Abstract
Introduction
More than 70 common alleles are known to be involved in breast cancer (BC) susceptibility, and several exhibit significant heterogeneity in their associations with different BC subtypes. Although there are differences in the association patterns between BRCA1 and BRCA2 mutation carriers and the general population for several loci, no study has comprehensively evaluated the associations of all known BC susceptibility alleles with risk of BC subtypes in BRCA1 and BRCA2 carriers.
Methods
We used data from 15,252 BRCA1 and 8,211 BRCA2 carriers to analyze the associations between approximately 200,000 genetic variants on the iCOGS array and risk of BC subtypes defined by estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and triple-negative- (TN) status; morphologic subtypes; histological grade; and nodal involvement.
Results
The estimated BC hazard ratios (HRs) for the 74 known BC alleles in BRCA1 carriers exhibited moderate correlations with the corresponding odds ratios from the general population. However, their associations with ER-positive BC in BRCA1 carriers were more consistent with the ER-positive associations in the general population (intraclass correlation (ICC) = 0.61, 95% confidence interval (CI): 0.45 to 0.74), and the same was true when considering ER-negative associations in both groups (ICC = 0.59, 95% CI: 0.42 to 0.72). Similarly, there was strong correlation between the ER-positive associations for BRCA1 and BRCA2 carriers (ICC = 0.67, 95% CI: 0.52 to 0.78), whereas ER-positive associations in any one of the groups were generally inconsistent with ER-negative associations in any of the others. After stratifying by ER status in mutation carriers, additional significant associations were observed. Several previously unreported variants exhibited associations at P <10−6 in the analyses by PR status, HER2 status, TN phenotype, morphologic subtypes, histological grade and nodal involvement.
Conclusions
Differences in associations of common BC susceptibility alleles between BRCA1 and BRCA2 carriers and the general population are explained to a large extent by differences in the prevalence of ER-positive and ER-negative tumors. Estimates of the risks associated with these variants based on population-based studies are likely to be applicable to mutation carriers after taking ER status into account, which has implications for risk prediction.
Electronic supplementary material
The online version of this article (doi:10.1186/s13058-014-0492-9) contains supplementary material, which is available to authorized users.
Introduction
Women who carry pathogenic mutations in BRCA1 or BRCA2 have markedly increased risks of developing breast cancer. The distributions of breast cancer tumor characteristics differ between BRCA1 mutation carriers, BRCA2 mutation carriers and those arising in the general population. The majority of breast tumors arising in BRCA1 carriers show low or absent expression of estrogen receptor (ER) [1]-[3], whereas the majority of BRCA2-associated tumors are ER-positive [1],[4],[5].
Many common breast cancer susceptibility alleles identified through population-based genome-wide association studies (GWASs) have also been associated with breast cancer risk in BRCA1 and BRCA2 carriers [6],[7]. Several of these variants are specifically associated with the ER status of the breast cancer in the general population [8],[9]. Among the single-nucleotide polymorphisms (SNPs) that have been evaluated in mutation carriers so far, the variants found to be associated with breast cancer risk for BRCA1 carriers largely overlap with loci for which stronger associations with ER-negative breast cancer have been reported in the general population [8]-[12]. An important question for risk modelling and prevention studies is whether the effects of common variants on breast cancer risk in mutation carriers are mediated through a generic influence on the development of particular hormone receptor subtypes of breast cancer or through epistatic interaction with the BRCA1/2 mutation itself.
Previous studies by the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) described the impact of 29 breast cancer susceptibility variants from non-hereditary breast cancer studies on ER-positive and ER-negative breast cancer risk in BRCA1 and BRCA2 carriers [6],[7],[13]-[15]. These analyses demonstrated that, despite the lack of an association between some susceptibility variants and overall breast cancer risk for BRCA1 or BRCA2 carriers, residual associations exist with specific disease subtypes. In addition, the ER-specific associations in BRCA1 and BRCA2 carriers were mainly in the same direction and of a magnitude similar to the associations observed with breast cancer stratified by ER expression status in the general population. However, these studies were conducted on smaller numbers of mutation carriers than currently available and evaluated only a subset of the currently known breast cancer susceptibility alleles for their associations with ER-specific subtypes in carriers. Recently, 45 additional SNPs have been found to be associated with breast cancer risk in the general population [8]-[10],[16]. Eighteen of these SNPs showed evidence of association with ER-positive breast cancer, but not with ER-negative breast cancer, and four loci (1q32.1 LGR6, 2p24.1, 16q12 and 20q11) were associated only with ER-negative breast cancer in the general population. These 45 newly discovered loci have not yet been evaluated for their associations with breast cancer risk for mutation carriers.
In the present study, we assessed the disease subtype-specific associations of all 74 previously reported breast cancer susceptibility variants in 15,252 BRCA1 and 8,211 BRCA2 carriers. We evaluated whether differences in associations of known breast cancer susceptibility variants between BRCA1 carriers, BRCA2 carriers and the general population are mediated by tumor ER status in mutation carriers. We also analyzed the associations of about 200,000 variants on the iCOGS genotyping array with subtype-specific breast cancer risk in carriers in an attempt to uncover previously unreported subtype-specific associations in women with BRCA1 and BRCA2 mutations. In addition to ER and progesterone receptor (PR) status, we report, for the first time to our knowledge, associations by HER2 status and with triple-negative disease (TN, referring to ER-, PR- and human epidermal growth factor receptor 2 (HER2)-negative), and we also describe associations with clinical features such as “ductal, no specified subtype” (hereafter referred to as ductal) and lobular morphologic subtypes, nodal status and histological grade.
Methods
Study subjects
Data were obtained from 47 studies in 27 different countries in CIMBA [17]. Eligible study subjects were women who carry pathogenic mutations in BRCA1 or BRCA2. The majority were recruited through cancer genetics clinics and enrolled into national or regional studies. Written informed consent was obtained from all subjects. Each of the host institutions recruited under ethically approved protocols. A list of the local institutional review boards that provided ethical approval for this study is given in Additional file 1: Table S1. Eligibility was restricted to mutation carriers who were 18 years of age or older at recruitment. Data collected included year of birth, age at cancer diagnosis, personal history of bilateral prophylactic mastectomy and/or bilateral salpingo-oophorectomy, mutation description, tumor pathology and ethnicity.
Tumor pathology data
Breast tumor pathology data were gathered from a range of sources, specifically patient pathology reports, pathology review data, tumor registry records and tissue microarray results. These included information on ER, PR and HER2 status; morphologic subtype; lymph node involvement; and histological grade. For ER, PR and HER2, status was classified as negative or positive, with supplementary immunohistochemistry scoring or biochemical data and methodology provided when available. The vast majority of centers employed a cutoff of either ≥10% or ≥1% of tumor nuclei staining positive to define ER and PR positivity. Additional file 1: Table S2 lists the subtype definitions used by each study, which were not centrally reclassified, owing to the low proportion of records with supporting staining data. Similarly, HER2 status was determined using immunohistochemistry to detect strong complete membrane staining (with a score of 3+ considered positive) and/or in situ hybridization to detect HER2 gene amplification. To ensure consistency across studies, when information on the cells stained was available, we used the same cutoff to define ER-, PR- and HER2-positive tumors. The cutoffs used for the small number of cases where composite scoring methods based on the proportion and intensity of staining were available (Allred score, immunoreactive Remmele score) are given in Additional file 1: Table S2. Consistency checks were performed to validate receptor data against supplementary scoring information, if provided. Each cancer was assigned to a morphologic subgroup (ductal, lobular, medullary, other), which we confirmed using the World Health Organization International Classification of Diseases for Oncology (ICD-O) code for the classification of tumor type when sufficient information was provided [18]. Lymph node status, along with the number of nodes showing metastatic carcinoma, was provided when available. Histologic grade was assigned as grade 1, 2 or 3 by local pathologists who used a modified Scarff-Bloom-Richardson malignancy grading system.
Genotyping and quality control
Genotyping was carried out using the iCOGS custom array. The array development and details of the genotyping and quality control for the CIMBA samples are described in detail elsewhere [6],[7]. Briefly, genotyping for BRCA2 carriers was conducted at McGill University and Génome Québec Innovation Centre (Canada) and for BRCA1 carriers at the Mayo Clinic (USA). SNPs were excluded if they were located on the Y chromosome, if they were monomorphic, if they deviated significantly from Hardy-Weinberg equilibrium (P <10−7) or if they had call rates <95%. Samples were excluded if they had a call rate <95%, if they were of non-European ancestry or if they demonstrated extreme heterozygosity. After quality control, we had 200,720 SNPs available for analysis in 15,252 BRCA1 samples and 200,908 SNPs available for analysis in 8,211 BRCA2 samples.
Statistical methods
We evaluated the associations of each genotype with risks of developing breast cancer or breast cancer subtypes defined by the tumor characteristics or morphology. The analyses were carried out within a survival analysis framework. Individuals were censored at the first of the following events: breast cancer diagnosis, ovarian cancer diagnosis, bilateral mastectomy or age at last follow-up. In order to account for non-random ascertainment of mutation carriers with respect to their disease phenotype, we used a retrospective likelihood approach that models the probability of observing the genotypes conditional on the disease phenotype [19],[20]. It was assumed that the cancer incidence depends on the underlying SNP genotype through a Cox proportional hazards model:
where λ0(t i) is the baseline incidence and β is the logarithm of the per-allele hazard ratio (HR, under a multiplicative model). The association with overall breast cancer risk was evaluated by testing the hypothesis that β = 0 [20].
We evaluated the associations with the groups of each subtype class (for example, ER-positive and ER-negative), using an extension of the retrospective likelihood approach to model the simultaneous effect of each SNP on more than one tumor subtype [15]. Briefly, this involves modeling the conditional likelihood of the observed SNP genotypes and tumor subtypes, given the disease phenotypes. Within this framework, it is possible to estimate simultaneously the HRs for each tumor subtype and test for heterogeneity in the associations [15]. To maximize the available information, genotyped mutation carriers that were missing information on tumor characteristics were included in the analysis, and their disease subtype was assumed to be missing at random. In order to account for non-independence among relatives, a robust variance estimation approach was used [20]. Further details of the methods for evaluating the associations with overall breast cancer [20] and tumor subtypes have been described elsewhere [15]. We carried out association analyses by subtype for the following breast cancer characteristics: ER-positive and ER-negative, PR-positive and PR-negative, HER2-positive and HER2-negative, TN breast cancer (that is, negative for ER, PR and HER2) and non-TN (that is, tumor positive for at least one of the three receptors), ductal morphologic subtype, lobular morphologic subtype, nodal involvement (no involved lymph nodes and at least one involved lymph node) and histological grade (high grade (grade 3) and non-high grade (grades 1 and 2)). Only samples with complete information on ER, PR and HER2 expression were included in the analysis for TN as well as non-TN breast cancer. The SNP associations by tumor morphologic subtype were evaluated by comparing ductal tumors to all others and, in a separate analysis, lobular tumors to all others. We are not reporting association analyses for risk of medullary morphologic subtype, owing to sparse data as well as the difficulties in diagnosing medullary breast tumors reliably [21],[22]. All analyses were stratified by country of residence. The United States and Canada strata were further subdivided by reported Ashkenazi Jewish ancestry. For subtypes with small groups, strata of geographically close countries were combined to provide sufficiently large groups for estimation. All analyses used calendar year– and cohort-specific cancer incidences for BRCA1 and BRCA2. SNPs with minor allele frequencies <3% were excluded. The retrospective likelihood was modeled using custom-written functions implemented in the pedigree analysis software MENDEL [23].
When evaluating whether the known breast cancer susceptibility loci identified through population based studies also modify breast cancer risk in mutation carriers, a significance threshold of P <0.05 was used because of the strong prior evidence of association for these loci with disease risk. For the association analyses of all the approximately 200,000 variants on the iCOGS array with the breast cancer subtypes in mutation carriers, only associations with P < 5 × 10−8 were considered significant. The discussion of findings and the tables were extended to associations at P <10−6.
For variants associated with ER-positive or ER-negative breast cancer with P <0.01, we evaluated whether the associations may have been affected by a possible survival bias due to inclusion of prevalent breast cancer cases in the analysis. For the sensitivity analysis, the association analysis by ER status was repeated after excluding mutation carriers diagnosed with breast cancer ≥5 years prior to study recruitment.
We evaluated the consistency between the breast cancer association estimates of previously reported breast cancer susceptibility variants in the general population (using published data) and the association estimates in BRCA1 and BRCA2 carriers using the intraclass correlation (ICC). We estimated ICC as outlined by Shrout and Fleiss [24] based on a one-way random-effects model and tested for agreement in absolute values of log HR. The same approach was used to evaluate the agreement between associations with ER-positive and/or -negative breast cancer in the general population and associations with ER-positive and/or -negative breast cancer in BRCA1 and in BRCA2 carriers. Furthermore, we carried out the same comparisons between associations for BRCA1 and associations for BRCA2 carriers.
Results
Subtype patterns
The analyses included data from 15,252 BRCA1 carriers and 8,211 BRCA2 carriers. Among the breast cancer–affected BRCA1 carriers, we had data on at least one disease characteristic of interest for 4,619 (59%) of the 7,797 affected women (Table 1). Data were available on tumor characteristics for 2,570 (59%) of the 4,330 affected BRCA2 carriers. Of the individuals with pathology information, 74% of the BRCA1 carriers and 75% of the BRCA2 carriers had data on ER status.
Table 1.
Breast tumor characteristics of 7,797 affected BRCA1 mutation carriers and 4,330 affected BRCA2 mutation carriers a
| BRCA1 mutation carriers | BRCA2 mutation carriers | |||||
|---|---|---|---|---|---|---|
| Yes, n (%) | No, n (%) | Unknown status | Yes, n (%) | No, n (%) | Unknown status | |
| Predictive markers | ||||||
| ER-positive | 819 (24) | 2,639 (76) | 4,339 | 1,490 (77) | 434 (23) | 2,406 |
| PR-positive | 662 (21) | 2,485 (79) | 4,650 | 1,099 (65) | 591 (35) | 2,640 |
| HER2-positive | 182 (9) | 1,816 (91) | 5799 | 121 (13) | 847 (87) | 3,362 |
| Non-TN | 580 (31) | 1,310 (69) | 5907 | 760 (85) | 136 (15) | 3,434 |
| Morphology | 3,789 | 2,087 | ||||
| Ductal | 3,159 (82) | 1,770 (79) | ||||
| Lobular | 89 (2) | 188 (8) | ||||
| Medullary | 290 (6) | 39 (2) | ||||
| Other | 470 (10) | 246 (11) | ||||
| Grade | 4,645 | 3,840 | ||||
| Grade 1 | 81 (3) | 113 (7) | ||||
| Grade 2 | 574 (18) | 700 (42) | ||||
| Grade 3 | 2,497 (79) | 839 (51) | ||||
| Nodal involvement | 1,103 (33) | 2,274 (67) | 4,420 | 804 (43) | 1,068 (57) | 2,458 |
| Stage | 5,991 | 3,382 | ||||
| Stage 0b | 65 (4) | 121 (13) | ||||
| Stage 1 | 825 (46) | 327 (35) | ||||
| Stage 2 | 772 (43) | 390 (41) | ||||
| Stage 3 | 127 (7) | 96 (10) | ||||
| Stage 4 | 17 (1) | 14 (1) | ||||
aER, Estrogen receptor positive; HER2, Human epidermal growth factor receptor 2; PR, Progesterone receptor; TN, Triple-negative. bCarcinoma in situ.
Single-nucleotide polymorphism associations
After quality control, genotype data were available for analysis for 200,720 SNPs for BRCA1 carriers and for 200,908 SNPs for BRCA2 carriers. After adjusting for sample size and excluding SNPs chosen for inclusion on the genotyping array based on reported associations in subsets of the current sample, the inflation coefficient λ1000 values were 1.01 for ER-positive disease in BRCA1 carriers, 1.02 for ER-negative in BRCA1, 1.01 for ER-positive in BRCA2 and 1.02 for ER-negative in BRCA2 carriers (Additional file 1: Figure S1 and Figure S2). Similar patterns were observed for other tumor characteristics (results not shown). After excluding variants located at known breast cancer susceptibility loci, there was no evidence for an excess in associations by ER status beyond the number expected.
Associations of previously reported breast cancer susceptibility loci
Associations with overall breast cancer and by tumor estrogen receptor status
First, we considered the associations with risk for overall breast cancer and for tumor subtypes for the 74 breast cancer susceptibility variants that have been reported up to April 2013. In light of the strong prior evidence of association, we considered associations at P <0.05 as evidence that a previously reported breast cancer susceptibility allele also modifies overall or ER-specific breast cancer risk in mutation carriers. The associations with overall breast cancer risk and risk of breast cancer subtypes for all 74 variants are given in Table 2 and Additional file 1: Tables S4 to S10. Of the breast cancer susceptibility loci that had not previously been evaluated for an association in mutation carriers, SNPs at 5q33.3, 8q24.21, 11q24.3, 12q22, 16q12.1, 22q13.1 were associated with overall breast cancer risk for BRCA1 carriers, and SNPs at 6p23, 11q24.3 and 16q12.1 were associated with breast cancer risk for BRCA2 carriers at P <0.05 (Table 2). Overall, 15 breast cancer susceptibility variants were associated with ER-negative breast cancer in BRCA1 carriers and 8 variants in BRCA2 carriers at P <0.05 (Table 2). Ten significant associations with ER-positive breast cancer in BRCA1 carriers and fourteen in BRCA2 carriers were found. The strongest association with ER-positive breast cancer was observed for rs2981579 in FGFR2 at 10q26.12 for both BRCA1 and BRCA2 carriers. SNP rs10069690 in TERT at 5p15.33 displayed the strongest association with ER-negative breast cancer for BRCA1 carriers and rs9348512 at 6p24.3 for BRCA2 carriers. We found significant differences in the associations by ER status for rs3803662 in TOX3 at 16q12.1 (P = 2 × 10−4) and rs13387042 at 2q35 (P =0.002) for BRCA1 carriers, which were not previously seen. Both SNPs showed evidence of association with ER-positive breast cancer only. Similarly, six of the loci that did not show evidence of association with overall breast cancer were associated with ER-positive and two with ER-negative breast cancer in BRCA1 carriers. This included two of the loci not previously evaluated in mutation carriers: 3q26.1 and 6p25.3. In BRCA2 carriers, four of the variants lacking evidence of association with overall breast cancer were associated with ER-negative and three with ER-positive breast cancer. This included four loci not previously evaluated in mutation carriers: 2q24, 14q13.3, 19q13.31 and 22q12.2. Of the breast cancer susceptibility loci that had not yet been evaluated for an association with breast cancer in mutation carriers, rs1011970 at CDKN2A/B and rs1292011 at 12q24.21 had significantly different associations with ER-positive and ER-negative cancer for BRCA1 carriers (P het = 0.009 and P het = 0.004, respectively, for the difference between ER-positive and ER-negative). SNP rs2236007 at 14q13.3 displayed differences by ER status for BRCA2 carriers (P het = 0.008). These three SNPs had associations in different directions for ER-positive and ER-negative tumors.
Table 2.
Associations of susceptibility loci with overall and estrogen receptor-positive and -negative breast cancer a
| BRCA1 carriers | BRCA2 carriers | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | ER-negative | ER-positive | Overall | ER-negative | ER-positive | |||||||||||||||||
| Locus | SNP | Position b | Nearby gene | Ref c | Eff d | K e | MAF | HR (95% CI) | P -value | HR (95% CI) | P -value | HR (95% CI) | P -value | P het-value f | MAF | HR (95% CI) | P -value | HR (95% CI) | P -value | HR (95% CI) | P -value | P het-value f |
| 1p36.22 | rs616488 | 10566215 | PEX14 | A | G | No | 0.32 | 0.96 (0.92 to 1.01) | 0.10 | 0.95 (0.90 to 1.00) | 0.07 | 1.00 (0.90 to 1.10) | 0.92 | 0.47 | 0.33 | 0.98 (0.92 to 1.04) | 0.52 | 1.01 (0.88 to 1.16) | 0.94 | 0.97 (0.9 to 1.05) | 0.46 | 0.69 |
| 1p13.2 | rs12022378 | 114448389 | SYT6 | G | A | No | 0.16 | 1.03 (0.98 to 1.09) | 0.25 | 1.03 (0.97 to 1.10) | 0.31 | 1.03 (0.90 to 1.17) | 0.68 | 0.94 | 0.17 | 1.03 (0.95 to 1.12) | 0.42 | 1.16 (0.97 to 1.37) | 0.10 | 1.00 (0.91 to 1.1) | 1.00 | 0.15 |
| 1p11.2 | rs11249433 | 121280613 | FCGR1B | A | G | Yes | 0.41 | 0.99 (0.95 to 1.04) | 0.78 | 0.99 (0.94 to 1.03) | 0.55 | 1.02 (0.93 to 1.13) | 0.63 | 0.50 | 0.41 | 1.05 (0.99 to 1.12) | 0.09 | 1.04 (0.91 to 1.19) | 0.57 | 1.06 (0.99 to 1.13) | 0.12 | 0.84 |
| 1q32.1a | rs6678914 | 202187176 | LGR6 | G | A | No | 0.4 | 0.98 (0.94 to 1.02) | 0.39 | 0.96 (0.91 to 1.01) | 0.08 | 1.06 (0.97 to 1.17) | 0.21 | 0.07 | 0.41 | 1.04 (0.98 to 1.10) | 0.19 | 0.94 (0.82 to 1.08) | 0.39 | 1.07 (1.00 to 1.15) | 0.05 | 0.11 |
| 1q32.1b | rs4245739 | 204518842 | MDM4 | A | C | Yes | 0.28 | 1.10 (1.05 to 1.15) | 4.6 × 10 −5 | 1.12 (1.07 to 1.19) | 1.4 × 10 −5 | 1.02 (0.91 to 1.13) | 0.76 | 0.11 | 0.28 | 0.97 (0.91 to 1.04) | 0.38 | 1.09 (0.95 to 1.26) | 0.21 | 0.94 (0.87 to 1.01) | 0.10 | 0.07 |
| 2p24.1 | rs12710696 | 19320803 | OSR1 | G | A | No | 0.39 | 1.01 (0.97 to 1.06) | 0.51 | 1.01 (0.97 to 1.07) | 0.57 | 1.01 (0.92 to 1.12) | 0.77 | 1.00 | 0.38 | 1.00 (0.94 to 1.06) | 0.99 | 1.14 (1.00 to 1.30) | 0.05 | 0.96 (0.9 to 1.03) | 0.28 | 0.02 |
| 2q14.2 | rs4849887 | 121245122 | G | A | No | 0.11 | 1.02 (0.96 to 1.09) | 0.53 | 1.01 (0.94 to 1.09) | 0.78 | 1.05(0.90 to 1.23) | 0.49 | 0.64 | 0.11 | 0.98 (0.89 to 1.08) | 0.72 | 1.08 (0.87 to 1.33) | 0.49 | 0.96 (0.85 to 1.07) | 0.43 | 0.34 | |
| 2q31.1 | rs2016394 | 172972971 | DLX2 | G | A | No | 0.47 | 1.01 (0.97 to 1.06) | 0.54 | 1.03 (0.98 to 1.08) | 0.23 | 0.96 (0.87 to 1.06) | 0.39 | 0.21 | 0.46 | 0.99 (0.93 to 1.05) | 0.74 | 1.07 (0.94 to 1.22) | 0.32 | 0.97 (0.9 to 1.04) | 0.36 | 0.21 |
| 2q31.1 | rs1550623 | 174212894 | CDCA7 | A | G | No | 0.15 | 1.01 (0.95 to 1.07) | 0.72 | 1.02 (0.95 to 1.09) | 0.54 | 0.97 (0.85 to 1.12) | 0.72 | 0.57 | 0.15 | 0.97 (0.89 to 1.06) | 0.49 | 1.03 (0.87 to 1.23) | 0.70 | 0.95 (0.87 to 1.05) | 0.31 | 0.41 |
| 2q35 | rs13387042 | 217905832 | TNP1 | A | G | Yes | 0.47 | 0.98 (0.94 to 1.02) | 0.41 | 1.02 (0.97 to 1.07) | 0.37 | 0.86 (0.78 to 0.95) | 1.9 × 10 −3 | 2.3 × 10 −3 | 0.48 | 0.99 (0.93 to 1.05) | 0.68 | 0.98 (0.87 to 1.12) | 0.82 | 0.99 (0.93 to 1.06) | 0.74 | 0.96 |
| 2q35 | rs16857609 | 218296508 | DIRC3 | G | A | No | 0.26 | 1.04 (1.00 to 1.09) | 0.06 | 1.03 (0.98 to 1.09) | 0.23 | 1.08 (0.97 to 1.21) | 0.14 | 0.47 | 0.27 | 0.96 (0.90 to 1.03) | 0.26 | 0.87 (0.74 to 1.01) | 0.08 | 0.99 (0.92 to 1.07) | 0.82 | 0.14 |
| 3p26.1 | rs6762644 | 4742276 | ITPR1 | A | G | No | 0.36 | 1.04 (0.99 to 1.08) | 0.09 | 1.00 (0.95 to 1.05) | 0.92 | 1.16 (1.05 to 1.28) | 2.9 × 10 −3 | 0.01 | 0.37 | 0.98 (0.93 to 1.05) | 0.61 | 1.08 (0.94 to 1.24) | 0.27 | 0.96 (0.89 to 1.03) | 0.24 | 0.13 |
| 3p24.1 | rs4973768 | 27416013 | SLC4A7 | G | A | Yes | 0.49 | 1.01 (0.97 to 1.06) | 0.47 | 0.99 (0.95 to 1.04) | 0.81 | 1.09 (0.99 to 1.19) | 0.07 | 0.10 | 0.5 | 1.08 (1.02 to 1.15) | 7.8 × 10 −3 | 1.05 (0.92 to 1.20) | 0.46 | 1.10 (1.02 to 1.18) | 0.01 | 0.58 |
| 3p24.1 | rs12493607 | 30682939 | TGFBR2 | C | G | No | 0.35 | 0.99 (0.95 to 1.04) | 0.73 | 0.99 (0.94 to 1.04) | 0.63 | 1.01 (0.92 to 1.11) | 0.85 | 0.70 | 0.34 | 0.98 (0.92 to 1.04) | 0.54 | 1.07 (0.93 to 1.22) | 0.35 | 0.96 (0.89 to 1.03) | 0.23 | 0.17 |
| 4q24 | rs9790517 | 106084778 | TET2 | G | A | No | 0.23 | 0.98 (0.94 to 1.03) | 0.51 | 0.97 (0.92 to 1.03) | 0.30 | 1.03 (0.92 to 1.15) | 0.61 | 0.37 | 0.22 | 0.97 (0.91 to 1.05) | 0.48 | 0.91 (0.77 to 1.07) | 0.25 | 0.99 (0.91 to 1.08) | 0.88 | 0.35 |
| 4q34.1 | rs6828523 | 175846426 | EBF1 | C | A | No | 0.11 | 1.03 (0.96 to 1.1) | 0.41 | 1.03 (0.95 to 1.11) | 0.47 | 1.02 (0.88 to 1.19) | 0.78 | 0.94 | 0.1 | 0.98 (0.89 to 1.08) | 0.72 | 1.05 (0.85 to 1.30) | 0.64 | 0.96 (0.86 to 1.08) | 0.51 | 0.49 |
| 5p15.33 | rs10069690 | 1279790 | TERT | G | A | Yes | 0.28 | 1.21 (1.15 to 1.26) | 1.1 × 10 −15 | 1.24 (1.18 to 1.31) | 2.7 × 10 −15 | 1.09 (0.98 to 1.22) | 0.09 | 0.04 | 0.27 | 1.11 (1.04 to 1.19) | 1.6 × 10 −3 | 1.25 (1.08 to 1.44) | 3.2 × 10 −3 | 1.08 (1.00 to 1.17) | 0.06 | 0.09 |
| 5p15.33 | rs7725218 | 1282414 | TERT | G | A | Yes | 0.36 | 1.08 (1.04 to 1.13) | 3.2 × 10 −4 | 1.09 (1.04 to 1.15) | 6.4 × 10 −4 | 1.05 (0.95 to 1.15) | 0.37 | 0.47 | 0.36 | 1.06 (0.99 to 1.12) | 0.08 | 1.07 (0.93 to 1.23) | 0.35 | 1.05 (0.98 to 1.13) | 0.17 | 0.85 |
| 5p15.33 | rs2736108 | 1297488 | TERT | G | A | Yes | 0.29 | 0.89 (0.85 to 0.93) | 4.2 × 10 −7 | 0.86 (0.82 to 0.91) | 1.1 × 10 −7 | 0.98 (0.88 to 1.08) | 0.68 | 0.04 | 0.3 | 0.93(0.88 to 1.00) | 0.04 | 0.91 (0.78 to 1.05) | 0.20 | 0.94 (0.87 to 1.02) | 0.13 | 0.67 |
| 5p12 | rs10941679 | 44706498 | MRPS30 | A | G | Yes | 0.25 | 0.99 (0.94 to 1.04) | 0.61 | 0.99 (0.94 to 1.05) | 0.84 | 0.97 (0.86 to 1.08) | 0.54 | 0.66 | 0.24 | 1.07 (1.00 to 1.15) | 0.03 | 1.09 (0.94 to 1.27) | 0.24 | 1.07 (0.99 to 1.15) | 0.09 | 0.78 |
| 5q11.2 | rs889312 | 56031884 | MAP3K1 | A | C | Yes | 0.29 | 1.01 (0.97 to 1.06) | 0.52 | 0.99 (0.94 to 1.05) | 0.82 | 1.09 (0.98 to 1.21) | 0.12 | 0.15 | 0.3 | 1.04 (0.98 to 1.11) | 0.21 | 0.99 (0.86 to 1.14) | 0.90 | 1.06 (0.98 to 1.14) | 0.14 | 0.44 |
| 5q11.3 | rs10472076 | 58184061 | RAB3C | A | G | No | 0.37 | 1.00 (0.96 to 1.05) | 0.83 | 0.98 (0.94 to 1.03) | 0.52 | 1.08 (0.98 to 1.18) | 0.13 | 0.11 | 0.38 | 0.99 (0.93 to 1.05) | 0.78 | 1.02 (0.89 to 1.17) | 0.78 | 0.98 (0.92 to 1.05) | 0.64 | 0.66 |
| 5q11.3 | rs1353747 | 58337481 | PDE4D | A | C | No | 0.09 | 0.98 (0.91 to 1.05) | 0.53 | 0.95 (0.88 to 1.04) | 0.28 | 1.06 (0.90 to 1.24) | 0.50 | 0.29 | 0.09 | 0.95 (0.86 to 1.05) | 0.35 | 1.02 (0.81 to 1.27) | 0.88 | 0.94 (0.83 to 1.05) | 0.26 | 0.52 |
| 5q33.3 | rs1432679 | 158244083 | A | G | No | 0.44 | 1.05 (1.00 to 1.09) | 0.03 | 1.03 (0.98 to 1.08) | 0.25 | 1.10 (1.01 to 1.21) | 0.04 | 0.21 | 0.45 | 1.01 (0.95 to 1.07) | 0.79 | 0.92 (0.81 to 1.04) | 0.16 | 1.04 (0.97 to 1.11) | 0.30 | 0.09 | |
| 6p25.3 | rs11242675 | 1318878 | FOXQ1 | A | G | No | 0.35 | 0.96 (0.92 to 1) | 0.06 | 0.94 (0.90 to 0.99) | 0.03 | 1.01 (0.91 to 1.11) | 0.90 | 0.28 | 0.36 | 1.01 (0.95 to 1.07) | 0.79 | 1.02 (0.89 to 1.18) | 0.75 | 1.00 (0.93 to 1.08) | 0.90 | 0.83 |
| 6p24.3 | rs9348512 | 10456706 | TFAP2A | C | A | Yes | 0.34 | 1.00 (0.95 to 1.04) | 0.87 | 1.00 (0.95 to 1.05) | 0.88 | 1.00 (0.90 to 1.10) | 0.95 | 1.00 | 0.34 | 0.85 (0.8 to 0.9) | 9.2 × 10 −8 | 0.79 (0.69 to 0.91) | 1.1 × 10 −3 | 0.86 (0.8 to 0.92) | 3.4 × 10 −5 | 0.32 |
| 6p23 | rs204247 | 13722523 | RANBP9 | A | G | No | 0.44 | 1.00 (0.96 to 1.04) | 0.98 | 1.00 (0.95 to 1.04) | 0.84 | 1.01 (0.92 to 1.12) | 0.75 | 0.72 | 0.44 | 1.09 (1.03 to 1.15) | 3.4 × 10 −3 | 1.08 (0.94 to 1.24) | 0.25 | 1.09 (1.02 to 1.17) | 9.7 × 10 −3 | 0.93 |
| 6q14 | rs17530068 | 82193109 | FAM46A | A | G | Yes | 0.25 | 1.03 (0.98 to 1.08) | 0.23 | 1.03 (0.97 to 1.09) | 0.29 | 1.03 (0.92 to 1.14) | 0.63 | 0.95 | 0.25 | 1.10 (1.03 to 1.18) | 7.2 × 10 −3 | 1.07 (0.92 to 1.25) | 0.40 | 1.11 (1.02 to 1.2) | 0.01 | 0.70 |
| 6q25.1 | rs3757318 | 151914113 | ESR1 | G | A | Yes | 0.08 | 1.20 (1.11 to 1.29) | 1.1 × 10 −6 | 1.24 (1.14 to 1.35) | 5.6 × 10 −7 | 1.06 (0.89 to 1.26) | 0.51 | 0.12 | 0.09 | 1.15 (1.03 to 1.28) | 0.01 | 1.33 (1.07 to 1.65) | 9.1 × 10 −3 | 1.09 (0.96 to 1.24) | 0.17 | 0.12 |
| 6q25.1 | rs2046210 | 151948366 | ESR1 | G | A | Yes | 0.37 | 1.16 (1.12 to 1.21) | 2.4 × 10 −12 | 1.20 (1.15 to 1.26) | 2.8 × 10 −13 | 1.04 (0.94 to 1.15) | 0.42 | 0.01 | 0.37 | 1.06 (0.99 to 1.12) | 0.07 | 1.15 (1.00 to 1.33) | 0.04 | 1.03 (0.96 to 1.1) | 0.43 | 0.17 |
| 7q35 | rs720475 | 144074929 | ARHGEF5 | G | A | Yes | 0.26 | 0.98 (0.93 to 1.03) | 0.36 | 0.98 (0.93 to 1.04) | 0.54 | 0.96 (0.86 to 1.08) | 0.53 | 0.79 | 0.26 | 0.99 (0.92 to 1.06) | 0.71 | 0.94 (0.80 to 1.09) | 0.41 | 1.00 (0.93 to 1.08) | 0.96 | 0.45 |
| 8p12 | rs9693444 | 29509616 | DUSP4 | C | A | Yes | 0.33 | 1.01 (0.96 to 1.05) | 0.80 | 1.01 (0.96 to 1.06) | 0.69 | 0.99 (0.90 to 1.09) | 0.83 | 0.72 | 0.33 | 0.98 (0.93 to 1.05) | 0.61 | 0.92 (0.80 to 1.07) | 0.28 | 1.00 (0.94 to 1.07) | 0.94 | 0.32 |
| 8q21.11 | rs6472903 | 76230301 | A | C | Yes | 0.17 | 1.01 (0.96 to 1.07) | 0.74 | 1.01 (0.95 to 1.08) | 0.77 | 1.01 (0.89 to 1.14) | 0.89 | 0.99 | 0.16 | 0.97 (0.90 to 1.05) | 0.48 | 0.95 (0.79 to 1.14) | 0.56 | 0.98 (0.89 to 1.07) | 0.66 | 0.75 | |
| 8q21.11 | rs2943559 | 76417937 | HNF4G | A | G | Yes | 0.08 | 1.06 (0.98 to 1.14) | 0.12 | 1.07 (0.98 to 1.17) | 0.12 | 1.02 (0.86 to 1.22) | 0.78 | 0.68 | 0.09 | 1.10 (1.00 to 1.22) | 0.05 | 1.14 (0.91 to 1.43) | 0.27 | 1.09 (0.97 to 1.23) | 0.13 | 0.77 |
| 8q24.21 | rs11780156 | 129194641 | MYC | G | A | No | 0.19 | 0.95 (0.9 to 1) | 0.05 | 0.96 (0.90 to 1.02) | 0.15 | 0.93 (0.82 to 1.05) | 0.23 | 0.70 | 0.19 | 0.97 (0.90 to 1.04) | 0.44 | 0.95 (0.81 to 1.12) | 0.56 | 0.98 (0.9 to 1.06) | 0.60 | 0.79 |
| 8q24.21 | rs13281615 | 128355618 | A | G | Yes | 0.43 | 1.02 (0.98 to 1.06) | 0.44 | 1.01 (0.96 to 1.06) | 0.73 | 1.04 (0.95 to 1.15) | 0.38 | 0.55 | 0.43 | 1.03 (0.97 to 1.09) | 0.33 | 1.05 (0.92 to 1.20) | 0.44 | 1.02 (0.96 to 1.1) | 0.51 | 0.71 | |
| 9p21.3 | rs1011970 | 22062134 | CDKN2B | C | A | Yes | 0.17 | 1.02 (0.97 to 1.08) | 0.47 | 1.07 (1.00 to 1.14) | 0.05 | 0.87 (0.76 to 0.99) | 0.04 | 9.3 × 10 −3 | 0.17 | 1.03 (0.95 to 1.11) | 0.50 | 1.11 (0.94 to 1.32) | 0.21 | 1.00 (0.92 to 1.1) | 0.95 | 0.29 |
| 9q31.2 | rs10759243 | 110306115 | KLF4 | C | A | No | 0.31 | 0.98 (0.94 to 1.02) | 0.39 | 0.98 (0.93 to 1.03) | 0.49 | 0.98 (0.88 to 1.08) | 0.64 | 0.93 | 0.29 | 1.00 (0.94 to 1.06) | 0.94 | 0.94 (0.81 to 1.09) | 0.40 | 1.01 (0.94 to 1.09) | 0.69 | 0.37 |
| 9q31.2 | rs865686 | 110888478 | KLF4 | A | C | Yes | 0.36 | 0.99 (0.95 to 1.04) | 0.77 | 1.02 (0.97 to 1.07) | 0.52 | 0.92 (0.83 to 1.02) | 0.11 | 0.10 | 0.36 | 0.99 (0.93 to 1.05) | 0.75 | 1.16 (1.01 to 1.34) | 0.04 | 0.94 (0.88 to 1.01) | 0.12 | 0.01 |
| 10p12.31 | rs7072776 | 22032942 | MLLT10 | G | A | No | 0.31 | 0.99 (0.94 to 1.03) | 0.52 | 0.96 (0.91 to 1.01) | 0.11 | 1.08 (0.98 to 1.20) | 0.12 | 0.04 | 0.3 | 0.99 (0.92 to 1.05) | 0.67 | 0.88 (0.77 to 1.02) | 0.08 | 1.02 (0.94 to 1.09) | 0.66 | 0.09 |
| 10p12.31 | rs11814448 | 22315843 | DNAJC1 | A | C | No | 0.02 | 1.10 (0.94 to 1.29) | 0.22 | 1.25 (0.93 to 1.69) | 0.15 | 0.50 | ||||||||||
| 10q21.2 | rs10995190 | 64278682 | ZNF365 | G | A | Yes | 0.15 | 0.99 (0.93 to 1.05) | 0.70 | 1.02 (0.96 to 1.09) | 0.49 | 0.88 (0.76 to 1.00) | 0.06 | 0.05 | 0.15 | 0.94 (0.86 to 1.02) | 0.16 | 0.92 (0.76 to 1.12) | 0.43 | 0.95 (0.86 to 1.04) | 0.26 | 0.84 |
| 10q22.3 | rs704010 | 80841148 | ZMIZ1 | G | A | Yes | 0.37 | 1.01 (0.97 to 1.06) | 0.48 | 0.99(0.94 to 1.04) | 0.61 | 1.12 (1.01 to 1.23) | 0.03 | 0.03 | 0.38 | 1.01 (0.95 to 1.07) | 0.86 | 1.02 (0.90 to 1.16) | 0.78 | 1.00 (0.93 to 1.07) | 0.96 | 0.83 |
| 10q25.2 | rs7904519 | 114773927 | TCF7L2 | A | G | Yes | 0.47 | 1.09 (1.05 to 1.14) | 1.6 × 10 −5 | 1.09 (1.04 to 1.14) | 6.4 × 10 −4 | 1.12 (1.02 to 1.23) | 0.02 | 0.59 | 0.47 | 1.02 (0.96 to 1.08) | 0.56 | 0.89 (0.78 to 1.01) | 0.07 | 1.06 (0.99 to 1.13) | 0.10 | 0.02 |
| 10q26.12 | rs2981579 | 123337335 | FGFR2 | G | A | Yes | 0.42 | 0.99 (0.95 to 1.04) | 0.81 | 0.92 (0.87 to 0.96) | 6.9 × 10 −4 | 1.29 (1.17 to 1.43) | 3.1 × 10 −7 | 7.5 × 10 −9 | 0.44 | 1.24 (1.16 to 1.31) | 5.4 × 10 −12 | 1.05 (0.92 to 1.20) | 0.44 | 1.29 (1.21 to 1.38) | 2.2 × 10 −13 | 7.3 × 10 −3 |
| 10q26.12 | rs11199914 | 123093901 | FGFR2 | G | A | No | 0.33 | 1.02 (0.98 to 1.07) | 0.27 | 1.04 (0.99 to 1.10) | 0.09 | 0.96 (0.86 to 1.06) | 0.43 | 0.17 | 0.33 | 0.95 (0.90 to 1.02) | 0.15 | 0.90 (0.78 to 1.03) | 0.13 | 0.97 (0.9 to 1.04) | 0.44 | 0.33 |
| 11p15.5 | rs3817198 | 1909006 | LSP1 | A | G | Yes | 0.33 | 1.08 (1.03 to 1.13) | 6.5 × 10 −4 | 1.08 (1.03 to 1.14) | 2.9 × 10 −3 | 1.07 (0.97 to 1.18) | 0.17 | 0.89 | 0.34 | 1.11 (1.04 to 1.18) | 9.3 × 10 −4 | 0.98 (0.85 to 1.13) | 0.80 | 1.15 (1.07 to 1.24) | 1.1 × 10 −4 | 0.06 |
| 11q13.1 | rs3903072 | 65583066 | SNX32 | C | A | No | 0.47 | 0.99 (0.95 to 1.03) | 0.69 | 1.00 (0.95 to 1.05) | 0.94 | 0.96 (0.87 to 1.05) | 0.39 | 0.44 | 0.47 | 0.97 (0.91 to 1.03) | 0.34 | 0.89 (0.78 to 1.01) | 0.07 | 1.00 (0.93 to 1.07) | 0.91 | 0.12 |
| 11q13.3 | rs554219 | 69331642 | CCND1 | C | G | Yes | 0.12 | 1.03 (0.97 to 1.09) | 0.37 | 1.00 (0.93 to 1.08) | 0.96 | 1.12 (0.97 to 1.29) | 0.12 | 0.20 | 0.13 | 1.09 (1.00 to 1.19) | 0.05 | 0.90 (0.73 to 1.10) | 0.31 | 1.15 (1.04 to 1.26) | 5.0 × 10 −3 | 0.04 |
| 11q13.3 | c11_pos69088342 | 69379161 | C | A | Yes | 0.06 | 1.03 (0.95 to 1.13) | 0.45 | 1.00 (0.90 to 1.12) | 0.95 | 1.14 (0.93 to 1.39) | 0.20 | 0.30 | 0.06 | 1.07 (0.95 to 1.21) | 0.28 | 0.85 (0.64 to 1.14) | 0.29 | 1.14 (0.99 to 1.3) | 0.08 | 0.09 | |
| 11q13.3 | rs494406 | 69344241 | G | A | Yes | 0.25 | 1.02 (0.97 to 1.07) | 0.46 | 1.02 (0.97 to 1.08) | 0.42 | 1.00 (0.90 to 1.12) | 0.97 | 0.74 | 0.26 | 1.05 (0.98 to 1.12) | 0.16 | 1.03 (0.89 to 1.19) | 0.66 | 1.05 (0.98 to 1.14) | 0.18 | 0.80 | |
| 11q24.3 | rs11820646 | 129461171 | BARX2 | G | A | No | 0.39 | 0.93 (0.89 to 0.97) | 5.9 × 10 −4 | 0.94 (0.90 to 0.99) | 0.03 | 0.88 (0.79 to 0.97) | 8.9 × 10 −3 | 0.20 | 0.39 | 0.92 (0.86 to 0.97) | 4.2 × 10 −3 | 0.95 (0.83 to 1.09) | 0.45 | 0.91 (0.84 to 0.97) | 5.9 × 10 −3 | 0.56 |
| 12p13.1 | rs12422552 | 14413931 | ATF7IP | G | C | No | 0.27 | 1.01 (0.97 to 1.06) | 0.63 | 1.01 (0.96 to 1.07) | 0.68 | 1.01 (0.91 to 1.13) | 0.85 | 0.99 | 0.28 | 1.00 (0.93 to 1.06) | 0.92 | 1.02 (0.88 to 1.19) | 0.79 | 0.99 (0.92 to 1.07) | 0.79 | 0.73 |
| 12p11.22 | rs10771399 | 28155080 | PTHLH | A | G | Yes | 0.10 | 0.85 (0.80 to 0.91) | 1.9 × 10 −6 | 0.83 (0.77 to 0.9) | 1.2 × 10 −5 | 0.91 (0.78 to 1.06) | 0.24 | 0.36 | 0.1 | 0.89 (0.81 to 0.98) | 0.02 | 0.72 (0.56 to 0.91) | 6.6 × 10 −3 | 0.94 (0.84 to 1.05) | 0.25 | 0.05 |
| 12q22 | rs17356907 | 96027759 | NTN4 | A | G | No | 0.29 | 0.95 (0.91 to 1.00) | 0.03 | 0.94 (0.90 to 1.00) | 0.03 | 0.98 (0.88 to 1.09) | 0.71 | 0.56 | 0.3 | 0.99 (0.93 to 1.06) | 0.78 | 0.95 (0.83 to 1.10) | 0.51 | 1.00 (0.93 to 1.08) | 0.96 | 0.55 |
| 12q24.21 | rs1292011 | 115836522 | TBX3 | A | G | Yes | 0.41 | 1.00 (0.96 to 1.05) | 0.82 | 1.04 (0.99 to 1.10) | 0.08 | 0.88 (0.80 to 0.97) | 0.01 | 4.39 × 10 −3 | 0.41 | 0.92 (0.87 to 0.98) | 0.01 | 1.07 (0.94 to 1.23) | 0.29 | 0.88 (0.82 to 0.95) | 5.7 × 10 −4 | 0.01 |
| 13q13.1 | rs11571833 | 32972626 | BRCA2 | T | A | No | 0.01 | 1.02 (0.83 to 1.26) | 0.83 | 1.05 (0.83 to 1.34) | 0.66 | 0.92 (0.53 to 1.59) | 0.75 | 0.66 | 0.03 | 0.95 (0.78 to 1.16) | 0.62 | 0.91 (0.58 to 1.41) | 0.66 | 0.97 (0.78 to 1.2) | 0.76 | 0.80 |
| 14q13.3 | rs2236007 | 37132769 | PAX9 | G | A | No | 0.21 | 0.97 (0.92 to 1.02) | 0.19 | 0.95 (0.90 to 1.01) | 0.13 | 1.01 (0.90 to 1.14) | 0.87 | 0.42 | 0.21 | 0.99 (0.92 to 1.07) | 0.83 | 1.20 (1.03 to 1.40) | 0.02 | 0.94 (0.86 to 1.02) | 0.13 | 8.5 × 10 −3 |
| 14q24.1 | rs2588809 | 68660428 | G | A | No | 0.19 | 0.96 (0.91 to 1.01) | 0.09 | 0.95 (0.89 to 1.01) | 0.13 | 0.97 (0.86 to 1.09) | 0.56 | 0.85 | 0.2 | 1.01 (0.94 to 1.09) | 0.82 | 0.95 (0.80 to 1.13) | 0.55 | 1.03 (0.94 to 1.12) | 0.55 | 0.44 | |
| 14q24.1 | rs999737 | 69034682 | RAD51L1 | G | A | Yes | 0.21 | 0.96 (0.91 to 1.01) | 0.09 | 0.98 (0.92 to 1.04) | 0.43 | 0.90 (0.80 to 1.01) | 0.07 | 0.22 | 0.22 | 0.97 (0.91 to 1.05) | 0.48 | 0.95 (0.81 to 1.11) | 0.53 | 0.98 (0.91 to 1.06) | 0.65 | 0.72 |
| 14q32.11 | rs941764 | 91841069 | CCDC88C | A | G | No | 0.34 | 1.03 (0.98 to 1.07) | 0.23 | 1.02 (0.97 to 1.07) | 0.47 | 1.05 (0.95 to 1.17) | 0.34 | 0.62 | 0.34 | 1.03 (0.97 to 1.09) | 0.39 | 1.00 (0.88 to 1.14) | 0.97 | 1.04 (0.97 to 1.11) | 0.32 | 0.61 |
| 16q12.1a | rs3803662 | 52586341 | TOX3 | G | A | Yes | 0.29 | 1.06 (1.01 to 1.11) | 0.02 | 1.01 (0.96 to 1.07) | 0.65 | 1.22 (1.10 to 1.35) | 1.5 × 10 −4 | 2.39 × 10 −3 | 0.29 | 1.24 (1.16 to 1.32) | 6.2 × 10 −11 | 1.12 (0.97 to 1.30) | 0.11 | 1.27 (1.18 to 1.36) | 1.5 × 10 −10 | 0.15 |
| 16q12.1b | rs11075995 | 53855291 | FTO | A | T | No | 0.24 | 1.01 (0.96 to 1.06) | 0.61 | 0.98 (0.93 to 1.04) | 0.60 | 1.11 (0.99 to 1.24) | 0.07 | 0.07 | 0.24 | 1.02(0.95 to 1.09) | 0.59 | 1.01 (0.86 to 1.18) | 0.94 | 1.02 (0.94 to 1.11) | 0.57 | 0.85 |
| 16q12.1b | rs17817449 | 53813367 | FTO | A | C | No | 0.41 | 0.95 (0.91 to 0.99) | 0.02 | 0.94 (0.89 to 0.98) | 9.8 × 10 −3 | 1.00 (0.91 to 1.10) | 0.99 | 0.26 | 0.41 | 0.94 (0.88 to 1.00) | 0.03 | 1.05 (0.91 to 1.21) | 0.49 | 0.91 (0.85 to 0.97) | 6.6 × 10 −3 | 0.08 |
| 16q23.2 | rs13329835 | 80650805 | CDYL2 | A | G | No | 0.23 | 1.04 (0.99 to 1.09) | 0.09 | 1.03 (0.97 to 1.09) | 0.29 | 1.08 (0.97 to 1.21) | 0.17 | 0.48 | 0.24 | 1.03 (0.96 to 1.11) | 0.35 | 0.94 (0.81 to 1.10) | 0.43 | 1.06 (0.98 to 1.15) | 0.13 | 0.17 |
| 17q22 | rs6504950 | 53056471 | COX11 | G | A | Yes | 0.27 | 0.98 (0.94 to 1.03) | 0.49 | 0.99 (0.94 to 1.04) | 0.70 | 0.96 (0.87 to 1.07) | 0.49 | 0.68 | 0.27 | 1.04 (0.97 to 1.11) | 0.24 | 1.08 (0.94 to 1.24) | 0.27 | 1.03 (0.95 to 1.11) | 0.46 | 0.54 |
| 18q11.2 | rs527616 | 24337424 | AQP4 | C | G | No | 0.37 | 0.99 (0.95 to 1.03) | 0.61 | 0.97 (0.93 to 1.02) | 0.30 | 1.04 (0.94 to 1.15) | 0.42 | 0.26 | 0.37 | 0.96 (0.9 to 1.02) | 0.19 | 0.95 (0.83 to 1.08) | 0.41 | 0.96 (0.9 to 1.03) | 0.30 | 0.82 |
| 18q11.2 | rs1436904 | 22824665 | A | C | No | 0.39 | 0.99 (0.95 to 1.03) | 0.68 | 1.00 (0.95 to 1.05) | 0.86 | 0.98 (0.89 to 1.08) | 0.63 | 0.74 | 0.39 | 0.96 (0.9 to 1.02) | 0.14 | 1.02 (0.89 to 1.17) | 0.79 | 0.94 (0.87 to 1.01) | 0.07 | 0.30 | |
| 19p13.11 | rs8170 | 17389704 | BABAM1 | G | A | Yes | 0.19 | 1.19 (1.12 to 1.25) | 2.9 × 10 −10 | 1.22 (1.15 to 1.30) | 1.7 × 10 −10 | 1.06 (0.95 to 1.20) | 0.30 | 0.05 | 0.19 | 0.98 (0.91 to 1.06) | 0.62 | 1.06 (0.90 to 1.25) | 0.51 | 0.96 (0.88 to 1.05) | 0.37 | 0.33 |
| 19p13.11 | rs4808801 | 18571141 | ELL | A | G | No | 0.32 | 0.98 (0.94 to 1.02) | 0.40 | 0.99 (0.94 to 1.05) | 0.81 | 0.94 (0.85 to 1.04) | 0.23 | 0.35 | 0.33 | 0.97 (0.91 to 1.03) | 0.33 | 1.07 (0.93 to 1.23) | 0.33 | 0.94 (0.87 to 1.01) | 0.10 | 0.11 |
| 19q13.31 | rs3760982 | 44286513 | KCNN4 | G | A | No | 0.46 | 1.03 (0.99 to 1.08) | 0.10 | 1.03 (0.98 to 1.08) | 0.19 | 1.04 (0.95 to 1.14) | 0.41 | 0.90 | 0.46 | 1.05 (0.99 to 1.12) | 0.09 | 0.97 (0.86 to 1.11) | 0.69 | 1.08 (1.01 to 1.15) | 0.03 | 0.19 |
| 21q21.1 | rs2823093 | 16520832 | NRIP1 | G | A | Yes | 0.27 | 0.95 (0.91 to 1) | 0.04 | 0.96 (0.91 to 1.02) | 0.17 | 0.92 (0.82 to 1.02) | 0.11 | 0.44 | 0.27 | 0.94 (0.88 to 1.01) | 0.09 | 1.07 (0.92 to 1.25) | 0.36 | 0.91 (0.84 to 0.98) | 0.02 | 0.06 |
| 22q12.2 | rs132390 | 29621477 | EMID1 | A | G | No | 0.03 | 0.98 (0.87 to 1.1) | 0.75 | 0.93 (0.81 to 1.07) | 0.30 | 1.16 (0.91 to 1.46) | 0.23 | 0.13 | 0.04 | 1.17 (1.00 to 1.37) | 0.05 | 0.92 (0.61 to 1.39) | 0.70 | 1.24 (1.04 to 1.49) | 0.02 | 0.22 |
| 22q13.1 | rs6001930 | 40876234 | SGSM3 | A | G | No | 0.11 | 1.07 (1.00 to 1.14) | 0.03 | 1.06 (0.98 to 1.15) | 0.14 | 1.11 (0.96 to 1.30) | 0.17 | 0.60 | 0.10 | 1.02 (0.92 to 1.12) | 0.71 | 0.94 (0.76 to 1.18) | 0.61 | 1.04 (0.93 to 1.16) | 0.48 | 0.44 |
aCI, Confidence interval; ER, Estrogen receptor; HR, Hazard ratio; MAF, Mean allele frequency; SNP, Single-nucleotide polymorphism. Results represent BRCA1 and BRCA2 mutation carriers for 74 previously reported breast cancer (BC) susceptibility variants from population-based studies. P-values <0.05 are shown in bold. bPosition in build 37. cReference allele. cEffect allele. eAssociation in BRCA1 and BRCA2 carriers has been reported before. fP-value for the difference between the association with ER-positive BC and the association with ER-negative BC.
When association analyses for ER-positive and -negative disease were repeated after excluding prevalent breast cancer cases (Additional file 1: Table S3), the HR estimates were consistent with the estimates from the complete sample but were associated with larger confidence intervals. Therefore, it is unlikely that our results are influenced by survival bias.
Associations with other subtypes and clinical features
The pattern of associations of previously reported breast cancer susceptibility variants by PR and TN status were very similar to that by ER status (Additional file 1: Tables S4 and S6), but fewer associations were observed at P <0.01. SNP rs720475 at 7q35 was the only variant that was associated with HER2-positive disease (HR = 1.45 and P = 0.003 for HER2-positive, P het = 9 × 10−4 in BRCA2 carriers) (Additional file 1: Table S5).
For BRCA1 carriers, there were significant differences (P het <0.01) in the HR for high grade (grade 3) and grades 1 and 2 breast cancer for SNPs at 10q26.12 (FGFR2) and at 12q24.21 (Additional file 1: Table S9). SNP rs3803662 in TOX3 at 16q12.1 was associated exclusively with node-positive breast cancer (P = 2 × 10−4, P het = 0.005) (Additional file 1: Table S10). This was also the only variant associated with lobular cancer, as shown in Additional file 1: Table S8 (P = 8 × 10−6 for BRCA2 carriers). The HR for lobular cancer was larger than that for non-lobular cancer (lobular HR = 1.57, 95% CI: 1.29 to 1.92; non-lobular HR = 1.20, 95% CI: 1.13 to 1.28 for BRCA2 carriers; P het = 9 × 10−4). There was no evidence for differences in associations by histological grade and nodal involvement for BRCA2 carriers.
Comparison of patterns of associations by breast cancer estrogen receptor status between BRCA1 and BRCA2 carriers and the general population
We compared the log HR estimates for the breast cancer association of known breast cancer susceptibility variants for BRCA1 carriers, BRCA2 carriers, and for the general population using published data from the Breast Cancer Association Consortium (BCAC) [8]. The resulting ICC coefficients for log HR/OR estimates for all comparisons are shown in Table 3. Log HR estimates for overall breast cancer risk in BRCA2 carriers were very similar to the log odds ratios (ORs) from the general population (ICC = 0.63, 95% CI: 0.47 to 0.75) (Additional file 1: Figure S3B), whereas there was only moderate correlation between the log HR estimates for BRCA1 carriers and the log HR estimates from both other groups (ICC: BRCA1-BCAC estimates = 0.43, BRCA1-BRCA2 = 0.46) (Additional file 1: Figure S3A,C). When comparing ER-positive specific associations, we found stronger agreement between the log HR/OR estimates than for overall breast cancer. The ICC estimates ranged from 0.61 (95% CI: 0.45 to 0.74) (Figure 1B) for BCAC-BRCA1 to 0.69 (95% CI: 0.55 to 0.79) (Figure 1C) for BCAC-BRCA2. The ER-negative breast cancer log HR estimates in BRCA1 carriers and the corresponding BCAC estimates were strongly correlated (ICC = 0.59, 95% CI: 0.42 to 0.72) (Figure 1E). However, the ER-negative breast cancer log HR estimates in BRCA2 carriers were less strongly correlated with the corresponding estimates in BRCA1 carriers (ICC = 0.46, 95% CI: 0.25 to 0.62) (Figure 1D) and in BCAC (ICC = 0.28, 95% CI: 0.05 to 0.48) (Figure 1F). There was no evidence that the ICC was different from 0 for the comparison between ER-positive associations in BRCA1 carriers with ER-negative associations in BRCA2 carriers and vice versa (Additional file 1: Figure S4B,C). Similarly, there was no significant correlation between log OR estimates for ER-positive breast cancer in BCAC with log HR estimates for ER-negative breast cancer in BRCA1 and BRCA2 carriers (Additional file 1: Figures S5B and S6B). There was only moderate correlation between log OR estimates for ER-negative breast cancer in the general population and log HR estimates for ER-positive breast cancer in BRCA1 and BRCA2 carriers (ICC = 0.39 and ICC = 0.34, respectively) (Additional file 1: Figures S5C and S6C).
Table 3.
Comparisons between the associations of 74 breast cancer susceptibility loci in BRCA1 carriers, in BRCA2 carriers, and in population-based studies a
| BRCA1 carriers | BRCA2 carriers | ||||||
|---|---|---|---|---|---|---|---|
| Overall | ER-positive | ER-negative | Overall | ER-positive | ER-negative | ||
| BRCA2 carriers | Overall | 0.46 (0.25 to 0.62) | |||||
| ER-positive | 0.67 (0.52 to 0.78) | 0.13 (−0.10 to 0.35) | |||||
| ER-negative | 0.10 (−0.13 to 0.33) | 0.46 (0.25 to 0.62) | |||||
| BCAC | Overall | 0.43 (0.22 to 0.60) | 0.63 (0.47 to 0.75) | ||||
| ER-positive | 0.61 (0.45 to 0.74) | 0.16 (−0.07 to 0.38) | 0.69 (0.55 to 0.79) | 0.13 (−0.11 to 0.35) | |||
| ER-negative | 0.34 (0.12 to 0.53) | 0.59 (0.42 to 0.72) | 0.39 (0.17 to 0.57) | 0.28 (0.05 to 0.48) | |||
aER, Estrogen receptor. Data are intraclass correlation coefficients and 95% confidence intervals, which describe associations with overall breast cancer as well as by ER status. Data are derived from published studies by the Breast Cancer Association Consortium (BCAC).
Figure 1.
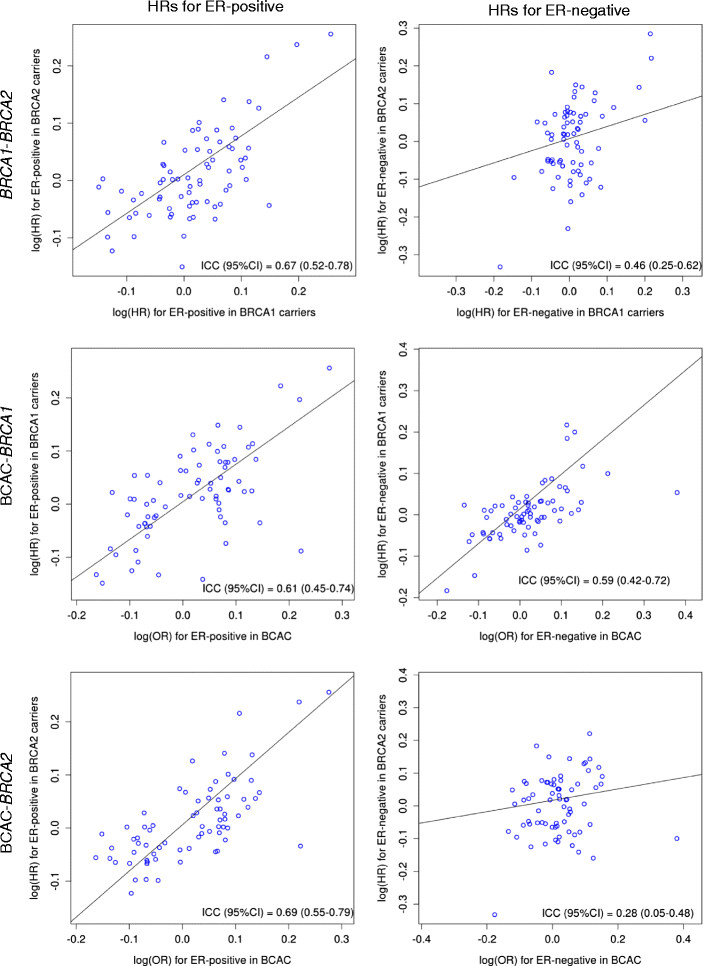
Estrogen receptor-positive and -negative log hazard ratio estimates in the general population and in BRCA1 and BRCA2 carriers. Dots represent the association of 74 previously reported breast cancer susceptibility single-nucleotide polymorphisms with estrogen receptor (ER)-positive breast cancer (A–C) and ER-negative breast cancer (D–F). (A) and (D) compare associations between BRCA1 and BRCA2 carriers, (B) and (E) between the general population and BRCA1 carriers and (C) and (F) between the general population and BRCA2 carriers. Association results for the general population were taken from reports published by the Breast Cancer Association Consortium (BCAC) [8]-[10],[16].
Associations by subtype with all single-nucleotide polymorphisms on iCOGS
Variants in or near the known breast cancer susceptibility loci TERT, ESR1 and 19p13.11 showed strong associations (P <10−6) with at least one of the categories in all subtype analyses in BRCA1 carriers. The same was true for SNPs in FGFR2 and TOX3 in BRCA2 carriers.
Variants on the iCOGS array that exhibited associations at P <10−6 with any of the breast cancer subtypes of ER-, PR-, HER2-positive or -negative and TN breast tumors are shown in Table 4. All the variants associated with ER-positive or ER-negative breast cancer at P <10−6 were located within known breast cancer susceptibility loci. Similar associations were observed with PR-positive and PR-negative breast cancer for BRCA1 carriers. A previously unreported SNP at 2p13.2 was associated with HER2-positive breast cancer in BRCA1 carriers. In BRCA2 carriers, two previously unreported variants showed evidence of association in the analysis by PR status. Furthermore, SNPs at 8q12.1 near TOX were associated with HER2-positive cancer in BRCA2 carriers. Only SNPs in the known breast cancer susceptibility loci FGFR2 and TOX3 were associated with non-TN breast cancer in BRCA2 carriers, and none were associated with TN breast cancer at P <10−6.
Table 4.
Associations with tumor subtypes a
| Affected by subtype, n (MAF) | Positive | Negative | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtype | Sample | SNP | Locus | Position b | Nearby gene | Ref c | Eff d | Reported e | Positive | Negative | Unknown | Number unaffected (MAF) | HR (95% CI) | P -value | HR (95% CI) | P -value | P het-value f |
| ER status | BRCA1 carriers | rs10069690 | 5p15.33 | 1332790 | TERT | G | A | Yes | 819 (0.27) | 2,635 (0.3) | 4,331 (0.3) | 7,449 (0.26) | 1.09 (0.98 to 1.22) | 0.09 | 1.24 (1.18 to 1.31) | 2.7 × 10−15 | 0.04 |
| rs2046210 | 6q25.1 | 151990059 | C6orf97 | G | A | Yes | 819 (0.36) | 2,639 (0.39) | 4,338 (0.38) | 7,453 (0.35) | 1.04 (0.94 to 1.15) | 0.42 | 1.2 (1.15 to 1.26) | 2.8 × 10−13 | 0.01 | ||
| rs2811708 | 9p21 | 21963422 | CDKN2A/B | C | A | Yes | 816 (0.24) | 2,634 (0.31) | 4,336 (0.28) | 7,445 (0.28) | 0.82 (0.73 to 0.91) | 2.9 × 10−4 | 1.13 (1.07 to 1.19) | 7.3 × 10−6 | 3.9 × 10−7 | ||
| rs45631579 | 10q26 | 123328965 | FGFR2 | A | G | Yes | 819 (0.48) | 2,639 (0.4) | 4,337 (0.42) | 7,455 (0.41) | 1.29 (1.17 to 1.43) | 2.6 × 10−7 | 0.92 (0.87 to 0.96) | 4.7 × 10−4 | 5.1 × 10−9 | ||
| rs2590275 | 12p12 | 28057334 | PTHLH | G | C | Yes | 819 (0.26) | 2,639 (0.25) | 4,338 (0.26) | 7,454 (0.28) | 0.92 (0.83 to 1.03) | 0.14 | 0.87 (0.82 to 0.92) | 5.3 × 10−7 | 0.34 | ||
| rs2363956 | 19p13.11 | 17255124 | ANKLE1 | C | A | Yes | 819 (0.47) | 2,635 (0.53) | 4,334 (0.52) | 7,447 (0.48) | 0.98 (0.89 to 1.08) | 0.71 | 1.25 (1.19 to 1.31) | 6.4 × 10−20 | 1.6 × 10−5 | ||
| BRCA2 carriers | rs2162540 | 10q26 | 123342126 | FGFR2 | A | G | Yes | 1450 (0.47) | 426 (0.41) | 2,348 (0.44) | 3,783 (0.38) | 1.34 (1.25 to 1.43) | 3.6 × 10−16 | 1.08 (0.94 to 1.23) | 0.27 | 6.1 × 10−3 | |
| rs17271951 | 16q12.1 | 51095541 | TOX3 | A | G | Yes | 1490 (0.32) | 434 (0.28) | 2,397 (0.3) | 3,879 (0.25) | 1.29 (1.2 to 1.39) | 8.4 × 10−12 | 1.14 (0.99 to 1.31) | 0.08 | 0.14 | ||
| PR status | BRCA1 carriers | rs10069690 | 5p15.33 | 1332790 | TERT | G | A | Yes | 662 (0.27) | 2,481 (0.29) | 4,642 (0.3) | 7,449 (0.26) | 1.10 (0.98 to 1.24) | 0.09 | 1.24 (1.17 to 1.3) | 1.3 × 10−14 | 0.11 |
| rs2046210 | 6q25.1 | 151990059 | C6orf97 | G | A | Yes | 662 (0.37) | 2,485 (0.39) | 4,649 (0.38) | 7,453 (0.35) | 1.09 (0.98 to 1.21) | 0.12 | 1.18 (1.13 to 1.24) | 2.0 × 10−11 | 0.19 | ||
| rs45631626 | 10q26 | 123327325 | FGFR2 | G | A | Yes | 662 (0.49) | 2,485 (0.4) | 4,647 (0.42) | 7,455 (0.42) | 1.33 (1.2 to 1.48) | 1.3 × 10−7 | 0.92 (0.88 to 0.97) | 9.6 × 10−4 | 5.3 × 10−9 | ||
| rs2590275 | 12p12 | 28057334 | PTHLH | G | C | Yes | 662 (0.26) | 2,485 (0.25) | 4,649 (0.26) | 7,454 (0.28) | 0.92 (0.82 to 1.03) | 0.16 | 0.87 (0.82 to 0.92) | 8.2 × 10−7 | 0.45 | ||
| rs8100241 | 19p13.11 | 17253894 | ANKLE1 | G | A | Yes | 657 (0.51) | 2,473 (0.47) | 4,628 (0.48) | 7,420 (0.52) | 0.98 (0.88 to 1.09) | 0.71 | 0.81 (0.78 to 0.85) | 1.8 × 10−17 | 2.4 × 10−3 | ||
| BRCA2 carriers | rs10017576 | 4q28.3 | 139799629 | G | A | No | 1099 (0.38) | 591 (0.43) | 2,639 (0.41) | 3,880 (0.43) | 0.83 (0.77 to 0.89) | 8.9 × 10−7 | 1.02 (0.92 to 1.14) | 0.67 | 2.7 × 10−3 | ||
| rs4376461 | 8p12 | 32822117 | NRG1 | C | A | No | 1099 (0.15) | 591 (0.10) | 2,640 (0.14) | 3,881 (0.15) | 1.00 (0.9 to 1.12) | 0.93 | 0.6 (0.5 to 0.73) | 2.4 × 10−7 | 1.4 × 10−5 | ||
| rs45631588 | 10q26 | 123341292 | FGFR2 | A | G | Yes | 1099 (0.47) | 591 (0.43) | 2,640 (0.45) | 3,881 (0.39) | 1.33 (1.23 to 1.44) | 2.7 × 10−13 | 1.15 (1.03 to 1.28) | 0.01 | 0.04 | ||
| rs17271951 | 16q12.1 | 51095541 | TOX3 | A | G | Yes | 1099 (0.32) | 591 (0.28) | 2,631 (0.3) | 3,879 (0.25) | 1.32 (1.21 to 1.43) | 4.3 × 10−11 | 1.14 (1.01 to 1.29) | 0.03 | 0.07 | ||
| HER2 status | BRCA1 carriers | rs17008885 | 2p13.2 | 73303647 | SMYD5 | T | A | No | 182 (0.19) | 1,816 (0.3) | 5,799 (0.3) | 7,455 (0.31) | 0.54 (0.43 to 0.69) | 7.8 × 10−7 | 0.99 (0.95 to 1.04) | 0.78 | 3.8 × 10−6 |
| rs10069690 | 5p15.33 | 1332790 | TERT | G | A | Yes | 181 (0.23) | 1,813 (0.29) | 5,791 (0.3) | 7,449 (0.26) | 0.9 (0.71 to 1.13) | 0.36 | 1.24 (1.18 to 1.3) | 3.4 × 10−17 | 0.01 | ||
| rs2046210 | 6q25.1 | 151990059 | C6orf97 | G | A | Yes | 182 (0.39) | 1,816 (0.4) | 5,798 (0.38) | 7,453 (0.35) | 1.12 (0.91 to 1.38) | 0.29 | 1.17 (1.11 to 1.22) | 1.5 × 10−10 | 0.71 | ||
| rs10843055 | 12p12 | 28063088 | PTHLH | A | C | Yes | 171 (0.06) | 1,743 (0.06) | 5,615 (0.06) | 7,234 (0.07) | 0.79 (0.52 to 1.2) | 0.26 | 0.79 (0.72 to 0.87) | 6.2 × 10−7 | 0.99 | ||
| rs4808616 | 19p13.11 | 17264033 | ANKLE1 | C | A | Yes | 176 (0.35) | 1,782 (0.32) | 5,706 (0.32) | 7,339 (0.28) | 1.37 (1.11 to 1.69) | 3.8 × 10−3 | 1.2 (1.14 to 1.26) | 6.8 × 10−13 | 0.25 | ||
| BRCA2 carriers | rs4305889 | 8q12.1 | 60352785 | TOX | A | G | No | 121 (0.18) | 845 (0.09) | 3,353 (0.11) | 3,874 (0.1) | 2.04 (1.53 to 2.71) | 8.7 × 10−7 | 0.96 (0.85 to 1.08) | 0.47 | 1.0 × 10−5 | |
| rs45631588 | 10q26 | 123341292 | FGFR2 | A | G | Yes | 121 (0.45) | 847 (0.46) | 3,362 (0.45) | 3,881 (0.39) | 1.27 (0.98 to 1.63) | 0.07 | 1.28 (1.19 to 1.37) | 2.1 × 10−12 | 0.95 | ||
| rs3817197 | 11p15.5 | 1862750 | LSP1 | G | A | Yes | 121 (0.55) | 847 (0.44) | 3,353 (0.45) | 3,877 (0.47) | 1.30 (1.02 to 1.64) | 0.03 | 0.84 (0.79 to 0.9) | 9.1 × 10−7 | 1.3 × 10−3 | ||
| rs17271951 | 16q12.1 | 51095541 | TOX3 | A | G | Yes | 121 (0.29) | 847 (0.31) | 3,353 (0.3) | 3,879 (0.25) | 1.10 (0.85 to 1.41) | 0.47 | 1.27 (1.18 to 1.37) | 6.9 × 10−11 | 0.3 | ||
| Triple-negativeg | BRCA1 carriers | rs10069690 | 5p15.33 | 1332790 | TERT | G | A | Yes | 579 (0.27) | 1,307 (0.3) | 5,899 (0.3) | 7,449 (0.26) | 1.06 (0.94 to 1.19) | 0.34 | 1.27 (1.2 to 1.36) | 5.2 × 10−14 | 0.02 |
| rs2046210 | 6q25.1 | 151990059 | C6orf97 | G | A | Yes | 580 (0.37) | 1,310 (0.41) | 5,906 (0.38) | 7,453 (0.35) | 1.01 (0.91 to 1.13) | 0.79 | 1.23 (1.16 to 1.31) | 5.5 × 10−12 | 6.8 × 10−3 | ||
| rs9512729 | 13q12.2 | 26974865 | LNX2 | G | A | No | 580 (0.34) | 1,309 (0.43) | 5,902 (0.41) | 7,454 (0.41) | 0.76 (0.69 to 0.85) | 9.1 × 10−7 | 1.08 (1.02 to 1.15) | 0.01 | 1.2 × 10−6 | ||
| rs8100241 | 19p13.11 | 17253894 | ANKLE1 | G | A | Yes | 577 (0.5) | 1,304 (0.46) | 5,877 (0.48) | 7,420 (0.52) | 0.94 (0.85 to 1.04) | 0.22 | 0.81 (0.76 to 0.86) | 2.4 × 10−13 | 0.03 | ||
| BRCA2 carriers | rs2162540 | 10q26 | 123342126 | FGFR2 | A | G | Yes | 735 (0.46) | 133 (0.36) | 3,356 (0.44) | 3,783 (0.38) | 1.39 (1.29 to 1.49) | 9.8 × 10−20 | 0.83 (0.67 to 1.03) | 0.10 | 4.0 × 10−5 | |
| rs1362548 | 16q12.1 | 51121452 | TOX3 | C | G | Yes | 760 (0.32) | 136 (0.29) | 3,434 (0.31) | 3,881 (0.26) | 1.26 (1.17 to 1.36) | 1.0 × 10−9 | 1.18 (0.94 to 1.49) | 0.16 | 0.63 | ||
aCI, Confidence interval; HR, Hazard ratio; MAF, Mean allele frequency. Single-nucleotide polymorphisms (SNPs) associated at P <10−6 with breast cancer by tumor estrogen receptor (ER) status, progesterone receptor (PR) status, human epidermal growth factor receptor 2 (HER2) status or triple-negative (negative for ER, PR and HER2) are shown. The most strongly associated SNP from each locus is reported. bPosition in build 36. cReference allele. dEffect allele. eVariant located in previously reported breast cancer susceptibility locus. fP-value for the difference in association between subtype positive and subtype negative breast cancer (for example, ER-positive vs ER-negative). gNon–triple negative was considered as “positive” and triple-negative as “negative.”
A SNP at 7q36 was associated with ductal subtype for BRCA1 carriers (Table 5). Two loci were associated with lobular breast cancer for BRCA2 carriers: 11q23.3 and Xp11.23.
Table 5.
Associations with ductal and lobular breast cancer a
| N tumors with morphology (MAF) | Other morphology | Tumors with morphology | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtype | Sample | SNP | Locus | Position b | Nearby gene | Ref c | Eff d | Reported e | Present | Other | Unknown | Number unaffected (MAF) | HR (95% CI) | P -value | HR (95% CI) | P -value | P het-value f |
| Ductal | BRCA1 carriers | rs10069690 | 5p15.33 | 1332790 | TERT | G | A | Yes | 3,155 (0.29) | 847 (0.30) | 3,783 (0.30) | 7,449 (0.26) | 1.23 (1.11 to 1.36) | 7.3 × 10−5 | 1.20 (1.14 to 1.26) | 2.0 × 10−11 | 0.67 |
| rs11155803 | 6q25.1 | 151987362 | C6orf97 | A | G | Yes | 3,159 (0.36) | 848 (0.35) | 3,786 (0.36) | 7,452 (0.32) | 1.13 (1.03 to 1.25) | 0.01 | 1.17 (1.11 to 1.23) | 4.6 × 10−10 | 0.58 | ||
| rs10252939 | 7q36 | 155587448 | A | G | No | 3,159 (0.28) | 849 (0.33) | 3,789 (0.29) | 7,455 (0.32) | 1.08 (0.98 to 1.19) | 0.14 | 0.87 (0.82 to 0.91) | 1.0 × 10−7 | 2.7 × 10−4 | |||
| rs8100241 | 19p13.11 | 17253894 | ANKLE1 | G | A | Yes | 3,137 (0.48) | 844 (0.49) | 3,777 (0.47) | 7,420 (0.52) | 0.87 (0.79 to 0.95) | 2.8 × 10−3 | 0.84 (0.8 to 0.88) | 1.5 × 10−13 | 0.51 | ||
| BRCA2 carriers | rs2162540 | 10q26 | 123342126 | FGFR2 | A | G | Yes | 1,728 (0.45) | 458 (0.44) | 2,038 (0.44) | 3,783 (0.38) | 1.24 (1.08 to 1.42) | 1.8 × 10−3 | 1.30 (1.21 to 1.39) | 3.1 × 10−14 | 0.55 | |
| rs1362548 | 16q12.1 | 51121452 | TOX3 | C | G | Yes | 1,770 (0.3) | 473 (0.32) | 2,087 (0.31) | 3,881 (0.26) | 1.32 (1.15 to 1.51) | 4.8 × 10−5 | 1.23 (1.15 to 1.32) | 7.3 × 10−9 | 0.36 | ||
| Lobular | BRCA2 carriers | rs2186703 | 11q23.3 | 115265717 | RPL15P15 | A | C | No | 188 (0.07) | 2,055 (0.03) | 2,087 (0.04) | 3,881 (0.03) | 1.09 (0.93 to 1.27) | 0.29 | 2.54 (1.78 to 3.62) | 2.8 × 10−7 | 1.5 × 10−5 |
| rs55998524 | Xp11.23 | 51082075 | NUDT10 | C | G | No | 188 (0.11) | 2,054 (0.05) | 2,086 (0.06) | 3,880 (0.05) | 0.96 (0.83 to 1.1) | 0.54 | 2.42 (1.77 to 3.32) | 3.6 × 10−8 | 7.0 × 10−8 | ||
aCI, Confidence interval; HR, Hazard ratio; MAF, Mean allele frequency. Single-nucleotide polymorphisms (SNPs) associated at P <10−6 with ductal carcinomas and, for BRCA2 mutation carriers, lobular carcinomas are shown. The most strongly associated SNP from each locus is reported. bPosition in build 36. cReference allele. dEffect allele. eVariant located in previously reported breast cancer susceptibility locus. fP-value for the difference in SNP association between ductal breast tumors and non-ductal breast tumors and for lobular breast tumors and non-lobular breast tumors.
There was one novel association with high-grade tumors in BRCA1 carriers (Table 6). Three previously unreported variants were associated with breast cancer nodal status in BRCA1 carriers: SNPs at 4q24 in the TET2 gene, at 5q32 in the SH3RF2 gene and at 7p22 in an intron of NXPH1. For BRCA2 carriers, only SNPs in FGFR2 and TOX3 exhibited associations at P <10−6 with breast cancer nodal status or histological grade.
Table 6.
Associations with grade and lymph node status a
| Affected by subtype, n (MAF) | Low-grade/no nodal involvement | High grade/nodal involvement | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtype | Sample | SNP | Locus | Position b | Nearby gene | Ref c | Eff d | Reported e | High-grade/node-positive | Low-grade/node-negative | Unknown | Number unaffected (MAF) | HR (95% CI) | P -value | HR (95% CI) | P -value | P het-value f |
| Grade 3 | BRCA1 carriers | rs17651413 | 2q33.1 | 202677054 | KIAA2012 | A | G | No | 2,495 (0.12) | 655 (0.11) | 4,644 (0.13) | 7,454 (0.11) | 1.01 (0.86 to 1.2) | 0.86 | 1.20 (1.12 to 1.29) | 8.4 × 10−7 | 0.08 |
| rs10069690 | 5p15.33 | 1332790 | TERT | G | A | Yes | 2,493 (0.3) | 654 (0.29) | 4,638 (0.3) | 7,449 (0.26) | 1.15 (1.02 to 1.29) | 0.02 | 1.22 (1.16 to 1.29) | 2.4 × 10−13 | 0.39 | ||
| c6_pos151989450 | 6q25.1 | 151989450 | C6orf97 | G | A | Yes | 2,497 (0.11) | 655 (0.08) | 4,645 (0.1) | 7,455 (0.08) | 1.01 (0.83 to 1.24) | 0.91 | 1.31 (1.21 to 1.42) | 2.2 × 10−11 | 0.02 | ||
| rs8100241 | 19p13.11 | 17253894 | ANKLE1 | G | A | Yes | 2,483 (0.48) | 653 (0.52) | 4,622 (0.48) | 7,420 (0.52) | 0.95 (0.86 to 1.05) | 0.36 | 0.82 (0.78 to 0.86) | 2.2 × 10−16 | 0.01 | ||
| BRCA2 carriers | rs45631588 | 10q26 | 123341292 | FGFR2 | A | G | Yes | 839 (0.44) | 813 (0.48) | 2,678 (0.44) | 3,881 (0.39) | 1.36 (1.24 to 1.49) | 7.4 × 10−11 | 1.19 (1.09 to 1.3) | 1.7 × 10−4 | 0.06 | |
| rs35850695 | 16q12.1 | 51131844 | TOX3 | G | A | Yes | 839 (0.32) | 813 (0.3) | 2,678 (0.3) | 3,881 (0.26) | 1.2 (1.09 to 1.33) | 3.0 × 10−4 | 1.31 (1.19 to 1.44) | 4.0 × 10−8 | 0.26 | ||
| Nodes | BRCA1 carriers | rs1498125 | 4q24 | 106412012 | TET2 | A | G | No | 1,103 (0.2) | 2,274 (0.23) | 4,419 (0.22) | 7,455 (0.2) | 1.18 (1.1 to 1.25) | 4.4 × 10−7 | 0.98 (0.89 to 1.09) | 0.74 | 5.7 × 10−3 |
| rs10069690 | 5p15.33 | 1332790 | TERT | G | A | Yes | 1,100 (0.3) | 2,271 (0.29) | 4,414 (0.3) | 7,449 (0.26) | 1.19 (1.12 to 1.26) | 6.2 × 10−9 | 1.24 (1.13 to 1.35) | 2.0 × 10−6 | 0.5 | ||
| rs11743632 | 5q32 | 145389263 | SH3RF2 | G | A | No | 1,103 (0.31) | 2,274 (0.37) | 4,418 (0.36) | 7,453 (0.37) | 1.05 (0.99 to 1.11) | 0.1 | 0.8 (0.74 to 0.87) | 2.3 × 10−7 | 6.9 × 10−7 | ||
| rs9383936 | 6q25.1 | 151986307 | C6orf97 | G | A | Yes | 1,103 (0.11) | 2,274 (0.09) | 4,420 (0.1) | 7,455 (0.08) | 1.16 (1.06 to 1.28) | 1.3 × 10−3 | 1.41 (1.24 to 1.6) | 1.3 × 10−7 | 0.03 | ||
| rs2349485 | 7p22 | 8517481 | NXPH1 | A | C | No | 1,091 (0.37) | 2,249 (0.34) | 4,374 (0.34) | 7,398 (0.37) | 0.87 (0.82 to 0.92) | 8.5 × 10−7 | 0.97 (0.90 to 1.06) | 0.53 | 0.03 | ||
| rs11669059 | 19p13.11 | 17261453 | ANKLE1 | A | G | Yes | 1,103 (0.41) | 2,273 (0.4) | 4,418 (0.39) | 7,452 (0.43) | 0.85 (0.81 to 0.9) | 2.9 × 10−9 | 0.86 (0.80 to 0.93) | 2.6 × 10−4 | 0.81 | ||
| BRCA2 carriers | rs2981578 | 10q26 | 123330301 | FGFR2 | A | G | Yes | 795 (0.44) | 1,047 (0.48) | 2,428 (0.47) | 3,819 (0.52) | 0.87 (0.8 to 0.94) | 7.4 × 10−4 | 0.75 (0.68 to 0.82) | 1.0 × 10−9 | 0.02 | |
| rs35850695 | 16q12.1 | 51131844 | TOX3 | G | A | Yes | 804 (0.31) | 1,068 (0.3) | 2,458 (0.3) | 3,881 (0.26) | 1.22 (1.12 to 1.33) | 8.5 × 10−6 | 1.30 (1.18 to 1.44) | 2.6 × 10−7 | 0.36 | ||
aCI, Confidence interval; HR, Hazard ratio; MAF, Mean allele frequency. Single-nucleotide polymorphisms (SNPs) associated at P <10−6 with high- or low-grade breast tumors and lymph node–positive or lymph node–negative breast cancer are shown. The most strongly associated SNPs from each locus are reported. bPosition in build 36. cReference allele. dEffect allele. eVariant located in previously reported breast cancer susceptibility locus. fP-value for the difference in association between high-grade breast cancer and low-grade breast cancer for grade 3 and for the association with lymph node–positive breast cancer and lymph node–negative breast cancer.
Discussion
This is the first comprehensive report, to our knowledge, of the associations of genetic variants with risk of developing breast cancer by tumor subtypes in BRCA1 and BRCA2 carriers. We evaluated the associations with ER, PR and HER2 status; morphologic subtype (ductal or lobular); histological grade; and lymph node status.
Prior to this study, variants at five loci (5p15.33, 6q25.1, 11p15.5, 12p11.22 and 16q12.1) had been shown to be associated with breast cancer risk for both BRCA1 and BRCA2 carriers; variants at four additional loci were known to be associated with breast cancer risk for BRCA1 carriers only (1q32.1, 10q25.2, 14q24.1 and 19p13.11); and variants at six additional loci were known to be associated with risk for BRCA2 carriers (3p24.1, 5p12, 6p24.3, 10q26.12, 11q13 and 12q24.21) [6],[7],[10]. Among the 43 breast cancer susceptibility variants that had not previously been evaluated in mutation carriers, we observed six associations with breast cancer at P <0.05 in BRCA1 carriers (5p33.3, 8q24.21, 11q24.3, 12q22, 16q12.1b and 22q13.1) and three in BRCA2 carriers (6p23, 11q24.3 and 16q12.1b).
After stratifying by ER status, we observed additional associations that were not seen for overall breast cancer. Among the 43 susceptibility variants that were evaluated in mutation carriers for the first time, we identified two additional associations in BRCA1 carriers when stratifying by ER status (3q26.1, 6p25.3) and four in BRCA2 carriers (2q24, 14q13.3, 19q13.3, 22q12.2). Population-based studies have shown that seven of the 74 breast cancer susceptibility variants display stronger associations with ER-negative disease in the general population [9]. Consistent with these findings, SNPs at 1q32.1 (MDM4), 5p15.33, 6q25.1 and 19p13 were associated with ER-negative breast cancer in BRCA1 carriers and SNPs at 2p24.1, 5p15.33 and 6q25.1 in BRCA2 carriers. No data were available for the SNP at 20q11.
We were able to confirm most of the associations with ER and PR subtypes of the 12 SNPs reported in the previous smaller CIMBA study [15]. In addition, two variants from that analysis now displayed evidence at P <0.05: rs13387042 at 2q35 with ER-positive breast cancer in BRCA1 carriers and rs2046210 at 6q25.1 with ER-negative breast cancer in BRCA2 carriers.
We also evaluated the associations of the 74 previously reported breast cancer susceptibility loci with other breast cancer subtypes. Variants at 5p15.33 (TERT), 6q25.1 (ESR1) and 19p13.11 showed associations in all subtype analyses for BRCA1 carriers. These are the most strongly associated loci for overall breast cancer in BRCA1 carriers, but they had not previously been investigated for their roles in subtypes other than ER. Variants at these three loci were associated with ER-, PR- and HER2-negative and TN subtypes. These variants were also associated with risk of high-grade tumors, with some suggestive evidence that this association was different from the association with grades 1 and 2 tumors for SNPs at ESR1 and 19p13. The three loci were associated with ductal as well as non-ductal subtypes and node-positive as well as node-negative breast cancer. For BRCA2 carriers, SNPs at loci 10q26 (FGFR2) and 16q12.1 (TOX3) were associated with all subtypes of breast cancer. SNPs at FGFR2 and TOX3 have consistently been associated with overall and with ER-positive breast cancer in population-based cases [25] as well as in BRCA1/2 carriers [15]. Furthermore, SNPs at these two loci were associated with PR-positive and HER2-negative disease. There was no evidence for a difference in HR estimates by tumor grade, nodal involvement or morphologic subtype (ductal).
It is important to note that for each of the 74 known loci considered, we evaluated only the associations for the specific SNPs that have been reported by the BCAC. We have not considered all genetic variants within a given region. Therefore, we cannot rule out the possibility that more strongly associated variants exist at these loci than the SNPs reported here. Future fine-mapping efforts in conjunction with BCAC analyses should clarify this. Such studies may also identify the causal variant and together with subsequent functional studies gather insights about the functional mechanisms causing these association signals. This in turn may yield insights about the etiology of breast cancer in BRCA1 and BRCA2 carriers.
We compared HRs for the association with overall breast cancer and ER-positive and ER-negative breast cancer for all the 74 known breast cancer susceptibility variants between BRCA1 carriers, BRCA2 carriers and population-based studies using published data from BCAC. Although only some of these variants were associated at P <0.05 with breast cancer in mutation carriers as outlined above, there was nevertheless strong correlation between the HRs for overall breast cancer from the general population and those from BRCA2 carriers, and moderate correlation between the HRs from BRCA1 carriers and from BCAC. These results suggest that many of these variants may also be associated with breast cancer risk for mutation carriers, but the power to detect statistically significant associations in mutation carriers is low. These variants could be employed in risk prediction models for mutation carriers. Future studies should be aimed at assessing the associations of the combined effects of the SNPs in mutation carriers in terms of polygenic risk scores.
We used these comparisons to assess the hypothesis that observed differences in the associations between BRCA1 and BRCA2 carriers and the general population are mediated by ER status. The smaller correlation between the association estimates for BRCA1 carriers and both BCAC and BRCA2 carriers compared with those between BCAC and BRCA2 carriers is consistent with this hypothesis. Moreover, we found stronger correlations between the HRs for ER-positive disease in all three two-way comparisons (ICC = 0.61 to 0.69). The correlation between ORs for ER-negative disease from BCAC and HRs for ER-negative disease from BRCA1 carriers was also strong (ICC = 0.59). Correlations diminished when comparing HR estimates for ER-positive with estimates for ER-negative breast cancer; most of them were not significantly different from zero. This finding suggests that, to a large extent, the difference in SNP association patterns is due to mediating effects of tumor ER status. Under such a model, the effects of common breast cancer susceptibility variants and of BRCA1 and BRCA2 mutations on breast cancer risk would be multiplicative, after taking into account tumor ER status. As BRCA1 carriers are more likely to develop ER-negative disease, SNPs associated with this subtype will be more informative in models to predict overall breast cancer risk in these women, whereas SNPs associated with ER-positive disease will be more useful in BRCA2 carriers. Furthermore, ER-specific SNP associations could be used to provide separate estimates of ER-negative and ER-positive breast cancer risk for mutation carriers. This understanding allows the development of more refined models and is critical to the provision of accurate information to women considering more targeted preventive options.
However, the fact that the correlations between the HR estimates matched for ER status were smaller than 1 implies that there were still some differences in the associations after accounting for ER status. This could be due to sampling error or to real differences in genetic associations between BRCA1 and BRCA2 carriers and the general population. There are examples for such differences: BRCA1 and BRCA2 carrier-specific modifiers, such as the recently identified variant at 6p24, which was associated with breast cancer risk only in BRCA2 carriers [6], and the ovarian cancer susceptibility locus 4q32, which appeared to modify ovarian cancer risk only for BRCA1 carriers [7]. In addition, an ovarian cancer susceptibility locus 17q11.2 identified through population-based data has been shown to display a consistent association in BRCA2 carriers, whereas an association of similar magnitude has been ruled out in BRCA1 carriers [26]. The extent to which genetic susceptibility to breast cancer in mutation carriers and in the general population is shared, as well as the extent to which it is mediated by ER status, need to be quantified systematically by future studies.
We also assessed the associations of over 200,000 SNPs on the iCOGS array. We identified several variants not previously reported that were associated with breast cancer at P <10−6 in the analyses by PR status, HER status, TN phenotype, histological grade, nodal involvement, ductal and lobular morphologic subtypes. In the absence of P-values at genome-wide significance levels for these associations to account for multiple testing, these associations require confirmation through by gathering additional data.
Although this is the largest study of its kind, the statistical power to detect associations of variants conferring small effects with specific tumor characteristics may be low, owing to the limited data available. Future studies of additional BRCA1 and BRCA2 carriers with detailed tumor pathology information on new and previously recruited mutation carriers are needed. In this study, tumor pathology information was retrieved primarily from medical records. Despite extensive efforts, it is difficult to control the quality of these data. If there is low reproducibility in the classification of tumor characteristics for some samples, this could potentially add to the sampling error and make it more difficult to detect subtype-specific associations.
Conclusions
We have identified additional genetic modifiers of breast cancer risk for mutation carriers among reported breast cancer susceptibility loci. Large differences in absolute risk are expected between mutation carriers who carry many and mutation carriers who carry few risk alleles of modifying variants [6],[7]. Therefore, in combination with previously identified modifiers, these variants may be of value for cancer risk prediction. Moreover, our results show that, to a large extent, the differences in breast cancer associations of known breast cancer susceptibility loci between BRCA1 and BRCA2 carriers and the general population are due to differences in the prevalence of tumor subtypes in BRCA1 and BRCA2 carriers. Estimates of the risks associated with these genetic variants based on large population-based association studies are likely to be applicable also to mutation carriers after taking ER status into account. Our results thus have implications for developing risk prediction models for breast cancer subtype-specific risks in mutation carriers that incorporate the effects of these SNPs.
Additional file
Electronic supplementary material
Additional file 1: Table S1.: Ethics committees that granted approval for the access and use of the data for this study. Table S2. Hormone receptor definitions used by the studies. Table S3. Associations of breast cancer susceptibility variants with breast cancer by tumor ER status after excluding prevalent cases. Table S4. Associations of breast cancer susceptibility variants with breast cancer by tumor PR status. Table S5. Associations of breast cancer susceptibility variants with breast cancer by tumor HER2 status. Table S6. Associations of breast cancer susceptibility variants with breast cancer by tumor triple-negative status. Table S7. Associations of breast cancer susceptibility variants with ductal breast cancer. Table S8. Associations of breast cancer susceptibility variants with lobular breast cancer. Table S9. Associations of breast cancer susceptibility variants with breast cancer by tumor grade. Table S10. Associations of breast cancer susceptibility variants with breast cancer by nodal involvement. Figure S1. QQ plots for SNP associations with ER-positive and -negative breast cancer in BRCA1 carriers. Figure S2. QQ plots for SNP associations with ER-positive and -negative breast cancer in BRCA2 carriers. Figure S3. Comparison of the breast cancer associations of breast cancer susceptibility loci in BCAC and in BRCA1 and BRCA2 carriers. Figure S4. Comparison of the associations of breast cancer susceptibility loci by ER status in BRCA1 and in BRCA2 carriers. Figure S5. Comparison of the associations of breast cancer susceptibility loci by ER status in population-based studies (BCAC) and in BRCA1 carriers. Figure S6. Comparison of the associations of breast cancer susceptibility loci by ER status in population-based studies (BCAC) and in BRCA2 carriers. (PDF 750 KB)
Below are the links to the authors’ original submitted files for images.
Acknowledgements
This study would not have been possible without the contributions of the following people: Per Hall (COGS); Paul Pharoah, Kyriaki Michailidou, Manjeet K Bolla, Qin Wang (BCAC), Andrew Berchuck (OCAC), Rosalind A Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Alison M Dunning, Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Anna Gonzalez-Neira and the staff of the CNIO genotyping unit, Daniel C Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of McGill University and Génome Québec Innovation Centre, Stig E Bojesen, Sune F Nielsen, Borge G Nordestgaard, and the staff of the Copenhagen DNA laboratory, Sharon A Windebank, Christopher A Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility; Maggie Angelakos, Judi Maskiell, Gillian Dite, Helen Tsimiklis; members and participants in the New York site of the Breast Cancer Family Registry; members and participants in the Ontario Familial Breast Cancer Registry; Vilius Rudaitis, Laimonas Griškevičius, Drs Janis Eglitis, Anna Krilova and Aivars Stengrevics; the families who contribute to the BMBSA study; Yuan Chun Ding and Linda Steele; Alicia Barroso, Rosario Alonso and Guillermo Pita; Monica Barile and Irene Feroce of the Istituto Europeo di Oncologia, Milan, Italy; Alessandra Viel of the Centro di Riferimento Oncologico, IRCCS, Aviano (PN), Laura Ottini of the “Sapienza” University, Rome, Italy; Antonella Savarese and Aline Martayan of the Istituto Nazionale Tumori Regina Elena, Rome, Italy; Stefania Tommasi and Brunella Pilato of the Istituto Nazionale Tumori “Giovanni Paolo II”, Bari, Italy; Loris Bernard and the personnnel of the Cogentech Cancer Genetic Test Laboratory, Milan, Italy. The EMBRACE Collaborating Centers are: Coordinating Centre, Cambridge: Debra Frost, Steve Ellis, Radka Platte, Jo Perkins; North of Scotland Regional Genetics Service, Aberdeen: Zosia Miedzybrodzka, Helen Gregory; Northern Ireland Regional Genetics Service, Belfast: Patrick Morrison, Lisa Jeffers; West Midlands Regional Clinical Genetics Service, Birmingham: Trevor Cole, Kai-ren Ong, Jonathan Hoffman; South West Regional Genetics Service, Bristol: Alan Donaldson, Margaret James; East Anglian Regional Genetics Service, Cambridge: Marc Tischkowitz, Joan Paterson, Sarah Downing, Amy Taylor; Medical Genetics Services for Wales, Cardiff: Alexandra Murray, Mark T Rogers, Emma McCann; St James’s Hospital, Dublin & National Centre for Medical Genetics, Dublin: M John Kennedy, David Barton; South East of Scotland Regional Genetics Service, Edinburgh: Mary Porteous, Sarah Drummond; Peninsula Clinical Genetics Service, Exeter: Carole Brewer, Emma Kivuva, Anne Searle, Selina Goodman, Kathryn Hill; West of Scotland Regional Genetics Service, Glasgow: Rosemarie Davidson, Victoria Murday, Nicola Bradshaw, Lesley Snadden, Mark Longmuir, Catherine Watt, Sarah Gibson, Eshika Haque, Ed Tobias, Alexis Duncan; South East Thames Regional Genetics Service, Guy’s Hospital London: Louise Izatt, Chris Jacobs, Caroline Langman; North West Thames Regional Genetics Service, Harrow: Huw Dorkins; Leicestershire Clinical Genetics Service, Leicester: Julian Barwell; Yorkshire Regional Genetics Service, Leeds: Julian Adlard, Gemma Serra-Feliu; Cheshire & Merseyside Clinical Genetics Service, Liverpool: Ian Ellis, Claire Foo; Manchester Regional Genetics Service, Manchester: D Gareth Evans, Fiona Lalloo, Jane Taylor; North East Thames Regional Genetics Service, NE Thames, London: Lucy Side, Alison Male, Cheryl Berlin; Nottingham Centre for Medical Genetics, Nottingham: Jacqueline Eason, Rebecca Collier; Northern Clinical Genetics Service, Newcastle: Fiona Douglas, Oonagh Claber, Irene Jobson; Oxford Regional Genetics Service, Oxford: Lisa Walker, Diane McLeod, Dorothy Halliday, Sarah Durell, Barbara Stayner; the Institute of Cancer Research and Royal Marsden NHS Foundation Trust: Ros Eeles, Susan Shanley, Nazneen Rahman, Richard Houlston, Elizabeth Bancroft, Elizabeth Page, Audrey Ardern-Jones, Kelly Kohut, Jennifer Wiggins, Elena Castro, Anita Mitra; North Trent Clinical Genetics Service, Sheffield: Jackie Cook, Oliver Quarrell, Cathryn Bardsley; South West Thames Regional Genetics Service, London: Shirley Hodgson, Sheila Goff, Glen Brice, Lizzie Winchester, Charlotte Eddy, Vishakha Tripathi, Virginia Attard; Wessex Clinical Genetics Service, Princess Anne Hospital, Southampton: Diana Eccles, Anneke Lucassen, Gillian Crawford, Donna McBride, Sarah Smalley; JoEllen Weaver and Dr Betsy Bove; National Cancer Genetics Network UNICANCER Genetic Group, France; GEMO collaborating groups: Coordinating Centres, Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Hospices Civils de Lyon, Centre Léon Bérard, & Equipe Génétique du cancer du sein, Centre de Recherche en Cancérologie de Lyon: Olga Sinilnikova, Sylvie Mazoyer, Francesca Damiola, Laure Barjhoux, Carole Verny-Pierre, Alain Calender, Sophie Giraud, Mélanie Léone; and Service de Génétique Oncologique, Institut Curie, Paris: Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Bruno Buecher, Claude Houdayer, Virginie Moncoutier, Muriel Belotti, Carole Tirapo, Antoine de Pauw; Institut Gustave Roussy, Villejuif: Brigitte Bressac-de-Paillerets, Olivier Caron; Centre Jean Perrin, Clermont-Ferrand: Yves-Jean Bignon, Nancy Uhrhammer; Centre Léon Bérard, Lyon: Christine Lasset, Valérie Bonadona, Sandrine Handallo. Centre François Baclesse, Caen: Agnès Hardouin, Pascaline Berthet; Institut Paoli Calmettes, Marseille: Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, Audrey Remenieras, François Eisinger; CHU Arnaud-de-Villeneuve, Montpellier: Isabelle Coupier, Pascal Pujol; Centre Oscar Lambret, Lille: Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin, Claude Adenis; Hôpital René Huguenin/Institut Curie, St Cloud: Etienne Rouleau, Rosette Lidereau, Liliane Demange, Catherine Nogues; Centre Paul Strauss, Strasbourg: Danièle Muller, Jean-Pierre Fricker; Institut Bergonié, Bordeaux: Emmanuelle Barouk-Simonet, Françoise Bonnet, Virginie Bubien, Nicolas Sevenet, Michel Longy; Institut Claudius Regaud, Toulouse: Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel; CHU Grenoble: Dominique Leroux, Hélène Dreyfus, Christine Rebischung, Magalie Peysselon; CHU Dijon: Fanny Coron, Laurence Faivre; CHU St-Etienne: Fabienne Prieur, Marine Lebrun, Caroline Kientz; Hôtel Dieu Centre Hospitalier, Chambéry: Sandra Fert Ferrer; Centre Antoine Lacassagne, Nice: Marc Frénay; CHU Limoges: Laurence Vénat-Bouvet; CHU Nantes: Capucine Delnatte; CHU Bretonneau, Tours: Isabelle Mortemousque; Groupe Hospitalier Pitié-Salpétrière, Paris: Florence Coulet, Chrystelle Colas, Florent Soubrier; CHU Vandoeuvre-les-Nancy: Johanna Sokolowska, Myriam Bronner; CHU Besançon: Marie-Agnès Collonge-Rame, Alexandre Damette; Creighton University, Omaha, NE, USA: Henry T Lynch, Carrie L Snyder; technical support of Ilse Coene en Brecht Crombez; Pedro Perez Segura, Atocha Romero, Paula Diaque and families; Drs Kristiina Aittomäki, Carl Blomqvist and Kirsimari Aaltonen, Taru A Muranen and registered nurses Irja Erkkilä and Virpi Palola. HEBON consists of the following collaborating centers: Coordinating center: Netherlands Cancer Institute, Amsterdam: MA Rookus, FBL Hogervorst, FE van Leeuwen, S Verhoef, MK Schmidt, JL de Lange, R Wijnands; Erasmus Medical Center, Rotterdam: JM Collée, AMW van den Ouweland, MJ Hooning, C Seynaeve, CHM van Deurzen, IM Obdeijn; Leiden University Medical Center, the Netherlands: CJ van Asperen, JT Wijnen, RAEM Tollenaar, P Devilee, TCTEF van Cronenburg; Radboud University Nijmegen Medical Center, the Netherlands: CM Kets, AR Mensenkamp; University Medical Center Utrecht, the Netherlands: MGEM Ausems, RB van der Luijt; Amsterdam Medical Center: CM Aalfs, TAM van Os; VU University Medical Center, Amsterdam: JJP Gille, Q Waisfisz, HEJ Meijers-Heijboer; University Hospital Maastricht, the Netherlands: EB Gómez-Garcia, MJ Blok; University Medical Center Groningen, the Netherlands: JC Oosterwijk, AH van der Hout, MJ Mourits, GH de Bock; The Netherlands Foundation for the Detection of Hereditary Tumours, Leiden: HF Vasen; The Netherlands Cancer Registry: S Siesling; The Dutch Pathology Registry (PALGA): LIH Overbeek; Hong Kong Sanatorium & Hospital; Hungarian Breast and Ovarian Cancer Study Group members: Janos Papp, Tibor Vaszko, Aniko Bozsik, Timea Pocza, Judit Franko, Maria Balogh, Gabriella Domokos, Judit Ferenczi, Department of Molecular Genetics, National Institute of Oncology, Budapest, Hungary, and the clinicians and patients; Oncogenetics Group and the High Risk and Cancer Prevention Unit of the University Hospital Vall d’Hebron led by Dr J Balmaña; ICO Hereditary Cancer Program team led by Dr Gabriel Capella; Dr Martine Dumont, Martine Tranchant. JS is Chairholder of the Canada Research Chair in Oncogenetics. JS and PS were part of the QC and Genotyping coordinating group of iCOGS (BCAC and CIMBA); Drs Ana Peixoto, Catarina Santos, Patrícia Rocha and Pedro Pinto; Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study and the many families who contribute to kConFab; Modifier Study of Quantitative Effects on Disease (MODSQUAD) acknowledges ModSQuaD members Csilla Szabo (National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA); Lenka Foretova and Eva Machackova (Department of Cancer Epidemiology and Genetics, Masaryk Memorial Cancer Institute, Brno, Czech Republic); and Michal Zikan, Petr Pohlreich and Zdenek Kleibl (Oncogynecologic Center and Department of Biochemistry and Experimental Oncology, First Faculty of Medicine, Charles University, Prague, Czech Republic); the NICCC National Familial Cancer Consultation Service team led by Sara Dishon, the lab team led by Dr Flavio Lejbkowicz, and the research field operations team led by Dr Mila Pinchev; Teresa Selander, Nayana Weerasooriya; Kevin Sweet, Caroline Craven, and Michelle O’Conor; The Ohio State University (OSU) Human Genetics Sample Bank; Yip Cheng Har, Nur Aishah Mohd Taib, Phuah Sze Yee, Norhashimah Hassan and all the research nurses, research assistants and doctors involved in the MyBrCa Study; Philip Iau, Sng Jen-Hwei and Sharifah Nor Akmal; Meirav comprehensive breast cancer center team at the Sheba Medical Center; Swedish scientists participating as SWE-BRCA collaborators are: from Lund University and University Hospital: Åke Borg, Håkan Olsson, Helena Jernström, Karin Henriksson, Katja Harbst, Maria Soller, Niklas Loman, Ulf Kristoffersson; from Gothenburg Sahlgrenska University Hospital: Anna Öfverholm, Margareta Nordling, Per Karlsson, Zakaria Einbeigi; from Stockholm and Karolinska University Hospital: Anna von Wachenfeldt, Annelie Liljegren, Annika Lindblom, Brita Arver, Gisela Barbany Bustinza, Johanna Rantala; from Umeå University Hospital: Beatrice Melin, Christina Edwinsdotter Ardnor, Monica Emanuelsson; from Uppsala University: Hans Ehrencrona, Maritta Hellström Pigg, Richard Rosenquist; from Linköping University Hospital: Marie Stenmark-Askmalm, Sigrun Liedgren; Cecilia Zvocec, Qun Niu, physicians, genetic counselors, research nurses and staff of the Cancer Risk Clinic, and the many families who contribute to our program; Joyce Seldon, MSGC, and Lorna Kwan, MPH; Dr Robert Nussbaum and the following genetic counselors for participant recruitment: Beth Crawford, Kate Loranger, Julie Mak, Nicola Stewart, Robin Lee, Amie Blanco and Peggy Conrad; Salina Chan; Paul Pharoah, Simon Gayther, Susan Ramus, Carole Pye, Patricia Harrington and Eva Wozniak; Geoffrey Lindeman, Marion Harris, Martin Delatycki of the Victorian Familial Cancer Trials Group; Sarah Sawyer, Rebecca Driessen and Ella Thompson.
Funding statement
The CIMBA data management and analysis are funded through Cancer Research UK grant C12292/A11174. ACA is a Senior Cancer Research UK Research Fellow. Funding for the iCOGS infrastructure came from: the European Community’s Seventh Framework Programme under grant agreement 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112: the Genetic Associations and Mechanisms in Oncology (GAME-ON) initiative), the US Department of Defense (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. This work was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US government or the BCFR. This work was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this article does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US government or the BCFR. This work was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US government or the BCFR. BFBOCC is partly supported by: Lithuania (BFBOCC-LT): Research Council of Lithuania grant LIG-07/2012; Latvia (BFBOCC-LV) is partly supported by LSC grant 10.0010.08 and in part by a grant from the ESF 2009/0220/1DP/1.1.1.2.0/09/APIA/VIAA/016 and Liepaja’s municipal council.
BIDMC is supported by the Breast Cancer Research Foundation. BRCA-gene mutations and breast cancer in South African women (BMBSA) was supported by grants from the Cancer Association of South Africa (CANSA) to Elizabeth J van Rensburg. SLN was partially supported by the Morris and Horowitz Families Endowed Professorship. This work was supported by the NEYE Foundation. The CNIO study was supported by Spanish Association against Cancer (AECC08), RTICC 06/0020/1060 and FISPI12/00070 and Mutua Madrileña Foundation (FMMA). City of Hope Clinical Cancer Genetics Community Network and the Hereditary Cancer Research Registry, supported in part by Award Number RC4CA153828 (Principal Investigator: J Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funds from Italian citizens who allocated the “5 × 1000” share of their tax payment, according to Italian laws, in support of the Fondazione IRCCS Istituto Nazionale Tumori (to SM), and of IRCCS AOU San Martino, IST Istituto Nazionale per la Ricerca sul Cancro (to LV); Italian Association for Cancer Research (AIRC) to GG; Associazione CAOS-Varese (to MGT). SH is supported by an NHMRC Program Grant (to GCT). GCT is an NHMRC Senior Principal Research Fellow. This research has been cofinanced by the European Union (European Social Fund (ESF)) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF), Research Funding Program of the General Secretariat for Research & Technology: ARISTEIA, investing in knowledge society through the European Social Fund. The DKFZ study was supported by the DKFZ. EMBRACE is supported by Cancer Research UK grants C1287/A10118, C1287/A16563 and C1287/A17523. D Gareth Evans and Fiona Lalloo are supported by an NIHR grant to the Biomedical Research Centre, Manchester. The investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Ros Eeles and Elizabeth Bancroft are supported by Cancer Research UK grant C5047/A8385. The authors acknowledge support from The University of Kansas Cancer Center (P30 CA168524) and the Kansas Bioscience Authority Eminent Scholar Program. AKG was funded by grants 5U01 CA113916, R01 CA140323 from the National Institutes of Health and by the Chancellors Distinguished Chair in Biomedical Sciences Professorship. The German Consortium of Hereditary Breast and Ovarian Cancer (GC-HBOC) is supported by the German Cancer Aid (grant no 109076, Rita K Schmutzler) and by the Center for Molecular Medicine Cologne (CMMC). The study was supported by the Ligue National Contre le Cancer; the Association “Le cancer du sein, parlons-en!” Award; and the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program. Tom Van Maerken is senior clinical investigator of FWO. This study was supported by National Cancer Institute grants to the Gynecologic Oncology Group (GOG) Administrative Office and Tissue Bank (CA 27469), the GOG Statistical and Data Center (CA 37517), and GOG’s Cancer Prevention and Control Committee (CA 101165). Drs Greene, Mai and Savage were supported by funding from the NCI Intramural Research Program. HCSC was supported by a grant RD12/00369/0006 and 12/00539 from ISCIII (Spain), partially supported by European Regional Development FEDER funds. The HEBCS was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society and the Sigrid Juselius Foundation. The HEBON study is supported by the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088, NKI2007-3756, the Netherlands Organization of Scientific Research grant NWO 91109024, the Pink Ribbon grant 110005 and the BBMRI grant NWO 184.021.007/CP46. HEBON thanks the registration teams of the Comprehensive Cancer Centre Netherlands and Comprehensive Centre South (together the Netherlands Cancer Registry) and PALGA (Dutch Pathology Registry) for part of the data collection. HRBCP is supported by The Hong Kong Hereditary Breast Cancer Family Registry and the Dr Ellen Li Charitable Foundation, Hong Kong.
The Hungarian Breast and Ovarian Cancer Study was supported by Hungarian Research Grants KTIA-OTKA CK-80745 and OTKA K-112228. ICO: Contract grant sponsor: Asociación Española Contra el Cáncer, Spanish Health Research Fund; Carlos III Health Institute; Catalan Health Institute and Autonomous Government of Catalonia. Contract grants ISCIIIRETIC RD06/0020/1051, RD12/0036/008, PI10/01422, PI10/00748, PI13/00285 and 2009SGR290. The IHCC was supported by Grant PBZ_KBN_122/P05/2004. The ILUH group was supported by the Icelandic Association “Walking for Breast Cancer Research” and by the Landspitali University Hospital Research Fund. This work was supported by the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program, Canadian Breast Cancer Research Alliance grant 019511 and Ministry of Economic Development, Innovation and Export Trade grant PSR-SIIRI-701. IOVHBOCS is supported by Ministero della Salute and “5 × 1000” Istituto Oncologico Veneto grant. This study was in part supported by Liga Portuguesa Contra o Cancro. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. GCT and ABS are supported by the NHMRC Research Fellowship scheme. KOHBRA is supported by a grant from the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs, Republic of Korea (1020350). MAYO is supported by NIH grant CA128978, an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a US Department of Defense Ovarian Cancer Idea award (W81XWH-10-1-0341) and a grant from the Breast Cancer Research Foundation.
Jewish General Hospital Weekend to End Breast Cancer, Quebec Ministry of Economic Development, Innovation and Export Trade. MODSQUAD was supported by MH CZ - DRO (MMCI, 00209805) and by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101) (to LF), and by Charles University in Prague project UNCE204024 (MZ). MSKCC is supported by grants from the Breast Cancer Research Foundation and Robert and Kate Niehaus Clinical Cancer Genetics Initiative 1R01 CA149429-01. The research of MH Greene and PL Mai was supported by the Intramural Research Program of the US National Cancer Institute, NIH, and by support services contracts N02-CP-11019-50 and N02-CP-65504 with Westat (Rockville, MD, USA). NICCC is supported by Clalit Health Services in Israel. Some of its activities are supported by the Israel Cancer Association and the Breast Cancer Research Foundation (BCRF), New York.
This work has been supported by the Russian Foundation for Basic Research (grant 13-04-92613).
This work was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US government or the BCFR. OSUCCG is supported by The Ohio State University Comprehensive Cancer Center. This work was supported by the ITT (Istituto Toscano Tumori) grants 2011–2013; the Ministry of Science, Technology and Innovation, Ministry of Higher Education (UM.C/HlR/MOHE/06); and the Cancer Research Initiatives Foundation. This project was partially funded through a grant by the Israel cancer association and the funding for the Israeli Inherited breast cancer consortium.
SWE-BRCA collaborators are supported by the Swedish Cancer Society. UCHICAGO is supported by NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA125183), R01 CA142996, 1U01 CA161032 and by the Ralph and Marion Falk Medical Research Trust, the Entertainment Industry Fund National Women’s Cancer Research Alliance and the Breast Cancer Research Foundation. OIO is an ACS Clinical Research Professor. Jonsson Comprehensive Cancer Center Foundation; Breast Cancer Research Foundation. UCSF Cancer Risk Program and Helen Diller Family Comprehensive Cancer Center. UKFOCR was supported by a project grant from CRUK to Paul Pharoah. National Institutes of Health (NIH) (R01 CA102776 and R01 CA083855; Breast Cancer Research Foundation; Susan G Komen Foundation for the cure, Basser Research Center for BRCA. Victorian Cancer Agency, Cancer Australia, National Breast Cancer Foundation. The Women’s Cancer Program (WCP) at the Samuel Oschin Comprehensive Cancer Institute is funded by the American Cancer Society Early Detection Professorship (SIOP-06-258-01-COUN).
Abbreviations
- BC
Breast cancer
- BCAC
Breast Cancer Association Consortium
- CIMBA
Consortium of Investigators of Modifiers of BRCA1/2
- ER
Estrogen receptor
- GWAS
Genome-wide association study
- HER2
Human epidermal growth factor receptor 2
- HR
Hazard ratio
- ICC
Intraclass correlation
- OR
Odds ratio
- PR
Progesterone receptor
- SNP
Single-nucleotide polymorphism
- TN
Triple-negative
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13058-014-0492-9) contains supplementary material, which is available to authorized users.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KBK, ACA, SLN, MR drafted the initial manuscript. KBK performed the statistical analyses. ACA, SLN, MR, AMM, DB, LM, KBK, ILA, ABS, MKS, RKS, CE, BW, HN, MT, MS, PR, SJR, SMD and FJC are members of the CIMBA pathology working group and participated in the design of the study. LM and DB are the CIMBA database managers. GCT initiated and coordinated CIMBA. All authors except KBK, DB, LM and AL acquired phenotypic data and DNA samples or performed SNP genotyping. All authors read and approved the final manuscript.
Contributor Information
Karoline B Kuchenbaecker, Email: kbk21@medschl.cam.ac.uk.
Susan L Neuhausen, Email: sneuhausen@coh.org.
Mark Robson, Email: robsonm@mskcc.org.
Daniel Barrowdale, Email: daniel@srl.cam.ac.uk.
Lesley McGuffog, Email: lesley@srl.cam.ac.uk.
Anna Marie Mulligan, Email: annamarie.mulligan@uhn.ca.
Irene L Andrulis, Email: andrulis@lunenfeld.ca.
Amanda B Spurdle, Email: amanda.spurdle@qimrberghofer.edu.au.
Marjanka K Schmidt, Email: mk.schmidt@nki.nl.
Rita K Schmutzler, Email: rita.schmutzler@uk-koeln.de.
Christoph Engel, Email: christoph.engel@imise.uni-leipzig.de.
Barbara Wappenschmidt, Email: barbara.wappenschmidt@uk-koeln.de.
Heli Nevanlinna, Email: heli.nevanlinna@hus.fi.
Mads Thomassen, Email: mads.thomassen@ouh.rsyd.dk.
Melissa Southey, Email: msouthey@unimelb.edu.au.
Paolo Radice, Email: paolo.radice@istitutotumori.mi.it.
Susan J Ramus, Email: sramus@usc.edu.
Susan M Domchek, Email: susan.domchek@uphs.upenn.edu.
Katherine L Nathanson, Email: knathans@exchange.upenn.edu.
Andrew Lee, Email: andrewl@srl.cam.ac.uk.
Sue Healey, Email: sue.healey@qimrberghofer.edu.au.
Robert L Nussbaum, Email: nussbaumr@humgen.ucsf.edu.
Timothy R Rebbeck, Email: rebbeck@exchange.upenn.edu.
Banu K Arun, Email: barun@mdanderson.org.
Paul James, Email: paul.james@petermac.org.
Beth Y Karlan, Email: beth.karlan@cshs.org.
Jenny Lester, Email: jenny.lester@cshs.org.
Ilana Cass, Email: ilana.cass@cshs.org.
Breast Cancer Family Registry, Email: esther.john@cpic.org.
Mary Beth Terry, Email: mt146@cumc.columbia.edu.
Mary B Daly, Email: mary.daly@fccc.edu.
David E Goldgar, Email: david.goldgar@hsc.utah.edu.
Saundra S Buys, Email: saundra.buys@hci.utah.edu.
Ramunas Janavicius, Email: ramunas.janavicius@santa.lt.
Laima Tihomirova, Email: laima@biomed.lu.lv.
Nadine Tung, Email: ntung@bidmc.harvard.edu.
Cecilia M Dorfling, Email: celmari.dorfling@up.ac.za.
Elizabeth J van Rensburg, Email: lizette.vanrensburg@up.ac.za.
Linda Steele, Email: lsteele@coh.org.
Thomas v O Hansen, Email: tvoh@rh.dk.
Bent Ejlertsen, Email: bent.ejlertsen@regionh.dk.
Anne-Marie Gerdes, Email: anne-marie.gerdes@regionh.dk.
Finn C Nielsen, Email: fcn@rh.dk.
Joe Dennis, Email: jgd29@cam.ac.uk.
Julie Cunningham, Email: cunningham.julie@mayo.edu.
Steven Hart, Email: hart.steven@mayo.edu.
Susan Slager, Email: slager.susan@mayo.edu.
Ana Osorio, Email: aosorio@cnio.es.
Javier Benitez, Email: jbenitez@cnio.es.
Mercedes Duran, Email: merche@ibgm.uva.es.
Jeffrey N Weitzel, Email: jweitzel@coh.org.
Isaac Tafur, Email: oncmd98@yahoo.com.
Mary Hander, Email: mhander@kh.org.
Paolo Peterlongo, Email: paolo.peterlongo@ifom.eu.
Siranoush Manoukian, Email: siranoush.manoukian@istitutotumori.mi.it.
Bernard Peissel, Email: bernard.peissel@istitutotumori.mi.it.
Gaia Roversi, Email: gaia.roversi@istitutotumori.mi.it.
Giulietta Scuvera, Email: giulietta.scuvera@istitutotumori.m.it.
Bernardo Bonanni, Email: bernardo.bonanni@ieo.it.
Paolo Mariani, Email: paolo.mariani@ifom.eu.
Sara Volorio, Email: sara.volorio@ifom.eu.
Riccardo Dolcetti, Email: rdolcetti@cro.it.
Liliana Varesco, Email: liliana.varesco@hsanmartino.it.
Laura Papi, Email: l.papi@dfc.unifi.it.
Maria Grazia Tibiletti, Email: mariagrazia.tibiletti@ospedale.varese.it.
Giuseppe Giannini, Email: giuseppe.giannini@uniroma1.it.
Florentia Fostira, Email: florentia@rrp.demokritos.gr.
Irene Konstantopoulou, Email: reena@rrp.demokritos.gr.
Judy Garber, Email: judy_garber@dfci.harvard.edu.
Ute Hamann, Email: u.hamann@dkfz-heidelberg.de.
Alan Donaldson, Email: alan.donaldson@uhbristol.nhs.uk.
Carole Brewer, Email: carole.brewer1@nhs.net.
Claire Foo, Email: claire.foo@lwh.nhs.uk.
D Gareth Evans, Email: gareth.evans@cmft.nhs.uk.
Debra Frost, Email: djsf2@medschl.cam.ac.uk.
Diana Eccles, Email: d.m.eccles@soton.ac.uk.
Fiona Douglas, Email: fiona.douglas@nuth.nhs.uk.
Angela Brady, Email: a.brady@imperial.ac.uk.
Jackie Cook, Email: jackie.cook@sch.nhs.uk.
Marc Tischkowitz, Email: mdt33@cam.ac.uk.
Julian Adlard, Email: julian.adlard@leedsth.nhs.uk.
Julian Barwell, Email: julian.barwell@uhl-tr.nhs.uk.
Kai-ren Ong, Email: kai-ren.ong@bwhct.nhs.uk.
Lisa Walker, Email: lisa.walker@ouh.nhs.uk.
Louise Izatt, Email: louise.izatt@gstt.nhs.uk.
Lucy E Side, Email: sidel@gosh.nhs.uk.
M John Kennedy, Email: jkennedy@stjames.ie.
Mark T Rogers, Email: mark.rogers@wales.nhs.uk.
Mary E Porteous, Email: mary.porteous@ed.ac.uk.
Patrick J Morrison, Email: patrick.morrison@belfasttrust.hscni.net.
Radka Platte, Email: rp356@medschl.cam.ac.uk.
Ros Eeles, Email: rosalind.eeles@icr.ac.uk.
Rosemarie Davidson, Email: rosemarie.davidson@ggc.scot.nhs.uk.
Shirley Hodgson, Email: shodgson@sgul.ac.uk.
Steve Ellis, Email: sde26@medscl.cam.ac.uk.
Andrew K Godwin, Email: agodwin@kumc.edu.
Kerstin Rhiem, Email: kerstin.rhiem@uk-koeln.de.
Alfons Meindl, Email: alfons.meindl@lrz.tu-muenchen.de.
Nina Ditsch, Email: nina.ditsch@med.uni-muenchen.de.
Norbert Arnold, nkarnold@email.uni-kiel.de.
Hansjoerg Plendl, Email: plendl@medgen.uni-kiel.de.
Dieter Niederacher, Email: niederac@uni-duesseldorf.de.
Christian Sutter, Email: c.sutter@med.uni-heidelberg.de.
Doris Steinemann, Email: steinemann.doris@mh-hannover.de.
Nadja Bogdanova-Markov, Email: nadja.bogdanova-markow@ukmuenster.de.
Karin Kast, Email: karin.kast@uniklinikum-dresden.de.
Raymonda Varon-Mateeva, Email: raymonda.varon-mateeva@charite.de.
Shan Wang-Gohrke, Email: shan.wang-gohrke@uniklinik.ulm.de.
Andrea Gehrig, Email: gehrig@biozentrum.uni-wuerzburg.de.
Birgid Markiefka, Email: birgid.markiefka@uk-koeln.de.
Bruno Buecher, Email: bruno.buecher@curie.net.
Cédrick Lefol, Email: cedrick.lefol@curie.net.
Dominique Stoppa-Lyonnet, Email: dominique.stoppa-lyonnet@curie.net.
Etienne Rouleau, Email: etienne.rouleau@curie.net.
Fabienne Prieur, Email: fabienne.prieur@chu-st-etienne.fr.
Francesca Damiola, Email: francesca.damiola@lyon.unicancer.fr.
Laure Barjhoux, Email: laure.barjhoux@lyon.unicancer.fr.
Laurence Faivre, Email: laurence.faivre@chu-dijon.fr.
Michel Longy, Email: longy@bergonie.org.
Nicolas Sevenet, Email: sevenet@bergonie.org.
Olga M Sinilnikova, Email: olga.sinilnikova@lyon.unicancer.fr.
Sylvie Mazoyer, Email: sylvie.mazoyer@lyon.unicancer.fr.
Valérie Bonadona, Email: valerie.bonadona@lyon.unicancer.fr.
Virginie Caux-Moncoutier, Email: virginie.moncoutier@curie.net.
Claudine Isaacs, Email: isaacsc@georgetown.edu.
Tom Van Maerken, Email: tom.vanmaerken@ugent.be.
Kathleen Claes, Email: kathleen.claes@ugent.be.
Marion Piedmonte, Email: mpiedmon@gogstats.org.
Lesley Andrews, Email: lesley.andrews@sesiahs.health.nsw.gov.au.
John Hays, Email: hays.161@osu.edu.
Gustavo C Rodriguez, Email: grodriguez@northshore.org.
Trinidad Caldes, Email: trinidad.caldes@salud.madrid.org.
Miguel de la Hoya, Email: mdhoya@hotmail.com.
Sofia Khan, Email: sofia.khan@helsinki.fi.
Frans BL Hogervorst, Email: f.hogervorst@nki.nl.
Cora M Aalfs, Email: c.m.aalfs@amc.uva.nl.
JL de Lange, Email: j.d.lange@nki.nl.
Hanne EJ Meijers-Heijboer, Email: h.meijers@vumc.nl.
Annemarie H van der Hout, Email: a.h.van.der.hout@umcg.nl.
Juul T Wijnen, Email: j.wijnen@lumc.nl.
KEP van Roozendaal, Email: kees.vanroozendaal@mumc.nl.
Arjen R Mensenkamp, Email: arjen.mensenkamp@radboudumc.nl.
Ans MW van den Ouweland, Email: a.vandenouweland@erasmusmc.nl.
Carolien HM van Deurzen, Email: c.h.m.vandeurzen@erasmusmc.nl.
Rob B van der Luijt, Email: r.b.vanderluijt@umcutrecht.nl.
Edith Olah, Email: e.olah@oncol.hu.
Orland Diez, Email: odiez@vhebron.net.
Conxi Lazaro, Email: clazaro@iconcologia.net.
Ignacio Blanco, Email: iblanco@iconcologia.net.
Alex Teulé, Email: ateule@iconcologia.net.
Mireia Menendez, Email: mmenendez@iconcologia.net.
Anna Jakubowska, Email: aniaj@pum.edu.pl.
Jan Lubinski, Email: lubinski@pum.edu.pl.
Cezary Cybulski, Email: cearycy@pum.edu.pl.
Jacek Gronwald, Email: jgron@pum.edu.pl.
Katarzyna Jaworska-Bieniek, Email: ka_jaworska@wp.pl.
Katarzyna Durda, Email: k.durda@onet.pl.
Adalgeir Arason, Email: adalgeir@landspitali.is.
Christine Maugard, Email: christinemaugard@gmail.com.
Penny Soucy, Email: penny.soucy@crchul.ulaval.ca.
Marco Montagna, Email: montagna@unipd.it.
Simona Agata, Email: simona.agata@ioveneto.it.
Manuel R Teixeira, Email: manuel.teixeira@ipoporto.min-saude.pt.
Curtis Olswold, Email: olswold.curtis@mayo.edu.
Noralane Lindor, Email: lindor.noralane@mayo.edu.
Vernon S Pankratz, Email: pankratz.vernon@mayo.edu.
Emily Hallberg, Email: hallberg.emily@mayo.edu.
Xianshu Wang, Email: wang.xianshu@mayo.edu.
Csilla I Szabo, Email: szaboc@mail.nih.gov.
Joseph Vijai, Email: josephv@mskcc.org.
Lauren Jacobs, Email: jacobsl@mskcc.org.
Marina Corines, Email: corinesm@mskcc.org.
Anne Lincoln, Email: lincolna@mskcc.org.
Andreas Berger, Email: andreas.berger@meduniwien.ac.at.
Anneliese Fink-Retter, Email: anneliese.fink-retter@meduniwien.ac.at.
Christian F Singer, Email: christian.singer@meduniwien.ac.at.
Christine Rappaport, Email: christine.rappaport@akhwien.at.
Daphne Gschwantler Kaulich, Email: daphne.gschwantler-kaulich@meduniwien.ac.at.
Georg Pfeiler, Email: georg.pfeiler@meduniwien.ac.at.
Muy-Kheng Tea, Email: muy-kheng.tea@meduniwien.ac.at.
Catherine M Phelan, Email: catherine.phelan@moffitt.org.
Phuong L Mai, Email: maip@mail.nih.gov.
Mark H Greene, Email: greenem@mail.nih.gov.
Gad Rennert, Email: rennert@tx.technion.ac.il.
Evgeny N Imyanitov, Email: evgeny@imyanitov.spb.ru.
Gord Glendon, Email: gord.glendon@cancercare.on.ca.
Amanda Ewart Toland, Email: amanda.toland@osumc.edu.
Anders Bojesen, Email: anders.bojesen@slb.regionsyddanmark.dk.
Inge Sokilde Pedersen, Email: isp@rn.dk.
Uffe Birk Jensen, Email: uffejens@rm.dk.
Maria A Caligo, Email: adelaide.caligo@do.unipi.it.
Eitan Friedman, Email: feitan@post.tau.ac.il.
Raanan Berger, Email: raanan.berger@sheba.health.gov.il.
Yael Laitman, Email: yael.laitman@sheba.health.gov.il.
Johanna Rantala, Email: johanna.rantala@karolinska.se.
Brita Arver, Email: brita.wasteson-arver@karolinska.se.
Niklas Loman, Email: niklas.loman@med.lu.se.
Ake Borg, Email: ake.borg@med.lu.se.
Hans Ehrencrona, Email: hans.ehrencrona@med.lu.se.
Olufunmilayo I Olopade, Email: folopade@medicine.bsd.uchicago.edu.
Jacques Simard, Email: jacques.simard@crchul.ulaval.ca.
Douglas F Easton, Email: douglas@srl.cam.ac.uk.
Georgia Chenevix-Trench, Email: georgia.trench@qimrberghofer.edu.au.
Kenneth Offit, Email: offitk@mskcc.org.
Fergus J Couch, Email: couch.fergus@mayo.edu.
Antonis C Antoniou, Email: antonis@srl.cam.ac.uk.
EMBRACE Study, Email: sde26@medscl.cam.ac.uk.
GEMO Study Collaborators, Email: dominique.stoppa-lyonnet@curie.net.
HEBON, Email: f.hogervorst@nki.nl.
KConFab Investigators, Email: heather.thorne@petermac.org.
References
- 1.Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, et al. Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) Cancer Epidemiol Biomarkers Prev. 2012;21:134–47. doi: 10.1158/1055-9965.EPI-11-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakhani SR, Jacquemier J, Sloane JP, Gusterson BA, Anderson TJ, van de Vijver MJ, et al. Multifactorial analysis of differences between sporadic breast cancers and cancers involving BRCA1 and BRCA2 mutations. J Natl Cancer Inst. 1998;90:1138–45. doi: 10.1093/jnci/90.15.1138. [DOI] [PubMed] [Google Scholar]
- 3.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–80. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bane AL, Beck JC, Bleiweiss I, Buys SS, Catalano E, Daly MB, et al. BRCA2 mutation-associated breast cancers exhibit a distinguishing phenotype based on morphology and molecular profiles from tissue microarrays. Am J Surg Pathol. 2007;31:121–8. doi: 10.1097/01.pas.0000213351.49767.0f. [DOI] [PubMed] [Google Scholar]
- 6.Gaudet MM, Kuchenbaecker KB, Vijai J, Klein RJ, Kirchhoff T, McGuffog L, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9:e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–8. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–84. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens KN, Fredericksen Z, Vachon CM, Wang X, Margolin S, Lindblom A, et al. 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 2012;72:1795–803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antoniou AC, Kuchenbaecker KB, Soucy P, Beesley J, Chen X, McGuffog L, et al. Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast Cancer Res. 2012;14:R33. doi: 10.1186/bcr3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couch FJ, Gaudet MM, Antoniou AC, Ramus SJ, Kuchenbaecker KB, Soucy P, et al. Common variants at the 19p13.1 and ZNF365 loci are associated with ER subtypes of breast cancer and ovarian cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2012;21:645–57. doi: 10.1158/1055-9965.EPI-11-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulligan AM, Couch FJ, Barrowdale D, Domchek SM, Eccles D, Nevanlinna H, et al. Common breast cancer susceptibility alleles are associated with tumour subtypes in BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Res. 2011;13:R110. doi: 10.1186/bcr3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French JD, Ghoussaini M, Edwards SL, Meyer KB, Michailidou K, Ahmed S, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013;92:489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE, et al. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA) Breast Cancer Res. 2007;9:104. doi: 10.1186/bcr1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Percy C, Vanholten V, Muir G, Sobin L. New International Classification of Diseases for Oncology (Icd-O, 2nd Edition) and the Neoplasm Chapter of the 10th Revision of the International Classification of Diseases (Icd-10) Laboratory Investigation. 1990;62:A113. [Google Scholar]
- 19.Antoniou AC, Sinilnikova OM, Simard J, Leone M, Dumont M, Neuhausen SL, et al. RAD51 135G-- > C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studie. Am J Hum Genet. 2007;81:1186–200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes DR, Lee A, Easton DF, Antoniou AC, Investigators E, kConFab I. Evaluation of association methods for analysing modifiers of disease risk in carriers of high-risk mutations. Genet Epidemiol. 2012;36:274–91. doi: 10.1002/gepi.21620. [DOI] [PubMed] [Google Scholar]
- 21.Gaffey MJ, Mills SE, Frierson HF, Jr, Zarbo RJ, Boyd JC, Simpson JF, et al. Medullary carcinoma of the breast: interobserver variability in histopathologic diagnosis. Mod Pathol. 1995;8:31–8. [PubMed] [Google Scholar]
- 22.Rigaud C, Theobald S, Noel P, Badreddine J, Barlier C, Delobelle A, et al. Medullary carcinoma of the breast: A multicenter study of its diagnostic consistency. Arch Pathol Lab Med. 1993;117:1005–8. [PubMed] [Google Scholar]
- 23.Lange K, Weeks D, Boehnke M. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–2. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 24.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86:420–8. doi: 10.1037/0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 25.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 26.Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015;47:164–71. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1.: Ethics committees that granted approval for the access and use of the data for this study. Table S2. Hormone receptor definitions used by the studies. Table S3. Associations of breast cancer susceptibility variants with breast cancer by tumor ER status after excluding prevalent cases. Table S4. Associations of breast cancer susceptibility variants with breast cancer by tumor PR status. Table S5. Associations of breast cancer susceptibility variants with breast cancer by tumor HER2 status. Table S6. Associations of breast cancer susceptibility variants with breast cancer by tumor triple-negative status. Table S7. Associations of breast cancer susceptibility variants with ductal breast cancer. Table S8. Associations of breast cancer susceptibility variants with lobular breast cancer. Table S9. Associations of breast cancer susceptibility variants with breast cancer by tumor grade. Table S10. Associations of breast cancer susceptibility variants with breast cancer by nodal involvement. Figure S1. QQ plots for SNP associations with ER-positive and -negative breast cancer in BRCA1 carriers. Figure S2. QQ plots for SNP associations with ER-positive and -negative breast cancer in BRCA2 carriers. Figure S3. Comparison of the breast cancer associations of breast cancer susceptibility loci in BCAC and in BRCA1 and BRCA2 carriers. Figure S4. Comparison of the associations of breast cancer susceptibility loci by ER status in BRCA1 and in BRCA2 carriers. Figure S5. Comparison of the associations of breast cancer susceptibility loci by ER status in population-based studies (BCAC) and in BRCA1 carriers. Figure S6. Comparison of the associations of breast cancer susceptibility loci by ER status in population-based studies (BCAC) and in BRCA2 carriers. (PDF 750 KB)


