphere were performed. Six different dilutions of cultures (10 cells to 107 cells, left to right) were spotted in triplicate on an agar plate before exposure to chemicals.
AcrAB Involvement on Olefin Production
Among olefins used for plastic production, styrene has been reported in the literature to be produced at levels that affect E. coli growth (McKenna and Nielsen, 2011). To investigate the involvement of the AcrAB-TolC in styrene production, we used the reported L-phenylalanine over-producing E. coli strain (E. coli NST74) (McKenna and Nielsen, 2011), in which we deleted acrAB, and introduced a plasmid encoding the reported styrene pathway (see methods). We made two separate production plasmids using two different IPTG-inducible promoters; ptrcSty, for strong induction and placSty, for weak induction. These plasmids, or the corresponding control plasmids (ptrc0 and plac0) were introduced in strains containing the plasmids encoding the pump (pABc) or the control (p0c). We found that the growth is affected by the absence of the AcrAB-TolC pump (Fig. 4) and consequently also production, in the styrene production strain. For the strain with styrene production genes under the control of a strong promoter (ptrcSty), we observed no growth in liquid culture, when the pump was not expressed (p0c) (data not shown). For the strain with ptrcSty and pABc, styrene production could be induced with IPTG but had a strong impact on growth (Fig. S1B). Additionally we observed a high standard deviation between replicates (Fig. S1B). When grown on solid agar plates with IPTG, we observed colonies with variable morphology and size (Fig. S1C) and were likely responsible for the high growth variation among liquid culture replicates. The DNA sequence of the plasmids, extracted from cells with a different morphology and size, revealed the presence of insertional sequences in the promoter or coding sequence of FDC1 or PAL2 (data not shown), inactivating styrene production. These observations indicated that use of a strong promoter affects the growth of the strain and results in a strong selective pressure against the production of styrene. To moderate the impact of styrene production, we used a weakly induced promoter, placSty. Correspondingly, we observed less growth variation between replicates (Fig. S1A) and in this case, the strain lacking acrAB was able to grow. We observed that the absence of the pump had a small negative impact on growth when styrene is not produced, and a strong negative impact when styrene is produced (Fig. 4A). Using the weaker promoter for the styrene production genes, we could measure and compare the quantity of styrene produced by the strain as a function of pump expression (Fig. 4B). The results show that the quantity of styrene produced was significantly higher when the pump expression is induced (Fig. 4B). Taken together these data show that the AcrAB-TolC pump is involved in tolerance towards toxic compounds such as styrene and important in reaching optimal production levels for a given biosynthetic pathway. However due to the high selective pressure against styrene production, we discontinued styrene as a model compound for the rest of this study.
Figure 4.
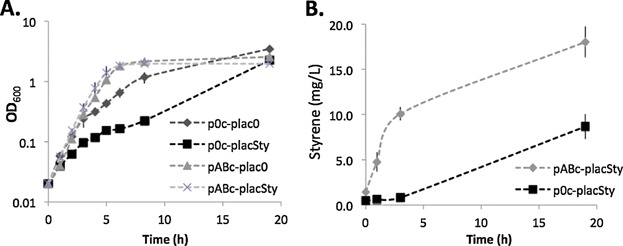
Impact of AcrAB expression on styrene production and growth. A: Growth of E. coli NST74 ΔacrAB containing pABc or p0c and either placSty or plac0 at 37 °C, in LB supplemented with the appropriate antibiotics and inducers. Bars represent standard deviation between duplicates. B: Styrene produced by E. coli NST74 ΔacrAB containing placSty and pABc or p0c. Bars represent standard deviation between duplicates and is representative of assays conducted multiple times.
Improvement of Tolerance Via Pump Overexpression
We tested if the overexpression of the AcrAB-TolC pump provides an advantage during 1-hexene exposure. We cloned pABc encoding acrAB (or p0c) and pTolCa (or p0a) into E. coli K12 wild type, and tested the survival of the strain in presence of 1-hexene. 1-hexene has a boiling point of 63 °C, and to address this volatility we used a saturated atmosphere survival assay (Fig. 1B). We observed that overexpressing acrA, acrB, and tolC in the wild type strain increases survival in 1-hexene (Fig. 5). Figure 5 shows that the overexpression of only tolC in wild type strains does not improve tolerance to 1-hexene. However, the overexpression of only acrAB improves 1-hexene tolerance relative to wild type. An explanation could be that TolC is not the limiting protein of this complex. This stoichiometry could be due to the involvement of TolC in other membrane complexes (Ramos et al., 2002). Our results also indicated that there is an upper limit to increasing pump expression. Increasing concentrations of inducer causes a reduction in growth (Fig. S2). Thus, while a limited increase in the levels of AcrAB-TolC improves tolerance to 1-hexene, further increase in tolerance to the olefin compound may only be achieved using a more efficient pump or one that has lower toxicity when overexpressed.
Figure 5.
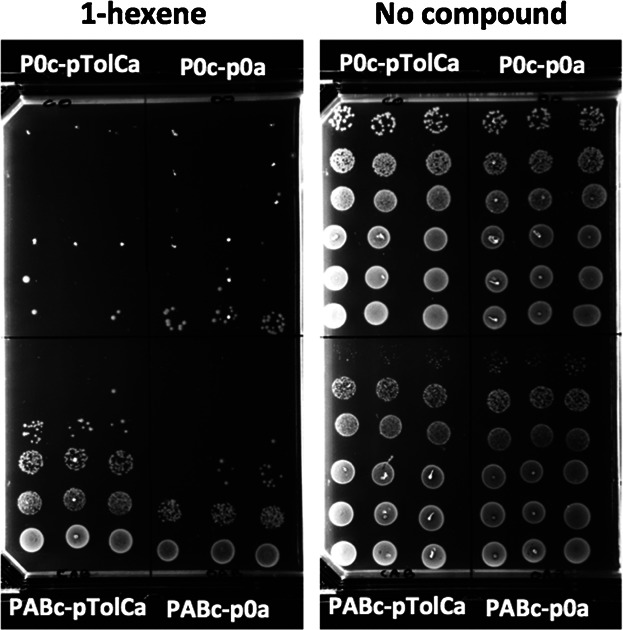
Impact of AcrA, AcrB, and TolC over-production on 1-hexene tolerance. A survival test of E. coli K12 wild type containing pABc or p0c and either pTolCa or p0a, in an 1-hexene saturated atmosphere was performed. Six different dilutions of cultures (10 cells to 107 cells, top to bottom in each of the four panels) were spotted in triplicate on an agar plate. Plates were then exposed to 1-hexene (Fig. 1B).
Directed Evolution of AcrB to Improve Tolerance to 1-Hexene
Next we sought to improve the AcrB protein function by using directed evolution. To identify AcrB variants that provide greater tolerance relative to the wild type, we developed a high throughput screening method using the 1-hexene saturated atmosphere assay (Fig. 1B). Using error prone PCR we generated a library of acrB variants. We cloned the acrB variants on a low copy plasmid, in an operon with the acrA gene to maintain stoichiometry of the pump complex, and introduced the library into E. coli K12 ΔacrAB. We individually tested 2500 clones expressing an acrB variant and observed that 1.1% of the clones had better survival in presence of 1-hexene relative to the strain encoding the wild type pump. We also used a second strategy, where selection pressure was applied directly after library transformation, by plating the transformed and recovered cells in a 1-hexene saturated atmosphere. In this second strategy, we found that only one cell out of eight survived, and among the 500 colonies tested in presence of 1-hexene, 9% were more tolerant than the strain expressing the wild-type pump. Of the total set of clones obtained, from both the strategies described above, we re-tested the 20 most tolerant variants in duplicate, and of these 16 (80%) colonies maintained increased survival relative to the strain expressing wild type acrB. Plasmids from these 16 strains were retransformed into the original background strain and 11 (63%) maintained the increased tolerance phenotype, confirming that in these cases the 1-hexene tolerant phenotype resulted from the acrB variant genes rather than background mutations in the genome. Since most variants contained more than one mutation, we reconstructed acrB gene variants with the individual single mutations to identify the ones responsible for the improved phenotype. We identified and confirmed six amino acid mutations in AcrB that provide improvement in tolerance over the wild type: A279T, Q584R, F617L, L822P, F927S, and F1033Y (Fig. 6A). These mutations are not localized in the same site but are distributed all along the protein sequence (Fig. 6B and C).
Figure 6.
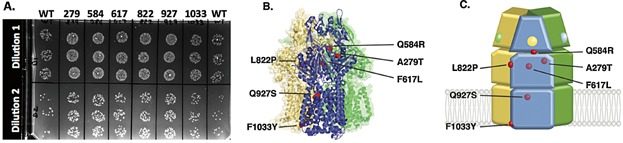
Impact of AcrB mutations on 1-hexene tolerance. A: Survival test of E. coli strain producing either the AcrB wild type or variants in a 1-hexene saturated atmosphere was performed. Two dilutions of cultures (Dilution 1: 105 cells and Dilution 2: 104 cells) were spotted in triplicate on an agar plate. B: Localization of the six beneficial mutations represented in red on the structure of the AcrB trimer or C: on the schematic representation of the AcrB trimer.
To determine if the mutations were synergistic, we combined selected mutations in a single acrB gene generating eight new variants with 2, 3, or 6 beneficial mutations (Table I). Of the combinations tested, strains containing AcrB with two mutations were found to be more tolerant to 1-hexene relative to an AcrB with a single mutation (Fig. S3A). However combining three or more mutations in the same protein did not result in additional tolerance. We compared the levels of the wild type AcrB protein to three of the variants using clarified cell lysates normalized for total protein (see methods). The tested AcrB variants were present at lower levels than the AcrB wild type and an increase in the number of beneficial mutations resulted in a decrease in protein quantity (Fig. S3B). Taken together, our data suggest that the tested mutations were potentially disruptive and destabilizing for the protein. Correspondingly, it is most likely that the improvements in tolerance are due to improvements in the efficiency or mode of action of the pump, rather than increased protein production or stability.
Table I.
AcrB variants
| AcrB variants with several mutations | Description |
|---|---|
| var1 | A279T, F617L |
| var2 | F617L, L822P |
| var3 | A279T, L822P |
| var4 | A279T, Q584R |
| var5 | A279T, F617L, and L822P |
| var6 | A279T, Q584R, and F617L |
| var7 | A279T, Q584R, and L822P |
| var8 | A279T, Q584R, F617L, L822P, F927S, and F1033Y |
We examined the confirmed evolved AcrB variants in liquid cultures containing 1-hexene and other industrially relevant olefins. We analyzed the growth of selected strains containing the AcrB variants in presence of different concentration of toxic olefins (Fig. 7). In presence of 1-hexene, we observed that several strains with evolved AcrB variants reached a higher optical density than the wild type after 2.5 h (Fig. 7A), consistent with the phenotype observed in the agar plate assay (Fig. 1B). In presence of bile salts, the mutation A269T and Q584R showed a minor but significant improvement in the growth. However none of the evolved AcrB variants provided growth benefit in presence of styrene (Fig. 7B). Thus, the mutations either provide no advantage against styrene or do not have measurable advantage.
Figure 7.
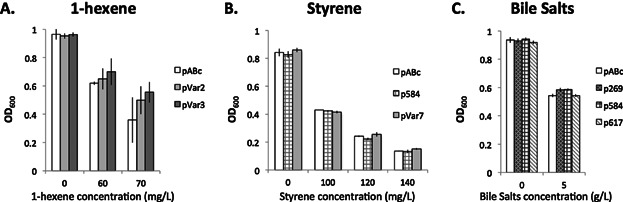
Impact of AcrB mutations on growth in the presence of 1-hexene, styrene, and bile salts. Different concentrations of 1-hexene (A), styrene (B), or bile salts (C) were added to an OD600 = 0.2 culture of E. coli K12 ΔacrAB containing either the AcrB wild type (pABc) or select AcrB variants. OD was monitored after 2.5 h, 2 h, and 1.5 h of culture at 37 °C for 1-hexene, styrene, and bile salts, respectively. Data show an average of biological replicates; triplicates for 1-hexene and bile salts, and duplicates for styrene.
Discussion
In this study we show that the E. coli efflux pump, AcrAB-TolC, plays a major role in tolerance to styrene and alpha-olefins, and may provide a significant advantage in reaching high production levels of toxic compounds. One of the first steps to improve the tolerance to alpha-olefins was to determine the benefit from increasing the levels of the AcrAB-TolC efflux pump. We observed that increasing the expression of this pump improved tolerance to 1-hexene but this strategy is limited by the toxicity known to be linked to overexpression of membrane proteins (Wagner et al., 2007). We thus decided to improve the pump function using directed evolution.
We show that engineering the AcrB subunit of this efflux pump improves tolerance to 1-hexene. Our goal was to find variants that are able to improve the tolerance to 1-hexene and to characterize the primary domains responsible for 1-hexene export. It has been shown that the entrance of substrates into the efflux pump and the amino acids involved in the substrate binding depends of the properties and the size of the compounds (Eicher et al., 2012; Takatsuka et al., 2010). To better understand the domain involved in 1-hexene export we analyzed several beneficial mutations obtained after a round of evolution. We identified several mutations (A279T, Q584R, F617L, L822P, F927S, and F1033Y) that resulted in improved tolerance to 1-hexene. We found that these mutations have an additive effect, but also established that some changes may be disruptive for protein solubility. Since all tested AcrB variants in our study resulted in decreased protein levels, it is more likely that the improvement in the tolerance conferred by these variants is due to an alternate mechanism, such as higher pump efficiency.
Crystallographic and site-directed mutagenesis studies of AcrB have determined the role of domains and of several amino acids in the protein (Husain and Nikaido, 2010; Murakami et al., 2002). Among the beneficial amino acid changes we identified, F927S and especially F1033Y were the most unexpected. F927S is localized in the transmembrane domain (Fig. 6B and C), at the top of the TM10 α-helix. This helix contains a crucial amino acid (K940) involved in proton transport (Su et al., 2006) and a mutation in this helix could impact the rotational movements leading to compound export. The mechanism of the F1033Y mutation is unclear but this amino acid could potentially interact with the newly discovered AcrZ protein (Hobbs et al., 2012). The improved tolerance with F1033Y and F927S could also be due to an indirect effect such as altering the membrane structure. However the mutations A279T, Q584R, F617L, and L822P were localized in positions known to be important for pump function. L822 is positioned between the two β-sheets Cβ13 and Cβ14 belonging to the pore subdomains PN1 and PC2, respectively (Fig. 6B and C). The mutation of a leucine to a proline is located between these 2 β-sheets at the “ceiling” of the vestibule, suggested to be a highly probable substrate entrance point, possibly altering the flexibility and/or the opening of the vestibule facilitating 1-hexene entrance into the pore. A279 is located in the binding pocket in which residues E273, N274, D276, I277, play important roles (Husain et al., 2011). Why the introduction of a polar amino acid at this position would improve the efflux of hydrophobic compound is unclear, but every substrate must eventually leave the binding site in order for its efflux to occur, so the substitution A279T (hydrophobic to polar residue) may help substrate release to the gate and the funnel. Q584 is located in a position potentially involved in trimer assembly. The AcrB subunit is reported to fold independently, and then assemble into a trimer (Yu et al., 2011). It has been shown that the P223 from one AcrB interacts with Q584 from another AcrB polypeptide, and is required for the assembly and stability of the trimer. The mutation Q584R could therefore impact trimer assembly and stability (Fig. 6B and C). This mutation also had a minor but significant improvement in E. coli tolerance to bile salts but not to styrene. Finally, the amino acid F617 has been shown to be located in the switch loop of the hydrophobic binding pocket. Reported crystal structures suggest that this amino acid could directly interact with various known substrates (Bohnert et al., 2008; Eicher et al., 2012; Vargiu et al., 2011). Mutating F617 to an alanine has been reported to have a direct impact on substrate uptake and was responsible for a substantial decrease in transport of novobiocin, but had a minor effect on the transport of oxacillin and various other macrolides (Bohnert et al., 2008). In our study, the mutation to a leucine at this position may have also improved transport of 1-hexene.
Our study, as well as other reports in the literature (Bohnert et al., 2008) suggest that AcrB is destabilized when more than three mutations are simultaneously introduced. However, additional rounds of evolution, starting with variants containing fewer mutations, could stabilize the protein and allow the introduction of more mutations to further improve function. Heterologous pumps from other organisms have been shown to bestow solvent tolerance in E. coli (Dunlop et al., 2011) and may be good candidates for future studies as well as directed evolution. Use of dynamic control systems to regulate transporter expression has been shown to enhance the final tolerance phenotype (Frederix et al., 2014) and provides another engineering route to explore. Additionally, analyses of more AcrB variants with increased tolerance to 1-hexene could provide a better understanding of the path(s) of this compound through the pump, allowing targeted engineering of the protein. Directed evolution of AcrB has also been conducted by other groups. AcrB variants with improved tolerance to n-butanol (Fisher et al., 2013) as well as α-pinene and n-octane are known (Foo and Leong, 2013), and the authors propose the improvements to have resulted from changes in the AcrB-TolC interaction, in the enlargement of the entrance to the cleft, in the facilitation of conformational changes or in the improvement of affinity for the substrate. Taken together with these previous studies, our results show that, within this pump's complex mode of action, several domains can be mutated and lead to a more efficient pump. Our study emphasizes the importance of an AcrAB-TolC type system for improved tolerance and export in host engineering, and shows that directed evolution remains a powerful tool to obtain improved variants of such systems.
We thank Huu Tran for his skillful technical assistance with the liquid handler robot, Dominque Loque and Fan Yang for providing the A. thaliana cDNA, James Kirby for help designing the GC-MS method, Thomas Ruegg for the Omnilog utilization, Marijke Frederix for helpful discussion and advice on various aspects of this study, and Nathan Hillson and Daniel Liu for reviewing the manuscript. Zosia Rostomian prepared Figure 1, 6B and C. The portion of the work conducted by the Lawrence Berkeley National Laboratory and the Joint BioEnergy Institute (JBEI) was supported by the U.S. DOE under Contract no. DE-AC02-05CH11231. This work, funded by Total, was performed as part of a collaborative program between JBEI and Total.
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology 2:2006 0008.
Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences 97(12):6640-6645.
Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6(5):343-5.McKenna R, Nielsen DR. 2011. Styrene biosynthesis from glucose by engineered E. coli. Metabolic engineering 13(5):544-54.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert JA, Schuster S, Seeger MA, Fahnrich E, Pos KM, Kern WV. Site-directed mutagenesis reveals putative substrate binding residues in the Escherichia coli RND efflux pump AcrB. J Bacteriol. 2008;190(24):8225–8229. doi: 10.1128/JB.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Ling H, Chang MW. Transporter engineering for improved tolerance against alkane biofuels in Saccharomyces cerevisiae. Biotechnol Biofuels. 2013;6(1):21. doi: 10.1186/1754-6834-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi R, Nguyen T, Chang G. Transporter-mediated biofuel secretion. Proc Natl Acad Sci USA. 2013;110(19):7642–7647. doi: 10.1073/pnas.1301358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509(7501):512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher T, Cha HJ, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, Kern WV, Verrey F, Grutter MG, Diederichs K. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci USA. 2012;109(15):5687–5692. doi: 10.1073/pnas.1114944109. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MA, Boyarskiy S, Yamada MR, Kong N, Bauer S, Tullman-Ercek D. Enhancing tolerance to short-chain alcohols by engineering the Escherichia coli AcrB efflux pump to secrete the non-native substrate n-butanol. ACS Synth Biol. 2013;3(1):30–40. doi: 10.1021/sb400065q. [DOI] [PubMed] [Google Scholar]
- Foo JL, Leong SS. Directed evolution of an E. coli inner membrane transporter for improved efflux of biofuel molecules. Biotechnol Biofuels. 2013;6(1):81. doi: 10.1186/1754-6834-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederix M, Hutter K, Leu J, Batth TS, Turner WJ, Ruegg TL, Blanch HW, Simmons BA, Adams PD, Keasling JD. Development of a native Escherichia coli induction system for ionic liquid tolerance. PLoS ONE. 2014;9(7):e101115. doi: 10.1371/journal.pone.0101115. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Ham TS, Dmytriv Z, Plahar H, Chen J, Hillson NJ, Keasling JD. Design, implementation and practice of JBEI-ICE: An open source biological part registry platform and tools. Nucleic Acids Res. 2012;40:e141. doi: 10.1093/nar/gks531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci USA. 2012;109(41):16696–16701. doi: 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain F, Bikhchandani M, Nikaido H. Vestibules are part of the substrate path in the multidrug efflux transporter AcrB of Escherichia coli. J Bacteriol. 2011;193(20):5847–5849. doi: 10.1128/JB.05759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain F, Nikaido H. Substrate path in the AcrB multidrug efflux pump of Escherichia coli. Mol Microbiol. 2010;78(2):320–330. doi: 10.1111/j.1365-2958.2010.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarboe LR, Zhang X, Wang X, Moore JC, Shanmugam KT, Ingram LO. Metabolic engineering for production of biorenewable fuels and chemicals: Contributions of synthetic biology. J Biomed Biotechnol. 2010;2010:761042. doi: 10.1155/2010/761042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J Biol Eng. 2011;5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna R, Nielsen DR. Styrene biosynthesis from glucose by engineered E. coli. Metab Eng. 2011;13(5):544–554. doi: 10.1016/j.ymben.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419(6907):587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H, Takatsuka Y. Mechanisms of RND multidrug efflux pumps. Biochimica et biophysica acta. 2009;1794(5):769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A. Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol. 2002;56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- Schubert P, Steinbuchel A, Schlegel HG. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol. 1988;170(12):5837–5847. doi: 10.1128/jb.170.12.5837-5847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313(5791):1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, Diederichs K, Pos KM. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struc Mol Biol. 2008;15(2):199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5(1):e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkema J, de Bont JA, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59(2):201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CC, Li M, Gu R, Takatsuka Y, McDermott G, Nikaido H, Yu EW. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J Bacteriol. 2006;188(20):7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka Y, Chen C, Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci USA. 2010;107(15):6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol. 2009;191(6):1729–1737. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka Y, Nikaido H. Site-directed disulfide cross-linking to probe conformational changes of a transporter during its functional cycle: Escherichia coli AcrB multidrug exporter as an example. Method Mol Biol. 2010;634:343–354. doi: 10.1007/978-1-60761-652-8_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Yamada Y, Zgurskaya HI. Sequential mechanism of assembly of multidrug efflux pump AcrAB-TolC. Chem Biol. 2011;18(4):454–463. doi: 10.1016/j.chembiol.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: An ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1(1):107–125. [PubMed] [Google Scholar]
- Vargiu AV, Collu F, Schulz R, Pos KM, Zacharias M, Kleinekathofer U, Ruggerone P. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: Explanation and rationale by molecular dynamics simulations. J Am Chem Soc. 2011;133(28):10704–10707. doi: 10.1021/ja202666x. [DOI] [PubMed] [Google Scholar]
- Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109(50):20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Baars L, Ytterberg AJ, Klussmeier A, Wagner CS, Nord O, Nygren PA, van Wijk KJ, de Gier JW. Consequences of membrane protein overexpression in Escherichia coli. Mol Cell Proteom. 2007;6(9):1527–1550. doi: 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- Yu L, Lu W, Wei Y. AcrB trimer stability and efflux activity, insight from mutagenesis studies. PloS ONE. 2011;6(12):e28390. doi: 10.1371/journal.pone.0028390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


