Abstract
The genetic rules that dictate legume-rhizobium compatibility have been investigated for decades, but the causes of incompatibility occurring at late stages of the nodulation process are not well understood. An evaluation of naturally diverse legume (genus Medicago) and rhizobium (genus Sinorhizobium) isolates has revealed numerous instances in which Sinorhizobium strains induce and occupy nodules that are only minimally beneficial to certain Medicago hosts. Using these ineffective strain-host pairs, we identified gain-of-compatibility (GOC) rhizobial variants. We show that GOC variants arise by loss of specific large accessory plasmids, which we call HR plasmids due to their effect on symbiotic host range. Transfer of HR plasmids to a symbiotically effective rhizobium strain can convert it to incompatibility, indicating that HR plasmids can act autonomously in diverse strain backgrounds. We provide evidence that HR plasmids may encode machinery for their horizontal transfer. On hosts in which HR plasmids impair N fixation, the plasmids also enhance competitiveness for nodule occupancy, showing that naturally occurring, transferrable accessory genes can convert beneficial rhizobia to a more exploitative lifestyle. This observation raises important questions about agricultural management, the ecological stability of mutualisms, and the genetic factors that distinguish beneficial symbionts from parasites.
In a specialized organ known as the root nodule, the legume-rhizobium symbiosis links the energy-harvesting process of photosynthesis with the energy-demanding process of biological nitrogen (N) fixation and is, thus, of great ecological and agricultural importance. The question of how plant-microbe compatibility is encoded in this symbiosis has been the subject of intense inquiry over the last three decades, with efforts focused on early recognition events that are required for the initiation of infection and nodule development (Gibson et al. 2008; Oldroyd and Downie 2008). In some instances, however, naturally occurring legume-rhizobium incompatibility (defined here as the inability of the symbiosis to support plant growth) becomes evident only after nodule induction, resulting in visible nodules that do not fix N (Béna et al. 2005; Bladergroen et al. 2003; Selbitschka and Lotz 1991; Simsek et al. 2007; Tirichine et al. 2000). This abortive nodulation phenomenon is particularly problematic for a host plant, as it invests in the assembly of nodules that will be of no use, while potentially foregoing opportunities to engage in symbiosis with more beneficial strains. Indeed, there is abundant documentation from agricultural settings of superior rhizobium inoculants being outcompeted for nodule occupancy by indigenous rhizobium strains that are less-effective N fixers (Dowling and Broughton 1986; Triplett and Sadowsky 1992). Abortive nodulation may represent an important transition from a mutualistic lifestyle to that of a parasite or vice versa. While evolutionary and ecological models for such transitions have been widely addressed (Foster and Wenseleers 2006; Jones et al. 2009; Nowak 2006; Oono et al. 2011; Sachs et al. 2004), the genetic mechanisms behind them have been difficult to resolve.
The N-fixing symbiosis between Sinorhizobium meliloti and members of the legume genus Medicago (alfalfa and its relatives) has become one of the best-understood symbioses at the level of molecular genetic mechanisms and signal exchange. Mutations in the standard S. meliloti laboratory strain (Rm1021) have proven useful for identifying genes that contribute to effective nodulation and N fixation on Medicago hosts with which it is naturally compatible. More recently, the genetic and phenotypic diversity of wild S. meliloti isolates has been investigated (Béna et al. 2005; Galardini et al. 2011; van Berkum et al. 2006). These studies have helped to develop a sufficient understanding of S. meliloti as a species to allow molecular-level analyses of naturally variable traits. Here, we used a large set of natural S. meliloti strains and a panel of diverse Medicago hosts to first identify symbiotically incompatible host-strain pairs and, then, to isolate S. meliloti derivatives that overcome the initial incompatible condition. We show that this phenotypic change is brought about by the loss of accessory plasmids that, in addition to limiting symbiotic host range, also confer a hypercompetitive phenotype with respect to nodule invasion.
RESULTS
Isolation of rhizobia with increased host range
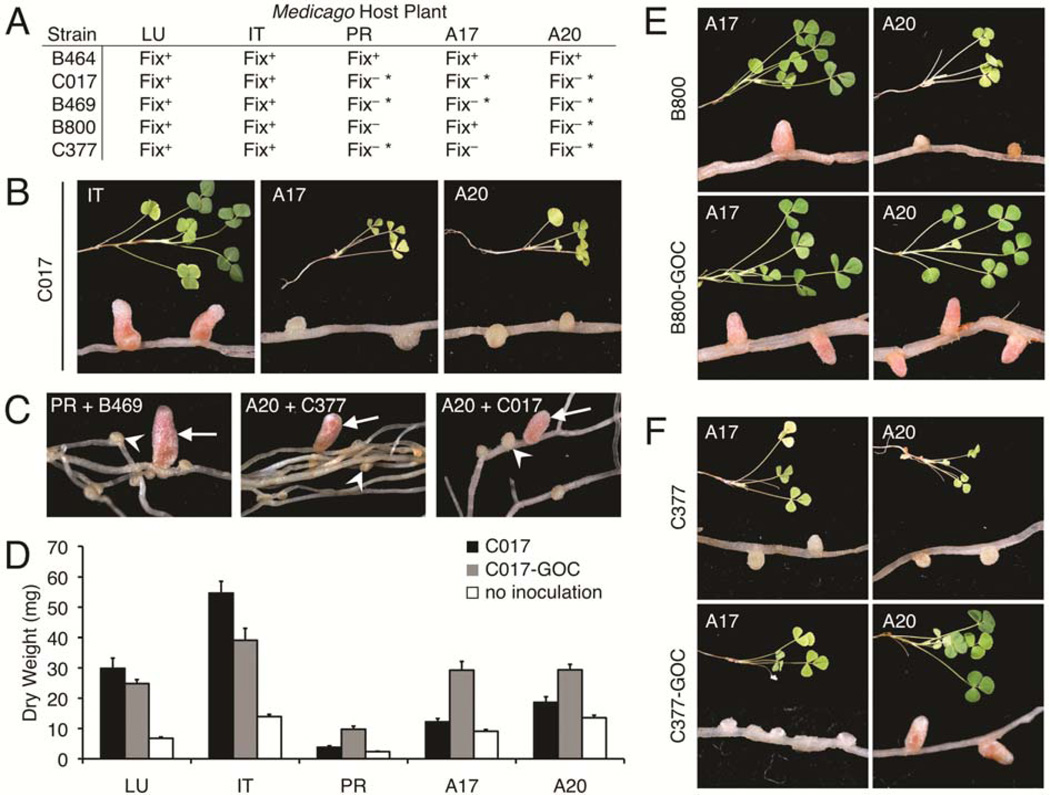
Using numerous S. meliloti isolates from the United States Department of Agriculture collection (van Berkum et al. 2006), we have evaluated symbiotic properties on several Medicago species, as well as multiple cultivars of the model plant Medicago truncatula (Fig. 1A; a more detailed data set may be found in Supplementary Table S1). Approximately half of all host-strain pairs resulted in an ineffective symbiosis, evidenced by chlorotic shoots and small, white nodules (Fig. 1B). In most cases, symbiotic incompatibility was host-conditioned, with strains exhibiting an effective N fixation phenotype (Fix+) on some Medicago hosts and an ineffective phenotype (Fix−) on others. This was apparent both for different species of Medicago (Fig. 1B) and different cultivars of M. truncatula (Fig. 1E and F). These strains, therefore, have the genetic capacity for N fixation, but that capacity is overridden by an unknown response by specific hosts that occurs after nodule initiation.
Fig. 1.
Host-range restriction in Sinorhizoboim meliloti is unstable. A, Strain-host combinations evaluated in this study. Symbiotic phenotypes were scored approximately 30 days postinoculation (dpi). Fix+, effective N-fixing pairs; Fix−, abortively nodulating pairs; Fix− *, pairs exhibiting suppressible incompatibility. LU = Medicago lupulina; IT = M. italica; PR = M. praecox; A17 = M. truncatula cv. A17; and A20 = M. truncatula cv. A20. B, Representative shoot and nodule images from compatible and incompatible pairs, 30 dpi. C, Examples of gain-of-compatibility (GOC) nodules (arrows) among incompatible nodules (arrowheads) on plants originally inoculated with single strains. D, Quantification of shoot dry mass 40 dpi with strain C017 (black; n = 8), C017-GOC (gray; n = 8), or no inoculation (white; n = 16). Error bars represent standard error of the mean. E, Representative shoot and nodule images of B800 and B800-GOC inoculated onto M. truncatula cvs. A17 and A20, 30 dpi. Note that both strains are Fix+ on A17 but only B800-GOC is Fix+ on A20. F, Representative shoot and nodule images of C377 and C377-GOC inoculated onto M. truncatula cvs. A17 and A20, 30 dpi. Note that both strains are Fix− on A17 but only C377-GOC is Fix+ on A20.
To better understand genetic mechanisms of host-conditioned incompatibility, several wild S. meliloti strains were screened on incompatible hosts in search of spontaneous mutants in which incompatibility was suppressed. These so-called gain-of-compatibility (GOC) mutants resulted in elongated, pink, N-fixing nodules among wild-type nodules that were small and white (Fig. 1C). Rhizobia isolated from GOC nodules showed a stable Fix+ phenotype after isolation and reinoculation (Supplementary Fig. S1). To confirm the host benefit brought about by GOC mutations, we measured shoot biomass from plants nodulated by S. meliloti C017 and its derivative C017-GOC. C017-GOC provided a greater host benefit on plants with which the C017 parent was incompatible (Fig. 1D, PR, A17, and A20). Conversely, on a host with which C017 was naturally compatible for N fixation (Fig. 1D, LU and IT), the C017-GOC strain did not confer additional benefit and may even have slightly reduced biomass.
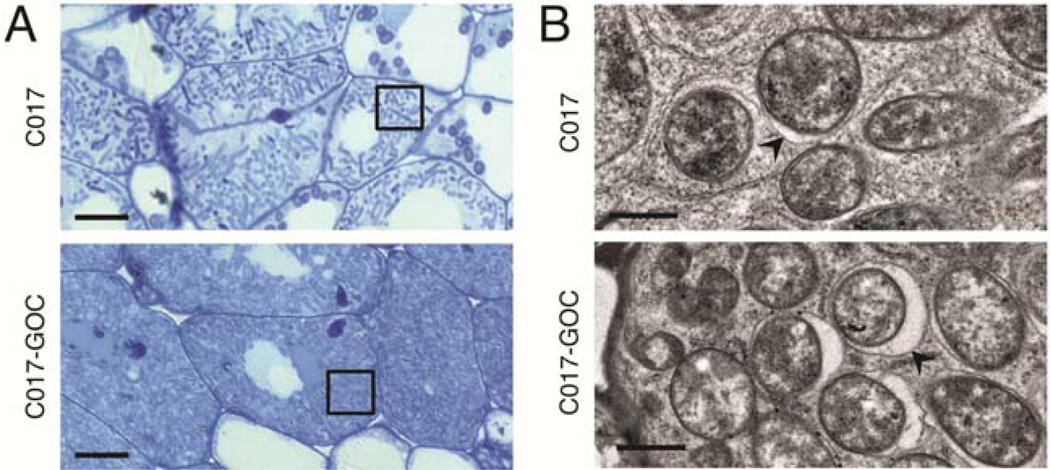
To assess how GOC derivatives affect nodule development on M. truncatula cv. A17, microscopic analysis was conducted on nodules that had been induced by either C017 (incompatible with A17) or C017-GOC (compatible with A17) (Fig. 2A and B). Under both conditions, bacteria successfully invaded the developing nodule, followed by morphological differentiation into swelled, intracellular bacteroids. It was consistently observed that C017-GOC attained higher bacteroid densities in the nodule cell cytoplasm than C017. It was therefore concluded that C017 incompatibility on A17 occurs very late in the establishment of the symbiosis, after bacterial entry and after at least partial bacteroid differentiation within nodule cells.
Fig. 2.
C017 and C017-GOC (a gain-of-compatibility derivative) occupy host plant cells but at different densities. A, Light micrographs of A17-infected nodule cells, 14 days postinoculation with strain C017 (top) or C017-GOC (bottom) from equivalent positions within the nodule. Inset boxes denote regions of nodule cell cytoplasm that are occupied by stained bacteroids. Scale bars = 10 µm. B, Transmission electron micrographs of nodules treated as in A, showing bacteroid cross-sections. Arrowheads indicate plant-derived peribacteroid membranes. Scale bars = 1 µm.
Large accessory plasmids can dictate symbiotic host range
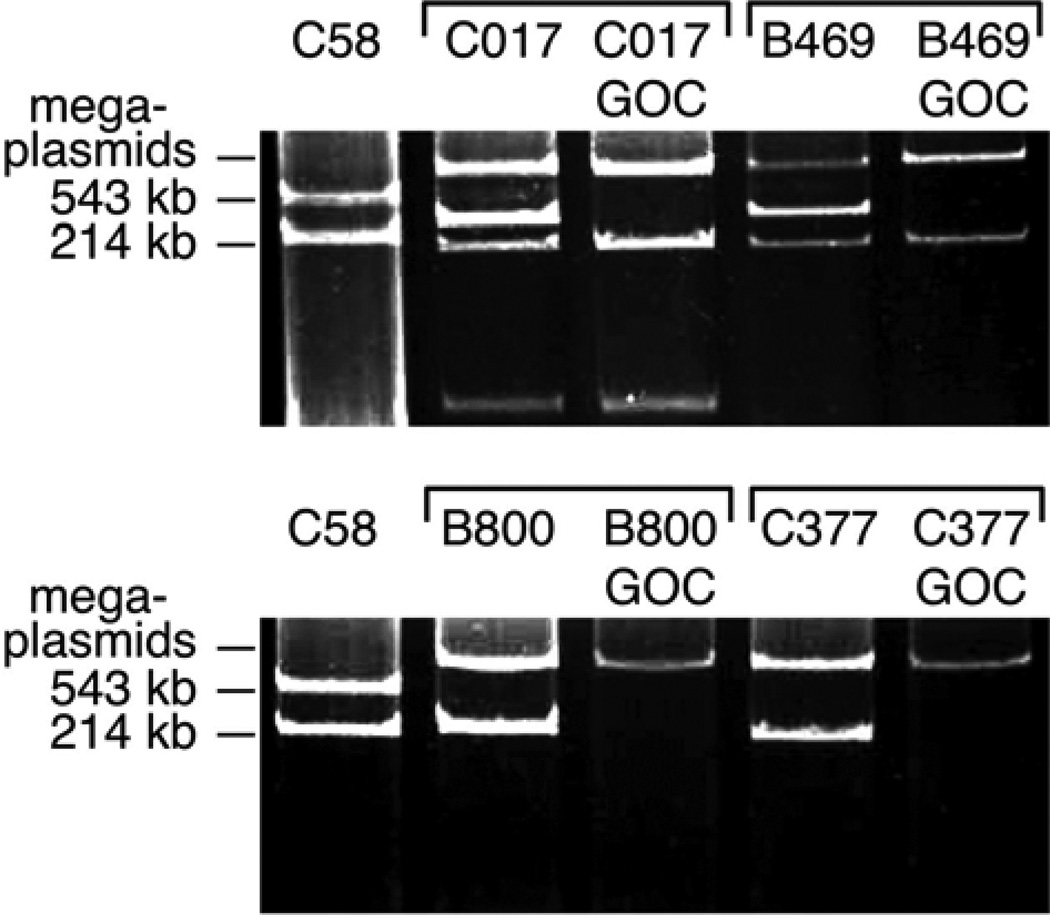
In hopes of finding a mutation responsible for the GOC phenotype, short-read, shotgun sequencing of strains C017 and C017-GOC was performed. Upon mapping reads to the reference Rm1021 genome using the GNUMAP algorithm (Clement et al. 2010), we discovered a significant amount of DNA in C017 that is absent in C017-GOC. Some of this DNA is similar to a known Sinorhizobium accessory plasmid, pSMeSM11a (Stiens et al. 2006). Subsequent Eckhardt gel analysis of all of our GOC derivatives and corresponding parent strains revealed that the GOC phenomenon correlates with the curing of accessory plasmids in the size range of 200 to 300 kb (Fig. 3). We generally refer to these as HR plasmids due to their effect on symbiotic host range (defined here as the range of plants benefitting from the symbiosis). Specific HR plasmid names assigned in this study (pHRC017, pHRB469, pHRB800, and pHRC377) are in reference to the parent strains in which they were initially discovered.
Fig. 3.
The gain-of-compatibility (GOC) phenomenon is associated with the loss of accessory plasmids. Eckhardt gels of parent strains and GOC derivatives. Agrobacterium tumefaciens C58 plasmids (543 and 214 kb) are included as molecular weight standards; S. meliloti symbiotic megaplasmids (>1 Mb) are indicated.
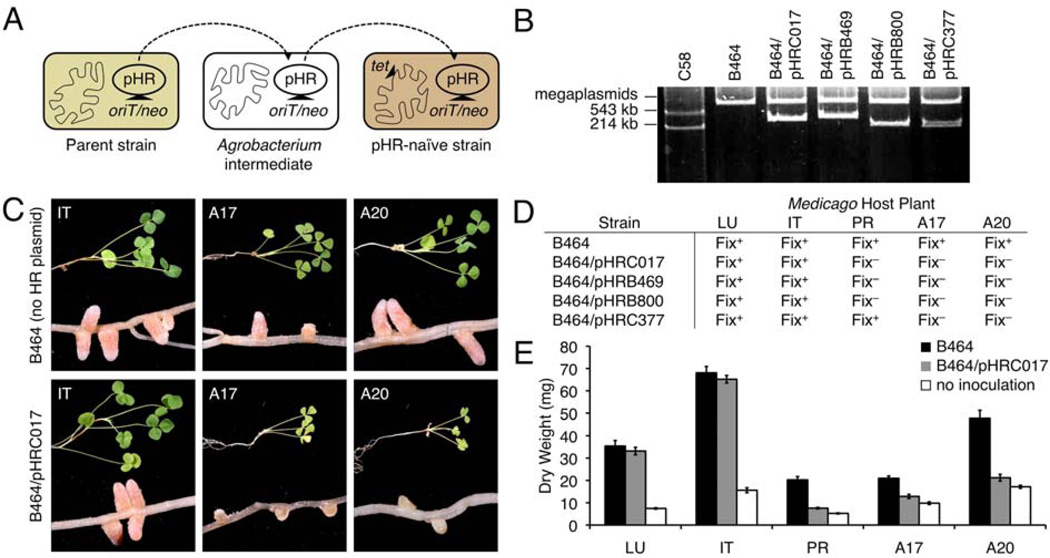
To confirm the role of HR plasmids in determining host compatibility, plasmid transfer experiments were performed. These experiments required modifying the plasmids with a genetic cassette containing a conjugative transfer origin (oriT) and a neomycin-resistance gene (neo). Recipient strains were modified with a tetracycline-resistance gene (tet) inserted at a neutral chromosomal location. This plasmid transfer procedure is diagrammed in Figure 4A and described below. In one experiment, pHRB469 was transferred from B469 into plasmid-free Agrobacterium tumefaciens UBAPF2 (Hynes et al. 1985) and, subsequently, into strain B469-GOCtet. The resulting transconjugant showed restored incompatibility on host plants with which B469 is incompatible (Supplementary Fig. S2).
Fig. 4.
Abortive nodulation is governed by autonomously functioning accessory plasmids. A, Diagram of the process used to introduce exogenous HR plasmids (large accessory plasmids affecting symbiotic host range) into plasmid-naive strains. B, Eckhardt gels showing B464 harboring different exogenous HR plasmids. C, Representative shoot and nodule phenotypes from plants inoculated with B464 with or without pHRC017 (30 days posinoculation [dpi]). D, Symbiotic outcomes on multiple hosts after inoculation with B464 harboring different HR plasmids. E, Quantification of shoot dry mass 40 dpi with strain B464 (black; n = 8), B464 and pHRC017 (gray; n = 8), or no inoculation (white; n = 16). Error bars represent standard error of the mean. All HR plasmids referred to in B, C, D, and E are modified with the oriT/neo cassette to permit transfer and selection. All recipient strains referred to in B, C, D, and E were modified with the tet cassette to permit selection of transconjugants.
We next considered the question of whether HR plasmids could be accommodated in a S. meliloti strain (B464) that is naturally devoid of accessory plasmids and is an effective N fixer on all of the Medicago hosts on our panel (Fig. 1A). After mobilization to the A. tumafaciens intermediate, all four HR plasmids were successfully transferred into B464tet (Fig. 4B). The HR plasmids conferred upon B464tet nearly the same host range restriction as they did to their parental S. meliloti strains (Fig. 4C to E). One exception was that of pHRB800, which did not restrict N fixation on M. truncatula A17 in parental strain B800 (Fig. 1E) but did in strain B464tet (Fig. 4D). This experiment shows that HR plasmids can restrict host range in a manner that is dominantly acting and largely (but not completely) independent of the genotype of the strain in which it resides. Further testing with all four HR plasmids in a variety of strain backgrounds revealed several other instances in which strain background can modulate the ultimate symbiotic consequence of harboring an HR plasmid (Supplementary Table S2).
HR plasmids can confer a competitive advantage for nodulation
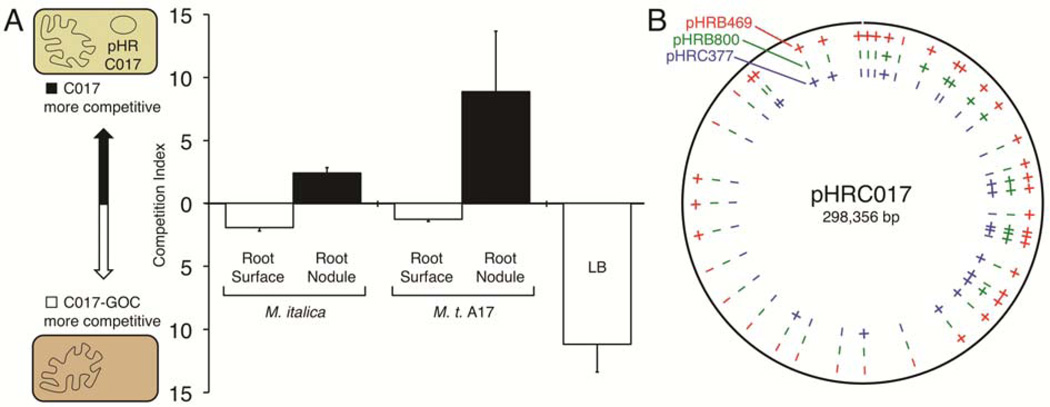
Because HR plasmids cause an abortive nodulation phenotype on certain Medicago hosts, we sought to better understand how these plasmids might provide a fitness advantage for bacterial cells in which they reside, even if they interfere with nodule development and N fixation. Supposing that HR plasmids might provide some growth advantage to free-living cells in the soil or rhizosphere, competition experiments were performed using mixed populations of strains C017 and C017-GOC. First, the competition index was determined for a C017 and C017-GOC mixed population grown for approximately 15 generations in Luria broth (LB). In this experiment, the GOC strain increased its representation in the population by approximately tenfold (Fig. 5A). This can be accounted for by the fact that C017-GOC has a faster doubling time than C017 in LB (5.5 versus 7.9 h). Next, mixed populations were grown on plants for three weeks. These bacteria were recovered both from the root surface (rhizosphere) and from within the nodules of surface-sterilized roots. In order to sample as many nodules as possible, tests for nodule occupancy were performed on a whole-plant basis. While pHRC017 imposed a modest disadvantage in the rhizosphere, it reproducibly provided a competitive advantage for nodule occupancy (Fig. 5A). Similar results were seen for competition between strains B469 and B469-GOC (data not shown).
Fig. 5.
pHRC017 confers a competitive advantage for nodule occupancy and is genetically similar to other HR plasmids (large accessory plasmids affecting symbiotic host range). A, The competitive indexes for C017 and C017-GOC (a gain-of-compatibility derivative) grown in competition when inoculated onto a compatible host (M. italica; n = 3), onto an incompatible host (M. truncatula [M. t.] A17; n = 6), or into LB (Luria broth; n = 6). Error bars represent standard error of the mean. The HR plasmid in C017 was modified with the oriT/neo cassette to permit transfer and selection. B, Diagram and annotation of pHRC017. Conservation of sequence in other HR plasmids (based on polymerase chain reaction tests) is indicated by + or −.
We have considered how it is possible that an incompatible strain can achieve a larger population than its effective GOC counterpart, which induces larger and more densely colonized nodules. In one model, the plasmid-harboring incompatible strain may be more aggressive at stimulating nodule initiation, even though the resulting nodules are less beneficial to the host. In a second model, the plasmid may permit higher recoverability of rhizobia from nodules, even if the number of nodules induced by each strain in competition is equivalent. These models are not mutually exclusive. The colony counts reported in Figure 5A reflect tallies of white and pink nodules on plants from the competition experiments, suggesting that the plasmid-harboring C017 strain is more competitive at the level of nodule establishment.
To investigate specific molecular mechanisms that could be responsible for HR plasmid control over host range determination and nodulation competitiveness, pHRC017 was sequenced. This was done by transferring pHRC017 into A. tumefaciens UBAPF2 and isolating total DNA, subjecting it to 454 sequencing. Sequence reads corresponding to A. tumefaciens were subsequently subtracted, and the remaining reads were assembled into a single circular molecule of 298,356 bp. An annotated record for pHRC017 has been submitted to GenBank (accession number JQ665880). Among the approximately 430 potential open reading frames on this plasmid are several genes that may encode proteins of symbiotic consequence, such as a probable 1-aminocyclopropane-1-carboxylate (ACC) deaminase. ACC is the direct precursor to the phytohormone ethylene, and bacterial ACC deaminases are known to influence plant-microbe interactions (Ma et al. 2004; Nonaka et al. 2008). Additionally, a type IV secretion system (T4SS) operon, which may be involved in horizontal plasmid transfer, is encoded on pHRC017. In tests to determine if HR plasmids are self-transmissible, plasmid-harboring strains were mixed with plasmid-naive strains. For pHRB469, clear evidence of transfer was observed (Supplementary Fig. S3) but transfer of pHRC017 was not. Transfer of the other HR plasmids was not tested.
Due to its similarity with pSMeSM11a, we performed BLAST analysis of pHRC017 in comparison with other sequenced Sinorhizobium accessory plasmids and observed only limited similarities (Supplementary Fig. S4). The plasmid pSINME01 from S. meliloti AK83 (Galardini et al. 2011) exhibited the highest similarity, matching approximately 25% of pHRC017, with regions of similarity being distributed throughout the pHRC017 sequence. We used polymerase chain reaction (PCR)-based markers to compare pHRC017 with the other HR plasmids, which have not yet been sequenced (Fig. 5B). Of 41 markers, 30 markers amplify from pHRB469, 12 amplify from pHRB800, and 15 amplify from pHRC377. Six of the 41 pHRC017 markers amplify from all four HR plasmids (Fig. 5B; additional details in Supplementary Table S3). This analysis confirms that the four HR plasmids are genetically related but that a limited number of gene clusters are conserved in all four plasmids. How these genes contribute to host-conditioned compatibility and competition for nodule invasion awaits further characterization.
DISCUSSION
We have presented the discovery and characterization of accessory plasmids in S. meliloti that block effective N fixation in a host-conditioned manner. Rhizobial symbiotic host range control is typically associated with Nod-factor signaling, which occurs at the earliest stages of nodule development. The HR plasmids described here appear to influence host range at a much later stage of symbiotic infection, and they act dominantly to limit host range. The idea that host range in the Sinorhizobium-Medicago symbiosis can be determined after Nod-factor perception is well documented (Béna et al. 2005; Simsek et al. 2007; Tirichine et al. 2000), but genetic mechanisms for this phenomenon have been difficult to resolve. In various rhizobial genera, correlations between accessory plasmids and nodulation competitiveness (Martínez-Romero and Rosenblueth 1990; Sanjuan and Olivares 1991) as well as connections between accessory plasmids and reduced host benefit (Pankhurst et al. 1986; Selbitschka and Lotz 1991; Velázquez et al. 1995) have been reported. Our work highlights the autonomy with which HR plasmids can function in multiple strains, their effect on competitiveness for nodule invasion, and the host-conditioned nature of the phenomenon.
Our observations suggest that HR plasmids may control the synthesis of a signal that somehow disrupts the symbiotic dialogue. Whether this disruptive signal elicits a host defense response or merely prevents a required host response is unclear. This model is reminiscent of observations first made in the soybean-rhizobium symbiosis, in which type III effectors (T3E) elicit a negative response from certain soybean cultivars, yet potentiate symbiosis with others (Deakin and Broughton 2009; Krishnan et al. 2003; Viprey et al. 1998). More recently, the soybean response to T3E has been linked to specific leucine-rich repeat resistance gene alleles, demonstrating that rules for rhizobium-legume compatibility are governed by pathways that are typically attributed to pathogen immunity (Yang et al. 2010). Unlike T3E-mediated incompatibility, which is evident at a very early stage of symbiotic infection (Yang et al. 2010), HR plasmid–mediated incompatibility blocks the symbiotic program at a very late stage, so that incompatible nodules can still harbor significant rhizobial populations.
As an alternative to the model described above, which involves the synthesis of a new compatibility-determining signal, HR plasmids may alter the normal synthesis of a signal encoded elsewhere in the rhizobial genome. In a previous study, it was suggested that differential succinylation of exopolysaccharide I (EPS I) could explain S. meliloti compatibility with M. truncatula cultivars A17 and A20 (Simsek et al. 2007). These M. truncatula cultivars were also used in our study. With these observations in mind, one can envision a model in which HR plasmids cause the modification of some common signal such as EPS I or lipopolysaccharide. Such a modification could give rise to the restricted host ranges reported here. It is notable that pHRB800 controls host range at the cultivar level in M. truncatula (Fig. 1E) in a manner similar to the differentially succinylating strains reported previously.
The evolutionary maintenance of HR plasmids is interesting to consider, since these plasmids appear to skew the exchange of benefits between host and symbiont, essentially converting a mutualistic relationship to a parasitic one. HR plasmids confer what may be thought of as a ‘cheater’ phenotype on S. meliloti cells that harbor them. If we simplistically assume that rhizobial populations possess some relatively stable balance of beneficial and nonbeneficial strains, then our observations relating to HR plasmids provide possible insights into mechanisms that maintain this balance. First, we observed that the negative effect of HR plasmids is host-conditioned, meaning that negative effects of cheating can be masked (Fig. 1D). Second, we found that HR plasmids may erode host benefit in some strain backgrounds but not in others; that is, HR plasmids function in a ‘strain-conditioned’ manner (Fig. 4D). Third, with cheater functions encoded on a curable and transferrable plasmid, rhizobial strains are not dedicated to a specific lifestyle; they may either gain or lose exploitative properties in single steps. Finally, we have shown that HR plasmids couple cheating functions with functions that increase competitiveness for nodule occupancy (Fig. 5A). In this way, exploitative genes keep a foothold in spite of the fitness disadvantage brought about by the cost incurred to the host. These mechanisms would tend to support the maintenance of exploitative functions, even under conditions that would select for mutualism. However, we do not dismiss the possibility that selection for HR plasmid maintenance may be largely driven by factors unrelated to the symbiosis.
This study points to a potential scenario in which commercial rhizobium inoculants that have been optimized for nitrogen fixation might be outcompeted by indigenous rhizobia or even neutralized by horizontal transfer of HR plasmids from coexisting native rhizobia. There is some evidence that HR plasmids may be naturally transmissible, based on the presence of T4SS genes in at least two of them (pHRC017 and pHRB469) and the observed transfer of pHRB469 from one strain to another. Future work should be dedicated to better defining the ecological and agricultural significance of HR plasmids and further characterizing molecular mechanisms of late-stage incompatibility brought about by these plasmids. We are currently developing tools for creating large-scale deletions in HR plasmids, which will assist in the genetic dissection of plasmid-mediated host range determination.
MATERIALS AND METHODS
Bacterial culture
Escherichia coli and S. meliloti cultures were grown at 37 and 30°C, respectively, in Luria broth (LB) supplemented with appropriate antibiotics as follows (in micrograms per milliliter): chloramphenicol, 30; kanamycin, 30; neomycin, 100; rifampicin, 100; streptomycin, 200; and tetracycline, 5. Rhizobia were extracted from nodules by surface sterilization and maceration (details below).
Plasmid and strain construction
Plasmids and strains used in this study are listed in Supplementary Table S4. Plasmids were constructed using standard techniques with enzymes purchased from New England Bio-labs (Ipswich, MA, U.S.A.) The high-fidelity polymerase Pfx50 (Invitrogen, Carlsbad, CA, U.S.A.) was used for insert amplification. All custom oligonucleotides were purchased from Invitrogen and are listed in Supplementary Table S5. Mobilization of plasmids was accomplished by triparental mating with helper E. coli B001 (DH5α harboring plasmid pRK600). pRK600 expresses trans-acting proteins required for mobilization of plasmids harboring the RK2 transfer origin (oriT). For modification of HR plasmids to enable pRK600-mediated transfer, an oriT/neo cassette was introduced by single-crossover homologous recombination using the pUC-based plasmid pJG194 (Griffitts and Long 2008). pJG194 was targeted (nondisruptively) to the acdS locus in pHRC017, pHRB469, and pHRB800 and the traA locus in pHRC377. Fragments corresponding to the regions upstream of acdS and traA were amplified using primer pairs oJG1127 and oJG1128 and oMC089 and oMC090, respectively, followed by ligation into pJG194 to yield integration plasmids pJG461 (for C017), pJG463 (for B469), pJG476 (for B800), and pJG499 (for C377). These modified pJG194 constructs were introduced into S. meliloti strains by triparental mating. When appropriate, S. meliloti was made tetracycline resistant by integration of pJG505 into the rhaK-icpA intergenic region of the chromosome. The rhaK-icpA fragment in pJG505 was amplified using primers oMC014 and oMC015. HR plasmid transfer experiments depicted in Figure 4 were accomplished by triparental mating, using the helper strain B001 (described above). Briefly, a parent S. meliloti strain carrying an oriT/neo-modified HR plasmid was mixed with A. tumefaciens UBAPF2 for approximately 12 h, followed by selection on neomycin and rifampicin (to counterselect the donor). The resulting strain was then mixed with the tetracycline-resistant S. meliloti recipient (e.g., B464tet) for approximately 12 h, followed by selection on neomycin and tetracycline (the latter to counterselect the A. tumefaciens donor). The plasmid transfer experiment was accomplished by biparental mating (i.e., by excluding helper strain B001) for approximately 12 h, followed by selection on neomycin and tetracycline (to counterselect the donor).
Plant growth and nodulation
Medicago species and cultivars used in this study are listed in Supplementary Table S6. Scarified and surface-sterilized seeds were allowed to germinate in petri plates, and 2-day-old seedlings were planted in sterile Turface clay particles (Turface Athletics, Buffalo Grove, IL, U.S.A.) and were allowed to grow for 4 days before being inoculated with 1 ml of S. meliloti cells suspended in 2.6 mM KH2PO4 (adjusted to pH 7.0 with 2N KOH) to an optical density at 600 nm (OD600) of approximately 0.1. Nodulation was allowed to proceed for two to six weeks, depending on the experiment. Plants were watered with standard nodulation medium (SNM). SNM contains (per liter) 0.35 g of KH2PO4, 0.25 g of MgSO4, 0.15 g of CaCl2·2H2O, and 2 ml of minor salts solution (500× minor salts [per liter] = 9.5 g of Na2-EDTA·2H2O, 7 g of FeSO4·7H2O, 1.5 g of ZnSO4·7H2O, 1.5 g of H3BO3, 1.5 g of MnSO4·H2O, 0.15 g of Na2MoO4·2H2O, 15 mg of CuSO4, and 15 mg of CoCl2). The pH of this solution was adjusted to 7.0 with 2N KOH, and the medium was sterilized by autoclaving. Plants were maintained at approximately 27°C under fluorescent lamps (2.7 klux intensity, 16 h day length). To determine dry weight of plants, samples were placed in an oven at 50°C for 48 h and were then weighed on a fine balance.
Microscopy
At 14 days after inoculation, whole nodules were excised, were fixed with 2% glutaraldehyde in 50 mM cacodylate (pH 7.2) for 2 h, were washed, were postfixed in 1% osmium tetroxide for 2 h, and were washed again with H2O. Samples were then stained in 0.5% uranyl acetate overnight, followed by dehydration in an acetone series. Samples were then embedded in Spurr’s resin and were cured for 48 h at 70°C. Sections were made using an ultramicrotome. For light microscopy, 600-nm sections were bound to a microscope slide (by heating at 70°C), were stained with toluidine blue, and were imaged with an Axio Imager.A1 microscope. For transmission electron microscopy, 80-nm sections were stained in Reynold’s lead citrate for 10 min, followed by washing with H2O. Sections were imaged with a Tecnai G2 T-12 transmission electron microscope.
Modified Eckhardt gel electrophoresis
Modified Eckhardt gels were performed as previously described (Hynes and McGregor 1990) with modifications. Briefly, bacteria were grown to an OD600 of 0.6 in LB. Culture (150 µl) was added to 500 µl of chilled 0.3% sarkosyl in 1× SBE (20× SBE = 500 ml of H2O, 4 g of NaOH, 3.72 g of Na2-EDTA·2H2O, pH to 8.0 with boric acid). Each sample was pelleted and resuspended in 20 µl of lysis solution (1× SBE, 10 mg of sucrose per milliliter, 1 mg of lysozyme per milliliter, 40 µg of RNase A per milliliter), followed immediately by loading into a sodium dodecyl sulfate (SDS)–SBE minigel (1× SBE, 0.8% agarose, 0.5% SDS). Each sample remained in the well for 2 min, followed by electrophoresis at 23 V for 10 min, followed by electrophoresis at 96 V for 90 min. The minigel was then stained for 1 h in 0.4 µg of ethidium bromide per milliliter and was destained for 10 min in H2O prior to imaging.
Competition experiments
Strains employed were C241 (C017 modified with a neo marker in pHRC017) and C017-GOC. Newly saturated cultures were diluted to an OD600 of 0.05 in 2.6 mM KH2PO4 (pH 7.0), were mixed at a 1:1 ratio, and were inoculated into LB broth (5 µl of cells per 4-ml culture) or onto plants (1 ml of suspension per plant). LB cultures were grown for 72 h, followed by dilution plating onto LB-streptomycin and LB-streptomycin-neomycin. Plants were harvested after 21 days. To evaluate rhizosphere colonization, whole-root systems were vortexed for 30 s in 5 ml of LB-10% glycerol and were dilution-plated onto LB with streptomycin (LB-Sm) and LB with streptomycin and neomycin (LB-Sm-Nm). To evaluate root nodule occupancy, the roots were then surface-sterilized in 75% ethanol for 30 s and in 1.5% hypochlorite for 30 s and were then washed in H2O. Surface-sterilized whole roots were macerated in LB and 10% glycerol and were dilution-plated onto LB-Sm and LB-Sm-Nm. The competition index (CI) was calculated according to Ellermeier and associates (2006) as follows. When C017-GOC yielded more colony-forming units (CFU) (LB and rhizosphere), CI = (C017-GOC recovered/C241 recovered)/(C017-GOC inoculated/C241 inoculated); when C241 yielded more CFU (whole roots), CI = (C241 recovered/C017-GOC recovered)/(C241 inoculated/C017-GOC inoculated).
Sequencing and annotation of pHRC017, and comparison with other HR plasmids
Original evidence of the existence of pHRC017 came from Illumina-sequencing of strains C017 and C017-GOC. DNA was purified in a manner similar to that described below. Sequence reads from both samples were mapped to the S. meliloti 1021 genome sequence using GNUMAP (Clement et al. 2010). DNA segments from strain C017 that were absent in strain 1021 and C017-GOC were analyzed by BLASTn.
For pyrosequencing of pHRC017, the plasmid was marked with an oriT/neo cassette and was transferred to plasmid-free A. tumefaciens UBAPF2 to yield strain C382. C382 was grown overnight in LB, and a total of 6 ml of saturated culture was pelleted and resuspended in 2.4 ml of TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). Cells were then treated with 0.6% SDS and 150 µg of proteinase K per milliliter and were incubated at 42°C for 20 min. The resulting lysate was brought to 0.7 M NaCl and 0.6 % cetyltrimethylammonium bromide, followed by vortexing and incubation at 65°C for 10 min. The sample was then extracted with phenol and chloroform. The aqueous phase was isolated and treated with 80 µg of RNase A per milliliter, was incubated at 37°C for 10 min, and was reextracted with chloroform. The aqueous phase was precipitated with isopropanol, was resuspended in TE buffer, and was further purified over a Nucelobond Midi column, according to manufacturer’s instructions. The 454 library preparation was performed according to the rapid library preparation protocol, followed by sequencing on the 454 Genome Analyzer FLX. Assembly into contigs was performed using Newbler (version 2.5.3) after subtracting reads that corresponded to A. tumefaciens. Contig edge ambiguities were resolved by PCR. The previously introduced oriT/neo cassette was then removed from the assembly to reconstitute the native pHRC017 sequence. Reads were remapped to this assembly, using Newbler (version 2.5.3) to verify the sequence. The final assembly was annotated using Glimmer (version 3.02), and predicted open reading frames were assigned putative functions after BLASTx analysis. This sequence can be accessed in GenBank (accession number JQ665880).
Genetic regions on pHRC017 were compared with the other three HR plasmids (pHRB469, pHRB800, and pHRC377) by PCR. Primers oMC73 and oMC74, oMC81 and oMC82, and oMC209 and oMC292 were used for this analysis. For most markers, primers were designed to amplify approximately 500-bp regions (Fig. 3B).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Brigham Young University (BYU) undergraduates S. Rogers, C. Crum, and A. Draper for assistance with experiments; P. Van Berkum of the United States Department of Agriculture-Agriculture Research Service program for providing many of the strains used in this study; M. Hynes for providing A. tumefaciens UBAPF2; I. Oresnik for advice relating to Eckhardt gels; and M. Standing for help with electron microscopy. This work was supported by a Helen Hay Whitney postdoctoral fellowship, BYU mentoring environment grant 425420, National Science Foundation grant IOS-1054980 (all to J. S. Griffitts), and NIH grant R01GM093628 (to S. R. Long). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors make the following declaration about their contributions. M. B. Crook, D. P. Lindsay, S. R. Long, and J. S. Griffitts conceived and designed the experiments. M. B. Crook, D. P. Lindsay, M. B. Biggs, and J. S. Bentley performed the experiments. M. B. Crook, D. P. Lindsay, J. C. Price, S. C. Clement, M. J. Clement, and J. S. Griffitts analyzed the data. M. J. Clement, S. R. Long, and J. S. Griffitts contributed reagents, materials, and analysis tools. M. B. Crook, S. R. Long, and J. S. Griffitts wrote the paper, and all the authors approved the manuscript.
Footnotes
Nucleotide sequence data is available in the GenBank database under accession number JQ665880.
LITERATURE CITED
- Béna G, Lyet A, Huguet T, Olivieri I. Medicago-Sinorhizobium symbiotic specificity evolution and the geographic expansion of Medicago. J. Evol. Biol. 2005;18:1547–1558. doi: 10.1111/j.1420-9101.2005.00952.x. [DOI] [PubMed] [Google Scholar]
- Bladergroen MR, Badelt K, Spaink HP. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant-Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- Clement NL, Snell Q, Clement MJ, Hollenhorst PC, Purwar J, Graves BJ, Cairns BR, Johnson WE. The GNUMAP algorithm: Unbiased probabilistic mapping of oligonucleotides from next-generation sequencing. Bioinformatics. 2010;26:38–45. doi: 10.1093/bioinformatics/btp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin WJ, Broughton WJ. Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat. Rev. Microbiol. 2009;7:312–320. doi: 10.1038/nrmicro2091. [DOI] [PubMed] [Google Scholar]
- Dowling DN, Broughton WJ. Competition for nodulation of legumes. Annu. Rev. Microbiol. 1986;40:131–157. doi: 10.1146/annurev.mi.40.100186.001023. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, González-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Foster KR, Wenseleers T. A general model for the evolution of mutualisms. J. Evol. Biol. 2006;19:1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x. [DOI] [PubMed] [Google Scholar]
- Galardini M, Mengoni A, Brilli M, Pini F, Fioravanti A, Lucas S, Lapidus A, Cheng JF, Goodwin L, Pitluck S, Land M, Hauser L, Woike T, Mikhailova N, Ivanova N, Daligault H, Bruce D, Detter C, Tapia R, Han C, Teshima H, Mocali S, Bazzicalupo M, Biondi EG. Exploring the symbiotic pangenome of the nitrogen-fixing bacterium Sinorhizobium meliloti. BMC Genomics. 2011;12:1–15. doi: 10.1186/1471-2164-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffitts JS, Long SR. A symbiotic mutant of Sinorhizobium meliloti reveals a novel genetic pathway involving succinoglycan biosynthetic functions. Mol. Microbiol. 2008;67:1292–1306. doi: 10.1111/j.1365-2958.2008.06123.x. [DOI] [PubMed] [Google Scholar]
- Hynes MF, McGregor NF. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 1990;4:567–574. doi: 10.1111/j.1365-2958.1990.tb00625.x. [DOI] [PubMed] [Google Scholar]
- Hynes MF, Simon R, Pühler A. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid. 1985;13:99–105. doi: 10.1016/0147-619x(85)90062-9. [DOI] [PubMed] [Google Scholar]
- Jones EI, Ferrière R, Bronstein JL. Eco-evolutionary dynamics of mutualists and exploiters. Am. Nat. 2009;174:780–794. doi: 10.1086/647971. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Lorio J, Kim WS, Jiang GQ, Kim KY, DeBoer M, Pueppke SG. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant-Microbe Interact. 2003;16:617–625. doi: 10.1094/MPMI.2003.16.7.617. [DOI] [PubMed] [Google Scholar]
- Ma W, Charles TC, Glick BR. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl. Environ. Microbiol. 2004;70:5891–5897. doi: 10.1128/AEM.70.10.5891-5897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Romero E, Rosenblueth M. Increased bean (Phaseolus vulgaris L.) nodulation competitiveness of genetically modified Rhizobium strains. Appl. Environ. Microbiol. 1990;56:2384–2388. doi: 10.1128/aem.56.8.2384-2388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Sugawara M, Minamisawa K, Yuhashi K, Ezura H. 1-Aminocyclopropane-1-carboxylate deaminase enhances Agrobacterium tumefaciens-mediated gene transfer into plant cells. Appl. Environ. Microbiol. 2008;74:2526–2528. doi: 10.1128/AEM.02253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA. Five rules for the evolution of cooperation. Science. 2006;314:1560–1563. doi: 10.1126/science.1133755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Oono R, Anderson CG, Denison RF. Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc. Biol. Sci. 2011;278:2698–2703. doi: 10.1098/rspb.2010.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankhurst CE, MacDonald PE, Reeves JM. Enhanced nitrogen fixation and competitiveness for nodulation of Lotus pedunculatus by a plasmid-cured derivative fo Rhizobium loti. J. Gen. Microbiol. 1986;132:2321–2328. [Google Scholar]
- Sachs JL, Mueller UG, Wilcox TP, Bull JJ. The evolu tion of cooperation. Q Rev. Biol. 2004;79:135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- Sanjuan J, Olivares J. NifA-NtrA regulatory system activates transcription of nfe, a gene locus involved in nodulation competitiveness of Rhizobium meliloti. Arch. Microbiol. 1991;155:543–548. doi: 10.1007/BF00245347. [DOI] [PubMed] [Google Scholar]
- Selbitschka W, Lotz W. Instability of cryptic plasmids affects the symbiotic effectivity of Rhizobium leguminosarum bv. viceae strains. Mol. Plant-Microbe Interact. 1991;4:608–618. [Google Scholar]
- Simsek S, Ojanen-Reuhs T, Stephens SB, Reuhs BL. Strain-ecotype specificity in Sinorhizobium meliloti-Medicago truncatula symbiosis is correlated to succinoglycan oligosaccharide structure. J. Bacteriol. 2007;189:7733–7740. doi: 10.1128/JB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiens M, Schneiker S, Keller M, Kuhn S, Pühler A, Schlüter A. Sequence analysis of the 144-kilobase accessory plasmid pSmeSM11a, isolated from a dominant Sinorhizobium meliloti strain identified during a long-term field release experiment. Appl. Environ. Microbiol. 2006;72:3662–3672. doi: 10.1128/AEM.72.5.3662-3672.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, de Billy F, Huguet T. Mtsym6, a gene conditioning Sinorhizobium strain-specific nitrogen fixation in Medicago truncatula. Plant Physiol. 2000;123:845–851. doi: 10.1104/pp.123.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett EW, Sadowsky MJ. Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 1992;46:399–428. doi: 10.1146/annurev.mi.46.100192.002151. [DOI] [PubMed] [Google Scholar]
- van Berkum P, Elia P, Eardly BD. Multilocus sequence typing as an approach for population analysis of Medicago-nodulating rhizobia. J. Bacteriol. 2006;188:5570–5577. doi: 10.1128/JB.00335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez E, Mateos PF, Pedrero P, Dazzo FB, Martínez-Molina E. Attenuation of symbiotic effectiveness by Rhizobium meliloti SAF22 related to the presence of a cryptic plasmid. Appl. Environ. Microbiol. 1995;61:2033–2036. doi: 10.1128/aem.61.5.2033-2036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 1998;28:1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- Yang SM, Tang F, Gao MQ, Krishnan HB, Zhu HY. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18735–18740. doi: 10.1073/pnas.1011957107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.