Abstract
The genus Plasmodium is a diversified group of parasites with more than 200 known species that includes those causing malaria in humans. These parasites use numerous proteins in a complex process that allows them to invade the red blood cells of their vertebrate hosts. Many of those proteins are part of multi-gene families; one of which is the merozoite surface protein-3 (msp3) family. The msp3 multi-gene family is considered important in the two main human parasites, Plasmodium vivax and Plasmodium falciparum, as its paralogs are simultaneously expressed in the blood stage (merozoite) and are immunogenic. There are large differences among Plasmodium species in the number of paralogs in this family. Such differences have been previously explained, in part, as adaptations that allow the different Plasmodium species to invade their hosts. To investigate this, we characterized the array containing msp3 genes among several Plasmodium species, including P. falciparum and P. vivax. We first found no evidence indicating that the msp3 family of P. falciparum was homologous to that of P. vivax. Subsequently, by focusing on the diverse clade of nonhuman primate parasites to which P. vivax is closely related, where homology was evident, we found no evidence indicating that the interspecies variation in the number of paralogs was an adaptation related to changes in host range or host switches. Overall, we hypothesize that the evolution of the msp3 family in P. vivax is consistent with a model of multi-allelic diversifying selection where the paralogs may have functionally redundant roles in terms of increasing antigenic diversity. Thus, we suggest that the expressed MSP3 proteins could serve as “decoys”, via antigenic diversity, during the critical process of invading the host red blood cells.
Keywords: Antigen, Homology, Merozoite surface protein 3, Multi-gene family, Plasmodium, Plasmodium vivax, Plasmodium falciparum
1. Introduction
Malaria parasites, of the genus Plasmodium, invade the red blood cells (RBCs) of a diverse range of vertebrate species, including humans. Due to the importance that this process has in disease and the completion of the parasite life cycle, the proteins involved have been the focus of research efforts aimed at developing an anti-malarial vaccine (Conway 2007). Many of those proteins belong to multi-gene families.
Previous comparative genomic studies (Cai et al., 2010; Liew et al., 2010; Neafsey et al., 2012; Tachibana et al., 2012) have shown that multi-gene families, especially those expressed by the parasite during RBC invasion, can be highly variable among Plasmodium lineages. Some antigen families are restricted to certain Plasmodium lineages, such as the SICAvar and kir families of Plasmodium knowlesi (Pain et al., 2008), the var genes of Plasmodium falciparum (Zilversmit et al., 2013), and the vir family of Plasmodium vivax (Bernabeu et al., 2012). These lineage-specific families have been associated with virulence and antigenic variation; however, they are not necessarily functionally equivalent as much remains to be known about their roles in invasion (Wasmuth et al., 2009; Frech and Chen, 2013). Others antigen families appear to have orthologs across many Plasmodium species, but exhibit wide variation in their numbers of paralogs among lineages; these include the serine-repeat antigen (SERA) (Arisue et al., 2011), reticulocyte binding protein (RBP) (Gunalan et al., 2013), merozoite surface protein-7 (MSP7) (Garzón-Ospina et al., 2010), and merozoite surface protein-3 (MSP3) families (Carlton et al., 2008). This variation in the number of paralogs has increasingly received attention since it is thought to be associated with the parasite’s host range (Arisue et al., 2011; Tachibana et al., 2012), although such studies have often been made using few or distantly related species and without reference to variation within species. Finer scale comparative studies could thus provide new insight about the evolution of these blood-stage antigen families of Plasmodium parasites. Here we sought to apply such an approach to the msp3 gene family.
The first protein termed “merozoite surface protein-3” (MSP3) was identified in P. falciparum (McColl et al., 1994; Oeuvray et al., 1994). Five years later, a new merozoite surface protein was discovered in P. vivax and likewise named MSP3 due to putative similarities to the one identified previously in P. falciparum (Galinski et al., 1999). Soon after, possible paralogs were identified in P. vivax and P. knowlesi (Galinski et al., 2001) that shared several protein characteristics such as the large, alanine-rich central domain predicted to form coiled-coil tertiary structure motifs. The MSP3 proteins were therefore grouped into a multi-gene family with putative orthologs in several Plasmodium species (Jiang et al., 2013). Subsequent sequencing of the first P. vivax genome (strain Salvador I) revealed a total of 12 msp3 paralogs clustered on P. vivax chromosome 10 (Carlton et al., 2008). These showed weak similarity to four msp3 gene family members on P. falciparum chromosome 10 and to two P. knowlesi msp3 genes located on different chromosomes. The authors speculated that there had been a significant expansion of the msp3 gene family in P. vivax, perhaps as a means to enhance immune evasion, as P. falciparum and P. vivax msp3 gene family members had been shown to be antigenic (Carlton et al., 2008). However, no further analyses supporting this finding were carried out at the time.
Extensive polymorphism in two P. vivax paralogs, msp3 “alpha” (PVX_097720) and “beta” (PVX_097680), was known and used in molecular epidemiologic applications (Bruce et al., 1999; Rayner et al., 2004; Rice et al., 2013), but with the availability of additional P. vivax genomes, extensive polymorphism was documented throughout the family (Neafsey et al., 2012). Such initial observations of polymorphism, together with the fact that these proteins are immunogenic (Bitencourt et al., 2013; Lima-Junior et al., 2011, 2012; Mourão et al., 2012), are thought to indicate that the msp3 family has an important function in the RBC invasion process, possibly through a role in immune evasion (Jiang et al. 2013).
Recent and ongoing genomic sequencing efforts for P. vivax, and the diverse clade of closely related parasites found in Southeast Asian cercopithecine hosts, allow for evolutionary genetic investigations such as the one carried out here (Carlton et al., 2013; Escalante et al., 2005). Thought to have originated 6.5–10.3 million years ago (Mya) (Pacheco et al., 2011; Pacheco et al., 2012b), this monophyletic group of parasites exhibits different host ranges and diversity in their RBC infection cycle phenotypes, such as periodicity (Coatney et al., 1971; Escalante et al., 1998, 2005). Thus, this clade offers an extraordinary model to better understand the complex invasion biology of malarial parasites and to better explore the role of the gene families involved.
In this investigation, we sought to evaluate two hypotheses: (1) that the msp3 gene family is orthologous among Plasmodium lineages, and (2) that expansion of the msp3 family is related to the species’ adaptation to the human host. Then, we proceeded to explore other factors that may have contributed to the evolution of this family by studying patterns of intraspecific and intergenic variation for these loci.
2. Materials and Methods
2.1. Synteny and homology
P. vivax msp3 family gene members lie clustered in an approximately 60 kilobase (kb) array on chromosome 10. Their encoded proteins were previously described to share several features: (1) a distinctively high alanine content; (2) a large central domain predicted to form coiled-coil tertiary motifs; and (3) a small peptide near the N-terminal that is considered characteristic of this gene family and has been termed a ‘signature’ peptide (Galinski et al., 2001; Jiang et al., 2013). We use a combination of criteria to identify putative orthologs and paralogs to these proteins that includes sequence similarity, synteny (relative genomic position), and the three protein characteristics mentioned above.
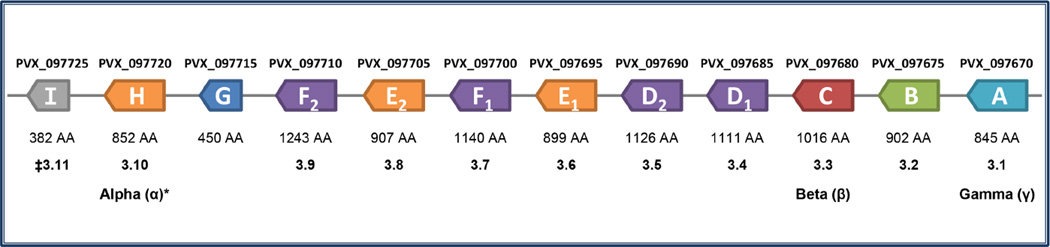
Following PlasmoDB version 8.2 (Aurrecoechea et al., 2009), the 12 genes in the P. vivax Salvador-1 strain reference genome annotated as putative msp3 paralogs were retrieved. In addition, the six open reading frames (ORFs) located in a cluster with P. falciparum merozoite surface protein 3 (PF3D7_1035400, also known as “SPAM”) were also retrieved; Singh et al. (2009) grouped these into a putative P. falciparum msp3 gene family previously. For the ease of reference of specific genes and their putative orthologs in species without an annotated genome, we will denote the 12 P. vivax Salvador-1 msp3 genes by giving them a letter from A to I following a 5’ to 3’ order in the array (Fig. 1). For example, msp3A in Plasmodium inui is an ortholog to msp3A of P. vivax. Paralogs found in one or few genomes that are likely the result of recent duplication events are indicated by adding numbers, for example, msp3E1 and E2 are paralogous copies of msp3E. This labeling is used since the names and genes included in the P. vivax msp3 family have changed previously (Galinski et al. 2001; Carlton et al. 2008; Jiang et al. 2013). All the different names used for the msp3 genes, along with their corresponding PlasmoDB gene IDs are also shown in Figure 1. Names for P. falciparum MSP3 proteins follow Singh et al. (2009).
Figure 1.
The 12 putative members of the P. vivax msp3 family. The twelve putative P. vivax msp3 genes in the PlasmoDB annotation of the Salvador I reference genome are shown with their gene IDs and the naming used here. Amino acid (AA) lengths and other aliases for the genes are shown below the genes. *See Galinski et al. (2001) for a description of α, β, and γ, the first 3 genes in the family identified, and Jiang et al. (2013) for their use of 3.1 through 3.11‡. Colors correspond to phylogenetic relationships, see Figure 5.
To find putative msp3 genes in additional species with complete genomes, conserved genes flanking the msp3 arrays were used as genomic landmarks. Both the P. vivax and P. falciparum msp3 arrays were flanked by genes with orthologs clearly identifiable throughout all Plasmodium species, allowing identification of the syntenic chromosomal regions in other Plasmodium species by BLAST (Altschul et al., 1997) and FASTA (Pearson and Lipman 1988) searches. For more details about these conserved flanking genes see Supplementary Material 1. All annotated genes in the region between the flanking marker genes were retrieved from the reference genomes of P. knowlesi, P. berghei, and P. chabaudi.
Specifically, we characterized the syntenic cluster in three closely related Asian macaque parasites, Plasmodium cynomolgi, P. inui and P. knowlesi, which together with P. vivax are within what we will refer to as the Asian primate Plasmodium clade (Escalante et al., 2005). Plasmodium vivax, P. cynomolgi, P. knowlesi have annotated genomes available, while preliminary genomic data from P. inui (Carlton et al., 2013) allowed us to perform targeted Sanger sequencing of its msp3 array (see below). We also extracted putative msp3 orthologs from locally assembled 454 (Roche, Applied Science, Basel, Switzerland) reads for Plasmodium gonderi, a parasite of Cercocebus and Mandrillus hosts in Africa (Coatney et al., 1971). Studying P. gonderi allowed us to infer ancestral states (Carlton et al., 2013) as this parasite is basal to the Asian Plasmodium clade (Escalante et al., 1998, 2005). Finally, from PlasmoDB, the syntenic arrays in two rodent malaria parasites, Plasmodium berghei (ANKA) and Plasmodium chabaudi chabaudi (AS), and the human parasite P. falciparum (3D7) were characterized for comparison.
The sets of flanking genes and msp3 paralogs from reference genomes were then used to search recently completed genomes of four additional P. vivax strains (Neafsey et al., 2012) and two P. cynomolgi strains (Tachibana et al., 2012). Putative ORFs homologous to the flanking genes or genes (paralogs and orthologs) within the msp3 arrays were identified by reciprocal BLAST and FASTA searches and the NCBI ORF Finder tool (available at ncbi.nlm.nih.gov/gorf/).
2.2. Sampling, amplification, cloning, and Sanger sequencing
To determine polymorphism and potential copy number variation within a species, sequences for each msp3 gene were also obtained from three to nine P. vivax isolates (in addition to the five strains with genomic data: India VII, Brazil I, Mauritania I, and North Korea), and from five to nine P. cynomolgi isolates (in addition to the two strains with whole genome data). Isolates for these two species were well characterized laboratory isolates from geographically diverse locations (Supplementary Material 1). Complete NCBI sequences from additional isolates were available for two of the P. vivax genes (msp3H “alpha” and msp3C “beta”) and were included in the analysis. In addition, between 10 and 17 clinical isolates from Thailand and Venezuela were also sampled to augment the sample size for three of the P. vivax genes (msp3H “alpha”, msp3E1, and E2). The number of P. vivax and P. cynomolgi samples for each gene are shown in Table 1 and Table 2. Supplementary Figure 1 shows the isolates sequenced for each gene in the msp3 arrays.
Table 1.
Polymorphism found in the msp3 gene family of Plasmodium vivax
| Gene (Population) |
n | Allele Range |
Aligned Length |
Alignable Sites |
Indel Haplotypes |
Aver age Indel Leng th |
d (SE) | dS | dN | dS-dN (SE) |
p (Z-stat) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| msp3A “γ” | 9 | 2538- 2955 |
3030 | 2427 | 9 | 35.5 | 0.136 (0.006) |
0.1 23 |
0.1 39 |
−0.016 (0.011) |
0.144 (1.469) |
| msp3B | 8 | 1770- 2748 |
2796 | 1701 | 8 | 50.1 | 0.150 (0.006) |
0.1 43 |
0.1 52 |
−0.008 (0.015) |
0.579 (0.556) |
| msp3C “β” | 1 1 |
2079- 3051 |
3084 | 2055 | 11 | 140. 5 |
0.067 (0.004) |
0.0 70 |
0.0 67 |
+0.003 (0.009) |
0.729 (− 0.348) |
| msp3D1 | 6 | 3336- 3397 |
3429 | 3276 | 6 | 17.8 | 0.096 (0.004) |
0.0 99 |
0.0 96 |
+0.003 (0.010) |
0.742 (− 0.330) |
| msp3D2 | 7 | 3369- 3462 |
3522 | 3297 | 7 | 15.8 | 0.098 (0.003) |
0.1 04 |
0.0 97 |
+0.007 (0.009) |
0.478 (− 0.713) |
| msp3E1-E2‡ | 4 0 |
2613- 2754 |
2808 | 2541 | ‡ | 14.8 | 0.087 (0.003) |
0.1 00 |
0.0 83 |
+0.017 (0.008) |
‡ |
|
msp3E (Thailand) |
1 5 |
2646- 2724 |
2808 | 2565 | ‡ | 13.0 | 0.088 (0.004) |
0.1 04 |
0.0 84 |
+0.020 (0.008) |
‡ |
|
msp3E (Venezuela) |
1 5 |
2643- 2754 |
2808 | 2583 | ‡ | 17.7 | 0.081 (0.004) |
0.0 92 |
0.0 79 |
+0.013 (0.009) |
‡ |
| msp3F1 | 1 1 |
3402- 4347 |
4614 | 3261 | 11 | 108. 6 |
0.108 (0.004) |
0.1 14 |
0.1 07 |
+0.007 (0.009) |
0.439 (− 0.777) |
| msp3F2 | 7 | 3387- 3885 |
3966 | 3303 | 7 | 44.3 | 0.145 (0.005) |
0.1 80 |
0.1 36 |
+0.044 (0.012) |
0.004 (− 3.677) |
| msp3G | 1 4 |
1281- 1389 |
1389 | 1239 | 12 | 57.8 | 0.015 (0.002) |
0.0 28 |
0.0 11 |
+0.017 (0.006) |
0.004 (− 2.908) |
| msp3H “α” | 4 8 |
1779- 2601 |
2646 | 1413 | 28 | 134. 9 |
0.027 (0.003) |
0.0 44 |
0.0 23 |
+0.021 (0.007) |
0.004 (− 2.936) |
|
msp3H (Thailand) |
1 7 |
1779- 2580 |
2646 | 1686 | 13 | 150. 9 |
0.033 (0.002) |
0.0 52 |
0.0 28 |
+0.024 (0.007) |
0.001 (− 3.403) |
|
msp3H (Venezuela) |
1 0 |
2232- 2295 |
2646 | 2214 | 4 | 21.8 | 0.031 (0.002) |
0.0 39 |
0.0 29 |
+0.010 (0.005) |
0.064 (− 1.870) |
| msp3I | 1 4 |
1149- 1161 |
1161 | 1068 | 6 | 21.2 | 0.008 (0.002) |
0.0 09 |
0.0 08 |
+0.002 (0.005) |
0.742 (− 0.331) |
Average of the pairwise comparisons within putative orthologs using the annotation of the Salvador I strain as a reference. Standard error (SE) estimates from 1000 bootstrap replicates and p-values from the codon based Z test are shown. Significant values (p<0.05) are bolded. Allele lengths and the number of alignable sites among putative orthologs are in base pairs. Indel haplotypes were counted by identifying sequences with unique associations of indel events.
Alleles for these two paralogs were not distinguishable and not recoverable from all isolates; all alleles were aligned and analyzed together. Therefore, this comparison likely includes multiple paralogs, making the haplotype frequency and Z test statistics inappropriate.
Table 2.
Polymorphism found in the msp3 gene family of Plasmodium cynomolgi
| Gene | n | Allele Range |
Aligned Length |
Alignable Sites |
Indel Haplotypes |
Avera ge Indel Lengt h |
d (SE) | dS | dN | dS-dN (SE) |
p (Z-stat) |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
msp3A “γ” |
9 | 1764- 1863 |
1899 | 1689 | 5 | 23.5 | 0.067 (0.004) |
0.0 63 |
0.0 68 |
−0.005 (0.010) |
0.619 (0.499) |
| msp3B1 -4‡ |
7 | 2671- 2952 |
3171 | 2334 | ‡ | 30.2 | 0.118 (0.004) |
0.1 45 |
0.1 11 |
+0.035 (0.011) |
‡ |
|
msp3C “β” |
9 | 2406- 2559 |
2571 | 2397 | 5 | 36.2 | 0.037 (0.002) |
0.0 46 |
0.0 34 |
+0.011 (0.006) |
0.065 (− 1.861) |
| msp3D | 5 | 2751- 3405 |
3447 | 2685 | 4 | 114.0 | 0.066 (0.004) |
0.0 87 |
0.0 61 |
+0.026 (0.010) |
0.012 (− 2.564) |
| msp3E1 | 9 | 2622- 2688 |
2727 | 2589 | 6 | 18.7 | 0.043 (0.003) |
0.0 67 |
0.0 36 |
+0.030 (0.008) |
0.0003 (− 3.721) |
| msp3F1 | 8 | 3048- 3135 |
3192 | 2979 | 6 | 23.6 | 0.097 (0.003) |
0.1 27 |
0.0 90 |
+0.037 (0.010) |
0.0004 (− 3.630) |
| msp3G | 8 | 1167- 1203 |
1221 | 1155 | 5 | 18.0 | 0.077 (0.006) |
0.0 76 |
0.0 78 |
−0.002 (0.013) |
0.878 (0.154) |
|
msp3H “α” |
9 | 1959- 1968 |
2028 | 1902 | 4 | 19.3 | 0.064 (0.004) |
0.0 83 |
0.0 58 |
+0.024 (0.010) |
0.020 (− 2.367) |
| msp3I | 9 | 576-708 | 729 | 573 | 6 | 36.0 | 0.081 (0.009) |
0.1 39 |
0.0 69 |
+0.070 (0.027) |
0.014 (− 2.496) |
Average of the pairwise comparisons within putative orthologs using the annotation of the strain B as a reference. Standard error (SE) estimates from 1000 bootstrap replicates and p-values from the codon based Z test are shown. Significant values (p<0.05) are bolded. Allele lengths and the number of informative sites are in base pairs. Indel haplotypes were counted by identifying sequences with unique associations of indel events.
The number of alleles recovered from isolates (putative paralogous copies) varied from 1 to 4 for this gene; all alleles and paralogs of msp3B were aligned and analyzed together. Therefore this comparison includes multiple paralogs, making the haplotype frequency and Z test statistics inappropriate.
In some instances, we cloned and sequenced genes from the four P. vivax strains with genomic sequencing data available; thus, we were able to confirm by Sanger sequencing some of the polymorphism observed from genomic data. We re-sequenced all putative msp3 ORFs from the genomic data for one P. cynomolgi strain (Berok), using overlapping clones of up to 6,000 base pairs (bp) to cover regions with breaks in genomic sequencing coverage (see Supplementary Figure 2). For P. inui strain OS, the entire 14,000 bp MSP3 array was also confirmed by amplifying it in four overlapping fragments that were cloned and sequenced (Supplementary Fig. 2). Using the preliminary P. inui strain OS genomic data, we were able to sequence the ORFs matching msp3 genes for the Leaf Monkey II and Leucophyrus strains (both from Peninsular Malaysia) as well. A total of eight isolates (OS, Leucophyrus, Perak, Celebes I, Celebes II, N-34, Taiwan I and Hawking) were also amplified, cloned and sequenced for the P. inui chimeric pseudogene (see results). Multiple clones (from two to nine) of each sample of a gene were sequenced for nearly all samples for all species. Reproducible clones from the same isolate that differed by more than 1% (e.g. 25 or more substitutions for a 2,500 bp gene) were considered outside the range of PCR and sequencing error and included in the analysis as possible paralogs or distinct alleles from mixed infections. In the case of P. gonderi, the putative msp3 ORFs identified from locally assembled 454 data were not confirmed by re-sequencing.
Amplification of complete or nearly complete coding sequences was performed by the polymerase chain reaction (PCR) using TaKaRa LA Taq (TaKaRa Mirus Bio Inc, Shiga, Japan) or AmpliTaq Gold (Applied Biosytems, Roche, USA) polymerase, depending on gene length. For more details about the PCR protocols see Supplementary Material 1 and Supplementary Table 1. PCR products were purified using the QIAquick DNA gel extraction kit (QIAgen, Hilden, Germany) and cloned using the pGEM-T Easy Vector System (Promega, WI, USA). Clones were sequenced using an Applied Biosystems 3730 capillary sequencer. Sequences reported in this study were deposited in GenBank under the accession numbers: KC907427 to KC907623. The accession codes and all the sequences analyzed here are listed in Supplementary Table 2.
2.3. Protein domain analysis
To determine if the distinctively high alanine content of the msp3 family was maintained among putative orthologs, amino acid composition was recorded for each msp3 gene in P. vivax, P. cynomolgi, P. knowlesi, P. inui, and P. gonderi. Tertiary protein structure motifs were predicted using the Paircoil2 (McDonnell et al., 2006) program and the locations of indels and unalignable sites were plotted using DnaSP v5.10.01 (Librado and Rozas 2009) and an Excel macro (available upon request). A multispecies alignment of 100 amino acids in the N-terminal among putative msp3 genes was used to identify the MSP3 ‘signature’ NLRNG.
2.4. Evolutionary genetic analyses
Msp3 interspecies family and individual gene alignments for each species were constructed using the MUSCLE algorithm (Edgar 2004) as implemented in SeaView4 (Gouy 2010), with manual editing. MSP3 proteins have a large central domain predicted to form coiled-coil tertiary motifs and many msp3 genes have multiple indels within that domain with high divergence among paralogs. Thus, three different sets of alignments were constructed using the alignable sites from the N and C terminals: (1) an alignment that included all the paralogs from Asian non-human primate malarias, as well as, P. vivax and P. gonderi; (2) a set of P. vivax and P. cynomolgi species-specific alignments for each paralog; and (3) interspecies alignment of both P. vivax and P. cynomolgi for different paralogs. The number of sites used in each analysis is reported in the results section. Phylogenetic analyses were performed using Maximum Likelihood (ML) methods as implemented in PhyML v3.0 (Guindon and Gascuel 2003) and Bayesian inference methods using MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003). Both methods used a general time reversible+gamma model (GTR+-Γ) because it best fit the data as estimated by MEGA v5. (Tamura et al. 2011). Bayesian support for nodes was inferred in MrBayes using between 30 and 55 × 106 Markov Chain Monte Carlo (MCMC) steps, depending on the alignment, with sampling every 100 generations. We discarded 50% of the samples as burn-in (Ronquist and Huelsenbeck 2003).
Genetic diversity among isolates for each P. vivax and P. cynomolgi paralog was estimated using the overall mean distance, d (the average number of pairwise nucleotide differences per site) in MEGA5. Since the loci are highly divergent within this multi-gene family, genetic diversity was estimated solely on the alignable sites for each locus. The Jukes-Cantor model of nucleotide substitution was used to correct estimates of pairwise distance. To investigate possible signatures of selection, the diversity at synonymous (dS) and nonsynonymous (dN) sites was also calculated, using the Nei and Gojobori (1986) method with the Jukes-Cantor correction. The standard errors of dS and dN estimates were calculated with 1000 bootstrap replicates and the significance of the difference dS-dN was tested using a two-tailed Z test (Nei and Kumar 2000), an evolutionary genetic test used repeatedly for malaria antigens (e.g.: Escalante et al., 2004; Pacheco et al., 2012a; Weedall and Conway 2010). The null hypothesis was that synonymous and nonsynonymous positions would have similar substitution rates under neutrality (dS = dN); a significant deviation from our null hypothesis was thus interpreted as possible evidence of natural selection. Divergence between paralogs and orthologs was estimated using net between group mean divergences (Tamura et al., 2011).
The RDP3 program (Martin et al., 2010) was used to screen for recombination and gene conversion events, using the RDP (Martin and Rybicki 2000) algorithm. Events where the recombination donor and recipient sequences could be identified were recorded, using default parameters and a significance cut-off of 0.05.
3. Results and Discussion
3.1. No evidence that Pfmsp3 and Pvmsp3 are homologous
Comparisons, along with references to their suggested relatedness, have been made repeatedly between the msp3 genes of P. vivax and those of P. falciparum (Bitencourt et al., 2013; Jiang et al., 2013; Mourão et al., 2012). However, explicit investigation of their homology has not been performed. Accordingly, we used a combination of synteny, sequence similarity (see Supplementary Table 3), and protein characteristics to address this question of homology. The use of multiple criteria provides a standard for ortholog discovery (Maguire et al., 2013).
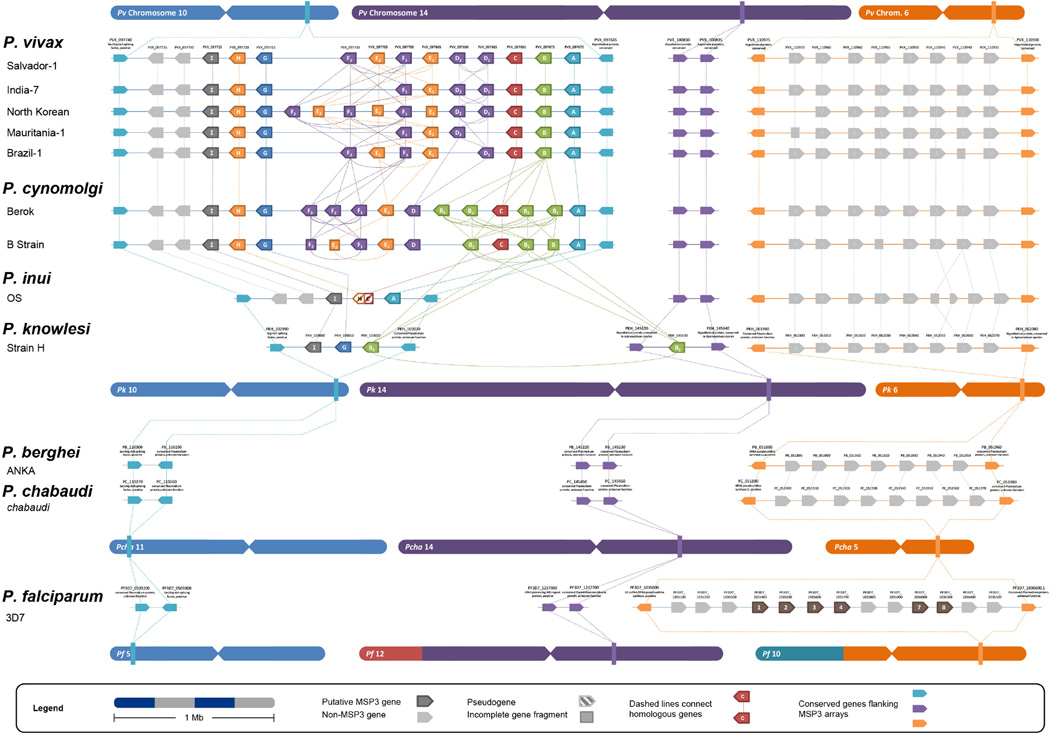
Figure 2 shows the characterized arrays with their chromosomal positions and the homologous chromosomal regions coded by color. Interestingly, the region in the genomes of P. berghei, P. chabaudi, and P. falciparum that was syntenic to the P. vivax msp3 (Pvmsp3) array (shown in blue) did not contain any genes (Fig. 2). Furthermore, the msp3 family in P. falciparum (PfMSP3) is not syntenic to that of P. vivax either, instead lying in an array syntenic to a region on chromosome 6 (shown in orange) in P. vivax and the Asian primate Plasmodium clade. This contrasts with other erythrocyte stage antigens such as MSP1, MSP4/5, MSP8, MSP9, MSP10, and AMA1 that are present and readily identified using synteny across the species studied (Carlton et al., 2008). Other tandemly clustered antigen families not found in the hyper-variable sub-telomeric regions, such as SERA, are arranged in synteny across Plasmodium as well. Indeed, upon completion of the first P. vivax genome it was found that 99% of the 3,000 plus orthologs identified, including 29 out of the 30 GPI-anchored surface proteins, were positioned syntenically among P. vivax, P. knowlesi, the rodent malaria parasites, and P. falciparum (Carlton et al., 2008). This provides an initial indication that the respective msp3 families of P. vivax and P. falciparum are not homologous.
Figure 2.
The PvMSP3 and PfMSP3 families are not syntenic among Plasmodium species. Chromosomes and position of the gene arrays are drawn approximately to scale; centromeres are arbitrarily placed in the middle of chromosomes. Homologous synteny blocks are colored the same on the chromosomes, such that for example, the region syntenic to P. vivax chromosome 14 (purple) in the P. falciparum genome (in this case, on P. falciparum chromosome 12) is colored similarly (see Frech and Chen (2011) for more about synteny blocks). P. falciparum surface proteins clustering with PfMSP3 (PfMSP3.1) in the PfMSP3 array (orange) include MSP6 (PfMSP3.2), MSP-H101 (PfMSP3.3), Duffy Binding-Like MSP (PfMSP3.4), MSP11 (PfMSP3.7), and MSP/PF10_0355 (PfMSP3.8). Additional P. falciparum antigens not originally considered part of the Pf msp3 family are also present within the array, nluding S-antigen (PF3D7_1036400), liver stage antigen 1 (PF3D7_1036400), and GLURP (PF3D7_1036400).
The clustering of these P. falciparum antigens in an array with no homologous counterpart in the P. vivax genome is worth noting. As discussed before (Frech and Chen 2011), the P. vivax array on chromosome 6 (shown in orange in Figure 2) syntenic to the P. falciparum msp3 array does not contain apparent orthologs to Pfmsp3. Evidence of homology between protein coding sequences in Pvmsp3 and Pfmsp3 arrays was also missing as reciprocal BLAST searches failed to identify significant similarity (data not shown).
After being unable to find Pvmsp3 homologs outside the Asian primate Plasmodium clade using synteny and sequence similarity, we examined the domain architecture of the PvMPS3 family and made comparisons to that of PfMSP3. Domain architecture for the P. falciparum msp3 family has been explored previously (Gondeau et al., 2009; Imam et al., 2011). Although one P. falciparum MSP3 protein (PfMSP3.1) shares a region of heptad repeats resulting in predicted coiled-coil tertiary motifs with P. vivax MSP3, the other PfMSP3 proteins lacked the coiled-coil motifs (Jiang et al., 2013). Additionally, all the PfMSP3 proteins have a leucine zipper motif at the extreme C-terminal end of the protein (Jiang et al., 2013), two of the genes (Pfmsp3.3 and Pfmsp3.4) contain a central Duffy-binding like domain, and others have proline rich regions (Singh et al., 2009).
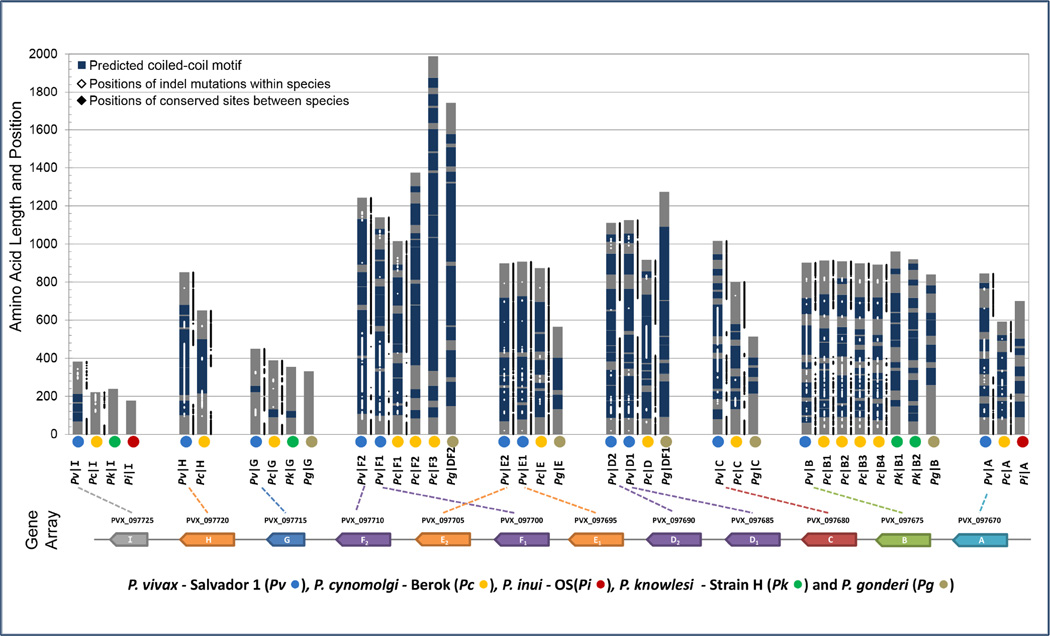
Characterizing the MSP3 proteins of P. vivax using amino acid composition, coiled-coil motif prediction and the so-called ‘signature’ peptide revealed that the msp3 family in P. vivax again markedly differed from that of P. falciparum. The first three members of the MSP3 family known in P. vivax were initially characterized by the presence of a large, hallmark, alanine rich (>20%) central domain predicted to form coiled-coil tertiary protein structure motifs (Galinski et al., 1999, 2001). We find here that, with two exceptions (MSP3G and MSP3I), this high alanine content (Supplementary Figure 3) and coiled-coil (Fig. 3) domain was maintained across proteins in the Pvmsp3 family and across their orthologs in the species of the Asian primate Plasmodium clade. Large coiled-coil motifs were also predicted for the P. gonderi ORFs identified from draft genomic sequencing data (Fig. 3). The first predicted coiled-coil motif began most often within the first 100 amino acids of a PvMSP3 and, though with small breaks in between, the motifs continued to until within 100 to 150 amino acids of the C-terminus (of the approximately 600–1300 amino acid proteins). This large coiled-coil motif-containing domain located in the center of the protein predicted for the P. vivax msp3 genes and their putative orthologs in P. cynomolgi, P. inui, P. knowlesi and P. gonderi contrasts that regions of coiled-coils were small or absent for P. falciparum. Any Duffy-binding like domains or leucine zipper motifs were also absent in PvMSP3 (Jiang et al., 2013).
Figure 3.
Domain architecture of the Pvmsp3 family and its orthologs in related species. Vertical gray bars represent putative MSP3 proteins, with their amino acid lengths drawn to scale. The positions of coiled-coil tertiary motifs as predicted by PairCoil2 are shown in blue. Positions of sites within indels from intraspecies alignments of P. vivax and P. cynomolgi isolates are plotted onto the proteins in white along with the positions of sites conserved and alignable between P. vivax and P. cynomolgi (in black). The P. vivax Salvador-1 gene array is shown for reference along with the abbreviations used for the nonhuman primate malaria species analyzed.
For P. vivax and the other members of the Asian primate Plasmodium clade, the size of the individual predicted coiled-coil motifs and the positions of breaks were not always consistent between isolates and orthologs from other species (Fig. 3), though it is unclear if this is due to the high levels of amino acid polymorphism observed (discussed later) or to noise introduced by the automated prediction. The gene encoding MSP3G was the first exception to this pattern in PvMSP3, as although it had an alanine rich central domain in P. vivax, alanine richness was due to the presence of simple repetition of 8–17 units of AN(N/K)A that was unique to this gene, and did not result in the prediction of coiled-coil motifs (Fig. 3). The second exception, MSP3I, was also alanine rich in P. vivax, but the entire central domain was missing in its putative P. cynomolgi, P. inui, and P. knowlesi msp3I orthologs. Despite these two exceptions in P. vivax, which may indicate that the two putative msp3 genes msp3G and msp3I are not functionally related to the rest of the family, it is clear that the protein domain organization of PvMSP3 is not similar to that of PfMSP3.
Although a majority of PvMSP3 proteins shared a similar predicted structure and a highly biased amino acid composition in the central domain (alanine, lysine, and glutamic acid together constituted approximately 60% of residues, Supplementary Fig. 3), low levels of sequence similarity (less than 30% protein identity) were observed between the central regions of those proteins containing the domain. Sequence complexity was clearly reduced due to the compositional bias in the region, but simple repeats and homopolymers were absent. P. falciparum MSP3 proteins, on the other hand, lacked the alanine rich central domain and did not share this bias towards alanine, lysine, and glutamic acid.
Given this divergence among Pvmsp3 genes, the maintenance of a specific, short protein motif near the N-terminus (NLRNG) that has been termed an MSP3 ‘signature’ motif has received some attention (Jiang et al., 2013). Despite the general conservation among paralogs at this short peptide, we observed population polymorphism and fixed divergence in some putative msp3 genes. Sequences of MSP3G (PVX_097715, which was excluded from the family by Jiang et al. (2013) for this reason) had a motif of N(I/M)RN(E/D) instead. P. gonderi MSP3G diverged even further in having the sequence GARNN. MSP3I had the NLRNG consensus sequence in P. vivax, but a sequence of NLR(I/N)E fixed in P. cynomolgi. As discussed above, MSP3G and MSP3I also lacked the central domain containing coiled-coil motifs; this divergence from the conserved peptide sequence seems to yield further support to questioning their relatedness to the rest of the family. However, several PvMSP3 proteins that shared the other characteristic MSP3 protein features diverged from this NLRNG sequence as well; MSP3H, the MSP3 “alpha” of Galinski et al. (2001), had a histidine residue fixed at the first position of this NRLNG peptide for all P. vivax and P. cynomolgi sequences, while MSP3B, C, and E had at least one isolate with a mutation in this region.
Interestingly, the P. falciparum msp3 family, despite not being syntenic with Pvmsp3, differing in domain architecture, and lacking sequence similarity elsewhere in the gene, shares a similar peptide near the N-terminal (Singh et al., 2009). Of further interest, the NLR(N/K)(A/G) peptide shared by the six ORFs of P. falciparum is situated within a highly active binding peptide (Rodriguez et al., 2008), thought to interact with a glycoprotein-like receptor on the erythrocyte surface. Regardless of this intriguing similarity, it is unlikely that these five amino acids are sufficient evidence for a common evolutionary origin for all MSP3 proteins. Indeed, it is found in more than 40 other proteins in P. falciparum (data not shown). The presence of this peptide in both the P. vivax - P. gonderi clade and P. falciparum msp3 arrays could be explained by parallel evolution and warrants further investigation of the binding activity of this region in Pvmsp3. Therefore, considering that both MSP3 families (in the P. falciparum and P. vivax clades) are expressed and immunogenic, as well as taking into account that there is no evidence for homology, we hypothesize that these families represent a case of convergent evolution in the proteins involved in the RBC invasion process.
3.2. No evidence that the number of paralogs in P. vivax is an adaptation to the human host or correlates with host range
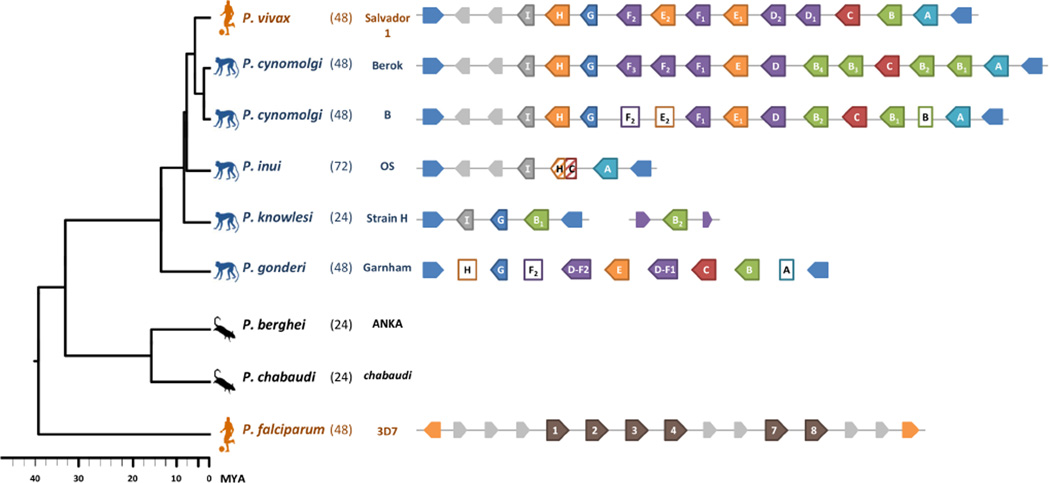
We then focused on the Asian primate Plasmodium clade, which includes P. vivax, where the homology of the msp3 family was evident (see Supplementary Table 3). First, we tested the hypothesis that the increased number of msp3 genes in P. vivax represented a lineage specific expansion. To do so, we identified putative msp3 homologs in an increased set of Plasmodium species. Figure 4 shows the number of genes recovered, hosts, and a well-known RBC cycle phenotype, eruption periodicity, for the eight species studied. If an increased number of msp3 genes in P. vivax reflected an expansion specific to that parasite lineage, we would expect the closely related species that infect Southeast Asian macaques (Escalante et al., 1998, 2005) to have a reduced msp3 family. However, a similar number of genes was found in both P. vivax (12 genes) (Carlton et al., 2008) and P. cynomolgi (12–14 genes) (Tachibana et al., 2013). By analysis of genomic assemblies and targeted re-sequencing of the msp3 array where assembly had failed, we were able to identify and confirm ORFs homologous to all 12 P. vivax genes in P. cynomolgi. In addition to the two P. cynomolgi strains with whole genome sequence data (Berok and B strains), we amplified and sequenced ORFs showing sequence similarity to each of the P. vivax msp3 genes in an additional three to seven P. cynomolgi isolates (Fig. S2 and Supplementary Table 3). This provides strong evidence that the high number of msp3 paralogs observed in P. vivax is also present among multiple P. cynomolgi strains. Thus, the so-called expansion of the msp3 family at least predates the P. vivax-P. cynomolgi split (estimated at 2.36–5.27 Mya, Pacheco et al., 2012b) and predates the P. vivax lineage’s adaptation to the human host.
Figure 4.
The msp3 gene family size in Plasmodium species. Msp3 gene family size recovered for the species studied along with their host (human, cercopithecine, or rodent) and merozoite eruption periodicities. 24, 48, and 72 hour cycles are referred to as quotidian, tertian, and quartan, respectively. The species phylogeny and time axis in millions of years ago (Mya) are from Pacheco et al. (2012b). The P. gonderi array was not fully assembled or re-sequenced and therefore gene order is not known; rectangles not filled in with color represent partial gene fragments. Putative msp3 genes are outlined in color and larger than non-msp3 genes (gray) and the conserved flanking genes used as markers.
Whereas P. vivax and P. cynomolgi had a comparable number of paralogs overall, their similar numbers were reached by differential expansions in the number of highly similar (65–82% protein identity) paralogs within four out of seven msp3 paralog lineages. Indeed, msp3 copy number variants are segregating in both P. vivax and P. cynomolgi populations (Fig. 2). Specifically, we were unable to recover 12 complete ORFs corresponding to the 12 P. vivax Salvador I paralogs in all four of the recently sequenced P. vivax strains (Neafsey et al., 2005), even after targeted Sanger re-sequencing (Fig. 2). Likewise, we identified multiple ORFs (msp3B3, B4, and F3) in the Berok strain of P. cynomolgi, but failed to find them in the B strain. PCR amplification, cloning and sequencing of the regions containing these three Berok ORFs produced sequences identical to that seen in the draft assembly and two of these three additional ORFs, msp3B4 and F3, were also successfully amplified, cloned and sequenced in the Gombak strain. This yields confidence that these additional P. cynomolgi paralogs were not assembly or PCR artifacts in just the Berok strain. As a consequence, a simple count of the number of msp3 coding genes in P. cynomolgi and P. vivax was not sufficient to compare these two species. With evidence of differential retention and expansion of msp3 lineages among and between species, inferring adaptation based simply on differences in the number of copies between species seems an oversimplification; a finding likely to apply in general for the comparative study of gene families in Plasmodium and other organisms.
Next, we characterized this family in two species that have similar host ranges to P. cynomolgi: P. inui and P. knowlesi. Both are largely sympatric with P. cynomolgi and all three species have been isolated multiple times from the same host species (Macaca fascicularis, M. nemestrina, M. mulatta, and M. hecki) (Coatney et al., 1971). Figure 4 shows the number of msp3 genes that we were able to identify in these parasites. Amplifying, cloning and re-sequencing of the P. inui array and re-analysis of the P. knowlesi Strain H reference genome revealed that these two species had substantially reduced MSP3 families, with three and four genes, respectively. As seen in Figure 3, the P. inui msp3A and P. knowlesi msp3B putative orthologs exhibited the large, central stretch of coiled-coil motifs characteristic of the msp3 family in P. vivax.
For P. knowlesi, one gene, PKH_145630 “merozoite surface protein”, on chromosome 14 and not within the syntenic msp3 array showed strong sequence similarity (e-value = 0, Supplementary Table 3) to a putative P. knowlesi msp3 gene (MSP3B1, PKH_103020) within the syntenic msp3 array and was included for later analysis. In addition to these two genes showing strong sequence similarity to P. vivax and P. cynomolgi msp3B, we identified putative orthologs to msp3G and msp3I in P. knowlesi as well.
In the case of P. inui, we found an msp3 pseudogene, a 1,064 bp region in the center of the P. inui OS strain msp3 array that appeared to be a “chimera”, with its N and C terminal halves showing strong sequence similarity (e-value < e-140) to the respective N and C termini of two different P. vivax and P. cynomolgi msp3 genes (msp3C and H respectively, Fig. 2). Targeted amplification and Sanger sequencing of the locus and 300–500 base pairs of the upstream and downstream noncoding sequence from eight P. inui isolates confirmed the msp3 pseudogene found in the local assembly. Additionally, among all these eight isolates sampled from a broad geographic distribution, stop codons were fixed at position 90 and 211, along with a fixed frame shift mutation at codon 166 that broke the reading frame in all three frames. This leaves P. inui with only two intact msp3 reading frames (msp3A and msp3I), and the msp3C–H pseudogene (Fig. 2).
The presence of msp3C and H fragments in the pseudogene suggests that the ancestor of the P. inui lineage previously contained complete msp3C and H genes and a larger msp3 family. Indeed, an unequal crossing over event in the P. inui lineage that deleted the segment of genes between msp3C and H (msp3D, E, F, and G) and interrupted the reading frame when joining the C and H fragments could explain the presence of the chimeric pseudogene and the msp3 family size in P. inui.
Likewise, the presence of msp3G in P. knowlesi, as well as P. vivax and P. cynomolgi, indicates that their common ancestor (Fig. 4) had msp3G. Moreover, despite being absent in the P. inui msp3 array, the presence of two genes in P. knowlesi for which their reciprocal best BLAST hit was to msp3B in P. vivax and P. cynomolgi is evidence that msp3B orthologs predate the divergence of P. inui from the P. vivax - P. cynomolgi lineage. These data indicate that msp3B and msp3G were lost independently in the P. inui lineage, while retained in P. knowlesi. Thus, the reduced MSP3 families of P. inui and P. knowlesi are most parsimoniously explained by separate, independent deletions from a larger msp3 family.
It is worth noting that the observed gene turnover in these arrays for P. knowlesi and P. inui contrasts with the relative conservation observed in the large syntenic blocks across the genomes of the Plasmodium genus (Frech and Chen 2011). We were able to identify orthologs to all but 18 of the other 308 intratelomeric P. vivax genes on chromosome 10 in P. cynomolgi and P. knowlesi, making the intrasyntenic contraction/expansion seen for msp3 substantial in comparison (from 3 to 12–14 genes for these species). The rate of gene death in the msp3 arrays for P. knowlesi and P. inui appears to be exceptionally fast in comparison to the hundreds of other Plasmodium genes on the same region of chromosome 10.
3.3. An earlier origin for an expanded msp3 family in P. vivax and related species
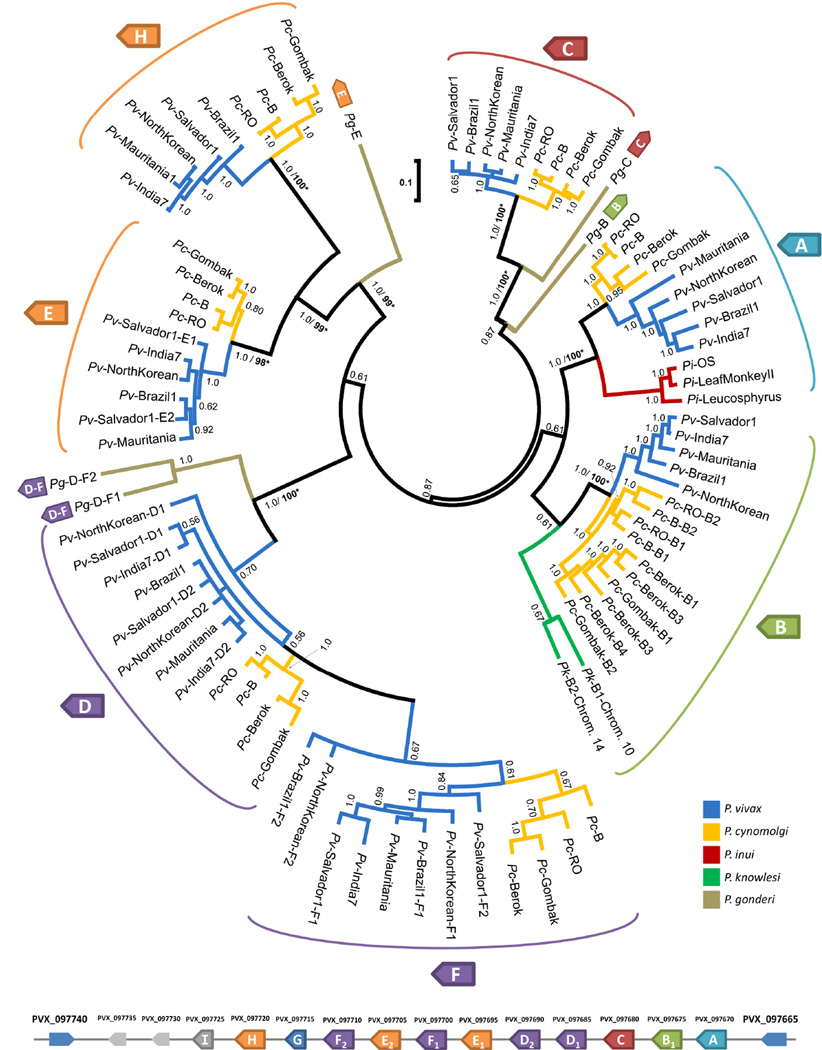
As stated above, the independent losses of msp3 paralogs in P. inui and P. knowlesi are better explained by an ancient higher number of paralogs in the msp3 array in the Asian primate Plasmodium clade. However, in order to further test this hypothesis, we analyzed the gene family in the African monkey parasite P. gonderi. This parasite is at the base of the Southeast Asian primate malaria clade (Escalante et al., 1998; Escalante et al., 2005; Pacheco et al., 2011, 2012b). Figure 4 also shows the six complete and three partial P. gonderi ORFs with significant similarity to P. vivax msp3 genes that were identified from draft genomic data using BLAST and by considering the domain architecture (supplementary Table 3 and Fig. 3). The ability to identify putative homologs for many of the Pvmsp3 lineages in the African monkey parasite P. gonderi, provide additional evidence for an ancient origin for a relative high number of msp3 paralogs. Specifically, we recovered four putative genes (msp3C, 2 mspD-F, and mspE) in P. gonderi that were absent in P. inui and P. knowlesi (supplementary Table 3 and Fig.3), thus providing additional support that these paralogs were present in the common ancestor of P. vivax, P. cynomolgi, P. inui and P. knowlesi (Fig. 4). It also indicates that P. vivax and P. cynomolgi actually conserved a high number of paralogs from the ancestral state, rather than had a recent expansion. This is evident in the phylogeny depicted in Figure 5 using the msp3 sequences identified in P. vivax, P. cynomolgi, P. inui, P. knowlesi and the basal species P. gonderi. Unfortunately, sequence similarity between genes in the msp3 family varied from 88% to 18% due to their highly divergent central region, leaving us with few of alignable sites (234 of 5940 bp). To improve the alignment the two genes, msp3I and G, that were substantially shorter (382 and 450 amino acids, respectively in P. vivax Salvador I) and lacked the characteristic coiled-coil motif domain (Fig. 3), were excluded from the phylogenetic analysis. However, bootstrap and posterior probability support at many nodes remained low after their exclusion, despite the number of alignable sites increasing to 947 bp. This could be explained, at least in part, by recombination-gene conversion.
Figure 5.
Bayesian phylogeny of the msp3 gene family of P. vivax and its closely related species. Nodes with greater than 50% posterior probability support from 55 × 106 MCMC generations are shown. Nodes with ML support based on a bootstrap with 100 pseudoreplications are indicated as * for 95–100%, nodes with less than 80% are not indicated. Branches are colored by species and clades are labeled by the gene from the Pvmsp3 array (shown below the phylogeny) they contain. A reduced set of 4 P. vivax, 4 P. cynomolgi, and 3 P. inui isolates for each gene was used for the unrooted phylogeny. Two P. cynomolgi genes, msp3F2 and msp3F3, which appeared to be recombinants of msp3F and msp3D (see results), were excluded in this tree. A Bayesian tree including these two genes is shown in the supplementary information (Supplementary Figure 4).
Nevertheless, as seen in Figure 5, msp3 homologs from the five species analyzed grouped into seven clades, corresponding to msp3A, B, C, D, E, F, and H. There was subgrouping among these seven clades such that msp3E and H were sister clades, and msp3F emerged from a paraphyletic clade shared with msp3D (msp3E and H, and msp3D and F, are colored similarly in the gene array to reflect this). A separate tree of just msp3F and msp3D sequences resolved the paraphyly (Supplementary Figure 4), with msp3F and msp3D sequences being sister clades. Plasmodium inui (branches shown in red in Fig. 5) and P. knowlesi (green) lacked all but one of the seven msp3 lineages. The two P. knowlesi genes, msp3B1 on chromosome 10 (PKH_103020) and msp3B2 on chromosome 14 (PKH_145630), seems to share a common ancestor with the P. vivax and P. cynomolgi msp3B lineage, reaffirming the homology of the non-syntenic P. knowlesi gene. Putative P. gonderi genes (brown) seem to share a common ancestor with the P. vivax (blue) and P. cynomolgi (yellow) genes.
3.4. Variation and recombination among and between genes in P. vivax and related species
We next explored intraspecies genetic diversity as it also reflects the processes driving the evolution of the msp3 family. To this end, we described the genetic polymorphism among P. vivax (Table 1), P. cynomolgi (Table 2), and P. inui isolates. This analysis was limited to the set of sites that could be confidently aligned. Nevertheless, substantial diversity at the nucleotide level was found among P. vivax (Table 1) and P. cynomolgi (Table 2) isolates. However, for several msp3 genes, and especially for P. cynomolgi, this diversity was biased toward synonymous sites. This may suggest that those aligned sites have been evolving under selective constraint in msp3 paralogs, an observation that has been made for other antigens (Pacheco et al., 2007; 2010; 2012a). This pattern, however, should be taken with caution since alignable sites may misrepresent the total polymorphism.
Indeed, the high frequency of indel events showed that changes to the protein coding sequence have occurred repeatedly (Table 1–Table 2). For example, 28 unique indel haplotypes were recovered from 48 isolates of Pvmsp3H. Moreover, the size of indels varied from single codons (e.g. a single serine insertion at amino acid position 20 in gene Pvmsp3D1) to events approaching 1,000 bp in length (e.g. isolates Indonesia I and North Korean lacked an 864 bp insert seen in 6 other isolates of Pvmsp3B). The range of allele sizes observed for several paralogs was wide as a result (Table 1). Small indels in regions of reduced sequence complexity are thought to be caused by polymerase slippage during replication, while large indel events are thought to be due to unequal crossing over during intragenic recombination events (Zilversmit et al., 2010). The frequency of indel events and their variance in size indicates that both mechanisms are commonly contributing to diversity among the MSP3 antigens.
We plotted the positions of indels and conserved sites within alignments onto the domain architecture of the msp3 family (using an Excel macro, available upon request, see Fig. 3). Indel and sequence diversity was high in the central domain, and mutations were observed at all positions within the heptad repeats described previously by Galinski et al. 2001. Haplotypes with large deletions in the N-terminal half of the central coiled-coil motif domain had been observed for msp3H (“alpha”) (Rayner et al., 2002) and msp3C (“beta”) (Rayner et al., 2004). We show here that this pattern extends to the majority of the rest of the msp3 family in P. vivax, especially genes msp3A, B, D, and F (Fig. 3). The sites conserved among isolates, under significant selective constraint for several paralogs, were largely restricted to the N and C termini, with indels and unalignable sites mostly mapping to the central domain (Fig. 3). Sites conserved between P. vivax and P. cynomolgi also mostly mapped to the N and C termini. Such functionally constrained segments may facilitate recombination. It is worth noting that, again, msp3G and msp3I differed from the other members of the family in having substantially less indel and nucleotide diversity among P. vivax isolates.
It is also worth noting that msp3H (“alpha”), the gene used for molecular genotyping in P. vivax due to its high polymorphism, was the least diverse (d = 0.027 ± 0.003) among those sharing the characteristic central domain (Table 1). Whereas other paralogs, such as msp3A (PVX_097670) or msp3B (PVX_097675), may provide additional options to those looking for highly polymorphic genetic markers to be used in epidemiologic investigations, their complex evolution limits their use in such studies (Rice et al., 2013).
Recombination seems to have taken place during the evolutionary history of this family as evidenced by the identification of multiple “chimeric” or recombinant ORFs. An example is the P. inui pseudogene that appeared to be a truncated chimera of msp3C and H. Evidence of intergenic recombination was also seen in P. gonderi msp3D–F2 and P. cynomolgi msp3F2 and F3. In each case, 125 to 150 amino acid segments at the N-terminal of these msp3F genes showed significant similarity, by alignment and BLAST searches (e-value < e-70) to the msp3D genes of P. vivax and P. cynomolgi. Furthermore, despite clearly seeing the gene turnover and paralog divergence characteristic of the birth and death model of gene family evolution (Garzón-Ospina et al., 2010), we also observed several cases where multiple paralogs within a species were more similar to each other than to their putative homologs in related species (Fig. 5). This would seem to indicate the action of gene conversion on pre-existing duplicates (Eirín-López et al., 2012), or repeated, recent duplications.
Assuming that the divergence between paralogs would be informative with regard to the time since their origin by duplication, we calculated the net group mean divergence (dA) between the P. vivax paralogs Pvmsp3D1-D2, Pvmsp3E1-E2, and Pvmsp3F1-F2, which were more closely related to a paralog in the P. vivax genome than to their putative ortholog in P. cynomolgi. An example of within species paralogs being more similar, P. vivax Salvador-1 genes Pvmsp3E1 and E2 were in the same terminal clade that was sister to their P. cynomolgi msp3E ortholog (Fig. 5). Table 3 shows that there was nearly equivalent divergence between each of these Pvmsp3 genes and their ortholog in P. cynomolgi. However, divergence between P. vivax paralogs was variable; with the two genes Pvmsp3E1 and E2 showing the least divergence between the two paralogs (Table 3). Indeed, during alignment of putative Pvmsp3E1 and E2 alleles, it was often unclear to which Salvador-1 reference gene (E1 or E2) an isolate’s allele was more similar. For the divergence calculations shown in Table 3, a pairwise distance matrix was used to assign Pvmsp3E sequences to the Salvador-1 gene they had minimal distance to. Moreover, as mentioned above, we were unable to recover two distinct Pvmsp3E copies in some of the strains of P. vivax with genomic sequencing available. This indicates a recent and independent gene duplication event in the Pvmsp3E lineage, or more frequent recombination/gene conversion between Pvmsp3E paralogs. The available evidence, however, favors that this pattern in Pvmsp3E is due to differential rates of gene conversion. First, an independent, more recent duplication of Pvmsp3E cannot properly account for the gene order observed in the Pvmsp3 array (see Figure 4). We would expect two independent, tandem duplications of msp3E1 and F2 to be more likely to produce an order of msp3E1-E2-F1-F2 with the duplicated genes each landing adjacent to their progenitor. A single, tandem duplication of the segment containing msp3E1 and F1, where the whole duplicated segment is inserted adjacently, seems a more parsimonious explanation for the msp3E1-F1-E2-F2 order seen. This supports that the paralogous copies of msp3E and F in P. vivax originated simultaneously and thus their different levels of divergence cannot be explained by different dates of duplication. Second, when we constructed a Bayesian phylogeny of all Pvmsp3E isolates and paralogs (Supplementary Figure 5), we failed to observe distinct clades that would correspond to diverged, distinct gene lineages. Rather, sequences of Pvmsp3E obtained from the same P. vivax isolate, and thus putative paralogs within the same genome (connected by red-dashed lines) were distributed throughout the phylogeny (Supplementary Figure 5). A potential source of error would be incorrectly assuming paralogy when distinct Pvmsp3E sequences were obtained from the same isolate. Clinical isolates could be of polyclonal infections; indeed, in one instance, five distinct msp3E sequences were obtained from a single Thailand isolate, likely evidence of a mixed infection. To alleviate this potential error, we performed the divergence analysis with only the three P. vivax strains that had been maintained in laboratory animals. In this smaller sample, where mixed infection could be ruled out, dA was even less (-0.006 ± 0.001), indicating more variation shared among sequences of Pvmsp3 than between supposed groups. This is consistent with the action of repeated inter-paralog recombination acting among Pvmsp3E paralogs, and indeed, recombination between Pvmsp3E sequences, as detected by the RDP algorithm, was observed throughout the phylogeny (recombination pairs connected by gray lines, Supplementary Figure 5). The observation of frequent recombination events and differing divergence between closely related paralog pairs for Pvmsp3 (Pvmsp3E versus F or D) is indicative that a simple model of the evolution of the msp3 family as a whole, such as a neutral birth and death process, may be inadequate. Work to better characterize the family’s function and the specific drivers of its evolution will have to account for this complexity. Here, we only have data from four species from a much-diversified clade (Pacheco et al 2011; 2012).
Table 3.
Paralog and ortholog divergence in the P. vivax msp3 gene family
| Gene | n | dA Between Pv Paralogs (SE) |
dA Between Pv and Pc Orthologs (SE) Orthologs (SE) |
dA Between Pv and Pc | |||
|---|---|---|---|---|---|---|---|
| msp3D | 18 | PvD1 vs. PvD2 | 0.038 (0.003) | PvD1 vs. PcD | 0.091 (0.005) | PvD2 vs. PcD | 0.092 (0.005) |
| msp3E | 25‡ | PvE1 vs. PvE2 | 0.007 (0.001) | PvE1 vs. PcE | 0.086 (0.005) | PvE2 vs. PcE | 0.085 (0.005) |
| msp3F | 26 | PvF1 vs. PvF2 | 0.024 (0.002) | PvF1 vs. PcF | 0.089 (0.005) | PvF2 vs. PcF | 0.091 (0.005) |
Divergence between P. vivax (Pv) isolates of the three highly similar paralog pairs msp3D1-D2, msp3E1-E2, and msp3F1-F2 and each paralog’s net divergence from its respective P. cynomolgi (Pc) ortholog are shown along with standard error (SE) estimates from 1000 bootstrap replicates. Sites with alignment gaps were excluded from analysis. The number of P. vivax and P. cynomolgi sequences (n) analyzed is shown.
For msp3E, only the 16 sequences from the 8 P. vivax isolates (Salvador I, Sumatra I, Thai III, two field isolates from Thailand, and three from Venezuela) for which exactly two divergent clones of msp3E were sequenced are included with the 9 P. cymomolgi msp3E sequences. Two Thai field isolates resulted in the amplification of more than two divergent clones of msp3E; these sequences are not included as it is not clear if this was a result of mixed infection.
Several models have been developed to explain gene duplication events that lead to multi-gene families. We evaluated these models (reviewed by Innan and Kondrashov (2010)) for their coherence with the observed pattern of variation and recombination among and between genes. Many of the models apply to gene families where paralogous genes differ with respect to specific amino acid substitutions that confer changes in function between the members that can lead to neofunctionalization or subfunctionalization. Additionally, some duplication events may serve to increase the amount of a circulating product such as an enzyme. However, here we find that variation between paralogs is constrained to the highly repetitive, low complexity region of the proteins, whose evolution is predominated by gene conversion events giving rise to several indel events. A suitable, flexible model that seems to fit the data and can accommodate putative gene conversion events is one of multi-allelic diversifying selection (Innan and Kondrashov 2010), where duplication increases the maximum level of heterozygosity possible in the population. This would be consistent with several patterns observed in terms of the number of indels and recent paralogs (e.g. Pvmsp3E1 and E2 alleles). Furthermore, it is compatible with the nature of the immune responses reported in some msp3 paralogs. Specifically, the central low complexity regions, precisely where the indels and highly level of divergence among paralogs occur, are highly immunogenic when compared with the more conserved N-C terminals (Lima-Junior et al. 2011). Thus, one could expect that variation will be maintained by selection in these low complexity regions since gene conversion and recombination between and among genes is beneficial as it creates new allelic combinations that are then are selected by diversifying or balancing selection, as expected by the host immune system. However, a formal test of this hypothesis will require an extended data set that includes a more detailed account of gene conversion and recombination events than the one reported here.
4. Conclusion
A role in immune evasion via antigenic diversity for the msp3 gene family could explain the complex patterns of variation observed among Plasmodium populations, in which its members are functionally (or at least structurally) redundant, yet present a highly diverse set of protein sequences with low complexity (Mendes et al., 2013; Schofield 1991) to the immune system. This pattern is consistent with the observation that low complexity regions seem to be more immunogenic in msp3 (Lima-Junior et al. 2011; 2012). Selection would be expected to place limits on the accumulation of variation in order to maintain functional redundancy, and we observed evidence of purifying selection primarily in codons at the N and C-termini of these proteins. We therefore suggest that the N and C-termini regions may be important to maintaining the family’s function in P. vivax and its related species. Nevertheless, we also observed redundancy in the diverse central domain, where despite high levels of intraspecific variation, they were still predicted to form large regions of coiled-coil tertiary protein structure motifs. Such a role in immune evasion could be particularly important and may explain the independent origin of an analogous group of proteins such as PfMSP3.
In summary, we find no evidence that Pfmsp3 and Pvmsp3 are actually homologous and hypothesize that they may be analogous, highlighting their functional importance in these two parasites. We find no evidence indicating that the number of genes in this gene family was either affected by the host switch to humans in the case of Pvmsp3 or that the number of paralogs is explained by host range. Finally, we hypothesize that a model of multi-allelic diversifying selection may fit the evolution of Pvmsp3, a model that is consistent with functional redundancy in terms of increasing antigenic diversity. However, population based investigations are still needed in order to test this hypothesis. Whereas msp3 paralogs are immunogenic, at least in P. vivax (Lima-Junior et al. 2011; 2012), there is no information indicating that they are associated with actual clinical protection. Thus, some of these paralogs could indeed act as “decoys” during the critical process of RBC invasion in tertian malarial parasites.
Supplementary Material
Highlights.
The array containing MSP3 genes was characterized among Plasmodium species.
The P. falciparum MSP3 proteins are analogous to those found in P. vivax.
The high number of paralogs has an earlier origin in the P. vivax clade.
Host range cannot explain the variation in the number of paralogs among species.
The MSP3 evolution is consistent with a multi-allelic diversifying selection model.
Acknowledgements
This work was supported by the US National Institutes of Health (R01 GM080586 and U19AI089688 to AAE). We thank the DNA laboratory at the School of Life Sciences for their technical support and David Fisher for computational support. BLR and MMA were supported in part by the US NIH: grant R25GM071798, ASU PREP for Biomedical Research. The content is solely the responsibility of the authors and does not represent the official views of the NIH or the Centers for Disease Control and Prevention.
Glossary
Abbreviations
- RBC
Red blood cell
- MSP3
merozoite surface protein-3
- Mya
millions of years ago
- dS
diversity at synonymous sites
- dN
diversity at non synonymous sites
- dA
net between group mean genetic divergence
Glossary
- Merozoite
the asexual, red blood cell infecting life stage of Plasmodium parasites
- synteny
shared relative genomic position
- host range
the set of host species that a parasitic organism colonizes
- periodicity
for malaria parasites, the length of the interval between fevers caused by synchronous release of merozoites in the bloodstream
Appendix A. Supplementary material
Supplementary Methods 1. Material and methods details (Microsoft Word File).
Supplementary Table 1. Primers, reaction conditions, and thermocycle programs for the P. vivax, P. cynomolgi, and P. inui msp3 genes or genome segments (Microsoft Excel file).
Supplementary Table 2. MSP3 isolates, sequences and accession numbers used for analysis (Microsoft Excel File).
Supplementary Table 3. BLASTn e-values from pairwise comparisons of MSP3 nucleotide sequences (Microsoft Excel File). Expected value of 10 used as a cut-off, comparisons in which no significant homology was found (the e-value score was 10 or above) are recorded as 10 in this table.
Supplementary Figure 1. Msp3 gene arrays sequenced for P. vivax, P. cynomolgi, P. inui and P. knowlesi. Gene arrays connected by a line represent isolates for which genomic sequencing was available and thus order on a chromosome or scaffold could be determined. Also, for those isolates with genomic sequencing data, rectangles represent incomplete or truncated ORFs, while genes filled in with color are those for which the genomic sequence was amplified, cloned and re-sequenced. Genes not filled in with color were not re-sequenced. The diagonal stripes of the msp3H-C ‘gene’ for P. inui signify it as a pseudogene. Gene length in amino acids (aa) and PlasmoDB gene ID codes are shown.
Supplementary Figure 2. P. cynomolgi strain Berok and P. inui strain OS sequencing schemes. Drawn to scale, with scale bars in kilobases (KB) and gene or cloned segment lengths in base pairs (bp) shown. Putative genes are labeled by the P. vivax gene to which they have most sequence similarity, either msp3 (see Figure 1) or noted as “O-” followed by the PlasmoDB code of the corresponding P. vivax gene. A.) The 17 P. cynomolgi segments cloned are shown in red. B.) The four overlapping fragments (P1-P4) for P. inui are shown with gray arrows.
Supplementary Figure 3. High alanine content in the msp3 family of P. vivax is maintained in orthologs. Amino acid composition for P. vivax (Pv) Salvador-1 genes (array shown below) and their respective orthologs in P. cynomolgi Berok strain (Pc), P. inui OS strain (Pi), P. knowlesi H strain (Pk), and P. gonderi (Pg), when present. Percentage alanine content is shown
Supplementary Figure 4. Additional Bayesian phylogenies of (A) the interspecies msp3 family and (B) msp3D and msp3E paralogs. Nodes with greater than 50% posterior probability support from 55 × 106 (A) or 31 × 106 (B) MCMC generations are shown. Branches are colored by species and clades are labeled by the gene from the Pvmsp3 array (shown below the phylogeny) they contain. Two P. cynomolgi genes, msp3F2 and msp3F3, which appeared to be recombinants of msp3F and msp3D (see results), were excluded from the phylogeny in Figure 5, but are included here in this figure.
Supplementary Figure 5. Phylogenetic relationships and recombination among P. vivax msp3E1 and msp3E2 paralogs. Nodes with greater than 50% bootstrap support are shown in the Bayesian tree of P. vivax (Pv) and P. cynomolgi (Pc) isolates. Alleles from the same isolate (putative paralogs) are connected with red-dotted lines. The recombination events detected by the RDP algorithm are connected with grey lines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
AAE, BLR, MAP, and MMA conceived and designed the study. AAE and MAP supervised the overall project. AAE, JMC, JWB contributed reagents, analytical tools, and the parasite material studied. BLR, JMC, MAP, and MMA generated the sequence data. AAE, BLR, MAP, and MMA analyzed the data. BLR wrote the initial draft of the manuscript; all authors contributed to and approved the final manuscript.
Contributor Information
Benjamin L. Rice, Email: Benjamin.Rice@asu.edu.
Mónica M. Acosta, Email: mmacost1@asu.edu.
M. Andreína Pacheco, Email: Maria.Pacheco@asu.edu.
Jane M. Carlton, Email: Jane.Carlton@nyu.edu.
John W. Barnwell, Email: wzb3@cdc.gov.
Ananias A. Escalante, Email: Ananias.Escalante@asu.edu.
References
- al-Khedery B, Barnwell JW, Galinski MR. Antigenic variation in malaria: a 3’ genomic alteration associated with the expression of a P. knowlesi variant antigen. Mol. Cell. 1999;3:131–141. doi: 10.1016/s1097-2765(00)80304-4. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amambua-Ngwa A, Tetteh KKA, Manske M, Gomez-Escobar N, Stewart LB, Deerhake ME, Cheeseman IH, Newbold CI, Holder AA, Knuepfer E, Janha O, Jallow M, Campino S, Macinnis B, Kwiatkowski DP, Conway DJ. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet. 2012;8:e1002992. doi: 10.1371/journal.pgen.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisue N, Kawai S, Hirai M, Palacpac NMQ, Jia M, Kaneko A, Tanabe K, Horii T. Clues to evolution of the SERA multigene family in 18 Plasmodium species. PLoS ONE. 2011;6:e17775. doi: 10.1371/journal.pone.0017775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr, Treatman C, Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann A, Magris MM, Urbaez M-L, Vivas-Martinez S, Durán R, Nieves T, Esen M, Mordmüller BG, Theisen M, Avilan L, Metzger WG. Naturally acquired immune responses to malaria vaccine candidate antigens MSP3 and GLURP in Guahibo and Piaroa indigenous communities of the Venezuelan Amazon. Malar. J. 2012;11:46. doi: 10.1186/1475-2875-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabeu M, Lopez FJ, Ferrer M, Martin-Jaular L, Razaname A, Corradin G, Maier AG, Del Portillo HA, Fernandez-Becerra C. Functional analysis of Plasmodium vivax VIR proteins reveals different subcellular localizations and cytoadherence to the ICAM-1 endothelial receptor. Cell. Microbiol. 2012;14:386–400. doi: 10.1111/j.1462-5822.2011.01726.x. [DOI] [PubMed] [Google Scholar]
- Bitencourt AR, Vicentin EC, Jimenez MC, Ricci R, Leite JA, Costa FT, Ferreira LC, Russell B, Nosten F, Rénia L, Galinski MR, Barnwell JW, Rodrigues MM, Soares IS. Antigenicity and immunogenicity of Plasmodium vivax merozoite surface protein-3. PLoS ONE. 2013;8:e56061. doi: 10.1371/journal.pone.0056061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce MC, Galinski MR, Barnwell JW, Snounou G, Day KP. Polymorphism at the merozoite surface protein-3alpha locus of Plasmodium vivax: global and local diversity. Am. J. Trop. Med. Hyg. 1999;61:518–525. doi: 10.4269/ajtmh.1999.61.518. [DOI] [PubMed] [Google Scholar]
- Cai H, Gu J, Wang Y. Core genome components and lineage specific expansions in malaria parasites plasmodium. BMC Genomics. 2010;(11 Suppl 3):S13. doi: 10.1186/1471-2164-11-S3-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RMR, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang’a S, Kooij TWA, Korsinczky M, Meyer EV-S, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax . Nature. 2008;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Das A, Escalante AA. Genomics, population genetics and evolutionary history of Plasmodium vivax . Adv. Parasitol. 2013;81:203–222. doi: 10.1016/B978-0-12-407826-0.00005-9. [DOI] [PubMed] [Google Scholar]
- Coatney RG, Collins WE, Warren M, Contacos PG. The Primate Malarias. US Government Printing Office; Washington, DC: 1971. [Google Scholar]
- Conway DJ. Molecular epidemiology of malaria. Clin. Microbiol. Rev. 2007;20:188–204. doi: 10.1128/CMR.00021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirín-López JM, Rebordinos L, Rooney AP, Rozas J. The birth-and-death evolution of multigene families revisited. Genome Dyn. 2012;7:170–196. doi: 10.1159/000337119. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Cornejo OE, Freeland DE, Poe AC, Durrego E, Collins WE, Lal AA. A monkey’s tale: the origin of Plasmodium vivax as a human malaria parasite. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1980–1985. doi: 10.1073/pnas.0409652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Cornejo OE, Rojas A, Udhayakumar V, Lal AA. Assessing the effect of natural selection in malaria parasites. Trends Parasitol. 2004;20:388–395. doi: 10.1016/j.pt.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech C, Chen N. Genome comparison of human and non-human malaria parasites reveals species subset-specific genes potentially linked to human disease. PLoS Comput. Biol. 2011;7:e1002320. doi: 10.1371/journal.pcbi.1002320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frech C, Chen N. Variant surface antigens of malaria parasites: functional and evolutionary insights from comparative gene family classification and analysis. BMC Genomics. 2013 Jun 27;14:427. doi: 10.1186/1471-2164-14-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski MR, Corredor-Medina C, Povoa M, Crosby J, Ingravallo P, Barnwell JW. Plasmodium vivax merozoite surface protein-3 contains coiled-coil motifs in an alanine-rich central domain. Mol. Biochem. Parasitol. 1999;101:131–147. doi: 10.1016/s0166-6851(99)00063-8. [DOI] [PubMed] [Google Scholar]
- Galinski MR, Ingravallo P, Corredor-Medina C, Al-Khedery B, Povoa M, Barnwell JW. Plasmodium vivax merozoite surface proteins-3beta and-3gamma share structural similarities with P. vivax merozoite surface protein-3alpha and define a new gene family. Mol. Biochem. Parasitol. 2001;115:41–53. doi: 10.1016/s0166-6851(01)00267-5. [DOI] [PubMed] [Google Scholar]
- Garzón-Ospina D, Cadavid LF, Patarroyo MA. Differential expansion of the merozoite surface protein (msp)-7 gene family in Plasmodium species under a birth-and-death model of evolution. Mol. Phylogenet. Evol. 2010;55:399–408. doi: 10.1016/j.ympev.2010.02.017. [DOI] [PubMed] [Google Scholar]
- Gondeau C, Corradin G, Heitz F, Le Peuch C, Balbo A, Schuck P, Kajava AV. The C-terminal domain of Plasmodium falciparum merozoite surface protein 3 self-assembles into alpha-helical coiled coil tetramer. Mol. Biochem. Parasitol. 2009;165:153–161. doi: 10.1016/j.molbiopara.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Gunalan K, Gao X, Yap SSL, Huang X, Preiser PR. The role of the reticulocyte-binding-like protein homologues of Plasmodium in erythrocyte sensing and invasion. Cell. Microbiol. 2013;15:35–44. doi: 10.1111/cmi.12038. [DOI] [PubMed] [Google Scholar]
- Howard RJ. Antigenic variation of bloodstage malaria parasites. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 1984;307:141–158. doi: 10.1098/rstb.1984.0115. [DOI] [PubMed] [Google Scholar]
- Imam M, Devi YS, Verma AK, Chauhan VS. Comparative Immunogenicities of full-length Plasmodium falciparum merozoite surface protein 3 and a 24-kilodalton N-terminal fragment. Clin. Vaccine Immunol. 2011;18:1221–1228. doi: 10.1128/CVI.00064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Jiang J, Barnwell JW, Meyer EVS, Galinski MR. Plasmodium vivax Merozoite Surface Protein-3 (PvMSP3): Expression of an 11 Member Multigene Family in Blood-Stage Parasites. PLoS ONE. 2013;8:e63888. doi: 10.1371/journal.pone.0063888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooij TW, Carlton JM, Bidwell SL, Hall N, Ramesar J, Janse CJ, Waters AP. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 2005;1:e44. doi: 10.1371/journal.ppat.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liew KJL, Hu G, Bozdech Z, Peter PR. Defining species specific genome differences in malaria parasites. BMC Genomics. 2010;11:128. doi: 10.1186/1471-2164-11-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Junior JC, Jiang J, Rodrigues-da-Silva RN, Banic DM, Tran TM, Ribeiro RY, Meyer VSE, De-Simone SG, Santos F, Moreno A, Barnwell JW, Galinski MR, Oliveira-Ferreira J. B cell epitope mapping and characterization of naturally acquired antibodies to the Plasmodium vivax merozoite surface protein-3α (PvMSP-3α) in malaria exposed individuals from Brazilian Amazon. Vaccine. 2011;29:1801–1811. doi: 10.1016/j.vaccine.2010.12.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Junior JC, Rodrigues-da-Silva RN, Banic DM, Jiang J, Singh B, Fabrício-Silva GM, Porto LCS, Meyer EVS, Moreno A, Rodrigues MM, Barnwell JW, Galinski MR, de Oliveira-Ferreira J. Influence of HLA-DRB1 and HLA-DQB1 alleles on IgG antibody response to the P. vivax MSP-1, MSP-3α and MSP-9 in individuals from Brazilian endemic area. PLoS ONE. 2012;7:e36419. doi: 10.1371/journal.pone.0036419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire SL, Ohéigeartaigh SS, Byrne KP, Schröder MS, O’Gaora P, Wolfe KH, Butler G. Comparative Genome Analysis and Gene Finding in Candida Species Using CGOB. Mol. Biol. Evol. 2013;30:1281–1291. doi: 10.1093/molbev/mst042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl DJ, Silva A, Foley M, Kun JF, Favaloro JM, Thompson JK, Marshall VM, Coppel RL, Kemp DJ, Anders RF. Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 1994;68:53–67. doi: 10.1016/0166-6851(94)00149-9. [DOI] [PubMed] [Google Scholar]
- McDonnell AV, Jiang T, Keating AE, Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006;22:356–358. doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- Mendes TAO, Lobo FP, Rodrigues TS, Rodrigues-Luiz GF, daRocha WD, Fujiwara RT, Teixeira SMR, Bartholomeu DC. Repeat-enriched proteins are related to host cell invasion and immune evasion in parasitic protozoa. Mol. Biol. Evol. 2013;30:951–963. doi: 10.1093/molbev/mst001. [DOI] [PubMed] [Google Scholar]
- Mourão LC, Morais CG, Bueno LL, Jimenez MC, Soares IS, Fontes CJ, Guimarães Lacerda MV, Xavier MS, Barnwell JW, Galinski MR, Braga EM. Naturally acquired antibodies to Plasmodium vivax blood-stage vaccine candidates (PvMSP-119 and PvMSP-3α359–798 and their relationship with hematological features in malaria patients from the Brazilian Amazon. Microbes Infect. 2012;14:730–739. doi: 10.1016/j.micinf.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Galinsky K, Jiang RHY, Young L, Sykes SM, Saif S, Gujja S, Goldberg JM, Young S, Zeng Q, Chapman SB, Dash AP, Anvikar AR, Sutton PL, Birren BW, Escalante AA, Barnwell JW, Carlton JM. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum . Nat. Genet. 2012;44:1046–1050. doi: 10.1038/ng.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Oeuvray C, Bouharoun-Tayoun H, Grass-Masse H, Lepers JP, Ralamboranto L, Tartar A, Druilhe P. A novel merozoite surface antigen of Plasmodium falciparum (MSP-3) identified by cellular-antibody cooperative mechanism antigenicity and biological activity of antibodies. Mem. Inst. Oswaldo Cruz. 1994;(89 Suppl 2):77–80. doi: 10.1590/s0074-02761994000600018. [DOI] [PubMed] [Google Scholar]
- Pacheco MA, Battistuzzi FU, Junge RE, Cornejo OE, Williams CV, Landau I, Rabetafika L, Snounou G, Jones-Engel L, Escalante AA. Timing the origin of human malarias: the lemur puzzle. BMC Evol. Biol. 2011;11:299. doi: 10.1186/1471-2148-11-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Elango AP, Rahman AA, Fisher D, Collins WE, Barnwell JW, Escalante AA. Evidence of purifying selection on merozoite surface protein 8 (MSP8) and 10 (MSP10) in Plasmodium spp. Infect. Genet. Evol. 2012a;12:978–986. doi: 10.1016/j.meegid.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Poe AC, Collins WE, Lal AA, Tanabe K, Kariuki SK, Udhayakumar V, Escalante AA. A comparative study of the genetic diversity of the 42kDa fragment of the merozoite surface protein 1 in Plasmodium falciparum and P. vivax . Infect. Genet. Evol. 2007;7:180–187. doi: 10.1016/j.meegid.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Reid MJC, Schillaci MA, Lowenberger CA, Galdikas BMF, Jones-Engel L, Escalante AA. The origin of malarial parasites in orangutans. PLoS ONE. 2012b;7:e34990. doi: 10.1371/journal.pone.0034990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, Ryan EM, Poe AC, Basco L, Udhayakumar V, Collins WE, Escalante AA. Evidence for negative selection on the gene encoding rhoptry-associated protein 1 (RAP-1) in Plasmodium spp. Infect. Genet. Evol. 2010;10:655–661. doi: 10.1016/j.meegid.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain A, Böhme U, Berry AE, Mungall K, Finn RD, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi . Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters W. Malaria of the orang-utan (Pongo pygmaeus) in Borneo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976;275:439–482. doi: 10.1098/rstb.1976.0089. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, McGee K, Keebler J, Awadalla P. Selection shapes malaria genomes and drives divergence between pathogens infecting hominids versus rodents. BMC Evol. Biol. 2008;8:223. doi: 10.1186/1471-2148-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Corredor V, Feldman D, Ingravallo P, Iderabdullah F, Galinski MR, Barnwell JW. Extensive polymorphism in the Plasmodium vivax merozoite surface coat protein MSP-3alpha is limited to specific domains. Parasitology. 2002;125:393–405. doi: 10.1017/s0031182002002317. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Huber CS, Feldman D, Ingravallo P, Galinski MR, Barnwell JW. Plasmodium vivax merozoite surface protein PvMSP-3 beta is radically polymorphic through mutation and large insertions and deletions. Infect. Genet. Evol. 2004;4:309–319. doi: 10.1016/j.meegid.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Rodriguez LE, Curtidor H, Urquiza M, Cifuentes G, Reyes C, Patarroyo ME. Intimate molecular interactions of P. falciparum merozoite proteins involved in invasion of red blood cells and their implications for vaccine design. Chem. Rev. 2008;108:3656–3705. doi: 10.1021/cr068407v. [DOI] [PubMed] [Google Scholar]
- Rice BL, Acosta MM, Pacheco MA, Escalante AA. Merozoite surface protein-3 alpha as a genetic marker for epidemiologic studies in Plasmodium vivax: a cautionary note. Malar. J. 2013;12:288. doi: 10.1186/1475-2875-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schofield L. On the function of repetitive domains in protein antigens of Plasmodium and other eukaryotic parasites. In: Regul, editor. Parasitol. Today. Vol. 7. 1991. pp. 99–105. [DOI] [PubMed] [Google Scholar]
- Singh S, Soe S, Weisman S, Barnwell JW, Pérignon JL, Druilhe P. A conserved multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum . PLoS ONE. 2009;4:e5410. doi: 10.1371/journal.pone.0005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana S-I, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, Arisue N, Palacpac NMQ, Honma H, Yagi M, Tougan T, Katakai Y, Kaneko O, Mita T, Kita K, Yasutomi VY, Sutton PL, Shakhbatyan R, Horii T, Yasunaga T, Barnwell JW, Escalante AA, Carlton JM, Tanabe K. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat. Genet. 2012;44:1051–1055. doi: 10.1038/ng.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborrini M, Stoffel SA, Westerfeld N, Amacker M, Theisen M, Zurbriggen R, Pluschke G. Immunogenicity of a virosomally-formulated Plasmodium falciparum GLURP-MSP3 chimeric protein-based malaria vaccine candidate in comparison to adjuvanted formulations. Malar. J. 2011;10:359. doi: 10.1186/1475-2875-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tyne D, Park DJ, Schaffner SF, Neafsey DE, Angelino E, Cortese JF, Barnes KG, Rosen DM, Lukens AK, Daniels RF, Milner DA, Jr, Johnson CA, Shlyakhter I, Grossman SR, Becker JS, Yamins D, Karlsson EK, Ndiaye D, Sarr O, Mboup S, Happi C, Furlotte NA, Eskin E, Kang HM, Hartl DL, Birren BW, Wiegand RC, Lander ES, Wirth DF, Volkman SK, Sabeti PC. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum . PLoS Genet. 2011;7:e1001383. doi: 10.1371/journal.pgen.1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth J, Daub J, Peregrín-Alvarez JM, Finney CA, Parkinson J. The origins of apicomplexan sequence innovation. Genome Res. 2009;19:1202–1213. doi: 10.1101/gr.083386.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedall GD, Conway DJ. Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol. 2010;26:363–369. doi: 10.1016/j.pt.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Zilversmit MM, Chase EK, Chen DS, Awadalla P, Day KP, McVean G. Hypervariable antigen genes in malaria have ancient roots. BMC Evol. Biol. 2013;13:110. doi: 10.1186/1471-2148-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilversmit MM, Volkman SK, DePristo MA, Wirth DF, Awadalla P, Hartl DL. Low-complexity regions in Plasmodium falciparum: missing links in the evolution of an extreme genome. Mol. Biol. Evol. 2010;27:2198–2209. doi: 10.1093/molbev/msq108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.