Abstract
The insulin-like growth factor (IGF) signaling pathway is thought to play a major role in the etiology of breast cancer. Although incidence rates of breast cancer overall are lower in African Americans than in Caucasians, African-American women have a higher incidence under age 40 years, are diagnosed with more advanced disease, and have poorer prognosis. We investigated the association of breast cancer and genetic variants in genes in the IGF signaling pathway in a population-based case–control study of African-American women. We found significant associations at a locus encompassing parts of the IGFBP2 and IGFBP5 genes on chromosome 2q35, which we then replicated in a case–control study of Nigerian women. Based on those initial findings, we genotyped a total of 34 single nucleotide polymorphisms (SNPs) across the region in both study populations. Statistically significant associations with breast cancer were observed across approximately 50 kb of DNA sequence encompassing three exons in the 3′ end of IGFBP2 and three exons in the 3′ end of IGFBP5. SNPs were associated with breast cancer risk with P values as low as P = 0.0038 and P = 0.01 in African-Americans and Nigerians, respectively. This study is the first to report associations between genetic variants in IGFBP2 and IGFBP5 and breast cancer risk.
Introduction
Breast cancer is the leading female cancer in the United States, with one in eight women predicted to be diagnosed during their lifetime. Overall, African-American women have a lower incidence of breast cancer than Caucasians, but they have a higher incidence before age 40 years, are diagnosed with more aggressive disease, and have poorer 5-year survival (Aziz et al. 1999; English et al. 2002; Marbella and Layde 2001; Ries et al. 2004; Swanson and Lin 1994; Trock 1996). The reasons for these racial differences are not known.
Extensive evidence from in vivo and in vitro model systems and human studies [reviewed in Jerome et al. (2003), Pollak et al. (2004), Sachdev and Yee (2001)] supports a major role for the insulin-like growth factor-I (IGF1) signaling pathway in breast cancer pathogenesis. Moreover, for pre-menopausal breast cancer, significant positive associations have been reported with serum levels of IGF1 (Bohlke et al. 1998; Bruning et al. 1995; Hankinson et al. 1998; Toniolo et al. 2000; Yu et al. 2002). Mammographic density, a strong risk factor for breast cancer, has also been positively associated with the ratio of IGF-1 to IGFBP-3 (insulin-like growth factor binding protein 3) in pre-menopausal women, but not in post-menopausal women (Byrne et al. 2000). Interestingly, in several studies, higher serum IGF-I levels have been reported in healthy African-Americans as compared to healthy Caucasians, among both males and females (Jernstrom et al. 2001; Wong et al. 1999; Yanovski et al. 2000). Thus we hypothesize that genetic variation in the IGF signaling pathway may contribute to racial/ethnic disparities in breast cancer incidence and disease progression. We focused on African-Americans, because they are more likely to be diagnosed with aggressive breast cancer, yet are under-represented in research investigating genetic risk factors for breast cancer.
As part of an ongoing study, we assessed genetic variants in the IGF1 signaling pathway, specifically in the IGF1, IGFBP1, IGFBP2, IGFBP3, and IGFBP5 genes, and their association with breast cancer in a case–control study of African-American women. We found significant associations with SNPs in a region that spanned the 3′ ends of IGFBP2 and IGFBP5. We then confirmed the results in a Nigerian case-control study. Based on these findings, we genotyped a total of 34 SNPS, including one candidate functional SNP, in both study populations to further investigate the association with breast cancer. The results of the analysis of the 34 SNPs are reported here.
Materials and methods
Study populations
All study participants provided written informed consent and the study was approved by the Institutional Review Boards of the participating institutions.
African-Americans
The African-American study population includes females aged 35–79 years who participated in a multiethnic population-based case-control study of breast cancer (John et al. 2003; Wang et al. 2005), and females aged <65 years who participated in the Northern California component of the Breast Cancer Family Registry (John et al. 2004). Both studies identified newly diagnosed, invasive breast cancer cases through the population-based Greater San Francisco Bay Area Cancer Registry, and unrelated population controls through random-digit dialing (RDD) that were frequency matched to cases on race/ethnicity and expected 5-year age distribution. For the Breast Cancer Family Registry, given limited funding for this activity, a 1:1 match of cases:controls was not the objective. However, the same proportion of controls and cases participated in the study, so that there was no apparent selection bias in controls. This analysis is based on cases diagnosed between May 1997 and April 1999 and matched controls from the multi-ethnic case–control study and cases diagnosed between January 1995 and September 1998 and matched controls enrolled in the family registry. Both studies administered a brief telephone-screening interview to assess study eligibility, family and personal history of breast cancer, and self-identified race/ethnicity. Eligible cases and controls were invited to participate in an in-person interview and collection of a blood sample. This analysis is based on 460 African-American cases and 279 African-American controls with available DNA samples.
Nigerians
The Nigerian study population includes women aged 24–90 years newly diagnosed with histologically confirmed, invasive breast cancer, identified at the Surgical Oncology and Radiotherapy Units of the University College Hospital (UCH) in Ibadan, Nigeria (Adebamowo et al. 2003). All consecutive cases diagnosed between March 1998 and December 2003 were eligible, and 94% participated. Unrelated controls aged 18–80 years were identified through population-based recruitment. A community adjoining the hospital was randomly selected from a list of all the communities in the area. This community is stable, socio-economically diverse and represents the diversity of patients seen at the UCH. Samples from this community were examined as part of the HapMap study for internal control, and there was no evidence of relatedness (unpublished data). Controls were randomly selected from the community population register and were eligible if a study nurse found no evidence of breast cancer by clinical breast examination. The primary objective of the collection was breast cancer cases and not 1:1 matching of cases and controls, so that far fewer controls were recruited than cases. Cases and controls completed an in-person interview and provided a blood sample. Ethnicity was based on self-identification and 77% of the participants were Yoruba, 14% Hausa, and 9% Ibo. This analysis is based on 461 cases and 163 controls with available DNA samples.
SNP selection
In total, 34 SNPs were genotyped in the African-American and Nigerian study populations. These included seven SNPs in IGFBP2 and 10 SNPs in IGFBP5, selected from the NIEHS Panel 1 (http://egp.gs.washington.edu/welcome.html), a panel of 90 individuals representing the ethnic diversity in the United States, including Caucasians, Hispanics, Asian Americans, and African-Americans. These 12 SNPs were selected with an r2 threshold of 0.80 using the program LDSelect (Carlson et al. 2004), and having minor allele frequencies greater than 0.05. In addition, 16 SNPs in the interval between rs9341177 and rs11575195 were subsequently selected and genotyped to increase the density of markers in the region showing the strongest evidence for association. These 16 SNPs included all SNPs with minor allele frequencies (MAF) >0.10 in the Yoruban population in the HapMap database (public release #20), for which we could design assays. An additional putative functional SNP in IGFBP5 was included.
SNP genotyping
Genotyping was performed by the MGB Taqman probe assay from Applied Biosystems Inc. (ABI) or MGB Eclipse™ probe assay from Nanogen Inc. Primer and probe sequences are available on request. Duplicates of 46 DNA samples and water controls were genotyped for quality control. The laboratory technician was blinded as to whether samples were duplicates, cases, or controls. The order of the DNA samples on 384-well plates was randomized in order to ensure the same study conditions for samples from cases and controls. Genotyping call rates ranged from 95 to 99% and duplicate concordance rates were higher than 99%.
For the MGB Taqman probe assays, reactions were carried out using 10 ng genomic DNA according to manufacturer’s protocols. The fluorescence profile was read on an ABI PRISM 7900 HT instrument and the results analyzed with Sequence Detection Software (ABI). For the MGB Eclipse™ probe assays, reactions were carried out using 10 ng genomic DNA according to manufacturer’s protocols. Endpoint dissociation melting curves were generated on the ABI PRISM 7900 HT instrument by monitoring fluorescence while heating the reactions from 30 to 80°C at a 10% rate. Genotypes were assigned using the Eclipse-MeltMacro_v2.328 program.
Statistical analysis
Hardy–Weinberg equilibrium (HWE) was assessed in cases and controls separately and the maximum likelihood estimate of the inbreeding coefficient (f) was reported if a statistically significant (P < 0.05) deviation from HWE was observed. The inbreeding coefficient ranges from −1 to 1, with −1 indicating complete heterozygosity (no homozygotes observed), 0 indicating genotypes in perfect HWE proportions, and 1 indicating complete homozygosity.
Each SNP was individually tested for the association with breast cancer using a logistic regression model with the genotype modeled as an additive variable, with levels 0, 1 and 2 representing the number of copies of the minor allele. This model is equivalent to the Armitage trend test (Armitage 1955) and has been shown to be robust to deviations from Hardy–Weinberg equilibrium and most powerful in the absence of dominance (Sasieni 1997). Deviation from additivity was tested using a second logistic regression model that included an additional parameter for deviation from additivity. If a SNP showed significant evidence for non-additivity, dominance and recessive models were also fit. Odds ratio estimates and 95% confidence intervals (CI) were obtained from the most statistically significant genetic model. This multiple model approach ensured that each SNP was tested with a model that provided maximum power (Chapman and Clayton 2007). All logistic regression models included age at diagnosis (cases) or age at enrollment (controls) as a covariate. Odds ratio estimates and 95% confidence intervals (CI) were obtained from the most statistically significant genetic model. A variable for self-identified ethnicity was included in the analysis of the Nigerian data for four SNPs that showed significant (P value <0.05) ethnic differences in allele frequencies. No adjustment for multiple testing was done. Given the correlation between SNPs due to linkage disequilibrium, and the increased prior probability of observing association at a candidate gene, a Bonferroni type of adjustment for multiple testing is not appropriate.
A simulation study was carried out to assess the statistical significance of the association findings. Data sets were generated under the null hypothesis of no association between SNP genotypes and breast cancer status by randomly assigning individuals to either case or control status, while maintaining the individuals’ genotypes and the study sample sizes. This simulation approach maintains the LD structure among the SNPs. Results were based on 10,000 simulated datasets.
Results
IGFBP2 and IGFBP5 are in a tail to tail configuration on chromosome 2q. Thirty-four SNPs were genotyped spanning from intron 1 of IGFBP2 to intron 1 of IGFBP5. The non-synonomous SNP, rs11575194 in the third exon of IGFBP5, was not included in the analysis because of the rarity of the minor allele (observed three times in African-Americans and not at all among Nigerians). Thus, 33 SNPs were analyzed for association with breast cancer in the two population samples. Deviations from Hardy–Weinberg equilibrium (P value < 0.01) only were observed for three SNPs in African-Americans (rs9341151, rs9341163 and rs2241193). None of the three SNPs were associated with breast cancer. Minor allele frequencies less than 0.05 were observed for one SNP in the African-American sample and for two SNPs in the Nigerian sample. These SNPs were included in the analysis, even though this study had very modest power to detect an association with an allele of such low frequency (Tables 1, 2).
Table 1.
Associations of genetic variants across IGFBP2 and IGFBP5 and breast cancer risk in African-American and Nigerian women
| SNP | African-Americans
|
Nigerians
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAFa
|
Association tests
|
Odds ratio | 95% CI
|
MAF+
|
Association tests
|
Odds ratio | 95% CI
|
|||||||
| Case | Control | Model | P value | Lower | Upper | Case | Control | Model* | P value | Lower | Upper | |||
| rs7603372 | 0.28 | 0.28 | Add | 0.94 | 0.99 | 0.78 | 1.25 | 0.18 | 0.19 | Add | 0.38 | 0.86 | 0.61 | 1.21 |
| rs9341134 | 0.15 | 0.10 | Add | 0.013 | 1.54 | 1.10 | 2.15 | 0.16 | 0.16 | Add | 0.88 | 1.03 | 0.70 | 1.51 |
| rs7600691 | 0.24 | 0.23 | Add | 0.71 | 1.90 | 0.92 | 3.95 | 0.33 | 0.32 | Add | 0.87 | 1.02 | 0.78 | 1.34 |
| rs9341145 | 0.04 | 0.03 | Add | 0.65 | 1.14 | 0.65 | 2.00 | 0.02 | 0.01 | Add | 0.25 | 2.15 | 0.58 | 8.00 |
| rs9341151 | 0.14 | 0.16 | Add | 0.56 | 0.92 | 0.69 | 1.22 | 0.26 | 0.21 | Add | 0.13 | 1.28 | 0.93 | 1.75 |
| rs9288521 | 0.11 | 0.10 | Rec | 0.04 | 0.24 | 0.06 | 0.93 | 0.10 | 0.10 | Add | 0.65 | 1.11 | 0.71 | 1.74 |
| rs9341157 | 0.12 | 0.11 | Add | 0.24 | 1.23 | 0.87 | 1.74 | 0.18 | 0.15 | Add | 0.53 | 1.13 | 0.78 | 1.64 |
| rs9341163 | 0.20 | 0.21 | Add | 0.77 | 0.96 | 0.75 | 1.24 | 0.27 | 0.31 | Rec | 0.016 | 0.47 | 0.26 | 0.87 |
| rs9341164 | 0.27 | 0.25 | Add | 0.38 | 1.12 | 0.87 | 1.42 | 0.33 | 0.31 | Add | 0.50 | 1.11 | 0.83 | 1.48 |
| rs9341165 | 0.08 | 0.07 | Add | 0.62 | 1.10 | 0.74 | 1.64 | 0.11 | 0.11 | Rec | 0.045 | 0.30 | 0.09 | 0.97 |
| rs9341177 | 0.15 | 0.16 | Add | 0.66 | 0.93 | 0.69 | 1.26 | 0.22 | 0.20 | Add | 0.29 | 1.19 | 0.86 | 1.65 |
| rs9341178 | 0.25 | 0.24 | Add | 0.63 | 1.06 | 0.83 | 1.36 | 0.34 | 0.30 | Rec | 0.01 | 2.62 | 1.26 | 5.47 |
| rs9341182 | 0.07 | 0.08 | Add | 0.83 | 0.96 | 0.64 | 1.44 | 0.13 | 0.08 | Dom | 0.013 | 1.95 | 1.15 | 3.31 |
| rs7567238 | 0.15 | 0.14 | Add | 0.49 | 1.11 | 0.82 | 1.50 | 0.20 | 0.15 | Dom | 0.01 | 1.76 | 1.14 | 2.70 |
| rs9288522 | 0.35 | 0.33 | Rec | 0.015 | 1.85 | 1.13 | 3.03 | 0.44 | 0.43 | Add | 0.52 | 1.09 | 0.83 | 1.44 |
| rs9341218 | 0.31 | 0.26 | Rec | 0.0067 | 2.34 | 1.27 | 4.33 | 0.39 | 0.39 | Add | 0.78 | 1.04 | 0.79 | 1.37 |
| rs9341223 | 0.26 | 0.23 | Rec | 0.0038 | 2.98 | 1.42 | 6.26 | 0.31 | 0.25 | Add | 0.035 | 1.38 | 1.02 | 1.87 |
| rs9341226 | 0.12 | 0.11 | Add | 0.60 | 1.09 | 0.79 | 1.51 | 0.17 | 0.10 | Add | 0.01 | 1.76 | 1.14 | 2.73 |
| rs9341227 | 0.05 | 0.06 | Add | 0.75 | 0.93 | 0.58 | 1.48 | 0.06 | 0.09 | Add | 0.07 | 0.62 | 0.37 | 1.04 |
| rs9288523 | 0.07 | 0.07 | Add | 0.98 | 1.00 | 0.68 | 1.50 | 0.09 | 0.07 | Add | 0.13 | 1.48 | 0.89 | 2.49 |
| rs2067041 | 0.42 | 0.41 | Add | 0.75 | 1.04 | 0.83 | 1.30 | 0.48 | 0.50 | Add | 0.60 | 0.93 | 0.72 | 1.22 |
| rs4674107 | 0.05 | 0.08 | Add | 0.023 | 0.62 | 0.41 | 0.94 | 0.01 | 0.01 | Add | 0.86 | 0.81 | 0.08 | 8.14 |
| rs3276 | 0.10 | 0.11 | Add | 0.60 | 0.91 | 0.65 | 1.28 | 0.11 | 0.08 | Add | 0.25 | 1.32 | 0.83 | 2.10 |
| rs11575213 | 0.29 | 0.28 | Rec | 0.013 | 2.24 | 1.18 | 4.25 | 0.36 | 0.28 | Add | 0.012 | 1.44 | 1.08 | 1.90 |
| rs11575212 | 0.14 | 0.14 | Add | 0.94 | 1.01 | 0.74 | 1.38 | 0.16 | 0.16 | Add | 0.58 | 1.11 | 0.77 | 1.60 |
| rs6746360 | 0.28 | 0.27 | Rec | 0.014 | 2.23 | 1.18 | 4.23 | 0.35 | 0.32 | Add | 0.28 | 1.16 | 0.88 | 1.53 |
| rs11575195 | 0.10 | 0.10 | Add | 0.72 | 1.07 | 0.75 | 1.51 | 0.10 | 0.10 | Add | 0.43 | 1.20 | 0.76 | 1.89 |
| rs7585038 | 0.10 | 0.07 | Add | 0.039 | 1.51 | 1.02 | 2.23 | 0.14 | 0.13 | Add | 0.65 | 1.10 | 0.74 | 1.63 |
| rs11575161 | 0.17 | 0.17 | Rec | 0.047 | 2.71 | 1.01 | 7.25 | 0.07 | 0.07 | Add | 0.96 | 0.99 | 0.59 | 1.65 |
| rs7420849 | 0.27 | 0.27 | Add | 0.93 | 1.01 | 0.80 | 1.27 | 0.21 | 0.19 | Add | 0.37 | 1.17 | 0.84 | 1.63 |
| rs7426116 | 0.20 | 0.27 | Add | 0.009 | 0.72 | 0.57 | 0.92 | 0.18 | 0.23 | Add | 0.030 | 0.70 | 0.51 | 0.97 |
| rs741384 | 0.46 | 0.44 | Add | 0.59 | 1.06 | 0.86 | 1.31 | 0.44 | 0.38 | Add | 0.057 | 1.30 | 0.99 | 1.69 |
| rs2241193 | 0.21 | 0.22 | Add | 0.51 | 0.92 | 0.72 | 1.18 | 0.18 | 0.19 | Add | 0.71 | 0.94 | 0.66 | 1.33 |
Minor allele frequency
Add additive, Dom dominant, Rec recessive
Table 2.
SNPs showing evidence for association with breast cancer in African-Americans
| SNP | Cases
|
f | Controls
|
f | Model | Obs. P value | Simul. P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | HWE | 1/1 | 1/2 | 2/2 | HWE | ||||||
| rs9341134 | 332 | 118 | 9 | 0.69 | NS | 216 | 47 | 4 | 0.46 | NS | Add | 0.013 | 0.011 |
| rs9288521 | 346 | 95 | 3 | 0.16 | NS | 226 | 40 | 7 | 0.01 | 0.18 | Rec | 0.040 | 0.008 |
| rs9288522 | 204 | 180 | 70 | 0.005 | 0.13 | 112 | 129 | 24 | 0.12 | NS | Rec | 0.015 | 0.013 |
| rs9341218 | 231 | 181 | 51 | 0.09 | NS | 144 | 116 | 14 | 0.12 | NS | Rec | 0.0067 | 0.0037 |
| rs9341223 | 256 | 157 | 41 | 0.02 | 0.11 | 156 | 110 | 9 | 0.04 | −0.12 | Rec | 0.0038 | 0.0021 |
| rs4674107 | 420 | 37 | 5 | 0.004 | 0.17 | 232 | 39 | 2 | 0.80 | NS | Add | 0.023 | 0.021 |
| rs11575213 | 243 | 174 | 45 | 0.10 | NS | 138 | 128 | 13 | 0.01 | −0.15 | Rec | 0.013 | 0.011 |
| rs6746360 | 245 | 174 | 45 | 0.09 | NS | 143 | 123 | 13 | 0.03 | −0.13 | Rec | 0.014 | 0.011 |
| rs7585038 | 373 | 84 | 6 | 0.62 | NS | 238 | 34 | 2 | 0.55 | NS | Add | 0.039 | 0.038 |
| rs11575161 | 324 | 114 | 23 | 0.005 | 0.26 | 185 | 84 | 5 | 0.17 | NS | Rec | 0.047 | 0.042 |
| rs7426116 | 297 | 144 | 22 | 0.41 | NS | 153 | 98 | 24 | 0.16 | NS | Add | 0.009 | 0.008 |
Associations with breast cancer risk in African-American women are shown in Table 1. Eleven of the 34 SNPs were associated at a P value less than 0.05, three of which showed P values less than 0.01. The strongest association was found for SNP rs9341223 (P value = 0.0038). SNPs rs9341218 (P value = 0.0067) and rs9288522 (P value = 0.015) were in linkage disequilibrium with rs9341223, showing r2 values of 0.63 and 0.58 between each respective pair. Seven SNPs showed significant non-additive effects, all of which showed the highest statistical significance under the recessive inheritance model. For all SNPs showing evidence for association, with the exception of rs9288521 and rs7426116, odds ratio estimates indicate that the minor allele confers increased risk. The significant associations observed at SNPs rs9341134, rs9341223, rs11575213, rs6746360 and rs11575161 in IGFBP2 and IGFBP5 led to the subsequent replication in the Nigerian study.
Associations with breast cancer risk in the Nigerian case–control study are shown in Table 1. Nine of the 33 SNPs analyzed showed evidence for association with P values less than 0.05, including three of the 11 SNPs associated with risk in African-American women. Notably, the SNP most strongly associated with risk in African-Americans, rs9341223, was also significantly associated with risk in Nigerians (P value = 0.035). For this SNP and for SNP rs11575213, different genetic models were significant for the two populations (recessive in African-Americans and additive in Nigerians).
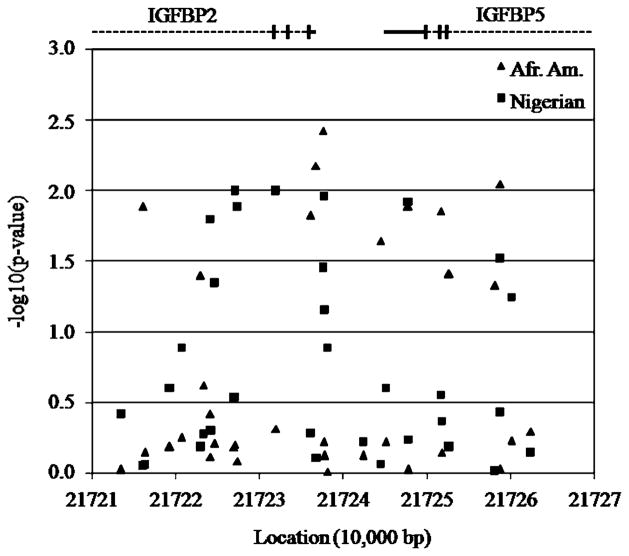
In Fig. 1, the locations and associations for the 33 SNPs tested across the IGFBP2 and IGFBP5 region in both populations are shown. The −1 × log 10 (P value) for the genetic model (additive, dominant or recessive) providing the strongest evidence for association is shown for each SNP. The locations of IGFBP2 and IGFBP5 are shown along the top of the figure, with bars indicating locations of exons. The two genes are oriented tail to tail with their 3′ untranslated regions (UTRs) separated by approximately 7,670 bp. The SNPs showing statistical significance are predominately clustered around the three 3′ exons and UTR of IGFBP2, the interval between the two genes, and the three 3′ exons and UTR of IGFBP5. The total region investigated spans 48,876 bp between SNPs rs7603372 and rs2241193. In the African-American women, all five of the SNPs showing evidence for association under the recessive model between SNPs rs9288522 and rs6746360 had significant r2 values between 0.58 and 0.63. The multiple significant associations observed at SNPs under the additive model or outside this region in the African-Americans cannot be explained by linkage disequilibrium. In the Nigerian women, all six SNPs showing evidence for association between SNPs rs9341178 and rs11575213 had significant r2 values between 0.28 and 0.64. All other SNPs showing evidence for association were not significantly correlated through linkage disequilibrium. Linkage disequilibrium patterns across the region with added marker density (between rs9341177 and rs11575195) were complex in both population samples and clear block patterns were not discernable. Haplotypes within two small regions of high linkage disequilibrium were no more informative than the individual SNPs constituting the haplotypes.
Fig. 1.
Associations of individual SNPs in the IGFBP2 and IGFBP5 region and breast cancer risk in African-American and Nigerian women
Tables 2 and 3 show the genotype counts for those SNPs showing evidence for association at P value < 0.05, in the African-American and Nigerian samples, respectively. HWE testing of SNPs using the combined case and control samples showed one SNP (rs4674107) with evidence for a deviation from equilibrium (P = 0.02). An excess of homozygous genotypes among cases and a deficiency of homozygous genotypes among controls could result from the disease association of a SNP that is acting under a recessive model of inheritance. It should be noted that the HWE test is highly sensitive to the number of homozygous genotype counts for rare alleles.
Table 3.
SNPs showing evidence for association with breast cancer in Nigerians
| SNP | Cases
|
f | Controls
|
f | Model | Obs. P value | Simul. P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | HWE | 1/1 | 1/2 | 2/2 | HWE | ||||||
| rs9341163 | 271 | 205 | 35 | 0.65 | NS | 77 | 51 | 20 | 0.02 | 0.19 | Rec | 0.016 | 0.012 |
| rs9341165 | 405 | 101 | 8 | 0.57 | NS | 121 | 21 | 6 | 0.003 | 0.28 | Rec | 0.045 | 0.053 |
| rs9341178 | 231 | 214 | 65 | 0.17 | NS | 69 | 67 | 10 | 0.23 | NS | Rec | 0.010 | 0.009 |
| rs9341182 | 390 | 116 | 6 | 0.40 | NS | 128 | 17 | 3 | 0.05 | NS | Dom | 0.013 | 0.014 |
| rs7567238 | 323 | 170 | 20 | 0.68 | NS | 111 | 30 | 6 | 0.06 | NS | Dom | 0.010 | 0.007 |
| rs9341223 | 248 | 239 | 48 | 0.37 | NS | 86 | 60 | 9 | 0.73 | NS | Add | 0.035 | 0.031 |
| rs9341226 | 355 | 142 | 17 | 0.55 | NS | 114 | 25 | 2 | 0.66 | NS | Add | 0.010 | 0.009 |
| rs11575213 | 223 | 240 | 71 | 0.61 | NS | 84 | 65 | 13 | 0.93 | NS | Add | 0.012 | 0.011 |
| rs7426116 | 351 | 141 | 21 | 0.17 | NS | 90 | 48 | 11 | 0.22 | NS | Add | 0.030 | 0.033 |
Using simulation, we assessed the probability of observing a P value less than that which was observed at each specific SNP. The last columns of Table 2 show the proportion of replicates showing P values less than observed at the individual SNP. The high correlation between the P values calculated from the real data and from the simulated data indicates that the individual results are robust. One possible exception is SNP rs9288521 that showed evidence for association under a recessive model but with very few genotypes homozygous for the minor allele.
We next analyzed the replicate datasets in order to assess the significance of our findings in the context of the 33 SNPs that were tested within each population. This analysis provides an estimate of the probability of observing results similar to those reported while taking into account the number of SNPs tested, the LD between the SNPs and the genetic models tested. There were three SNPs showing P values less than 0.01 in the African-American sample, as well as in the Nigerian sample. From the simulated data-sets, we assessed the probability of observing any three SNPs with P values less than 0.01 in each dataset. The simulated African-American data showed 152/10,000 replicates in which any three or more SNPs showed P values less than 0.01, providing a rough estimate of the probability of this observation as 0.015. Similarly, there was 149/10,000 simulated Nigerian datasets in which three or more SNPs showed P values less than 0.01. The African-American and Nigerian datasets both showed three or more SNPs with P values less than 0.01 for the same iteration of the datasets in only 3/10,000 replicates, providing a rough estimate of 0.0003 as the probability of such a replication occurring by chance.
Discussion
We initially found an association of breast cancer risk in African-American women with a locus on chromosome 2q35, encompassing parts of IGFBP2 and IGFBP5, and subsequently replicated this association in Nigerian women. The association was statistically significant in both populations. A total of 34 SNPs were genotyped in this region. Significant associations with individual SNPs at a common locus were observed in the two independent populations of African descent. Given the complex etiology of breast cancer, the genetic diversity within and between the two populations studied, and the relatively small sample size in both studies, the inconsistent nature of the results is not unexpected. It is unlikely that any of the genetic variants tested in this study are causal; the observed associations likely depend on the allele frequencies in the two populations, as well as the linkage disequilibrium patterns across the region. It is also possible that the associations are due to different etiological variants, or sets of variants, in the two populations, and that the effects are dependent on different genetic backgrounds and/or interactions with different environmental factors. Ideally, future association studies of this candidate region should be based on additional SNPs and larger sample sizes, accounting for population substructure, to reduce any random effects of sampling. Functional studies will need to be done to confirm the actual causal variant.
The IGF pathway is a plausible candidate pathway to investigate, because it is a key factor in the development and progression of breast cancer, based on evidence from more than 1,100 published papers, ranging from in vivo and in vitro studies in humans and mice to epidemiologic studies [reviewed in Jerome et al. (2003), Pollak et al. (2004) and Sachdev and Yee (2001)]. There have been a limited number of epidemiologic studies of the association of breast cancer risk and genetic variation in genes in the IGF pathway, focused on IGF1, IGFBP1, IGFBP3, and IGFALS. For IGF1, two reports showed significant associations (Canzian et al. 2006; Cleveland et al. 2006), whereas others found no association (Al-Zahrani et al. 2006; Gonzalez-Zuloeta Ladd et al. 2007; Missmer et al. 2002; Setiawan et al. 2006; Wagner et al. 2005). For IGFBP3, one study in Chinese women found a significantly elevated breast cancer risk associated with genetic variants in IGFBP3 (Ren et al. 2004), as did a study in German women (Wagner et al. 2005), whereas other studies found no associations (Al-Zahrani et al. 2006; Canzian et al. 2006; Cheng et al. 2006; Schernhammer et al. 2003). The inconsistent results may be due to many factors such as differences in genetic variants examined in the genes and/or in study design (e.g., restriction to post-menopausal or pre-menopausal breast cancers). No positive associations have been reported for IGFBP1 (Canzian et al. 2006; Cheng et al. 2006) and IGFALS (Canzian et al. 2006). A study investigating breast cancer survival among Chinese women found significant associations with two SNPs in IGFBP3 and one SNP in IGF1R, and no associations with SNPs in IGF1 or IGFALS (Deming et al. 2007). To our knowledge, no studies have been published investigating the association of genetic variation in IGFBP2 and IGFBP5 and breast cancer risk.
The insulin-like growth factor binding proteins (IGFBPs) serve as growth modulators, both independently and as regulators of IGFs (Mohan and Baylink 2002; Rosenzweig 2004). The different binding proteins may modulate IGF action differently, and the same binding protein may play a different role under different cellular conditions (Jones and Clemmons 1995). IGFBP-2 is expressed in invasive and in situ breast cancers, but not in normal tissues (Busund et al. 2005). It is involved in apoptosis by IGF signaling through the phosphoinositol-3-kinase (PI3K) pathway (Martin and Baxter 2007), and also independently of IGF (Frommer et al. 2006). IGFBP-2 also interacts with integrin to regulate tumor growth and migration (Pereira et al. 2004; Schutt et al. 2004). IGFBP-5 mRNA and protein are found in breast cancer tissues and in primary breast cancer cell cultures (Beattie et al. 2006), and are involved in apoptosis of the mammary gland, in both in vivo and in vitro mouse studies, through inhibition of IGF-signaling and independently of IGF through interactions with molecules in the extracellular matrix (Flint et al. 2003; Marshman et al. 2003). This same effect has been observed in human breast cancer cells (Butt et al. 2005, 2003). IGFBP-2 and IGFBP-5 are also associated with tamoxifen resistance (Becker et al. 2005; Maxwell and van den Berg 1999). IGFBP-2 and IGFBP-5 thus appear to have multiple roles in breast cancer, and variants in these genes could contribute to risk.
The causative genetic variants that underlay the observed associations across IGFBP2 and IGFBP5 must still be identified before it can be determined how these genes alter breast cancer risk and whether one or both genes are involved. Currently, one can only hypothesize that the genetic variant alters the expression of the IGFBP in the cell, the binding of IGF-1 to the IGFBP, the proteolytic action of the proteases, or the localization within the cell. Any alterations in the IGFBP protein could result in changes in amounts of IGF-1 to bind to the IGF receptor to activate the downstream phosphoinositol-3-kinase (PI3K) and the Rasmitogen-activated protein kinase (MAPK) pathways, or alter the IGFBP independent effects associated with apoptosis, cell adhesion, or cell migration.
In conclusion, the finding of a significant association in two independent populations of African descent, the consistent localization in the two populations, and the biological plausibility of the association between the candidate genes and the phenotype provide strong evidence for association with this locus. Further replication in larger populations of African descent, as well as in non-African populations, are required to increase confidence in the reported association and to localize the etiological component of the association.
Acknowledgments
We thank Jocelyn Koo for data management and Karen Kelly and Megan Rounds for genotyping, and Kavita Renduchintala, Ming Li, and Aileen Sy for help with the genotyping. We thank the women who participated in this research. This work was supported by the California Breast Cancer Program 9PB-0142 (to SLN). The multi-ethnic case-control study was supported by grant CA77305 from the National Cancer Institute (NCI) and grant 17-96-1-6071 by the U.S. Department of Defense (to EMJ). The Northern California Family Registry for Breast Cancer was supported by the NCI under RFA#CA95-003 through a cooperative agreement with the Northern California Cancer Center (to EMJ). Collection of the Nigerian case-control population was funded by NCI CA-RO1 89085-01A and the Ralph and Marion Falk Medical Research Trust (to OIO).
Contributor Information
Chad P. Garner, Department of Epidemiology, University of California Irvine, 224 Irvine Hall, Irvine, CA 92697-7550, USA
Yuan C. Ding, Department of Epidemiology, University of California Irvine, 224 Irvine Hall, Irvine, CA 92697-7550, USA
Esther M. John, Northern California Cancer Center, Fremont, CA, USA
Sue A. Ingles, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA
Olufunmilayo I. Olopade, Department of Medicine, University of Chicago, Chicago, IL, USA
Dezheng Huo, Department of Health Studies, University of Chicago, Chicago, IL, USA.
Clement Adebamowo, Department of Surgery, Division of Oncology, College of Medicine, University of Ibadan, Ibadan, Nigeria.
Temidayo Ogundiran, Department of Surgery, Division of Oncology, College of Medicine, University of Ibadan, Ibadan, Nigeria.
Susan L. Neuhausen, Email: sneuhaus@uci.edu, Department of Epidemiology, University of California Irvine, 224 Irvine Hall, Irvine, CA 92697-7550, USA
References
- Adebamowo CA, Ogundiran TO, Adenipekun AA, Oyesegun RA, Campbell OB, Akang EU, Rotimi CN, Olopade OI. Obesity and height in urban Nigerian women with breast cancer. Ann Epidemiol. 2003;13:455–461. doi: 10.1016/s1047-2797(02)00426-x. [DOI] [PubMed] [Google Scholar]
- Al-Zahrani A, Sandhu MS, Luben RN, Thompson D, Baynes C, Pooley KA, Luccarini C, Munday H, Perkins B, Smith P, Pharoah PD, Wareham NJ, Easton DF, Ponder BA, Dunning AM. IGF1 and IGFBP3 tagging polymorphisms are associated with circulating levels of IGF1, IGFBP3 and risk of breast cancer. Hum Mol Genet. 2006;15:1–10. doi: 10.1093/hmg/ddi398. [DOI] [PubMed] [Google Scholar]
- Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- Aziz H, Hussain F, Sohn C, Mediavillo R, Saitta A, Hussain A, Brandys M, Homel P, Rotman M. Early onset of breast carcinoma in African American women with poor prognostic factors. Am J Clin Oncol. 1999;22:436–440. doi: 10.1097/00000421-199910000-00002. [DOI] [PubMed] [Google Scholar]
- Beattie J, Allan GJ, Lochrie JD, Flint DJ. Insulin-like growth factor-binding protein-5 (IGFBP-5): a critical member of the IGF axis. Biochem J. 2006;395:1–19. doi: 10.1042/BJ20060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Sommer A, Kratzschmar JR, Seidel H, Pohlenz HD, Fichtner I. Distinct gene expression patterns in a tamoxifen-sensitive human mammary carcinoma xenograft and its tamoxifen-resistant subline MaCa 3366/TAM. Mol Cancer Ther. 2005;4:151–168. [PubMed] [Google Scholar]
- Bohlke K, Cramer DW, Trichopoulos D, Mantzoros CS. Insulin-like growth factor-I in relation to premenopausal ductal carcinoma in situ of the breast. Epidemiology. 1998;9:570–573. [PubMed] [Google Scholar]
- Bruning PF, Van Doorn J, Bonfrer JM, Van Noord PA, Korse CM, Linders TC, Hart AA. Insulin-like growth-factor-binding protein 3 is decreased in early-stage operable pre-menopausal breast cancer. Int J Cancer. 1995;62:266–270. doi: 10.1002/ijc.2910620306. [DOI] [PubMed] [Google Scholar]
- Busund LT, Richardsen E, Busund R, Ukkonen T, Bjornsen T, Busch C, Stalsberg H. Significant expression of IGFBP2 in breast cancer compared with benign lesions. J Clin Pathol. 2005;58:361–366. doi: 10.1136/jcp.2004.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AJ, Dickson KA, McDougall F, Baxter RC. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J Biol Chem. 2003;278:29676–29685. doi: 10.1074/jbc.M301965200. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Dickson KA, Jambazov S, Baxter RC. Enhancement of tumor necrosis factor-alpha-induced growth inhibition by insulin-like growth factor-binding protein-5 (IGFBP-5), but not IGFBP-3 in human breast cancer cells. Endocrinology. 2005;146:3113–3122. doi: 10.1210/en.2004-1408. [DOI] [PubMed] [Google Scholar]
- Byrne C, Hankinson SE, Pollak M, Willett WC, Colditz GA, Speizer FE. Insulin-like growth factors and mammographic density. Growth Horm IGF Res. 2000;10(Suppl A):S24–5. doi: 10.1016/s1096-6374(00)90011-x. [DOI] [PubMed] [Google Scholar]
- Canzian F, McKay JD, Cleveland RJ, Dossus L, Biessy C, Rinaldi S, Landi S, Boillot C, Monnier S, Chajes V, Clavel-Chapelon F, Tehard B, Chang-Claude J, Linseisen J, Lahmann PH, Pischon T, Trichopoulos D, Trichopoulou A, Zilis D, Palli D, Tumino R, Vineis P, Berrino F, Bueno-de-Mesquita HB, van Gils CH, Peeters PH, Pera G, Ardanaz E, Chirlaque MD, Quiros JR, Larranaga N, Martinez-Garcia C, Allen NE, Key TJ, Bingham SA, Khaw KT, Slimani N, Norat T, Riboli E, Kaaks R. Polymorphisms of genes coding for insulin-like growth factor 1 and its major binding proteins, circulating levels of IGF-I and IGFBP-3 and breast cancer risk: results from the EPIC study. Br J Cancer. 2006;94:299–307. doi: 10.1038/sj.bjc.6602936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J, Clayton D. One degree of freedom for dominance in indirect association studies. Genet Epidemiol. 2007;31:261–271. doi: 10.1002/gepi.20207. [DOI] [PubMed] [Google Scholar]
- Cheng I, Penney KL, Stram DO, Le Marchand L, Giorgi E, Haiman CA, Kolonel LN, Pike M, Hirschhorn J, Henderson BE, Freedman ML. Haplotype-based association studies of IGFBP1 and IGFBP3 with prostate and breast cancer risk: the multiethnic cohort. Cancer Epidemiol Biomarkers Prev. 2006;15:1993–1997. doi: 10.1158/1055-9965.EPI-06-0361. [DOI] [PubMed] [Google Scholar]
- Cleveland RJ, Gammon MD, Edmiston SN, Teitelbaum SL, Britton JA, Terry MB, Eng SM, Neugut AI, Santella RM, Conway K. IGF1 CA repeat polymorphisms, lifestyle factors and breast cancer risk in the Long Island Breast Cancer Study Project. Carcinogenesis. 2006;27:758–765. doi: 10.1093/carcin/bgi294. [DOI] [PubMed] [Google Scholar]
- Deming SL, Ren Z, Wen W, Shu XO, Cai Q, Gao YT, Zheng W. Genetic variation in IGF1, IGF-1R, IGFALS, and IGFBP3 in breast cancer survival among Chinese women: a report from the Shanghai Breast Cancer Study. Breast Cancer Res Treat. 2007;104:309–319. doi: 10.1007/s10549-006-9420-8. [DOI] [PubMed] [Google Scholar]
- English WP, Cleveland KE, Barber WH. There is no difference in survival between African-American and white women with breast cancer. Am Surg. 2002;68:594–597. [PubMed] [Google Scholar]
- Flint DJ, Beattie J, Allan GJ. Modulation of the actions of IGFs by IGFBP-5 in the mammary gland. Horm Metab Res. 2003;35:809–815. doi: 10.1055/s-2004-814164. [DOI] [PubMed] [Google Scholar]
- Frommer KW, Reichenmiller K, Schutt BS, Hoeflich A, Ranke MB, Dodt G, Elmlinger MW. IGF-independent effects of IG-FBP-2 on the human breast cancer cell line Hs578T. J Mol Endocrinol. 2006;37:13–23. doi: 10.1677/jme.1.01955. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Zuloeta Ladd AM, Liu F, Houben MP, Arias Vasquez A, Siemes C, Janssens AC, Coebergh JW, Hofman A, Janssen JA, Stricker BH, van Duijn CM. IGF-1 CA repeat variant and breast cancer risk in postmenopausal women. Eur J Cancer. 2007;43:1718–1722. doi: 10.1016/j.ejca.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Hankinson SE, Willett WC, Colditz GA, Hunter DJ, Michaud DS, Deroo B, Rosner B, Speizer FE, Pollak M. Circulating concentrations of insulin-like growth factor-I and risk of breast cancer. Lancet. 1998;351:1393–1396. doi: 10.1016/S0140-6736(97)10384-1. [DOI] [PubMed] [Google Scholar]
- Jernstrom H, Chu W, Vesprini D, Tao Y, Majeed N, Deal C, Pollak M, Narod SA. Genetic factors related to racial variation in plasma levels of insulin-like growth factor-1: implications for premenopausal breast cancer risk. Mol Genet Metab. 2001;72:144–154. doi: 10.1006/mgme.2000.3130. [DOI] [PubMed] [Google Scholar]
- Jerome L, Shiry L, Leyland-Jones B. Deregulation of the IGF axis in cancer: epidemiological evidence and potential therapeutic interventions. Endocr Relat Cancer. 2003;10:561–578. doi: 10.1677/erc.0.0100561. [DOI] [PubMed] [Google Scholar]
- John EM, Horn-Ross PL, Koo J. Lifetime physical activity and breast cancer risk in a multiethnic population: the San Francisco Bay area breast cancer study. Cancer Epidemiol Biomarkers Prev. 2003;12:1143–1152. [PubMed] [Google Scholar]
- John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, Ziogas A, Andrulis IL, Anton-Culver H, Boyd N, Buys SS, Daly MB, O’Malley FP, Santella RM, Southey MC, Venne VL, Venter DJ, West DW, Whittemore AS, Seminara D. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev. 1995;16:3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- Marbella AM, Layde PM. Racial trends in age-specific breast cancer mortality rates in US women. Am J Public Health. 2001;91:118–121. doi: 10.2105/ajph.91.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshman E, Green KA, Flint DJ, White A, Streuli CH, Westwood M. Insulin-like growth factor binding protein 5 and apoptosis in mammary epithelial cells. J Cell Sci. 2003;116:675–682. doi: 10.1242/jcs.00263. [DOI] [PubMed] [Google Scholar]
- Martin JL, Baxter RC. Expression of insulin-like growth factor binding protein-2 (IGFBP-2) by MCF-7 breast cancer cells is regulated through the PI3-KINASE/AKT/mTOR pathway. Endocrinology. 2007;148:2532–2541. doi: 10.1210/en.2006-1335. [DOI] [PubMed] [Google Scholar]
- Maxwell P, van den Berg HW. Changes in the secretion of insulin-like growth factor binding proteins-2 and -4 associated with the development of tamoxifen resistance and estrogen independence in human breast cancer cell lines. Cancer Lett. 1999;139:121–127. doi: 10.1016/s0304-3835(99)00009-9. [DOI] [PubMed] [Google Scholar]
- Missmer SA, Haiman CA, Hunter DJ, Willett WC, Colditz GA, Speizer FE, Pollak MN, Hankinson SE. A sequence repeat in the insulin-like growth factor-1 gene and risk of breast cancer. Int J Cancer. 2002;100:332–336. doi: 10.1002/ijc.10473. [DOI] [PubMed] [Google Scholar]
- Mohan S, Baylink D. IGF-binding proteins are multifunctional and act via IGF-dependent and -independent mechanisms. J Endocrinol. 2002;175:19–31. doi: 10.1677/joe.0.1750019. [DOI] [PubMed] [Google Scholar]
- Pereira JJ, Meyer T, Docherty SE, Reid HH, Marshall J, Thompson EW, Rossjohn J, Price JT. Bimolecular interaction of insulin-like growth factor (IGF) binding protein-2 with alphavbeta3 negatively modulates IGF-I-mediated migration and tumor growth. Cancer Res. 2004;64:977–984. doi: 10.1158/0008-5472.can-03-3056. [DOI] [PubMed] [Google Scholar]
- Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat Rev Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- Ren Z, Cai Q, Shu XO, Cai H, Li C, Yu H, Gao YT, Zheng W. Genetic polymorphisms in the IGFBP3 gene: association with breast cancer risk and blood IGFBP-3 protein levels among Chinese women. Cancer Epidemiol Biomarkers Prev. 2004;13:1290–1295. [PubMed] [Google Scholar]
- Ries L, Eisner M, Kosary C, Hankey B, Miller B, Clegg L, Mariotto A, Feuer E, Edwards B. SEER Cancer Statistics Review, 1975–2001. National Cancer Institute; Bethesda: 2004. [Google Scholar]
- Rosenzweig SA. What’s new in the IGF-binding proteins? Growth Horm IGF Res. 2004;14:329–336. doi: 10.1016/j.ghir.2004.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev D, Yee D. The IGF system and breast cancer. Endocr Relat Cancer. 2001;8:197–209. doi: 10.1677/erc.0.0080197. [DOI] [PubMed] [Google Scholar]
- Sasieni PD. From genotypes to genes: doubling the sample size. Biometrics. 1997;53:1253–1261. [PubMed] [Google Scholar]
- Schernhammer ES, Hankinson SE, Hunter DJ, Blouin MJ, Pollak MN. Polymorphic variation at the −202 locus in IGFBP3: Influence on serum levels of insulin-like growth factors, interaction with plasma retinol and vitamin D and breast cancer risk. Int J Cancer. 2003;107:60–64. doi: 10.1002/ijc.11358. [DOI] [PubMed] [Google Scholar]
- Schutt BS, Langkamp M, Rauschnabel U, Ranke MB, Elmlinger MW. Integrin-mediated action of insulin-like growth factor binding protein-2 in tumor cells. J Mol Endocrinol. 2004;32:859–868. doi: 10.1677/jme.0.0320859. [DOI] [PubMed] [Google Scholar]
- Setiawan VW, Cheng I, Stram DO, Penney KL, Le Marchand L, Altshuler D, Kolonel LN, Hirschhorn J, Henderson BE, Freedman ML. Igf-I genetic variation and breast cancer: the multiethnic cohort. Cancer Epidemiol Biomarkers Prev. 2006;15:172–174. doi: 10.1158/1055-9965.EPI-05-0625. [DOI] [PubMed] [Google Scholar]
- Swanson GM, Lin CS. Survival patterns among younger women with breast cancer: the effects of age, race, stage, and treatment. J Natl Cancer Inst Monogr. 1994:69–77. [PubMed] [Google Scholar]
- Toniolo P, Bruning PF, Akhmedkhanov A, Bonfrer JM, Koenig KL, Lukanova A, Shore RE, Zeleniuch-Jacquotte A. Serum insulin-like growth factor-I and breast cancer. Int J Cancer. 2000;88:828–832. doi: 10.1002/1097-0215(20001201)88:5<828::aid-ijc22>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Trock BJ. Breast cancer in African American women: epidemiology and tumor biology. Breast Cancer Res Treat. 1996;40:11–24. doi: 10.1007/BF01805999. [DOI] [PubMed] [Google Scholar]
- Wagner K, Hemminki K, Israelsson E, Grzybowska E, Soderberg M, Pamula J, Pekala W, Zientek H, Mielzynska D, Siwinska E, Forsti A. Polymorphisms in the IGF-1 and IGFBP 3 promoter and the risk of breast cancer. Breast Cancer Res Treat. 2005;92:133–140. doi: 10.1007/s10549-005-2417-x. [DOI] [PubMed] [Google Scholar]
- Wang W, John EM, Ingles SA. Androgen receptor and prostate-specific antigen gene polymorphisms and breast cancer in African-American women. Cancer Epidemiol Biomarkers Prev. 2005;14:2990–2994. doi: 10.1158/1055-9965.EPI-05-0310. [DOI] [PubMed] [Google Scholar]
- Wong WW, Copeland KC, Hergenroeder AC, Hill RB, Stuff JE, Ellis KJ. Serum concentrations of insulin, insulin-like growth factor-I and insulin-like growth factor binding proteins are different between white and African American girls. J Pediatr. 1999;135:296–300. doi: 10.1016/s0022-3476(99)70123-x. [DOI] [PubMed] [Google Scholar]
- Yanovski JA, Sovik KN, Nguyen TT, Sebring NG. Insulin-like growth factors and bone mineral density in African American and White girls. J Pediatr. 2000;137:826–32. doi: 10.1067/mpd.2000.109151. [DOI] [PubMed] [Google Scholar]
- Yu H, Jin F, Shu XO, Li BD, Dai Q, Cheng JR, Berkel HJ, Zheng W. Insulin-like growth factors and breast cancer risk in Chinese women. Cancer Epidemiol Biomarkers Prev. 2002;11:705–712. [PubMed] [Google Scholar]