Abstract
The purpose of this study was to develop a sensitive, rapid, and inexpensive immunofluorescence assay (IFA) using a recombinant porcine circovirus type 2 (PCV2) nucleocapsid protein for the serological detection of PCV2-specific antibodies in pig sera. The viral nucleocapsid protein encoded by the PCV2 ORF2 gene has recently been identified as the most immunoreactive viral protein that carries type-specific antigenic determinants. The ORF2 sequence of the IAF-2897 strain of PCV2 has been cloned into a pCEP5 eucaryotic expression vector under the control of the cytomegalovirus promoter, downstream of a polyhistidine sequence tag. The recombinant plasmid was used in transfection experiments with human epithelial kidney 293 cells that were further tested, and positive expression of the viral nucleocapsid protein was confirmed by IFA and Western blotting. Strong, specific fluorescence was observed in the nuclei of transfected cells. Test specificity to PCV2 was verified with several related infectious agents. Sensitivity was compared to that of standard IFA using PCV2-infected cells by evaluating the reactivities of 44 field serum samples from pigs on farms with a porcine population suffering from postweaning multisystemic wasting syndrome. The recombinant nucleocapsid-based test was able to detect 15 more positive-testing pigs than the PCV2-based IFA. Therefore, the relative sensitivity of the latter test was estimated at only 57.1% compared to that of the recombinant nucleocapsid-based test. The recombinant fusion protein has been purified by affinity chromatography and is being used to develop further sensitive serological tests.
Postweaning multisystemic wasting syndrome (PMWS) is an emerging swine disease first described in Canada in 1991 (7, 13); subsequent outbreaks have frequently struck pig farms in North and South America and in Europe and Asia (2, 3, 6, 15, 22, 28). The disease affects 5- to 12-week-old piglets and is characterized in part by weight loss, dyspnea, jaundice, and enlarged lymph nodes as well as by degeneration and necrosis of hepatocytes, multifocal lymphohistiocytic pneumonia, lymphocytic depletion, and multinucleated giant cell formation (13). Typically, morbidity rates can reach 5 to 50% in affected herds, and mortality is close to 100% in pigs that develop the full spectrum of symptoms associated with the disease (19). The rapid and simultaneous emergence of the disease in many different parts of the world as well as the uncertain mode of transmission and high rates of mortality in pigs suffering from PMWS has caused great concern throughout the swine industry. The agent thought to be mainly responsible for PMWS has been identified; a circovirus-like particle isolated from diseased pigs was associated with the syndrome and named recombinant (rec) porcine circovirus type 2 (PCV2) (2, 11, 12).
The first porcine circovirus, now known as PCV1, was discovered in 1974 as a nonpathogenic contaminant of the porcine kidney PK-15 cell line (27). It has since been characterized as a small nonenveloped single-stranded DNA virus with a 1.76-kb ambisense genome (25). Antibodies against the virus have been found in pigs worldwide, but no discernible pathogenic properties have been associated with PCV1 infection in swine (8, 10, 24). PCV2 isolated from pigs suffering from PMWS displays a 76 to 83% amino acid identity with nonpathogenic PCV1 and shares the same genomic organization, consisting of two major open reading frames (ORFs) coding for the replicase protein (Rep) and the nucleocapsid (NC) protein, respectively (12, 18, 19). The 702-nucleotide ORF2 sequence coding for the PCV2 NC protein is located on the complementary strand of the double-stranded replicative form of the PCV2 genome and shares only 66% amino acid identity with the sequence of the PCV1 ORF2, compared with an 85% identity between the ORF1s of both viruses (12). These data have led scientists to speculate that the PCV2 NC protein may be at least in part responsible for the pathogenicity associated with PCV2. Recently, several studies have suggested that the 28-kDa PCV2 NC protein is the major immunogenic protein of the virus and the principal bearer of type-specific epitopes (14, 20).
PMWS is most commonly diagnosed on the basis of the presence of histopathological lesions, its characteristic clinical symptoms, and on the detection of PCV2 in the lesions (23). Current methods most commonly used for the detection of PCV2 in pigs include indirect immunofluorescence assays (IFA) for PCV2-infected cells and PCR (21). Because the use of PCR requires several time-consuming steps, IFA is generally preferred for rapid and inexpensive diagnosis of PMWS. However, IFA requires the prior infection of porcine cells with live PCV2 virus, a procedure which can be very arduous because of its very slow replication cycle. The aim of this work was to develop a rapid, easy-to-use, and inexpensive ORF2-based serological diagnostic assay for PCV2 detection on the basis of a rec PCV2 NC protein produced in a eucaryotic expression system.
(This report was taken in part from a dissertation to be submitted by S. Racine to the INRS-Institut Armand-Frappier, Université du Québec, in partial fulfillment of the requirements for the M.Sc. degree.)
MATERIALS AND METHODS
Virus, cells, and antisera.
In a previous work, the IAF-2897 strain of PCV2 virus was isolated from lung tissue of young pigs with typical clinical signs of PMWS in Quebec farms and its genome was entirely sequenced (GenBank accession no. AF408635) (21). The virus was propagated in PCV1-free PK-15 porcine kidney (PKA) cells, which were graciously provided by A. Afshar (Animal Diseases Research Institute, Agriculture Canada, Nepean, Ontario, Canada). Cells were infected at a very low level of confluence with purified virus and grown in monolayers for 3 to 4 days in GIBCO minimal essential medium (Invitrogen, Burlington, Ontario, Canada) supplemented with 5% fetal bovine serum, 1 mM glutamine, 1% sodium pyruvate, and 100 μM penicillin-streptomycin. When cells reached confluence, supernatant was removed and cells were washed three times with phosphate-buffered saline (PBS). The virus was harvested by freezing and thawing the infected cells three times in a small volume of PBS. The resulting cell lysate was then clarified by centrifugation. Viral genomic DNA was extracted from stocks of virus with TriPure DNA isolation reagent (Roche Diagnostics, Laval, Quebec, Canada) according to the manufacturer's recommendations. Transfection experiments were carried out with the human embryonic kidney 293 cell line (ATCC CRL-1573). Cells were grown in GIBCO Dulbecco's modified Eagle medium (Invitrogen) supplemented with 5% fetal bovine serum, 1 mM glutamine, 1% sodium pyruvate, and 100 μM penicillin-streptomycin.
To produce a swine reference serum for use in serological tests of the IAF-2897 strain of PCV2, four 5-week-old specific-pathogen-free pigs were injected intranasally with 106 fluorescence units of semipurified PCV2 virus stock. Antisera from inoculated pigs were tested weekly for the presence of PCV2-specific antibodies until bleeding at 63 days postinfection (dpi). Negative-testing serum samples were collected from two noninfected specific-pathogen-free piglets that were maintained under the same conditions. A total of 44 field serum samples collected from different groups of pigs from southern Quebec farms with porcine populations suspected to be infected with PCV2 were provided by Biovet Inc. (St-Hyacinthe, Quebec, Canada).
Cloning of PCV2 ORF2 in a eucaryotic expression vector.
The PCV2 ORF2 sequence coding for the virus NC was amplified from the purified PCV2 DNA in two successive PCRs using the primers described in Table 1. These primers were designed to add a polyhistidine sequence to permit purification of the rec protein by use of an affinity column and an enterokinase site for the subsequent removal of the polyhistidine site. All primers overlap with each other or with the ORF2 sequence by 20 to 22 nucleotides. Both PCRs used Taq polymerase and consisted of an initial enzyme activation step at 95°C for 5 min followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 48°C for 1 min, and extension at 72°C for 3 min and a final extension at 72°C for 10 min. The first PCR amplified the ORF2 by use of the 6H-F2 no. 1 forward primer and the F2 reverse primer. The resulting PCR product was then purified using a QIAGEN PCR purification kit (QIAGEN Inc., Mississauga, Ontario, Canada). In the second PCR the 6H-F2 no. 2 forward primer was used with the same F2 reverse primer used previously. A 780-nucleotide DNA sequence of the expected size was confirmed by gel electrophoresis, and the final PCR product was then purified as previously described.
TABLE 1.
Oligonucleotide primers used for the construction and sequencing of the pCEP5-6H-ORF2 plasmid
| Primer | Sensea | Sequenceb | Length (bp) |
|---|---|---|---|
| 6H-F2 no. 1 | F | 5′-ACCATCACGGTGACGATGACGATAAGGATATGACGTATCCAAGGAGGCG-3′ | 49 |
| 6H-F2 no. 2 | F | 5′-CGCGGATCCGCGATGCATCACCATCACCATCACGGTGACGATGACGA-3′ | 47 |
| F2 | R | 5′-CGCGGATCCGCGCGCGGATCCGCGTTAGGGTTTAAGTGGGGGGT-3′ | 44 |
| SeqpCEP-F | F | 5′-GAGTGACAATGACATCCAC-3′ | 19 |
| SeqpCEP-R | R | 5′-GTCCTTCCGAGTGAGAGAC-3′ | 19 |
F, forward; R, reverse.
Nucleotide sequences corresponding to BamHI digestion sites are underlined, the polyhistidine sequences are boxed, the enterokinase digestion sites are in bold, and nucleotides corresponding to the ORF2 sequence are italicized.
The eucaryotic expression vector pCEP5 derived from the pCEP4 vector (9) was obtained from the laboratory of A. Kamen (BRI, NRC, Montreal, Canada). Both the vector and the final purified PCR product were digested with the BamHI enzyme for 2 h at 37°C. The enzyme was subsequently inactivated by incubation at 65°C for 20 min. The digested PCR product and plasmid were then ligated using T4 DNA ligase at 14°C for 14 h. Escherichia coli DH5α competent cells were transformed using the resulting pCEP-6H-ORF2 plasmid. Positive clones were screened by PCR, restriction enzyme digestion, and DNA sequencing using the SeqpCEP primers detailed in Table 1.
Expression of the rec nucleocapsid protein.
Transfection experiments were carried out in either 96-well plaques or 60-cm2 petri dishes, and 293 or PKA cells as well as polyethylenimine (PEI) gene transfer reagent (5) in its 25-kDa linear form (Polysciences, Warrington, Pa.) were used for all transfection experiments. Typically, 1 μg of linear 25-kDa PEI was mixed with 1 μg of pCEP5-6H-ORF2 DNA plasmid in a total volume of 100 μl of GIBCO serum-free Dulbecco's modified Eagle medium. This mix was incubated at 37°C for 10 min, and then 20 μl per well was added to the subconfluent 293 or PKA cell monolayers and incubated in standard culture conditions for 48 h.
Expression of the PCV2 ORF2 in transfected cells was verified by IFA using polyclonal PCV2 porcine sera from pigs experimentally infected with purified PCV2, as well as sera from PCV-free pigs. Briefly, transfected cells were washed with PBS and fixed with an acetone-methanol solution (1:1). Fixed cells were then rehydrated with PBS for 10 min. PCV2-positive and -negative pig sera were diluted at 1/100 in PBS with 0.15% Tween 20 and incubated with cells for 1 h at 37°C. Cells were washed three times with PBS and then incubated with rabbit anti-pig immunoglobulin G-fluorescein isothiocyanate conjugate (Sigma-Aldrich, Oakville, Ontario, Canada) diluted 1/100 in PBS during 45 min at 37°C. Cells were finally observed and photographed with an epifluorescence microscope (Leica; Leitz, Wetzlar, Germany). PCV2-infected PKA cells were also used as controls as described above.
Purification and characterization of rec protein.
293 cells transfected with pCEP5-6H-ORF2 expression vector in 100-mm-diameter petri dishes were lysed with a cell lysis buffer (150 mM NaCl, 10 mM Tris [pH 7.8], 1% Triton X, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing protease inhibitors. The cell lysate was then solubilized by repeated passage through a syringe and added to a HiTrap chelating affinity column (Amersham Pharmacia Biotech, Baie d'Urfé, Québec, Canada) per the manufacturer's instructions. Collected fractions were screened for the presence of the rec ORF2 protein by SDS-polyacrylamide gel electrophoresis and Western blotting.
Collected fractions of purified 6H-ORF2 protein as well as nontransfected 293 cell lysate and purified virus preparation were analyzed by Western blotting. Proteins were separated in SDS-12% polyacrylamide electrophoresis gels by use of the standard method and transferred onto a nitrocellulose membrane (Bio-Rad, Mississauga, Ontario, Canada), which was then incubated in blocking solution (4% skim milk, 0.05% Tween 20 in PBS). Polyclonal anti-PCV2 serum, as well as serum from PCV-free pigs, was diluted 1/500 in blocking solution and incubated with the membranes for 1 h at room temperature. Membranes were washed three times in PBS-Tween 20 and incubated for 1 h with a 1/1,000-diluted solution of goat anti-porcine immunoglobulin G-peroxidase conjugate (Sigma) in blocking solution. After three final washes, the blots were developed with 4-chloro-1-naphthol reagent (Bio-Rad).
Sensitivity and specificity of the IFA.
All field serum samples were tested by IFA on 96-well plaques of either 293 cells transfected with pCEP5-6H-ORF2 or PKA cells infected with purified PCV2 as described previously. Serum dilutions from 1/50 to 1/5,000 were used. Sera were considered positive for PCV2 at 1/50. Specificity of both diagnostic tests for PCV2 was assayed by testing serum samples from pigs infected with PCV1, porcine reproductive and respiratory syndrome virus, porcine hemagglutinating encephalomyelitis coronavirus, and Mycoplasma hyopneumoniae. The sensitivity values for both assays were determined using the following formulas, with the IFA performed on transfected 293 cells as a reference assay: (i) sensitivity = true positives/(true positives + false negatives) × 100 and (ii) specificity = true negatives/(true negatives + false positives) × 100.
RESULTS
Expression of the PCV2 NC protein in transfected cells.
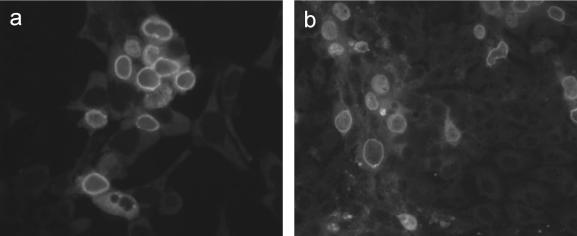
A mix of purified plasmid DNA and linear 25-kDa PEI was used to transfect subconfluent monolayers of 293 cells. Transfected cells showed no sign of cellular damage that could be attributed to the presence of the PCV2 NC protein or the PEI transfection reagent. The presence of the NC protein was determined by IFA with various types of antibodies. Intense and specific fluorescence was observed in the nuclei as well as in the perinuclear regions of transfected cells. The pattern of fluorescence was similar to that of PKA cells infected with wild-type PCV2 (Fig. 1).
FIG. 1.
Localization of the ORF2 protein in transfected cells. Fluorescence was localized in the peri- and intranuclear regions of 293 cells transfected with the pCEP5-6H-ORF2 plasmid (a). This specific fluorescence is similar to that observed in PCV2-infected PKA cells (b). To facilitate observation, the 293 cells were transfected with a suboptimal quantity of pCEP5-6H-ORF2 DNA.
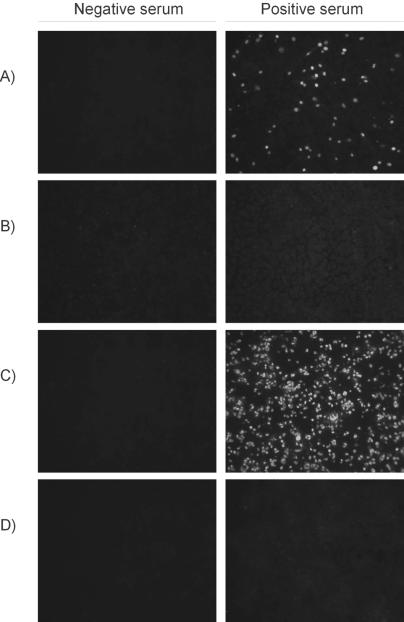
Transfected cells containing the recNC antigen reacted very strongly to polyclonal anti-PCV2 antibodies but did not cross-react with normal pig serum. The expression levels of the NC protein in in vitro infections of PKA cells and in 293 cells transfected with the pCEP-6H-ORF2 plasmid were compared by IFA (Fig. 2). The proportion of cells expressing the PCV2 NC antigen was much higher in cells transfected with the plasmid than in infected cells. Repeated experiments confirmed that, on average, the PCV2 recNC protein is expressed at levels roughly 9 times higher in cells transfected with the pCEP5-6H-ORF2 plasmid than in cells infected with the virus.
FIG. 2.
IFA comparison of PCV2 NC expression levels in PCV2-infected PKA cells and 293 cells transfected with pCEP5-6H-ORF2. (A) PKA cells infected with PCV2. (B) Mock-infected PKA cells. (C) 293 cells transfected with pCEP5-6H-ORF2. (D) Nontransfected 293 control cells. Cells were incubated with either polyclonal anti-PCV2 pig serum (positive serum) or PCV-negative pig serum (negative serum).
Characterization of the PCV2 recNC protein.
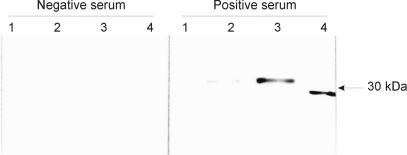
Transfection of subconfluent 293 cells in 60-mm-diameter petri dishes with the pCEP-6H-ORF2 plasmid resulted in the production of a significant quantity of PCV2 recNC which could then be recovered by cell lysis as described above. The resulting lysate containing both cellular proteins and the PCV2 recNC antigen was purified by passage through an Ni affinity column. Both the total protein from the cell lysate and the purified recNC reacted specifically in immunoblotting with polyclonal anti-PCV2 serum at a 1/500 dilution (Fig. 3, lanes 2 and 3). The recNC protein had an observed molecular mass of 32 kDa. Concentrated and purified PCV2 virus was used as a control antigen (Fig. 3, lane 4) and produced a specific reaction at a molecular mass of 29 kDa. Neither the recNC nor the purified PCV2 virus reacted with PCV2-negative swine serum.
FIG. 3.
Expression and purification of rec 6H-ORF2 protein and reactivity with polyclonal anti-PCV2 pig serum and PCV-negative serum in Western blotting. Lane 1, nontransfected 293 cell lysate; lane 2, total protein in cell lysate from 293 cells transfected with pCEP5-6H-ORF2; lane 3, 6H-ORF2 protein partially purified in a HiTrap chelating affinity column; lane 4, total protein in cell lysate from concentrated purified PCV2.
Accuracy of the rec ORF2-based diagnostic IFA.
The accuracy of the test was verified by comparing both the classic IFA diagnostic test using PCV2-infected PKA cells (classic IFA) and the recNC-based test (recNC IFA). All 44 porcine serum samples collected from PMWS-affected swine farms were tested using both IFA tests, and the observed results are summarized in Table 2. A total of 23 serum samples were positive for PCV2-specific antibodies by the PCV2-based test, while 35 serum samples were positive by the recNC-based test. The sensitivity of the classic IFA versus that of the recNC-based test is 57.1% when the latter is used as the reference assay. A total of 15 serum samples that were negative in the classic IFA were considered positive in the recNC IFA.
TABLE 2.
Comparison between IFA for the serologic detection of specific PCV2 antibodies in pigs
| Classic IFA result | No. (%) of serum samples with indicated result by recNC IFA
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 20 (45.5) | 3 (6.8) | 23 (52.3) |
| Negative | 15 (34.1) | 6 (13.6) | 21 (47.7) |
| Total | 35 (79.6) | 9 (20.4) | 44 (100.0) |
Among the 35 serum samples that tested positive for PCV2 antibodies in the recNC-based test, 10 showed antibody titers greater than 5,000 whereas only 3 showed antibody levels greater than 2,500 in the classic IFA. Serum samples from six pigs that tested negative in the classic IFA produced antibody titers over 2,500 with the new IFA test. Overall, 25 serum samples produced a significantly higher antibody titer with the recNC IFA than with the classic IFA. In addition, seven positive-testing serum samples gave roughly equal titers and nine were negative with both assays. Only three samples had antibody titers that were lower with the recNC IFA than with the classic IFA.
Positive-control sera from pigs infected with PCV1, porcine reproductive and respiratory syndrome virus, hemagglutinating encephalomyelitis coronavirus, or M. hyopneumoniae were also tested with both IFA (data not shown). Some nonspecific background fluorescence was observed among PCV2-infected PKA cells, but no reactivity whatsoever was observed in cells transfected with the pCEP5-6H-ORF2 plasmid. As a result, both IFA diagnostic tests had a specificity of 100%.
DISCUSSION
The dramatic increase in reported outbreaks of PMWS in pig farms in recent years has been an additional burden for pig producers already under pressure from the threat of other pig pathogens, increased public scrutiny, and intense competition in the marketplace. Early detection of PCV2 in pigs is essential to help contain the spread of the virus and of PMWS. The goal of the present study was to conceive a very specific and extremely sensitive but low-cost and easy-to-use test to help in the diagnosis of cases of early PCV2 infection in pigs. The IFA is probably the most commonly used method for the detection of PCV2 antibodies in pig serum, as it is faster and less expensive than PCR tests and does not require specialized equipment. However, the use of live PCV2 to infect cells used in IFA has always been problematic at best. Because the virus does not produce a visible cytopathic effect in infected cells and replicates only in cells currently undergoing mitosis, PCV2 infections yield very small amounts of virus (1). The use of d-glucosamine to stimulate the entry of cells in the S phase of mitosis to accelerate the replication of PCV2 has been suggested previously (26). However, it was also reported to be problematic because of its high level of cellular toxicity and therefore should generally be avoided when possible (1). There is also evidence of antigenic cross-reactivity between the Rep proteins of PCV1 and PCV2, which could generate false diagnostic results. Recent studies have demonstrated that the ORF1-encoded proteins in the two viruses were antigenically related whereas the ORF2 proteins were recognized differentially by polyclonal anti-PCV2 antibodies (4, 16, 17). No cross-reactivity was demonstrated between the NC proteins of the two viruses (29).
To circumvent the previous problems, the ORF2 gene from the PCV2 virus was cloned into a pCEP5 eucaryotic expression vector bearing a CMV2 promoter. The resulting pCEP5-6H-ORF2 plasmid was used to transfect cells that could then express the viral NC protein. Initial attempts to transfect PKA cells with the plasmid and either PEI or fuGENE6 transfection reagents (Roche Diagnostics) were a failure. No plasmid DNA was detected in the cytoplasm or the nuclei of transfected cells. Alternatively, 293 cells had previously been used successfully in conjunction with the pCEP5 vector and PEI (9). Subsequent transfection experiments with pCEP5-6H-ORF2 consistently produced clear and distinct fluorescence in the nucleus and the perinuclear area of the 293 cells (Fig. 1). This fluorescence pattern was identical to that observed in naturally infected PKA cells, but background fluorescence was markedly lower in transfected cells.
The proportion of transfected cells expressing the PCV2 recNC was roughly proportional to the quantity of plasmid DNA used in the transfection mix. Fluorescent cells may be observed with as little as 40 ng of plasmid DNA per well in a 96-well plate, the optimal amount of plasmid being about 200 to 300 ng per well. By use of these conditions, transfection of 293 cells with the plasmid generated on average 9 times more fluorescent cells expressing the PCV2 NC than transfection of PKA cells infected with the virus (Fig. 2). In addition, transfections of 293 cells in 60-mm-diameter petri dishes under the same transfection conditions yielded significantly larger quantities of NC product than transfection of PKA cells infected with PCV2 under the same culture conditions. The addition of a polyhistidine tag to the recNC protein allowed quick and easy purification and concentration of the protein by affinity chromatography with a metal chelate affinity column. Purified and nonpurified recNC from cell lysate analyzed by Western blotting reacted strongly with PCV2-specific antibodies (Fig. 3). As expected, the addition of the polyhistidine tag and the enterokinase restriction site to the NC amino acid sequence increased the molecular mass of the protein to roughly 32 kDa compared to about 29 kDa for the native protein. These results suggest that the recNC protein expressed in transfected cells is immunogenically identical to the native protein.
To determine whether recNC IFA using cells transfected with the pCEP-6H-ORF2 plasmid can be used as an accurate diagnostic tool for the detection of PCV2-specific antibodies in pigs, 44 swine serum samples from several porcine farms in southern Quebec that have had recurrent problems with PMWS outbreaks in the past years have been analyzed. Each serum sample was tested by classic IFA using a diagnostic protocol from local laboratories and by recNC IFA as described in the present study (Table 2). The results strongly indicate that the recNC IFA was able to detect much smaller amounts of antibodies in serum, as evidenced by comparing several serial serum dilutions in each test, and it is therefore a much more sensitive assay. The sensitivity of the classic IFA was only 57.1% compared to that of the recNC IFA. Of the 44 serum samples tested, only three samples that were considered to be positive using the classic IFA were shown to be negative with the recNC IFA. This could be explained by the high amount of background fluorescence often observed in classic IFA, particularly at low serum dilutions. However, fluorescence in transfected cells is strongly localized in the cell nucleus even under conditions of incubation with low serum dilutions. It is noteworthy that both IFA tests were also used with sera collected at different times postinfection from piglets experimentally infected with PCV2. The recNC IFA was able to detect the presence of PCV2-specific antibodies in serum as early as 7 dpi compared to 14 dpi for the classic IFA.
The results presented in this report indicate that an IFA diagnostic test using PCV2 recNC antigen expressed from transfected cells can quickly and accurately detect even small amounts of PCV2 antibodies in pig sera. The sensitivity of the assay is much improved compared to that of the classic IFA method using virus-infected cells. This system is particularly advantageous, because very large amounts of protein can be rapidly and inexpensively produced compared to the results seen with PCV2 infections, which can be relatively difficult to perform and produce only small quantities of virus. In addition, the recNC protein can be immediately purified and concentrated by affinity chromatography and can subsequently be used as an antigen for enzyme-linked immunosorbent assays and Western blot analyses and for further characterization studies. Additional work is necessary to determine whether the protein might be used as a subunit vaccine in pigs to prevent infection by PCV2.
Acknowledgments
We thank A. Kamen (BRI, NRC, Montreal, Canada) and Biovet Inc. (St-Hyacinthe, Québec, Canada) for providing, respectively, the pCEP5 eucaryotic expression vector and the swine sera from farms with pigs suffering from PMWS. We are grateful to Mourad Ouardani for his technical help as well as to Peter Tijssen for helpful discussions.
This work was partly supported by the Québec Ministry of Agriculture (MAPAQ), the Conseil de Recherche en Pêche et Agro-Alimentaire du Québec (CORPAQ) (grant 102055), and Biovet Inc. (St-Hyacinthe, Québec, Canada).
REFERENCES
- 1.Allan, G. M., and J. A. Ellis. 2000. Porcine circoviruses: a review. J. Vet. Diagn. Investig. 12:3-14. [DOI] [PubMed] [Google Scholar]
- 2.Allan, G. M., F. McNeilly, S. Kennedy, B. Daft, E. G. Clarke, J. A. Ellis, D. M. Haines, B. M. Meehan, and B. M. Adair. 1998. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 10:3-10. [DOI] [PubMed] [Google Scholar]
- 3.Allan, G. M., F. McNeilly, B. M. Meehan, S. Kennedy, D. P. Mackie, J. A. Ellis, E. G. Clark, E. Espuna, N. Saubi, P. Riera, A. Botner, and C. E. Charreyre. 1999. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet. Microbiol. 66:115-123. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard, P., D. Mahe, R. Cariolet, C. Truong, M. Le Dimna, C. Arnauld, N. Rose, E. Eveno, E. Albina, F. Madec, and A. Jestin. 2003. An ORF2 protein-based ELISA for porcine circovirus type 2 antibodies in post-weaning multisystemic wasting syndrome. Vet. Microbiol. 94:183-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boussif, O., F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix, and J. P. Behr. 1995. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA 92:7297-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, C., C. Chae, and E. G. Clark. 2000. Porcine postweaning multisystemic wasting syndrome in Korean pig: detection of porcine circovirus 2 infection by immunohistochemistry and polymerase chain reaction. J. Vet. Diagn. Investig. 12:151-153. [DOI] [PubMed] [Google Scholar]
- 7.Clark, E. 1997. Post-weaning multisystemic syndrome. Proc. Am. Assoc. Swine Prac. 28:499-501. [Google Scholar]
- 8.Dulac, G. C., and A. Afshar. 1989. Porcine circovirus antigens in PK-15 cell line (ATCC CCL-33) and evidence of antibodies to circovirus in Canadian pigs. Can. J. Vet. Res. 53:431-433. [PMC free article] [PubMed] [Google Scholar]
- 9.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, S., and J. J. Sands. 1994. Evidence of circovirus infection in British pigs. Vet. Rec. 134:680-681. [DOI] [PubMed] [Google Scholar]
- 11.Ellis, J., L. Hassard, E. Clark, J. Harding, G. Allan, P. Willson, J. Strokappe, K. Martin, F. McNeilly, B. Meehan, D. Todd, and D. Haines. 1998. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 39:44-51. [PMC free article] [PubMed] [Google Scholar]
- 12.Hamel, A. L., L. L. Lin, and G. P. Nayar. 1998. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72:5262-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harding, J. 1997. Post-weaning multisystemic wasting syndrome: preliminary epidemiology and clinical presentation. Proc. Am. Assoc. Swine Prac. 28:503. [Google Scholar]
- 14.Liu, Q., S. K. Tikoo, and L. A. Babiuk. 2001. Nuclear localization of the ORF2 protein encoded by porcine circovirus type 2. Virology 285:91-99. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Q., L. Wang, P. Willson, B. O'Connor, J. Keenliside, M. Chirino-Trejo, R. Melendez, and L. Babiuk. 2002. Seroprevalence of porcine circovirus type 2 in swine populations in Canada and Costa Rica. Can. J. Vet. Res. 66:225-231. [PMC free article] [PubMed] [Google Scholar]
- 16.Magar, R., P. Muller, and R. Larochelle. 2000. Retrospective serological survey of antibodies to porcine circovirus type 1 and type 2. Can. J. Vet. Res. 64:184-186. [PMC free article] [PubMed] [Google Scholar]
- 17.Mahe, D., P. Blanchard, C. Truong, C. Arnauld, P. Le Cann, R. Cariolet, F. Madec, E. Albina, and A. Jestin. 2000. Differential recognition of ORF2 protein from type 1 and type 2 porcine circoviruses and identification of immunorelevant epitopes. J. Gen. Virol. 81:1815-1824. [DOI] [PubMed] [Google Scholar]
- 18.Meehan, B. M., F. McNeilly, D. Todd, S. Kennedy, V. A. Jewhurst, J. A. Ellis, L. E. Hassard, E. G. Clark, D. M. Haines, and G. M. Allan. 1998. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 79:2171-2179. [DOI] [PubMed] [Google Scholar]
- 19.Morozov, I., T. Sirinarumitr, S. D. Sorden, P. G. Halbur, M. K. Morgan, K. J. Yoon, and P. S. Paul. 1998. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 36:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nawagitgul, P., I. Morozov, S. R. Bolin, P. A. Harms, S. D. Sorden, and P. S. Paul. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81(Pt. 9):2281-2287. [DOI] [PubMed] [Google Scholar]
- 21.Ouardani, M., L. Wilson, R. Jette, C. Montpetit, and S. Dea. 1999. Multiplex PCR for detection and typing of porcine circoviruses. J. Clin. Microbiol. 37:3917-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plana-Duran, J., M. Balasch, J. Segalés, C. Rosell, G. M. Rodriguez-Arrioja, M. Domingo, J. M. Folch, A. Sanchez, and A. Mankertz. 1999. Post-weaning multisystemic wasting syndrome in Spain. Vet. Rec. 145:87-88. [PubMed] [Google Scholar]
- 23.Rosell, C., J. Segalés, J. Plana-Duran, M. Balasch, G. M. Rodriguez-Arrioja, S. Kennedy, G. M. Allan, F. McNeilly, K. S. Latimer, and M. Domingo. 1999. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J. Comp. Pathol. 120:59-78. [DOI] [PubMed] [Google Scholar]
- 24.Tischer, I., L. Bode, D. Peters, S. Pociuli, and B. Germann. 1995. Distribution of antibodies to porcine circovirus in swine populations of different breeding farms. Arch. Virol. 140:737-743. [DOI] [PubMed] [Google Scholar]
- 25.Tischer, I., H. Gelderblom, W. Vettermann, and M. A. Koch. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64-66. [DOI] [PubMed] [Google Scholar]
- 26.Tischer, I., D. Peters, R. Rasch, and S. Pociuli. 1987. Replication of porcine circovirus: induction by glucosamine and cell cycle dependence. Arch. Virol. 96:39-57. [DOI] [PubMed] [Google Scholar]
- 27.Tischer, I., R. Rasch, and G. Tochtermann. 1974. Characterization of papovavirus- and picornavirus-like particles in permanent pig kidney cell lines. Zentbl. Bakteriol. [Orig. A] 226:153-167. [PubMed] [Google Scholar]
- 28.Trujano, M., G. Iglesias, J. Segalés, and J. M. Palacios. 2001. PCV-2 from emaciated pigs in Mexico. Vet. Rec. 148:792. [PubMed] [Google Scholar]
- 29.Truong, C., D. Mahe, P. Blanchard, M. Le Dimna, F. Madec, A. Jestin, and E. Albina. 2001. Identification of an immunorelevant ORF2 epitope from porcine circovirus type 2 as a serological marker for experimental and natural infection. Arch. Virol. 146:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]