Abstract
Objective
To our knowledge, there is no broad genomic analysis comparing skin and synovium in psoriatic arthritis (PsA). Also, there is little understanding of the relative levels of cytokines and chemokines in skin and synovium. The purpose of this study was to better define inflammatory pathways in paired lesional skin and affected synovial tissue in patients with PsA.
Methods
We conducted a comprehensive analysis of cytokine and chemokine activation and genes representative of the inflammatory processes in PsA. Paired PsA synovial tissue and skin samples were obtained from 12 patients on the same day. Gene expression studies were performed using Affymetrix HGU133 Plus 2.0 arrays. Confirmatory quantitative real-time polymerase chain reaction (PCR) was performed on selected transcripts. Cell populations were assessed by immunohistochemistry and immunofluorescence.
Results
Globally, gene expression in PsA synovium was more closely related to gene expression in PsA skin than to gene expression in synovium in other forms of arthritis. However, PsA gene expression patterns in skin and synovium were clearly distinct, showing a stronger interleukin-17 (IL-17) gene signature in skin than in synovium and more equivalent tumor necrosis factor (TNF) and interferon-γ gene signatures in both tissues. These results were confirmed with real-time PCR.
Conclusion
This is the first comprehensive molecular comparison of paired lesional skin and affected synovial tissue samples in PsA. Our results support clinical trial data showing that PsA skin and joint disease are similarly responsive to TNF antagonists, while IL-17 antagonists have better results in PsA skin than in PsA joints. Genes selectively expressed in PsA synovium might direct future therapies for PsA.
Psoriatic arthritis (PsA) is an inflammatory joint disease associated with psoriasis. Up to 30% of patients with psoriasis develop PsA (1). The pathogeneses of both the skin disease and the joint inflammation of PsA are not well defined. Early studies designated psoriasis and PsA as Th1-mediated diseases with a focus on interferon-γ (IFNγ) and interleukin-2 (IL-2) (2). More recent studies identify IL-17 as the most critical cytokine for sustaining skin disease, with important interactions between IL-17 and tumor necrosis factor (TNF) within skin cells (3,4). IL-17 has also been implicated in PsA, with an increased number of Th17 cells in the peripheral blood, synovial fluid, and synovial tissue of PsA patients (5–7). In addition, synoviocytes of PsA patients show increased expression of IL-17 receptor (IL-17R) compared with the synoviocytes of patients with osteoarthritis (OA) (7).
There is little understanding of the relative levels of cytokines and chemokines within skin and synovium in PsA. Moreover, to our knowledge, there is no broad genomic analysis comparing skin and synovium in PsA. The purpose of this study was to better define the inflammatory pathways of PsA in both skin and joint pathogenesis in matched lesional skin and affected synovial tissue specimens in patients with PsA. We conducted a comprehensive analysis of the cytokine and chemokine activation that defines Th1, Th2, Th9, Th22, and Th17 T cell subsets as well as genes representative of the inflammatory processes that are seen in psoriatic skin and joint disease. Our results establish marked within-patient differences in gene expression between lesional skin and affected synovium in PsA patients. Specifically, IL-17 expression is significantly higher in skin than in synovium, while IL-6 expression is higher in synovium.
Patients and Methods
Twelve patients (10 women and 2 men) who fulfilled the Moll and Wright criteria for PsA (8) were enrolled at the Arthritis Treatment Center, Frederick, MD (see Supplementary Table 1, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract). All patients had active inflammatory arthritis and active psoriatic skin lesions, were negative for rheumatoid factor, and received stable doses of medications including nonsteroidal antiinflammatory drugs and methotrexate. Patients receiving biologic agents, such as TNF inhibitors, were excluded unless they had undergone a therapeutic washout for at least 2 weeks. Samples of lesional psoriatic skin tissue and synovial tissue from inflamed joints were obtained from the same patient on the same day. The study was approved by the Institutional Review Board of The Rockefeller University, and all patients gave informed and written consent to participate in the study. The study was performed in accordance with the ethics principles of the Declaration of Helsinki. We chose not to use 6 scalp samples because our laboratory has described significant differences in gene expression between scalp and nonscalp samples mostly due to hair follicle–related genes (Suárez-Fariñas M, Krueger JG: unpublished observations).
Arthroscopy and skin biopsy
Synovial tissue samples from the most actively inflamed and accessible joint were recovered using arthroscopically guided synovial biopsy procedures. Psoriatic lesional skin was obtained from all patients by 4-mm punch biopsies. All tissue samples were snap-frozen in liquid nitrogen, stored at −80°C, and shipped on dry ice to the Laboratory of Investigative Dermatology at The Rockefeller University.
RNA extraction and microarray
One nanogram of total RNA was subjected to 2-cycle complementary DNA synthesis according to the Affymetrix protocol with a slight modification (9). Labeling of complementary RNA transcripts with biotin was performed using a GeneChip IVT Labeling kit (Affymetrix). Fifteen micrograms of biotin-labeled RNA was fragmented and hybridized to HGU133 Plus 2.0 arrays (Affymetrix), washed, stained, and scanned according to the manufacturer's protocol.
Quantitative real-time polymerase chain reaction (PCR)
The preamplification quantitative real-time PCR technique was used for measuring various genes in total RNA according to the manufacturer's protocol (Applied Biosystems) and as described previously (10). The sequences of primers and probes (Applied Biosystems) used in this study are shown in Supplementary Table 2, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract.
Immunohistochemistry and immunofluorescence
Immunohistochemical and immunofluorescence analyses were performed on cryostat tissue sections (4 PsA skin samples and 4 PsA synovium samples) with the antibodies listed in Supplementary Table 3 (available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract) as previously described (10).
Statistical analysis
Quality control of microarray data was conducted using a Harshlight package (http://asterion.rockefeller.edu/Harshlight/index2.html) (11) and ArrayQuality Control packages in R (http://www.r-project.org/). A GC-RMA algorithm was used to calculate expression values (12). Expression values across diseases for skin and synovium were adjusted by organ using a linear model. Changes in gene expression were modeled using mixed-effects models, which simultaneously account for across-group differences and within-patient correlation across skin–synovium samples. Hypotheses of interest were tested in an R Limma package framework using contrasts. P values were adjusted for multiple hypotheses using the Benjamini-Hochberg approach, which controls the false discovery rate (FDR). Contrasts were also used to obtain the transcriptomics estimates adjusted by organ.
Clustering
Unsupervised clustering of expression profiles for differentially expressed genes was carried out using Pearson's correlation and an average agglomeration algorithm.
Gene set variation analysis (GSVA)
To summarize per-patient differences across pathways, we also used GSVA (13). GSVA is a per-sample generalization of commonly used gene set enrichment analysis in which a pathway/gene set score is produced for each sample, thus allowing statistical modeling of functional pathways as done in gene-based analysis. GSVA estimates variation of a set of genes over the sample population. It uses a Z score, which is a statistical measurement of a score's relationship to the mean in a group of scores.
Upstream Regulator Analysis
An Ingenuity Pathway Analysis (IPA) (Ingenuity Systems; http://www.ingenuity.com) Upstream Regulator Analysis tool was used to identify upstream regulators that may be responsible for expression changes in the data set.
Results
Global comparison of transcriptional profiles of PsA skin and synovium
To gain a global view of the gene expression profiles of the PsA skin samples and PsA synovium samples, a principal components plot was used in which we also included normal skin, normal synovium, rheumatoid arthritis (RA) synovium, OA synovium, and systemic lupus erythematosus (SLE) synovium gene sets obtained from GEO (GEO accession nos. GSE7307 and GSE14195) and previously reported data (14,15). The skin and synovium are inherently different organs and the profiles were clearly separate (see Supplementary Figure 1, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract).
We then adjusted for organ-specific genes, which allowed us to focus on disease-specific genes by comparing diseased tissue with normal tissue from the same organ. Gene expression profiles in PsA skin (the remaining 6 nonscalp samples), PsA synovium, normal skin, normal synovium, RA synovium, OA synovium, and SLE synovium gene sets were again visualized on a principal components plot (Figure 1). There were many differences in gene expression data between the different forms of arthritis. For example, one clear difference between RA synovium and PsA synovium was the greater B cell signature in RA (Belasco J, Suárez-Fariñas M, Krueger JG: unpublished observations). Gene expression in PsA synovium was much more closely related to gene expression in PsA skin than to gene expression in synovium in other forms of arthritis, after adjustment for organ (skin and synovium) differences. However, even after accounting for organ-specific genes, while the skin and synovium of PsA patients were closely related, they were clearly distinct, showing disease-related differences. When we directly compared PsA skin with PsA synovium (prior to adjustment for organ differences), 2,590 probe sets were up-regulated in PsA skin and 3,057 probe sets were down-regulated in PsA skin. After adjusting for organ-specific differences, we compared the PsA skin and PsA synovium transcriptomes and still found many differentially expressed genes, with 809 probe sets up-regulated in PsA skin and 2,008 probe sets down-regulated in PsA skin (see Supplementary Table 4, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract).
Figure 1.

Principal components analysis showing that gene expression in psoriatic arthritis (PsA) synovium was much more closely related to gene expression in PsA skin than to gene expression in synovium in other forms of arthritis, after adjustment for organ (skin and synovium) differences. The principal components plot shows gene expression profiles in lesional skin from PsA patients, synovium from PsA patients, synovium from osteoarthritis (OA) patients, synovium from rheumatoid arthritis (RA) patients, synovium from systemic lupus erythematosus (SLE) patients, skin from healthy controls, and synovium from healthy controls after adjustment for organ differences. Lines connecting samples of skin and synovium from PsA patients indicate matched pairs. While skin and synovium from PsA patients are closely related, they are clearly distinct even after accounting for organ-specific genes, showing that there are disease-related differences between the 2 tissues. PC = principal component.
We also compared PsA synovium with normal synovium (the PsA synovium transcriptome), PsA skin with normal skin (the PsA skin transcriptome), and the PsA synovium transcriptome with the PsA skin transcriptome (Supplementary Table 4 and Figure 2A). It must be noted that even if probe sets are “shared” by both groups, the expression is not necessarily present in the same direction or magnitude. Our organ difference–adjusted comparison of the PsA skin and PsA synovium transcriptomes accounts for this as described in Patients and Methods. For example, integrin beta 8 (ITGB8), a shared gene, had a fold change of 4.99 in the PsA synovium transcriptome but a fold change of −3.76 in the PsA skin transcriptome. This indicates a different direction of differential expression between the transcriptomes. Kynureninase (KYNU), also a shared gene, had a fold change of 4.59 in the PsA synovium transcriptome and a fold change of 21.56 in the PsA skin transcriptome. This indicates the same direction of differential expression but a different magnitude. Another shared gene, TGFBR1, had a fold change of 2.22 in the PsA skin transcriptome and a fold change of 2.04 in the PsA synovium transcriptome, indicating the same direction and magnitude of differential expression between the transcriptomes.
We generated a heatmap of the top 50 shared genes with the same direction and similar magnitude of differential expression between the PsA synovium transcriptome and the PsA skin transcriptome (Figure 2B). Figure 2C shows a heatmap of all the genes that were shared but had a different magnitude of differential expression between these transcriptomes. Lesional skin from patients with psoriasis vulgaris was included as a positive control for disease activity in PsA skin. Normal skin, normal synovium, and lesional skin gene sets from patients with psoriasis vulgaris were obtained from previous reports or GEO (GEO accession no. GSE7307) (15,16).
Figure 2.
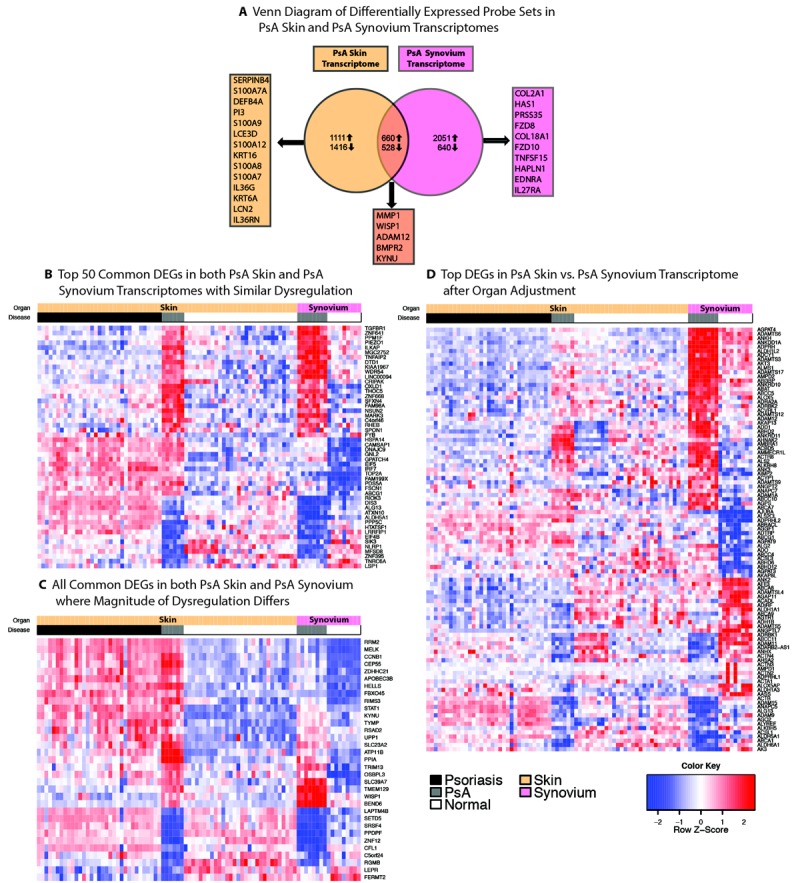
A, Venn diagram showing numbers of differentially expressed probe sets in psoriatic arthritis (PsA) skin (PsA skin versus normal skin) and PsA synovium (PsA synovium versus normal synovium) transcriptomes prior to adjustment for organ differences. Genes listed in boxes indicate biologically relevant genes in the top 60 differentially expressed genes (DEGs) (fold change of >2, false discovery rate [FDR] of <0.05) uniquely or commonly expressed by each tissue. B, Heatmap showing the top 50 genes shared by PsA skin and PsA synovium transcriptomes in which no difference in dysregulation is observed. C, Heatmap showing all the genes shared by PsA skin and PsA synovium transcriptomes in which the magnitude of dysregulation differs between skin and synovium (fold change of >2, FDR of <0.05). D, Heatmap comparing the 50 most up-regulated and 50 most down-regulated genes in PsA skin versus PsA synovium after adjustment for organ-specific genes. Heatmaps show PsA synovium (n = 12), normal synovium (n = 9), PsA skin (n = 6), psoriatic lesional skin (n = 33), and normal skin (n = 30) tissue samples.
To further highlight the major differences in the transcriptomes of these diseases, we generated a heatmap of the genes most differentially expressed (after adjustment for organ differences) between PsA skin and PsA synovium (using normal synovium and lesional skin from patients with psoriasis vulgaris as controls) (Figure 2D). Overall, the pattern of gene expression in PsA skin lesions was similar to that in psoriasis patients without arthritis. Even after adjustment for organ differences, the top genes included genes that were intrinsic to skin and that were highly up-regulated inflammatory genes. Many were keratinocyte specific, such as S100 calcium binding protein A7A (S100A7A), S100A9, lipocalin 2 (LCN2), and involucrin (IVL). Some were immunologically active genes often seen at epithelial barriers such as interleukin 36 gamma (IL36G) and interleukin 36 receptor antagonist (IL36RN), which are known to be increased in psoriasis (17). A small number of genes were highly up-regulated in PsA synovium but were overall down-regulated or showed no change in PsA skin, psoriatic skin, normal skin, or normal synovium. These included genes specific to synovium such as Frizzled family receptor 8 (FZD8), cytokine-like 1 (CYTL1), and collagen type II, alpha 1 (COL2A1); these genes may indicate the destruction and repair that are occurring in this tissue (18–21).
Gene expression in PsA skin
Many of the top genes differentially expressed in PsA skin compared with normal skin were terminal differentiation genes such as S100A7A, S100A9, S100A8, and S100A7 (fold change of >19) as well as peptidase inhibitor 3 (PI3) and small proline-rich protein 2C (SPRR2C) with fold changes of 74 and 24, respectively (see Supplementary Table 5, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract). Defensin, beta 4A (DEFB4A), a psoriasis hallmark gene, was increased, with a fold change of 80. In addition, IL36G, IL36RN, IL19, LCN2, and CXCL1 were all increased, with fold changes of >2.
Gene expression in PsA synovium
The top genes differentially expressed in PsA synovium compared with normal synovium were structural in nature and related to formation and breakdown of joint tissue. These included matrix metalloproteinase 1 (MMP1), COL2A1, WNT1 inducible signaling pathway protein 1 (WISP1), hyaluronan synthase I (HAS1), integrin-binding sialoprotein (IBSP), FZD8, and bone morphogenetic protein receptor, type II (BMPR2). In addition, collagen type XVIII alpha 1 (COL18A1) (an antiangiogenic protein) and coagulation factor V (F5) were among the top 50 differentially expressed genes (22) (see Supplementary Table 6, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract).
Analysis of disease-related transcripts in skin versus synovium by functional pathways
Our gene lists were subsequently interrogated using IPA Upstream Regulator Analysis. TNF was predicted to be an upstream regulator in both synovium and skin; however, TNF was much more strongly activated in the PsA synovium transcriptome (Z = 3.767) than in the PsA skin transcriptome (Z = 0.540). IFNG was also predicted to be activated in both synovium and skin (Z = 2.099 and Z = 2.293, respectively). IL17-related upstream regulators were not detected in synovium, while IL17R was activated in skin (Z = 2.795) (Figure 3A). VEGF and TGFB1, factors involved in increased vascularization (23), were activated upstream regulators in synovium (Z = 3.938 and Z = 2.047, respectively) but not in skin. WNT3A, which is antiosteoclastogenic (18), was predicted to be an upstream regulator only in synovium (Z = 2.379). Upstream Regulator Analysis also identified several transcription regulators (see Supplementary Tables 7 and 8, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract). The transcription factor v-ets erythroblastosis virus E26 oncogene homolog 1 (ETS1) was predicted to be activated only in synovium and not in skin. Ets-1 regulates genes involved in development, differentiation, and cell proliferation including angiogenesis, cartilage regeneration, and inhibition of Th17 cell development (24,25).
Figure 3.
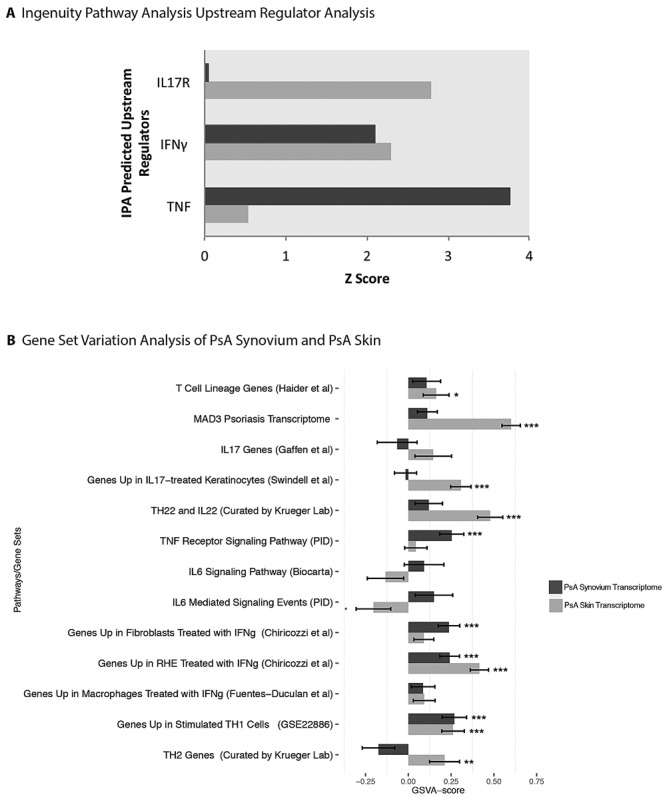
A, Ingenuity Pathway Analysis (IPA) Upstream Regulator Analysis showing that IL17 upstream regulators are present only in skin, while IFNγ and TNF are present in both skin and synovium. B, Gene set variation analysis (GSVA) of curated gene sets in psoriatic arthritis (PsA) synovium (PsA synovium versus normal synovium) and PsA skin (PsA skin versus normal skin) transcriptomes. Values are the mean ± SD of the Z score of the given set of genes. P values indicate significance or a trend of each transcriptome (PsA synovium or PsA skin) independently. ∗ = P < 0.1; ∗∗ = P < 0.05; ∗∗∗ = P < 0.01. IL-17 = interleukin-17; TNF = tumor necrosis factor; IFNγ = interferon-γ; RHE = reconstructed human epidermis.
To evaluate the differences between the PsA skin transcriptome and the PsA synovium transcriptome at the pathway level, we used GSVA (Figure 3B). We evaluated relative genomic scores within various gene sets that represent T cells, T cell axes, epidermis, keratinocytes, macrophages, fibroblasts, and curated groups of genes that we have used in prior studies or retrieved from the Broad Institute database (26–32). Overall, both PsA skin and PsA synovium had an IFNγ gene signature. While the IL-6 signaling pathway was not significantly expressed in either transcriptome, there was a trend toward positive IL-6 signaling in synovium but not in skin. The TNF receptor signaling pathway was significantly expressed only in synovium and was also expressed, although not significantly, in skin. The Th22 cell/IL-22 and IL-17 axes were only significantly positive in the skin and the IL-17 axes trended toward being negative in the synovium.
Analysis of inflammatory gene expression by real-time PCR
Gene arrays provide a means to analyze complex expression pathways, but their limited sensitivity does not allow reliable quantification of most primary cytokine transcripts. We used real-time PCR as a more sensitive measure and to further compare the immunologic similarities and differences between PsA skin and PsA synovium (Figure 4). Expression of messenger RNA (mRNA) for many cytokines and chemokines was observed, but this expression did not differ significantly between psoriatic skin and synovium as determined by real-time PCR. Messenger RNA was expressed for IFNγ (defining Th1 cells), IL-4 and IL-13 (defining Th2 cells), IL-22 (defining Th22 cells), IL-9 (defining Th9 cells), and FoxP3 (defining Treg cells). Messenger RNA for TNFα, IL-1β, IL-8, and matrix metalloproteinase 1 (MMP-1) was also expressed, but this expression did not differ significantly between psoriatic skin and synovium. However, expression of mRNA for IL-6 and CXCL2 was significantly increased in synovium compared with lesional skin (P ≤ 0.05), and the gene DEFB4A and mRNA for IL-1α were significantly increased in skin compared with synovium (P < 0.0005). Of the Th17-polarizing cytokines, mRNA for IL-23A was present in both skin and synovium with no significant difference in expression; expression of mRNA for both IL-17A and IL-17F was significantly higher in skin than in synovium (P = 0.01 and P = 0.001, respectively).
Figure 4.

Confirmation of microarray results by quantifying mRNA expression of biologically significant genes by real-time polymerase chain reaction and normalizing expression values to the housekeeping gene hARP. There is significant elevation of mRNA for interleukin-17A (IL-17A) and IL-17F in psoriatic lesional skin compared with inflamed psoriatic synovium (P = 0.01 and P = 0.001, respectively). In addition, the gene DEFB4A and mRNA for IL-1α are significantly increased in psoriatic skin compared with affected synovium (both P < 0.0005). Messenger RNA for IL-6 and CXCL2 is significantly increased in synovium (P < 0.05 and P = 0.05, respectively). Values are the mean ± SD. ∗ = P ≤ 0.05; ∗∗∗ = P ≤ 0.001. IFNγ = interferon-γ; IL-2RA = IL-2 receptor antagonist; TNFα = tumor necrosis factor α; MMP-1 = matrix metalloproteinase 1.
To verify that the low levels of cytokines or the absence of cytokines based on microarray data were not due to a lack of inflammatory cells, immunohistochemistry was performed to identify T cells (CD3+), macrophages (CD163+), and myeloid dendritic cells (DCs) (CD11c+) (see Supplementary Figure 2A, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract). The staining indicated that T cells were present in both skin and synovium, although there seemed to be fewer T cells in synovium. Macrophages appeared abundant in both skin and synovium. Myeloid DCs were also present in both tissues, but fewer myeloid DCs were present in synovium. Immunofluorescence performed with anti-CD11c and anti-TNF showed greater overall CD11c in skin than in synovium, and TNF appeared to be present in skin and synovium in relatively equal amounts (see Supplementary Figure 2B). Many CD11c+ DCs expressed high levels of TNF, as reported previously for inflammatory DCs in psoriatic lesions (33).
Immunohistochemistry was also performed for IL-17, IL-17R, and IL-6 (see Supplementary Figure 3, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.38995/abstract). There was generally more IL-17 and IL-17R staining in PsA skin than in PsA synovium. Also, there was generally more IL-17R staining than IL-17 staining in both PsA skin and PsA synovium. In PsA skin, IL-17 staining appeared more localized to the upper regions of the epidermis (stratum granulosum), while IL-17R staining was panepidermal. IL-6 staining was apparent at similar levels in both skin and synovium.
Discussion
To our knowledge, this study provides the first comprehensive genomic and molecular comparison of matched lesional skin and affected synovium in PsA. Our global gene expression analysis indicates that synovial tissue in PsA is much more similar to psoriatic lesional skin than to synovial tissue in other forms of arthritis such as RA, OA, and SLE. Given this information, the idea that treatments of other forms of inflammatory arthritis can be equally effective in PsA is likely flawed. This analysis clarifies the importance of differentiating PsA from other forms of arthritis.
One might think that there are similar pathomechanisms of disease between the involved skin and synovium in PsA. However, our comparative gene array analysis revealed that thousands of genes are differentially expressed between paired skin and synovium in PsA. Skin and synovium are very different organs, and it could be predicted that there would be numerous differences in gene expression. However, even with adjustment for organ-specific genes, there is still a great deal of differential gene expression.
Indeed, our study showed that there are clear differences in inflammatory gene expression between skin and synovium. We found a strong IL-17–related gene signature in skin relative to synovium. Many up-regulated genes in skin were IL-17 signature genes. For example, the genes S100A7, S100A8, and S100A9 have been shown to be up-regulated in vitro by IL-22, IL-17, or both cytokines (29,34–36). PI3 and SPRR2C were found to be IL-17 signature genes in IL-17–induced keratinocytes (4). IL-17A induces IL-36 cytokines, and IL-36 and IL-17 are synergistic for the production of S100A7 and TNFα (37). IL-17 promotes expression of CXCL1 in epithelial cell populations such as keratinocytes and synoviocytes (4,38,39). IL19, DEFB4A, LCN2, and CCL20 are also known to be classic IL-17 target genes (4,28).
The top differentially expressed genes in synovium were not generally related to an IL-17 gene signature. Instead, many of the top genes and predicted upstream regulators were related to cartilage and bone breakdown and formation (MMP1, COL2A1, WISP1, HAS1, IBSP, FZD8, BMPR2, WNT3A) or the angiogenesis that is present in PsA (COL18A1, F5, VEGF, TGFB1) (23). IL-17 can induce MMPs, but very few other IL-17–related genes were expressed in psoriatic or normal synovium, and expression of mRNA for MMP-1 was not significantly different between psoriatic skin and synovium by real-time PCR (28). Indeed, the expression of all the detectable IL-17–related genes in PsA synovium was not significantly different from that in normal synovium. For example, there were no significant differences in CXCL1, S100A7A, and LCN2, which are all considered to be IL-17–regulated products (28).
IPA identified IL17R as an upstream regulator for the PsA skin versus normal skin gene set, but not for the PsA synovium versus normal synovium gene set. In addition, GSVA showed a general trend for IL-17–related pathways to be more highly expressed in skin than in synovium, which further supports our IPA findings. It should be noted that many of the curated gene sets used for GSVA were developed in skin. However, the gene sets developed in skin contain many genes seen in a variety of tissues. In general, a strong signal can override the few genes that are specific for skin. For instance, even in gene sets developed in skin for IFNγ and TNF, it is possible to see a significantly elevated genomic score in synovium compared with skin. When possible, we included gene sets curated from connective tissue such as fibroblasts, and we also included the Gaffen gene set (28), which more generally includes genes from various cells and conditions. The Gaffen IL-17 gene set showed a positive trend in skin and a negative trend in synovium. As further confirmation of a stronger IL-17 gene signature in skin, mRNA for IL-17A and IL-17F was significantly elevated in skin compared with synovium by real-time PCR. Our data point to an overall stronger IL-17 gene signature in PsA skin, reflecting the IL-17 activation pathway that is seen and well understood in psoriasis in general.
Another upstream regulator identified by IPA only in PsA synovium was ETS1, a transcription factor involved in angiogenesis, cartilage regeneration, and inhibition of Th17 cell development (24,25). Overexpression of ETS1 has been shown to increase COL2A1 promoter activity. We found a significant increase in COL2A1 (fold change of 10.48) in PsA synovium compared with normal synovium. In addition, Ets-1 plays a role in angiogenesis and is associated with increased expression of VEGF (40). In our study, VEGF was predicted to be an activated upstream regulator in synovium but not in skin. It is possible that the decreased level of mRNA for IL-17 in PsA synovium compared with PsA skin is due to the inhibition of Th17 cell differentiation by an increase in Ets-1 in T cells in the synovium. This hypothesis needs further confirmation.
Unlike IL-17, TNF appears to have a more similar gene signature in skin and synovium. TNF was predicted by IPA to be an upstream regulator in both skin and synovium, and GSVA also suggested that TNF may play a role in both organs. In addition, expression of mRNA for TNFα was not significantly different between skin and synovium by real-time PCR. TNF is not transcriptionally regulated, so analysis of downstream genes is essential. IL1 is downstream of TNF and was up-regulated in both skin and synovium, while expression of IL6 (regulated by bioactive IL1) was even higher in synovium.
It should be considered that we compared only skin and synovium in our study. Enthesitis and bone erosion/proliferation play important roles in PsA (41,42). It is possible that our findings would be further elucidated by also analyzing entheses and bone in a similar group of patients. If IL-17 contributes to arthritic inflammation as suggested by some recent studies, the effect might be mediated by high levels of IL-17 in the circulation with effects on target cells that are not in synovial tissues (e.g., osteoclasts or other inflammatory cells at joint entheses) (5,6). Our immunohistochemical staining does show a higher amount of IL-17R compared with IL-17. This may indicate that there are many receptors on cells in the synovium, but not a great deal of IL-17. This suggests the potential for IL-17 effects in synovium, with the proviso that IL-17 is coming from the circulation or other nearby cells. In addition, IL-23 appears to be expressed in both skin and synovium. Again, it is possible that IL-23 could be produced and influence Th17 cells outside the synovium and that we are just not seeing a synovial response. Also, it is possible that TNF antagonists may work by modulating circulating IL-17.
In addition to comparing only skin and synovium in this study, there are other limitations that should be considered. Our sample size was relatively small, and a larger confirmatory study would be worthwhile. Also, the severity of skin lesions was not addressed, as we did not use any index of lesional severity at the time of recruitment. However, our group has investigated thin-plaque and thick-plaque psoriasis, and the gene profile is not very different across severity (Suárez-Fariñas M, Krueger JG: unpublished observations). Also, we included lesional skin from patients with moderate-to-severe psoriasis vulgaris as a positive control. Even if PsA lesional skin had less extensive involvement than typical psoriatic lesions, the characteristics of PsA lesional skin seem similar overall to those of psoriatic lesional skin. The top differentially expressed genes in the gene expression profile we present in the PsA skin transcriptome are very typical for psoriasis in general and include terminal differentiation genes and the psoriasis hallmark gene DEFB4A. Also, we were not able to make in-depth comparisons of many of the tissues mentioned in our report as this was outside the scope of this study. For example, we included data on psoriatic lesional skin, but we did not provide a thorough description of the comparison of psoriatic lesional skin and PsA lesional skin. This comparison deserves a deeper evaluation and will be addressed in future research. For the purposes of this study, psoriatic lesional skin was included only as a positive control for skin disease activity.
Our findings indicate that treating both skin lesions and arthritis with the same modalities may show different patterns of efficacy between the skin and the joint. Although TNF antagonists are somewhat effective treatments for both psoriasis without arthritis and skin/joint manifestations of PsA, treatment failures occur in at least 30–40% of the overall PsA population, and alternative treatments are needed (43,44). IL-17 has emerged as the most critical cytokine for sustaining skin disease. In psoriasis vulgaris trials, the emerging IL-17 antagonists yielded response rates that reflect virtual elimination of disease (45,46). IL-12/IL-23 antagonists, which ultimately suppress IL-17 signaling, show marked lesional skin improvement in psoriasis but less robust results for PsA joint symptoms (47–49). It is not clear whether IL-17 will play a major role in PsA. The lower responses of PsA to ustekinumab and brodalumab (compared with TNF antagonists) seem to predict that this cytokine will not have the dominant role in arthritis that it has in controlling the skin phenotype (47,50). In addition, some clinical trials have shown flares of arthritis in a small number of PsA patients whose skin disease was well controlled with ustekinumab (51,52).
Our findings of a relatively stronger IL-6 gene signature in synovium may point toward another target in PsA. GSVA showed a trend toward positive expression of IL-6 pathways in synovium and negative expression in skin. In addition, real-time PCR further confirmed our GSVA IL-6 pathway findings, with mRNA for IL-6 significantly increased in synovium compared with skin. Tocilizumab, a humanized monoclonal antibody directed against the IL-6 receptor, has been used in a small number of PsA patients with mixed results (53,54). A larger study would be needed to show more definitive results.
Our data provide insight into the outcomes of the most recent clinical trials of TNF antagonists, IL-12/IL-23 antagonists, and IL-17 antagonists in psoriasis and PsA. These clinical trials make it clear that the better the match between pathogenic molecules and therapeutic antagonists, the more effective the therapeutic agent may be. In PsA this is complicated by the differences in immunopathology of the skin and joint symptoms. We believe that our current expectations for treatment of PsA need to be reconsidered in light of emerging data on the treatment of psoriasis without arthritis. IL-17 appears to be a critical molecule for driving psoriatic skin disease. An equivalent pivotal molecule remains to be identified for PsA joint disease.
Author Contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Belasco had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Louie, Wei, Nograles, Suárez-Fariñas, Krueger.
Acquisition of data. Belasco, Louie, Wei, Nograles, Fuentes-Duculan, Krueger.
Analysis and interpretation of data. Belasco, Gulati, Fuentes-Duculan, Mitsui, Suárez-Fariñas, Krueger.
Role of the Study Sponsor
Amgen had no role in the study design or in the collection, analysis, or interpretation of the data, the writing of the manuscript, or the decision to submit the manuscript for publication. Publication of this article was not contingent upon approval by Amgen.
References
- 1.Zachariae H. Prevalence of joint disease in patients with psoriasis: implications for therapy. Am J Clin Dermatol. 2003;4:441–7. doi: 10.2165/00128071-200304070-00001. [DOI] [PubMed] [Google Scholar]
- 2.Cauli A, Mathieu A. Th17 and interleukin 23 in the pathogenesis of psoriatic arthritis and spondyloarthritis. J Rheumatol. 2012;89(Suppl):15–8. doi: 10.3899/jrheum.120234. [DOI] [PubMed] [Google Scholar]
- 3.Nograles KE, Davidovici B, Krueger JG. New insights in the immunologic basis of psoriasis. Semin Cutan Med Surg. 2010;29:3–9. doi: 10.1016/j.sder.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiricozzi A, Guttman-Yassky E, Suarez-Farinas M, Nograles KE, Tian S, Cardinale I. Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol. 2011;131:677–87. doi: 10.1038/jid.2010.340. [DOI] [PubMed] [Google Scholar]
- 5.Jandus C, Bioley G, Rivals JP, Dudler J, Speiser D, Romero P. Increased numbers of circulating polyfunctional Th17 memory cells in patients with seronegative spondylarthritides. Arthritis Rheum. 2008;58:2307–17. doi: 10.1002/art.23655. [DOI] [PubMed] [Google Scholar]
- 6.Leipe J, Grunke M, Dechant C, Reindl C, Kerzendorf U, Schulze-Koops H. Role of Th17 cells in human autoimmune arthritis. Arthritis Rheum. 2010;62:2876–85. doi: 10.1002/art.27622. [DOI] [PubMed] [Google Scholar]
- 7.Raychaudhuri SP, Raychaudhuri SK, Genovese MC. IL-17 receptor and its functional significance in psoriatic arthritis. Mol Cell Biochem. 2012;359:419–29. doi: 10.1007/s11010-011-1036-6. [DOI] [PubMed] [Google Scholar]
- 8.Moll JM, Wright V. Psoriatic arthritis. Semin Arthritis Rheum. 1973;3:55–78. doi: 10.1016/0049-0172(73)90035-8. [DOI] [PubMed] [Google Scholar]
- 9.Mitsui H, Suarez-Farinas M, Belkin DA, Levenkova N, Fuentes-Duculan J, Coats I. Combined use of laser capture microdissection and cDNA microarray analysis identifies locally expressed disease-related genes in focal regions of psoriasis vulgaris skin lesions. J Invest Dermatol. 2012;132:1615–26. doi: 10.1038/jid.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulati N, Suarez-Farinas M, Fuentes-Duculan J, Gilleaudeau P, Sullivan-Whalen M, Correa da Rosa J. Molecular characterization of human skin response to diphencyprone at peak and resolution phases: therapeutic insights. J Invest Dermatol. 2014;134:2531–40. doi: 10.1038/jid.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suarez-Farinas M, Pellegrino M, Wittkowski KM, Magnasco MO. Harshlight: a “corrective make-up” program for microarray chips. BMC Bioinformatics. 2005;6:294. doi: 10.1186/1471-2105-6-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu ZI, Rafael A, Gentleman R, Murillo FM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–17. [Google Scholar]
- 13.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nzeusseu Toukap A, Galant C, Theate I, Maudoux AL, Lories RJ, Houssiau FA. Identification of distinct gene expression profiles in the synovium of patients with systemic lupus erythematosus. Arthritis Rheum. 2007;56:1579–88. doi: 10.1002/art.22578. [DOI] [PubMed] [Google Scholar]
- 15.Yao Y, Richman L, Morehouse C, de los Reyes M, Higgs BW, Boutrin A. Type I interferon: potential therapeutic target for psoriasis? PLoS One. 2008;3:e2737. doi: 10.1371/journal.pone.0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman-Yassky E, Suarez-Farinas M, Chiricozzi A, Nograles KE, Shemer A, Fuentes-Duculan J. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J Allergy Clin Immunol. 2009;124:1235–44.e58. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 17.Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med. 2007;204:2603–14. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albers J, Keller J, Baranowsky A, Beil FT, Catala-Lehnen P, Schulze J. Canonical Wnt signaling inhibits osteoclastogenesis independent of osteoprotegerin. J Cell Biol. 2013;200:537–49. doi: 10.1083/jcb.201207142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenberg J, Ruetschi U, Skioldebrand E, Karrholm J, Lindahl A. Quantitative proteomics reveals regulatory differences in the chondrocyte secretome from human medial and lateral femoral condyles in osteoarthritic patients. Proteome Sci. 2013;11:43. doi: 10.1186/1477-5956-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oldberg A, Franzen A, Heinegard D. The primary structure of a cell-binding bone sialoprotein. J Biol Chem. 1988;263:19430–2. [PubMed] [Google Scholar]
- 21.Arheden K, Mandahl N, Heim S, Mitelman F. In situ hybridization localizes the human type II α1 collagen gene (COL2A1) to 12q13. Hereditas. 1989;110:165–7. doi: 10.1111/j.1601-5223.1989.tb00437.x. [DOI] [PubMed] [Google Scholar]
- 22.O'Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–85. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 23.Fearon U, Griosios K, Fraser A, Reece R, Emery P, Jones PF. Angiopoietins, growth factors, and vascular morphology in early arthritis. J Rheumatol. 2003;30:260–8. [PubMed] [Google Scholar]
- 24.Peng H, Tan L, Osaki M, Zhan Y, Ijiri K, Tsuchimochi K. ESE-1 is a potent repressor of type II collagen gene (COL2A1) transcription in human chondrocytes. J Cell Physiol. 2008;215:562–73. doi: 10.1002/jcp.21338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007;204:2825–35. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haider AS, Lowes MA, Suarez-Farinas M, Zaba LC, Cardinale I, Blumenberg M. Cellular genomic maps help dissect pathology in human skin disease. J Invest Dermatol. 2008;128:606–15. doi: 10.1038/sj.jid.5701067. [DOI] [PubMed] [Google Scholar]
- 27.Tian S, Krueger JG, Li K, Jabbari A, Brodmerkel C, Lowes MA. Meta-analysis derived (MAD) transcriptome of psoriasis defines the “core” pathogenesis of disease. PLoS One. 2012;7:e44274. doi: 10.1371/journal.pone.0044274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen F, Gaffen SL. Structure-function relationships in the IL-17 receptor: implications for signal transduction and therapy. Cytokine. 2008;41:92–104. doi: 10.1016/j.cyto.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nograles KE, Zaba LC, Guttman-Yassky E, Fuentes-Duculan J, Suarez-Farinas M, Cardinale I. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159:1092–102. doi: 10.1111/j.1365-2133.2008.08769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiricozzi A, Nograles KE, Johnson-Huang LM, Fuentes-Duculan J, Cardinale I, Bonifacio KM. IL-17 induces an expanded range of downstream genes in reconstituted human epidermis model. PloS One. 2014;9:e90284. doi: 10.1371/journal.pone.0090284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swindell WR, Johnston A, Voorhees JJ, Elder JT, Gudjonsson JE. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics. 2013;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowes MA, Chamian F, Abello MV, Fuentes-Duculan J, Lin SL, Nussbaum R. Increase in TNF-α and inducible nitric oxide synthase-expressing dendritic cells in psoriasis and reduction with efalizumab (anti-CD11a) Proc Natl Acad Sci U S A. 2005;102:19057–62. doi: 10.1073/pnas.0509736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boniface K, Bernard FX, Garcia M, Gurney AL, Lecron JC, Morel F. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol. 2005;174:3695–702. doi: 10.4049/jimmunol.174.6.3695. [DOI] [PubMed] [Google Scholar]
- 35.Gutowska-Owsiak D, Schaupp AL, Salimi M, Taylor S, Ogg GS. Interleukin-22 downregulates filaggrin expression and affects expression of profilaggrin processing enzymes. Br J Dermatol. 2011;165:492–8. doi: 10.1111/j.1365-2133.2011.10400.x. [DOI] [PubMed] [Google Scholar]
- 36.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–9. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrier Y, Ma HL, Ramon HE, Napierata L, Small C, O'Toole M. Inter-regulation of Th17 cytokines and the IL-36 cytokines in vitro and in vivo: implications in psoriasis pathogenesis. J Invest Dermatol. 2011;131:2428–37. doi: 10.1038/jid.2011.234. [DOI] [PubMed] [Google Scholar]
- 38.Zrioual S, Toh ML, Tournadre A, Zhou Y, Cazalis MA, Pachot A. IL-17RA and IL-17RC receptors are essential for IL-17A-induced ELR+ CXC chemokine expression in synoviocytes and are overexpressed in rheumatoid blood. J Immunol. 2008;180:655–63. doi: 10.4049/jimmunol.180.1.655. [DOI] [PubMed] [Google Scholar]
- 39.Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF) Nat Immunol. 2011;12:853–60. doi: 10.1038/ni.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oettgen P. The role of Ets factors in tumor angiogenesis. J Oncol. 2010;2010:767384. doi: 10.1155/2010/767384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGonagle D, Gibbon W, Emery P. Classification of inflammatory arthritis by enthesitis. Lancet. 1998;352:1137–40. doi: 10.1016/S0140-6736(97)12004-9. [DOI] [PubMed] [Google Scholar]
- 42.McGonagle D, Lories RJ, Tan AL, Benjamin M. The concept of a “synovio-entheseal complex” and its implications for understanding joint inflammation and damage in psoriatic arthritis and beyond. Arthritis Rheum. 2007;56:2482–91. doi: 10.1002/art.22758. [DOI] [PubMed] [Google Scholar]
- 43.Mease PJ, Goffe BS, Metz J, VanderStoep A, Finck B, Burge DJ. Etanercept in the treatment of psoriatic arthritis and psoriasis: a randomised trial. Lancet. 2000;356:385–90. doi: 10.1016/S0140-6736(00)02530-7. [DOI] [PubMed] [Google Scholar]
- 44.Mease PJ, Gladman DD, Ritchlin CT, Ruderman EM, Steinfeld SD, Choy EH Adalimumab Effectiveness in Psoriatic Arthritis Trial Study Group. Adalimumab for the treatment of patients with moderately to severely active psoriatic arthritis: results of a double-blind, randomized, placebo-controlled trial. Arthritis Rheum. 2005;52:3279–89. doi: 10.1002/art.21306. et al, for the. [DOI] [PubMed] [Google Scholar]
- 45.Papp KA, Leonardi C, Menter A, Ortonne JP, Krueger JG, Kricorian G. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N Engl J Med. 2012;366:1181–9. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- 46.Leonardi C, Matheson R, Zachariae C, Cameron G, Li L, Edson-Heredia E. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N Engl J Med. 2012;366:1190–9. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 47.Gottlieb A, Menter A, Mendelsohn A, Shen YK, Li S, Guzzo C. Ustekinumab, a human interleukin 12/23 monoclonal antibody, for psoriatic arthritis: randomised, double-blind, placebo-controlled, crossover trial. Lancet. 2009;373:633–40. doi: 10.1016/S0140-6736(09)60140-9. [DOI] [PubMed] [Google Scholar]
- 48.Papp KA, Langley RG, Sigurgeirsson B, Abe M, Baker DR, Konno P. Efficacy and safety of secukinumab in the treatment of moderate-to-severe plaque psoriasis: a randomized, double-blind, placebo-controlled phase II dose-ranging study. Br J Dermatol. 2013;168:412–21. doi: 10.1111/bjd.12110. [DOI] [PubMed] [Google Scholar]
- 49.McInnes IB, Sieper J, Braun J, Emery P, van der Heijde D, Isaacs JD. Efficacy and safety of secukinumab, a fully human anti-interleukin-17A monoclonal antibody, in patients with moderate-to-severe psoriatic arthritis: a 24-week, randomised, double-blind, placebo-controlled, phase II proof-of-concept trial. Ann Rheum Dis. 2014;73:349–56. doi: 10.1136/annrheumdis-2012-202646. [DOI] [PubMed] [Google Scholar]
- 50.Mease PJ, Genovese MC, Greenwald MW, Ritchlin CT, Beaulieu AD, Deodhar A. Brodalumab, an anti-IL17RA monoclonal antibody, in psoriatic arthritis. N Engl J Med. 2014;370:2295–306. doi: 10.1056/NEJMoa1315231. [DOI] [PubMed] [Google Scholar]
- 51.De Souza A, Ali-Shaw T, Reddy SM, Fiorentino D, Strober BE. Inflammatory arthritis following ustekinumab treatment for psoriasis: a report of two cases. Br J Dermatol. 2013;168:210–2. doi: 10.1111/j.1365-2133.2012.11206.x. [DOI] [PubMed] [Google Scholar]
- 52.Stamell EF, Kutner A, Viola K, Cohen SR. Ustekinumab associated with flares of psoriatic arthritis. JAMA Dermatol. 2013;149:1410–3. doi: 10.1001/jamadermatol.2013.5728. [DOI] [PubMed] [Google Scholar]
- 53.Hughes M, Chinoy H. Successful use of tocilizumab in a patient with psoriatic arthritis. Rheumatology (Oxford) 2013;52:1728–9. doi: 10.1093/rheumatology/kes432. [DOI] [PubMed] [Google Scholar]
- 54.Ogata A, Umegaki N, Katayama I, Kumanogoh A, Tanaka T. Psoriatic arthritis in two patients with an inadequate response to treatment with tocilizumab. Joint Bone Spine. 2012;79:85–7. doi: 10.1016/j.jbspin.2011.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


