Abstract
The release of nascent RNA from transcribing RNA polymerase complexes is required for all further functions carried out by RNA molecules. The elements and processing machinery involved in 3′ end formation therefore represent key determinants in the biogenesis and accumulation of cellular RNA. While these factors have been well-characterized for messenger RNA, recent work has elucidated analogous pathways for the 3′ end formation of other important cellular RNA. Here, we discuss four specific cases of non-mRNA 3′ end formation—metazoan small nuclear RNA, Saccharomyces cerevisiae small nuclear RNA, Schizosaccharomyces pombe telomerase RNA, and the mammalian MALAT1 large noncoding RNA—as models of alternative mechanisms to generate RNA 3′ ends. Comparison of these disparate processing pathways reveals an emerging theme of evolutionary ingenuity. In some instances, evidence for the creation of a dedicated processing complex exists; while in others, components are utilized from the existing RNA processing machinery and modified to custom fit the unique needs of the RNA substrate. Regardless of the details of how non-mRNA 3′ ends are formed, the lengths to which biological systems will go to release nascent transcripts from their DNA templates are fundamental for cell survival.
The 3′ end formation of mRNA molecules plays a central role in the termination of RNA polymerase II (RNAPII) transcription, is important for mRNA export and stability, and by the addition of the poly(A) tail determines their translational efficiency. Not surprisingly, a highly conserved set of demarcating elements and protein machinery guarantees that this critical RNA processing event takes place with precision and reproducible accuracy. For poly(A) mRNA, members of the Cleavage and Polyadenylation Specificity (CPSF) and Cleavage Stimulation Factor (CstF) complexes are vital to 3′ end formation. In contrast, replication-dependent histone mRNAs, the only mRNAs that lack a poly(A) tail, utilize distinct 3′ end formation machinery in addition to CPSF/CstF factors to yield a stable and translatable message characterized by a unique terminal secondary structure. Both of these processes have been well-characterized and will not be discussed further here having been reviewed extensively elsewhere 1, 2.
With the recent appreciation for the true size of the transcriptome, the spectrum of non-mRNA molecules has increased dramatically beyond that of the abundant ribosomal RNA (rRNA) and transfer RNA (tRNA). In addition to the small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA), there is an array of other nuclear partitioned RNA playing vital roles in coordinating gene expression. These RNAs include, but are not limited to, the RNA component of RNAseP involved in tRNA processing, the RNA component of the telomerase RNP required for the maintenance of telomeres, the Xist RNA involved in dosage compensation, and the ever-increasing number of long noncoding RNA (lncRNA) that play a wide range of functions from modulating chromatin modification to regulating alternative splicing (see 3, 4 for review). Proper biosynthesis of non-mRNA ensures their structural and functional integrity and is essential for the homeostasis of the cell. While the machinery involved in the 3′ end processing of non-mRNA is distinct from their mRNA counterparts, the role it plays in their accumulation is equally important. Reviewing four specific cases, we intend to demonstrate that either by using a dedicated processing complex or by borrowing from the existing RNA processing machinery, non-mRNA 3′ end formation rivals in complexity and ingenuity with mRNA end formation.
U snRNA 3′ end processing in metazoans
Demarcating Elements and Protein Factors
Uridine-rich small nuclear RNAs (U snRNAs) are a family of short (60–200 nucleotides) intronless non-coding RNAs. They form the RNA moiety of the major (U1, U2, U4, U5 and U6) and minor (U11, U12, U4atac, U5 and U6atac) spliceosomes (reviewed in5) with the exception of U7 that is involved in the 3′-end processing of the replication-dependent histone mRNAs (reviewed in 2).
Most of the U snRNAs are transcribed by RNAPII (U1, U2, U4, U4atac, U5, U7, U11 and U12) with the exception of the U6 snRNA (and U6atac) that is transcribed by the RNA polymerase III (RNAPIII). Regardless of their dependency on different RNA polymerases, the promoters of U snRNA genes share remarkable similarities. About 250 bp in length, they contain a distal sequence element (DSE) recruiting the Oct1 and Sp1 transcription factors and a proximal sequence element (PSE) that is bound by the snRNA activating protein complex (SNAPc), a five subunit complex specific to U snRNA transcription (reviewed in 6). The only distinction between the two types of promoters resides in the presence of a TATA box between the transcription start site and the PSE of the U6 promoter that confers its specificity for RNAPIII7.
Despite numerous similarities in U snRNA structure, promoter architecture, and ultimate function, the mechanism of 3′ end processing of RNAPII- and RNAPIII-dependent snRNAs is drastically different. U6 and U6atac are processed as typical RNAPIII transcripts. A stretch of thymidines at their 3′ end drives transcription termination 8. The U6 primary transcript is further processed by two antagonistic enzymatic activities. The U6-specific uridyl terminal transferase (TUT1 or U6-TUTase) adds a poly(U) tail to the 3′ end of the U6 snRNA 9, 10 while the 3′-to-5′ exonuclease Mpn1 trims the poly(U) tail to an average of five Us and generates the final 2′–3′ cyclic phosphate characteristic of the mature U6 snRNA 11.
The 3′-end processing of the RNAPII-transcribed U snRNAs is more complex and requires at least two independent steps. The first step, the endonucleolytic cleavage of the primary transcript (detailed below), takes place in the nucleus and leaves 2 to 10 nucleotides beyond the mature 3′-end 12–14. This intermediate transcript is then exported to the cytoplasm through the nuclear pore complex by the RNA adaptor PHAX and the CRM1/RanGTP complex15. In the cytoplasm, the pre-U snRNA undergoes a 3′ to 5′ exonucleolytic trimming step to generate the mature end of the U snRNA 12–14. Whereas in Saccharomyces cerevisiae the exonucleases involved in 3′ to 5′ digestion of the pre-U snRNA are largely known (see section below), the 3′ to 5′ exonuclease responsible for the cytoplasmic trimming of the metazoan U snRNA has not been identified yet.
Extensive work has shed light on the nuclear phase of the U snRNA 3′-end processing, which requires the presence of a U snRNA-specific promoter (as described above) and a conserved GTTTN0-3AAAPuNNAGA sequence, referred to as the 3′ box, located 9-19 nucleotides downstream from the mature end of the U snRNA 16–19. A third element, the carboxy-terminal domain (CTD) of the largest RNAPII subunit, Rpb1, and its phosphorylation by pTEFb (CDK9) were identified later as equally important for the 3′ end processing of the U snRNAs 20, 21. The CTD of the RNAPII is a domain formed by multiple YSPTSPS heptamer repeats (52 in human, 42 in fly and 26 in Saccharomyces cerevisiae (hereafter referred to as yeast) with various degree of conservation). Through the transcription cycle, the phosphorylation pattern on the different serines of the heptamer repeats drives the recruitment of various factors involved in transcription and RNA processing 22–24. Taken together, the different requirements for the 3′ end processing of the U snRNA (U snRNA promoter, 3′ box, and RNAPII CTD phosphorylation) has suggested a co-transcriptional mechanism where a processing factor is recruited at the U snRNA promoter and loaded onto the CTD of the RNAPII to cleave the nascent U snRNA after the transcription of the 3′ box element.
The Integrator Complex: the snRNA Processing Machinery
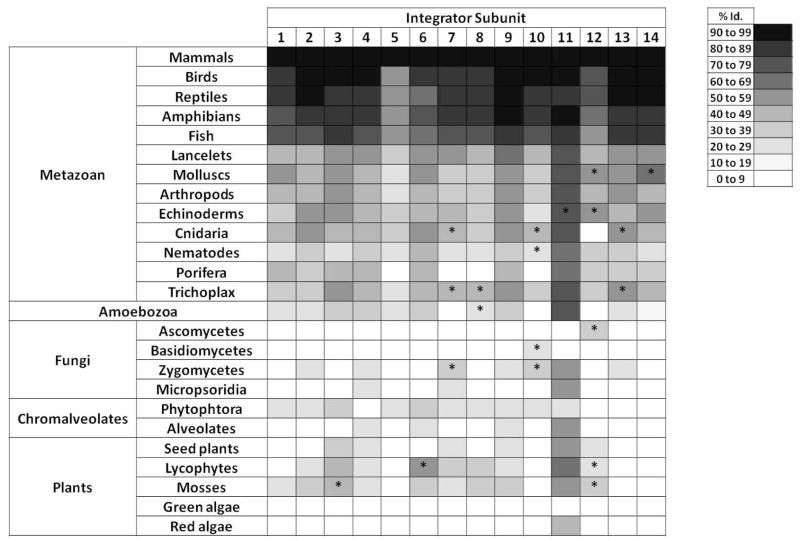
This hypothesis was validated by the purification and characterization of the Integrator complex 25. Integrator was first isolated as a large (>1 MDa) RNAPII-associated complex, containing at least 12 subunits. Recent studies have identified at least two new core subunits 26 as well as several potential protein partners 26–28. This complex appears to be broadly conserved through evolution (Figure 1). The majority of the Integrator subunits can be identified in metazoans and in plants, with the exception of green and red algae. Some of the complex subunits are also present in many unicellular eukaryotes including amoebas and chromalveolates, suggesting an early evolutionary origin. In fungi, the situation is more nuanced. The complex is absent from yeasts, in agreement with their distinct U snRNA biogenesis pathway (described below). However, some Integrator subunits can be identified, although with less conservation, in certain fungal phyla suggesting that the Integrator complex might have been lost through evolution in yeasts. Such a loss in yeast is not unprecedented, a similar evolutionary mechanism was advanced to explain the absence of the minor spliceosome (U11, U12 snRNAs and their associated spliceosomal proteins) in yeast despite its presence in other fungi and in several unicellular eukaryotes 29. Strikingly, herpesvirus saimiri coopts the cellular integrator complex during infection to generate U snRNA like molecules that function as microRNA precursor used to modulate ultimately host gene expression 30.
Figure 1. Phylogenetic distribution of the Integrator complex subunits.
Each column represents a subunit of the Integrator complex. Int13 and Int14 correspond to the recently characterized subunits Asunder and CG4785, respectively 26. For each subunit, a search was performed against the corresponding genome(s) using Blastp with default parameters. The shading of each cell represents the level of identity between the human sequence and the considered organism(s). A white cell indicates that the search failed to return a sequence with significant homology. When a homolog is identified, it was verified that a reciprocal Blastp search using the identified subunit against the Metazoan taxon returned the corresponding Integrator subunit as the most significant hit. Asterisks indicate that a significant homology was detected only on a portion of the protein sequence.
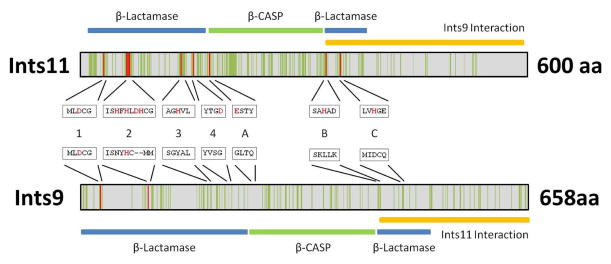
Unlike the mRNA 3′ end processing factors, few of the Integrator subunits present sequence or domain homology with any known proteins that could point toward a predicted function within the complex 31. However, two subunits in particular, IntS9/11, do have domains that readily suggest function. IntS11 is believed to be the endonuclease responsible for the pre-U snRNA 3′ end cleavage and forms a stable dimer with IntS9 through their C-terminal domains 32. Both IntS11 and IntS9 belong to the β-CASP (after the name of its representative members CPSF, Artemis, SNM1 and PSO2) family of metallo-β-lactamase fold nucleases 33, 34. Their closest homologs in this family are CPSF73 and CPSF100, two subunits of the cleavage and polyadenylation specificity factor complex (Figure 2) involved in the 3′ end cleavage of pre-mRNAs 1, 35. Moreover, depletion of IntS11 or the expression of a catalytic mutant of IntS11 results in misprocessing of the 3′ end of the pre-U snRNAs 25, 36.
Figure 2. Map of the Ints11 and Ints9 subunits of the Integrator complex.
The β-lactamase and β-CASP domains as well as the interaction domain between Ints11 and Ints9 are represented schematically. The vertical green bars represent conserved residues between Ints11 and CPSF73 and between Ints9 and CPSF100. The sequences of the motifs characteristic of the β-lactamase and β-CASP family (1 to 4 and A to C) are given for each protein 33. The amino acids responsible for the coordination of the zinc ions in the catalytic center of the proteins are represented in red. Note that most are absent in Ints9/CPSF100.
Mechanism of metazoan snRNA 3′ end formation
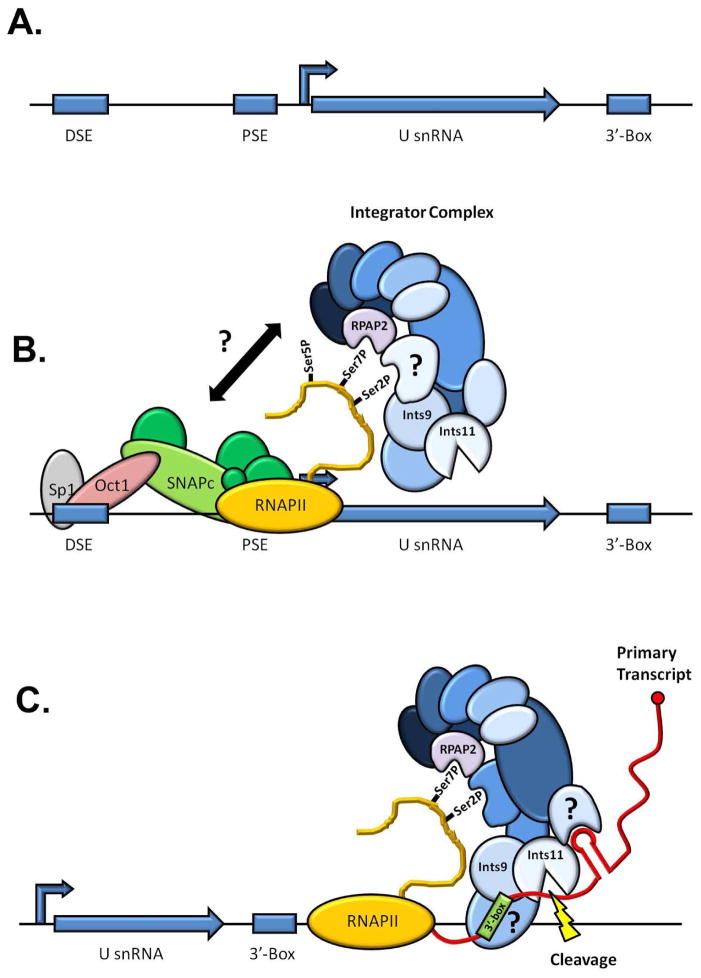
The precise molecular mechanism of the pre-U snRNA 3′ end processing is still unknown but a tentative mechanism has emerged (Figure 3) 37. Upon transcription initiation of the U snRNA genes, the CTD of the RNAPII is phosphorylated on both serine5 (ser5P) and serine7 (ser7P) of the heptamer repeats, most likely by CDK7 38, 39. The ser7P mark would then be recognized by the putative serine-5 phosphatase RPAP2. This interaction leads in turn to the recruitment of at least a subset of Integrator subunits that includes IntS4 and IntS5. Later during the transcription cycle, the phosphorylation of serine2 (ser2P) by the CDK9 kinase of the pTEFb complex and the dephosphorylation of ser5P by RPAP2 would enable the binding of the remaining Integrator subunits (including IntS11) culminating in the 3′ end cleavage of the pre-U snRNA.
Figure 3. Model for metazoan U snRNA 3′ end processing.
A. Organization of a typical U snRNA gene. The distal element (DSE) and the proximal element (PSE) are located approximately 200 and 50 bp upstream of the start site respectively. The 3′ box is located 9-19 bp downstream of the mature end of the U snRNA. B. Transcription initiation of U snRNA genes. The CTD of the polymerase is phosphorylated by CDK7 (ser5P and ser7P) and CDK9 (ser2P). RPAP2 binds specifically the ser7P mark. The Integrator complex is in turn recruited to the CTD through its interaction with RPAP2 and ser2P. The exact mechanism driving Integrator complex specificity for U snRNA genes is unknown, as is the Integrator subunit responsible for binding ser2P. C. U snRNA 3′-end recognition and cleavage. After transcription of the 3′ box, the Integrator complex recognizes the sequence of the 3′ box in the nascent transcript and most probably the terminal stem-loop of the U snRNA, triggering the cleavage by Ints11 of the pre-U snRNA between these two elements. The identity of the proteins contacting the nascent U snRNA is currently unknown.
This model posited above is attractive because it explains the long known mechanism of transcriptional coupling between the U snRNA promoter and the 3′ end processing. It also reaffirms the crucial role of the CDK9 activity 40 and emphasizes the importance of the double ser2P/ser7P CTD phosphorylation mark 41, 42. Yet it leaves some questions unanswered. First, neither the ser7P mark nor the presence of RPAP2 are specific to U snRNA genes 37, 43, raising the issue of the specificity of the Integrator recruitment to the U snRNA genes. Second, RPAP2 was initially identified as part of a larger RNAPII-associated complex 27, 44, and recent structural and biochemical evidence demonstrated that recombinant RPAP2 lacked ser5P phosphatase activity in vitro 45. Finally, it was recently reported that RPAP2 is dispensable for U snRNA processing, at least in fly 26. Altogether, these data indicate that RPAP2 cannot be the only determinant for Integrator recruitment and might act in conjunction with other factors yet to be characterized. In that line of thought, an RNAi screen in fly recently identified CycC/CDK8, a cyclin-dependent kinase pair normally present in the kinase module of the Mediator complex 46, as necessary for U snRNA processing 26. The finding that another kinase besides CDK7 and CDK9 plays a role in U snRNA processing suggests that all the protagonists involved in U snRNA processing have not yet been accounted for and that other layers of regulation might be at play.
To explain U snRNA promoter specificity, one hypothesis is that the Integrator complex may recognize a specific DNA sequence in the U snRNA promoter, even though no identified Integrator subunit has homology with a known DNA binding motif 31. As an alternative, Integrator could interact with a U snRNA specific transcription factor like the transcription activator SNAPc. Even though this factor was never detected in association with Integrator 25, 27, 44 it cannot be ruled out that this interaction might be transient and context dependent, thereby escaping its detection by conventional biochemical approaches.
Another open question is the mechanism of the 3′-end cleavage site recognition. No RNA-protein interactions have been demonstrated so far between the pre-U snRNA and the Integrator complex. The 3′ box, essential for 3′ end processing, is a tentative candidate to drive this interaction. Yet, this sequence is highly tolerant to mutations 47, suggesting that other determinants could be participating in the cleavage site recognition to form a bipartite motif that stabilizes the RNA/Integrator interaction. Such an element could be the terminal 3′ stem-loop that is present in all U snRNAs. This structure was proven to be important for U snRNA processing in some models 36, but dispensable in others 18 reaffirming that the recognition of the cleavage site is probably not dependent on a single motif but likely depends on the presence of several elements cooperating for the interaction between the Integrator complex and the pre-U snRNA.
After cleavage, the mechanism of transcription termination is unclear and might depend on the context of each particular U snRNA gene. For example, U2 transcription extends up to a kilobase beyond the 3′ box while U1 transcription stops shortly after cleavage 48. In the case of U2, transcription termination involves the boundary protein CTCF and the negative elongation factor NELF 49, while in vivo footprinting has revealed that an unidentified protein and potential terminator is bound immediately downstream from the 3′ box of the U1 snRNA gene 48. There have been no reports demonstrating that Xrn2 is involved in snRNA termination suggesting that either unique factors carry out this process or that IntS11 may function as an exonuclease similar to what has been shown for CPSF7350.
3′ end formation of snRNA in Saccharomyces cerevisiae
Rnt1 and exonucleolytic trimming
The U snRNA gene structure and cis requirements of Saccharomyces cerevisiae (hereafter referred to as yeast) for 3′ end formation differ significantly from their metazoan counterparts. There is no identifiable 3′ box downstream of the cleavage site and processing is not contingent on the type of RNAPII promoter that initiates transcription. These criteria are consistent with the lack of Integrator subunits in yeast and suggest that general factors are utilized to form the snRNA 3′ end. The two characterized types of cis-acting elements that direct 3′ end formation of yeast snRNAs are a stem loop (SL) sequence located downstream of the 3′ end of the mature product and a termination element located even further downstream. The SL is a target for the Rnt1 nuclease and the termination element utilizes the Nrd1/Nab3/Sen1 complex in the case of U1, U4, and U5 or possibly the CPF complex in the case of the U2 snRNA. These redundant pathways (SL and termination element) are likely in place to ensure accumulation of mature snRNA but also, given the more compact size of the yeast genome relative to humans, to prevent read through into neighboring genes.
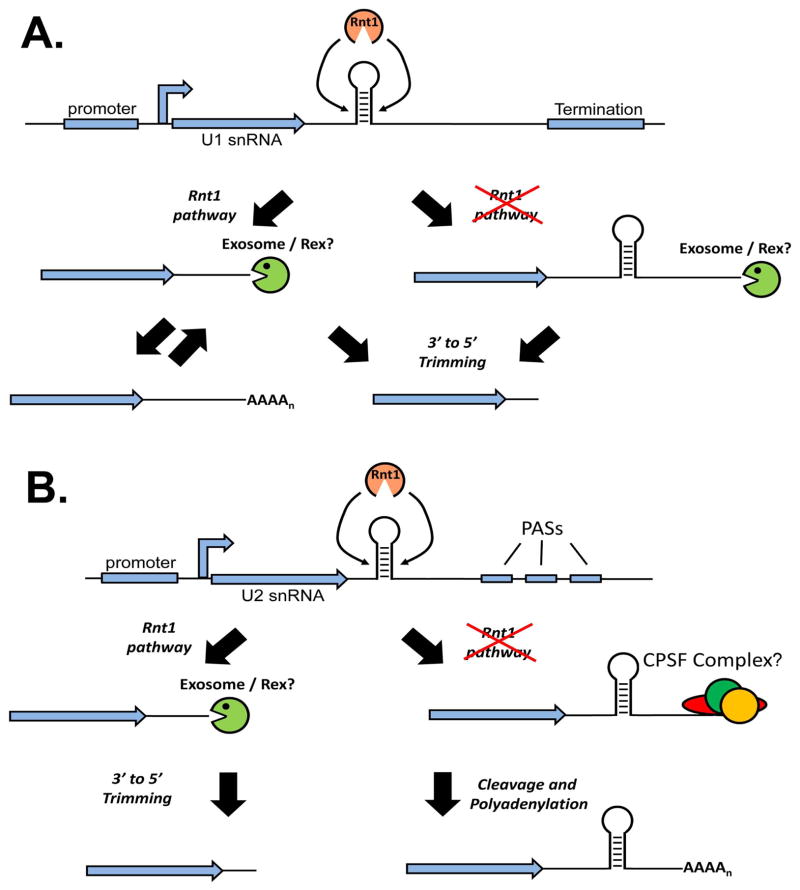
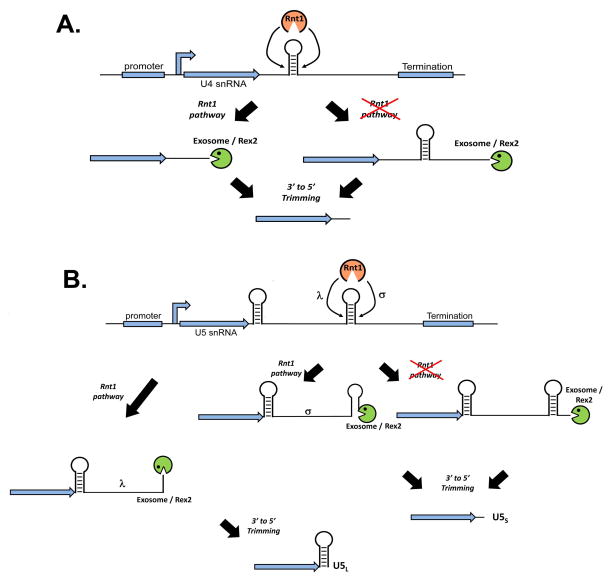
The initial generation of an rnt1 temperature-sensitive allele implicated this RNase III enzyme as essential for pre-rRNA processing 51. Upon further analysis, stem loop target sites within the 5′ and 3′ external transcribed spacers were identified as a specific cleavage site for Rnt1. Given that all four yeast U snRNAs have similar stem loops present downstream of their mature 3′ ends and have been shown to be substrates in vitro for recombinant Rnt1 enzyme, it is evident that Rnt1 is involved in the cleavage of precursor snRNA52–54. These Rnt1 cleavage sites, however, are not present at the ultimate 3′ end of the mature U snRNA demonstrating that additional processing is required.
In the case of U1 snRNA, the Rnt1-dependent cleavage product is processed further through the activity of the exosome 53 or possibly through the activity of alternative exonucleases called Rex (RNA exonuclease) proteins 55 (Figure 4A). Moreover, the Rnt1 U1 snRNA cleavage intermediate may cycle between a polyadenylated and non-polyadenylated form, although the dynamics of this putative toggling have not been examined56. In rnt1-Δ strains, elongated U1 snRNA transcripts are detectable, likely created through an alternative termination pathway (described in more detail below). These intermediates may be processed by the exosome given that their abundance is increased in rnt1, exosome double mutants 53. While the yeast U2 snRNA may use a similar Rnt1-dependent pathway that involves subsequent exonucleolytic trimming, it also utilizes a slightly different Rnt1-independent pathway that involves the use of polyadenylation sequences (PASs) located downstream of the stem loop cleavage site (Figure 4B) 52. Remarkably, yeast whose endogenous U2 snRNA gene is replaced with a U2 gene lacking the Rnt1 stem loop cleavage site possess exclusively lengthened and polyadenylated forms of U2 snRNA and are without any detectable phenotype. This demonstrates that poly(A)+ U2 snRNA is capable of incorporation into a snRNP and functional in catalysing the splicing reaction. The mechanism U2 snRNA poly(A) tails are added remains to be understood as the use of a PAS suggests involvement of the CPF machinery, however, other reports have not detected any U2 snRNA misprocessing phenotype when CPF proteins are mutated57.
Figure 4. Schematic of Saccharomyces cerevisiae U1 and U2 snRNA 3′ end formation pathways.
A. U1 snRNA utilizes both Rnt1-dependent and Rnt-independent mechanisms to achieve 3′ end formation. The RNAseIII, Rnt1, cleaves at a stem loop region generating a substrate for 3′ to 5′ exonucleases giving rise to the mature product, which can also be generated through termination followed by exonucleolytic trimming. A polyadenylated intermediate may also be formed that may cycle back to an exonuclease substrate. B. U2 snRNA uses both a Rnt1-dependent and Rnt1-independent mechanism to generate its 3′ end. The Rnt1-dependent is similar to the U1 snRNA 3′ end pathway but the Rnt1-independent utilizes Polyadenylation Sequences (PASs) located downstream of the Rnt1 cleavage site. This generates a cleavage and polyadenylation substrate and ultimately poly(A)+ U2 snRNA.
The 3′ end formation of the U4 snRNA utilizes redundant pathways related to U1 but distinct from U2 snRNA as no active PASs have been identified downstream of the stem loop (Figure 5A) 53, 58. Extended forms of U4 snRNA are present in yeast harbouring Rex2 deletions or temperature sensitive mutations in exosome subunits suggesting that these two factors are required for biogenesis of the mature U4 snRNA 55. Moreover, larger U4 snRNA intermediates are observed in exosome or Rex2 mutants that are also rnt1 null, demonstrating that the products of transcriptional termination are also subject to exonucleolytic trimming.
Figure 5. Schematic of Saccharomyces cerevisiae U4 and U5 snRNA 3′ end formation pathways.
A. U4 snRNA utilizes both Rnt1-dependent and independent pathways to generate its 3′ end. The Rnt1-dependent is related to U1 snRNA where the Rnt1 cleavage product acts as a substrate for the exosome and/or Rex2. The Rnt1-independent pathway uses a termination site followed by exosome and/or Rex2 trimming. B. The U5 snRNA utilizes the most complex Rnt1-dependent and independent pathways. Cleavage by Rnt1 at two alternative sites (L or S) leads to the exosome/Rex2-mediated trimming to either the U5L or U5S products. In the Rnt1-independent pathway, a downstream termination element is used and the long U5 precursor is processed to the U5S form.
The most complex 3′ end snRNA processing in yeast involves the biosynthesis of the U5 snRNA (Figure 5B). Two mature forms of U5 snRNA are detectable in yeast, the U5 Long (U5L) and U5 Short (U5S), which differ in length by ~33 nucleotides at the 3′ end 54. These two mature U5 isoforms are generated from the initial production of two distinct Rnt1 intermediate cleavage products termed U5σ and U5λ. Cleavage at the U5λ site is followed by subsequent exosome and/or Rex2-mediated trimming to form the U5L product while Rnt1 cleavage at the U5σ site is trimmed similarly to form the U5S product 53, 55. In the absence of Rnt1, transcription termination products are then trimmed by the exosome and/or Rex2 to form the U5S product. Both the U5S and U5L appear functionally equivalent in their capacity to facilitate splicing.
The Nrd1/Sen1/Nab3 complex
An alternative pathway to generate yeast snRNA 3′ ends that may act either redundantly or in parallel utilizes the Nrd1 termination complex, which was discovered through careful yet serendipitous experimentation (reviewed recently in59). In their continuing efforts to study U6 snRNA function, Steinmetz and Brow placed the U6 snRNA gene within the intron of a well-characterized splicing reporter comprised of an Actin-Cup1 fusion whose proper splicing results in copper resistance 60. Unexpectedly, positioning the U6 snRNA gene in the antisense orientation led to the initial discovery of a self-autonomous element capable of downregulating this reporter RNA leading to copper sensitivity 60. The downregulation was later proven to be due to premature termination of RNAPII transcription. Spontaneously arising suppressor mutations that caused transcriptional read through of the termination sequence were found leading to the ultimate identification of two genes whose expression is required for the function of this element: an uncharacterized gene that was termed Nrd1 (for nuclear pre-mRNA down-regulation) and the previously identified Sen1 RNA helicase 61.
The Nrd1 gene encodes a 575 amino acid nuclear protein in yeast that contains an N-terminal CTD-interacting domain (CID) as well as a C-terminal RNA recognition motif (RRM) 62. Both of these domains are required for the antisense U6 snRNA function within the CUP1 reporter suggesting that recruitment via RNAPII and specific interaction with the nascent transcript are required events to promote 3′ end formation 62. Subsequently, it was determined that the function of Nrd1 is not limited to the U6 antisense terminator element but rather is utilized by cells naturally and in a genome-wide scale 63. Expression profiling using poly(A)+ RNA revealed that ORFs downstream of many snoRNA genes were upregulated after Nrd1 depletion due to termination failure and transcriptional readthrough, which produces chimeric transcripts containing the snoRNA and mRNA fused together 63. Importantly, a Nrd1 terminator was identified downstream of the U4 snRNA Rnt1 cleavage site connecting this complex to snRNA biogenesis. Subsequent studies have shown the requirement of Nrd1 in the termination of several short RNAPII transcripts, most notably cryptic unstable transcripts (CUTs) 64, 65. Consistent with these observations, structural analysis of the Nrd1 CID-RNAPII CTD interaction has demonstrated a strong preference for serine 5 phosphorylation, which is most commonly found proximal to promoters66.
Nrd1 exists as a heterodimer with the nuclear polyadenylated RNA-binding 3 (Nab3) protein, which also contains an RRM thereby increasing the RNA binding capacity and specificity of the complex 67. Several studies indicate a sequence preference for Nrd1 of GUAR and for Nab3 of UCUU, both of which are enriched in terminator elements found downstream of snRNA 68–70. The emerging model of Nrd1-dependent termination downstream of snRNA integrates the numerous properties of Nrd1 that may be differentially required depending on the gene context: CTD interaction via Nrd1, heterodimeric association with Nab3, and individual RNA recognition through the Nrd1 and Nab3 RRMs (reviewed in more detail in 71. This combination of factors likely brings about high specificity to the complex. In addition Nrd1 and Nab3, this termination event likely involves members of the APT complex (Associated with Pta1), which includes: Pta1, Ref2, Pti1, Swd2, Glc7, Ssu72, and Syc172. In particular, the APT member Ssu72 was also identified genetically as required for U4 snRNA 3′ end formation and essentially phenocopies the transcriptional readthrough observed in Nrd1 mutants73. Once these recognition events transpire, the Sen1 helicase is thought to destabilize the RNA polymerase complex much in the same way that the Rho helicase stimulates termination in prokaryotes (Figure 6). It isn’t yet understood how the Nrd1 complex collaborates with members of the APT complex or how the Sen1 helicase activity leads to the dissociation of polymerase. What is likely, however, is that the longer snRNA precursors generated through Nrd1-dependent termination are trimmed by the exonucleases to form the mature snRNA. Indeed, recent cross-linking studies demonstrate localization overlap between Nrd1, Nab3, and the TRAMP complex74, supporting a model whereby termination is coupled to 3′ to 5′ exonucleolytic trimming.
Figure 6. Mechanism of Nrd1/Nab3/Sen1-dependent termination.
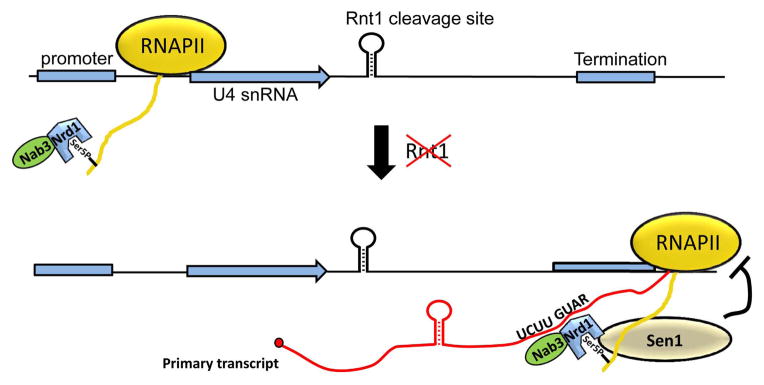
The Nrd1/Nab3 hetereodimer recognizes the CTD of Rpb1 that is phosphorylated at serine 5 within the RNAPII complex. Nrd1 contains the CTD interaction domain that recognizes this specific mark. In the absence of Rnt1, Nrd1 and Nab3 binding sites are exposed in the nascent snRNA transcript, which then facilitates termination by the Sen1 helicase. The termination product produces a free 3′ OH that is used as a substrate for the exosome or other 3′ to 5′ exonucleases.
TER1 3′ end formation
Ter1 characteristics
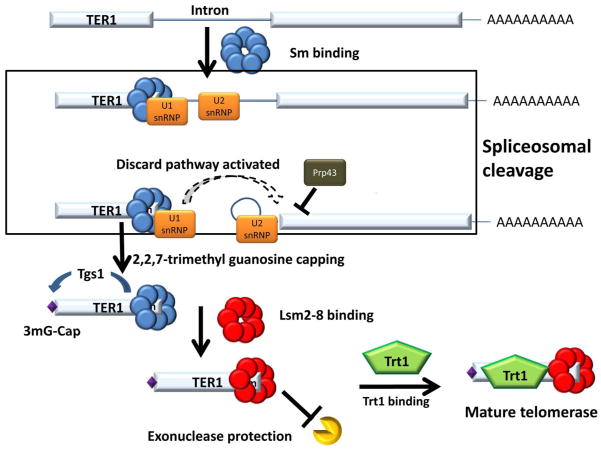
Telomerase, the ribonucleoprotein enzyme responsible for the maintenance of the chromosome ends in eukaryotic cells, contains a highly conserved protein component and a more evolutionarily flexible RNA component 75. In Schizosaccharomyces pombem, the RNA component is encoded by the TER1 gene. The approximately 1400 nucleotide long S. pombe TER1 primary transcript is formed initially through cleavage and polyadenylation but undergoes subsequent RNA processing steps resulting in the production of a mature 1200 nucleotide long non-polyadenylated form. Similar to snRNAs, the mature TER1 transcript contains an Sm binding motif as well as a 2,2,7-trimethylguanosine (TMG) cap 76, 77. Initial studies of the longer polyadenylated form of TER1 revealed the presence of 5′ splice site, branch point, and 3′ splice site consensus sequences located near its 3′ end. Sequencing revealed that the corresponding 56 nucleotide intron was removed, indicating that splicing does occur at these sites 78. Furthermore, the 5′ splice site, which overlaps with the Sm binding site, is located near the 3′ end of the mature TER1 in S. pombe (Figure 7). While the Sm site is conserved in Saccharomyces cerevisiae telomerase RNA (TCL1), it only plays a role in stabilizing the transcript and no evidence for a nearby splice site exists 79, 80.
Figure 7. Schizosaccharomyces pombe telomerase RNA slicing.
The primary TER1 transcript is produced by cleavage and polyadenylation and contains an intron near its 3′ end. Association of the Sm complex near the 5′ splice site promotes the first step of splicing, however, the second step of splicing does not occur. The high affinity of the U2snRNP for the bp leads to a splicing discard pathway through the involvement of Prp43. The association of the Sm complex stimulates the hypermethylation of the TER1 5′ cap by Tgs1 and the complex is ultimately replaced with the Lsm complex, which both protects the 3′ terminus of the TER1 transcript and facilitates recruitment of the Trt1 protein component of the telomerase RNP.
The Spliceosome as a 3′ end formation machine
The existence of splicing at the 3′ end of the TER1 transcript was unexpected; moreover, the exact role of the spliceosome in the biosynthesis of the transcript, once elucidated, was unprecedented. Mutations of the splice sites within the TER1 precursor RNA inhibit its splicing but only alterations of the 5′ splice site or branch point reduce the amount of mature TER1 product 78. Importantly, compensatory mutations to a mutant 5′ splice site in the U1 snRNA were able to restore TER1 3′ end processing, unequivocally implicating components of the spliceosome in transcript cleavage. Taken together, these results indicate a role only for the first step of the splicing reaction while there is a blockade in the second splicing step of exon ligation. Consistent with this model, replacement of the TER1 intron with a heterologous intron results in a proper cleavage only if the canonical 3′ splice site is mutated 78. Reciprocally, mutations in the TER1 intron which improved or facilitated the completion of splicing fail to generate a mature TER1 transcript. The resulting model of TER1 3′ end formation entails the utilization of the endonucleolytic activity of the spliceosome to execute the first step of splicing and subsequent release of the “5′ exon”-that becomes the mature TER1. This new function, in stark contrast to the typical function of the spliceosome in the removal of introns, was dubbed “spliceosomal cleavage” (Figure 7). The spliceosomal cleavage of the 3′ end of TER1 raises several questions. The most obvious pertains to how the TER1 intron is able to direct the first step of splicing yet inhibit its completion. Second, extending the former, regards what spliceosomal subunits are involved in the 3′ end formation of TER1. Finally, once the TER1 3′ end is formed, how is it stabilized?
Recent investigations have determined that the TER1 intron contains all of the elements necessary for spliceosomal cleavage and that the native promoter is not required81. Closer inspection of cis-acting splicing elements revealed a modest polypyrimidine tract, suboptimal positional of the branch point (too far upstream), and higher than typical complementarity between the branch point and U2 snRNA. Coexistence of these intronic features are statistically under-represented in the S. pombe genome and provide clues as to why this intron has unique properties81. Normal functioning introns similar to the TER1 intron can be converted into a spliceosomal cleavage-directing intron through increasing the number of base-pairs formed between the branch point and U2snRNA. This leads to a model suggesting that the reduced kinetics of U2 snRNA removal/remodelling within the spliceosome formed on the TER1 intron triggers a discard pathway preventing exon ligation. In support of this model, others have shown the involvement of the Prp43 splicing factor in the spliceosome discard pathway82, 83 and, indeed, temperature sensitive mutations in Prp43 disrupt TER1 3′ end formation81. Finally, the Sm binding site that binds to the Sm complex is not only required to facilitate spliceosomal cleavage but also is important for TER1 stability. The Sm ring is also thought to recruit the Tgs1 methylase, leading to the formation of the TMG cap 84. Once TER1 cap is hypermethylated, a switch from the Sm complex to the Lsm2-8 complex ensues, which not only provides protection from 3′ to 5′ exonucleases but is also important to recruit the protein component of the telomerase RNP (Figure 7).
Altogether, the spliceosomal cleavage of the S. pombe TER1 transcript represents an alternative use of many cellular components in an innovative and unusual manner. It remains to be seen if the human telomerase RNA component utilizes a similar machinery to form its 3′ end but a lack of identifiable splice sites suggests an alternative mechanism may be involved.
NEAT transcripts
Genomic Structure and Function
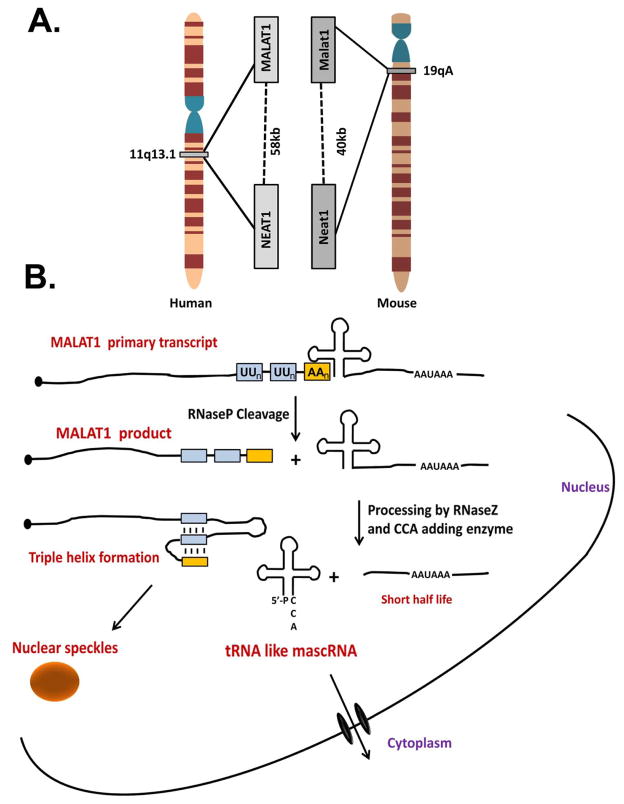
Two novel, RNAPII-transcribed, noncoding nuclear enriched abundant transcripts (NEAT): NEAT1 and NEAT2 (also known as MALAT1-Metastasis associated lung adenocarcinoma transcript-1) were discovered in a screen designed to identify nuclear retained polyadenylated RNA that may play a role in regulating gene expression 85. The gene loci of these two non-coding RNAs (ncRNAs) were found to be in close proximity to one another on human chromosome 11 and genomic synteny demonstrates that the mouse orthologs (Neat1 and Malat1) are organized in an analogous manner (Figure 8A). While NEAT1 is conserved in only small regions, MALAT1 is highly conserved throughout the length of the transcript between human and mouse. Recent studies revealed that both NEAT1 and MALAT1 are key players in nuclear architecture86–88. NEAT1 is associated with and forms an integral part of paraspeckles as RNAi-mediated depletion of NEAT1 results in their disintegration89, 90. Similar studies with MALAT1 demonstrate that it preferentially localizes to nuclear speckles91. Although their exact function within these nuclear organelles is not completely understood, there is a general consensus that they may behave as scaffolds for RNA binding proteins.
Figure 8. MALAT1 noncoding RNA processing.
A. Schematic of genomic organization of MALAT1 and NEAT1 in human and mouse. B. Schematic of the pathway of RNA processing events that lead to the generation of the MALAT1 mature transcript. The primary MALAT1 transcript contains a PAS as well as a cloverleaf structure, which is a substrate for RNAseP. The downstream product is further processed by RNAseZ and the CCA-adding enzyme preceding its export into the cytoplasm. The upstream MALAT1 transcript resulting from RNAseP digestion forms a triple helix at its 3′ end and is then localized to nuclear speckles.
3′ end processing of MALAT1 and NEAT1
Human and mouse MALAT1 full length transcripts have been reported to be ~8.7kb and ~7kb in length, respectively, and utilize canonical cleavage and polyadenylation signals directing the addition of a poly(A) tail 92, 93. However, careful inspection of these transcripts revealed that their most abundant forms are slightly shorter (~7kb vs. ~8.7kb and ~6.7kb vs ~7kb) and that there are no recognizable poly(A) sites directing cleavage in these regions. Rather, it was found that sequences immediately downstream of the abundant MALAT1 3′ end are predicted to adopt a cloverleaf structure similar to tRNA (Figure 8B). This sequence was subsequently proven to act as a substrate for RNaseP-mediated endonucleolytic processing. This cleavage event results in the simultaneous generation of two RNA species: the 3′ end of the mature MALAT1 nonpolyadenylated transcript and the 5′ end of its downstream cleavage product 94. Next, the 3′ end of the downstream product is recognized by the tRNA processing machinery (RNaseZ and the CCA-adding enzyme) generating a small tRNA-like RNA called the mascRNA (MALAT1 associated small cytoplasmic RNA) that is exported to the cytoplasm. The functional role of mascRNA is not yet clear; however, there is no evidence that it is charged with an amino acid or implicated in translation.
The NEAT1 locus has its own RNAPII promoter and encodes two transcripts that share the same 5′ end but have unique 3′ ends. The shorter isoform is a 3.7 kb (3.2 kb in mouse) unspliced and polyadenylated transcript termed NEAT1_1/MENε. The longer 23kb (~20kb in mouse) isoform, called NEAT1_2/MENβ, is also unspliced but is non-polyadenylated 85. Unlike the shorter MENε transcript, the MENβ 3′ end lacks canonical cleavage/polyadenylation signals indicating that another mechanism is required to generate the 3′ end of the MENβ transcript. Similar to MALAT1, a tRNA-like cloverleaf structure was reported to be present at the 3′ end of MENβ locus suggesting a possible RNaseP dependant 3′ end cleavage event. Subsequent analysis demonstrated that the MENβ ncRNA 3′ end is formed through a similar pathway as the MALAT1 ncRNA. However, the level of a “mascRNA-like” downstream cleavage product was poorly detected possibly due to an extremely short half-life 91.
Stability of NEAT transcripts through a 3′ triple helix
The presence of a poly(A) tail for RNAPII transcripts is an established feature that results in increased mRNA half-life and higher translation efficiency 95. Although not polyadenylated, the MALAT1 transcript is resistant to degradation and accumulates in the nucleus at very high levels 94. A recent report identified a mechanism to account for this unexpected stability 96. At the very 3′ terminus of the MALAT1 transcript, just upstream from the RNAseP processing site, two short U-rich regions interrupted by a conserved stem loop precede an encoded poly(A) rich tract (Figure 8B). Additionally, Brown et al demonstrated that Menβ also has enhanced stability through the likely formation of a triple helix97. While a structure has yet to be solved, extensive mutagenesis coupled with structural modelling strongly supports the prediction that these three runs of nearly identical nucleotides form of a stable triple helix. This structure likely provides protection from 3′ to 5′ exonucleases and functionally replaces a poly(A) tail to promote MALAT1 stability.
Conclusion
The diversity of the pathways in which non-mRNAs form their 3′ end is as compelling as it has been unexpected. The case studies described here exemplify the inherent fluidity of non-mRNA 3′ end formation research. Our understanding is still being advanced by a continually growing body of publications. While progress has been made, many questions are yet unanswered. In the case of metazoan U snRNAs, it is still unclear how the Integrator Complex is specifically recruited to the snRNA promoters and the details of CTD modification recognition are still unclear. In yeast snRNA 3′ end formation, it is still undetermined how the 3′ exonucleases recognize longer snRNA precursors. As for TER1 3′ formation, it remains to be seen if this mechanism is in place for other RNA transcripts. Also, whether or not triple helix formation is restricted to MALAT1 or represents a more general feature of noncoding RNAs is unknown. These are the types of questions that will undoubtedly be addressed in future work as they represent compelling questions in the field of non-mRNA 3′ end processing.
Acknowledgments
We thank Todd Albrecht and John Hagan for critically reading the manuscript. Work in the Wagner laboratory is supported from the following sources: National Institute of Health CA166274 (E.J.W) and MDA 202141 (E.J.W.).
Contributor Information
Natoya Peart, Department of Biochemistry and Molecular Biology, The University of Texas Medical School at Houston; The University of Texas Graduate School of Biomedical Sciences at Houston.
Anupama Sataluri, Department of Biochemistry and Molecular Biology, The University of Texas Medical School at Houston; The University of Texas Graduate School of Biomedical Sciences at Houston.
David Baillat, Email: David.Baillat@uth.tmc.edu, Department of Biochemistry and Molecular Biology, The University of Texas Medical School at Houston.
Eric J. Wagner, Email: Eric.J.Wagner@uth.tmc.edu, Department of Biochemistry and Molecular Biology, The University of Texas Medical School at Houston; The University of Texas Graduate School of Biomedical Sciences at Houston
References
- 1.Mandel C, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cellular and Molecular Life Sciences. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zong X, Tripathi V, Prasanth KV. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 5.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 6.Jawdekar GW, Henry RW. Transcriptional regulation of human small nuclear RNA genes. Biochim Biophys Acta. 2008;1779:295–305. doi: 10.1016/j.bbagrm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobo SM, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 8.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. Rna. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trippe R, Sandrock B, Benecke BJ. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shchepachev V, Wischnewski H, Missiaglia E, Soneson C, Azzalin CM. Mpn1, mutated in poikiloderma with neutropenia protein 1, is a conserved 3′-to-5′ RNA exonuclease processing U6 small nuclear RNA. Cell Rep. 2012;2:855–865. doi: 10.1016/j.celrep.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 12.Madore SJ, Wieben ED, Kunkel GR, Pederson T. Precursors of U4 small nuclear RNA. J Cell Biol. 1984;99:1140–1144. doi: 10.1083/jcb.99.3.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madore SJ, Wieben ED, Pederson T. Eukaryotic small ribonucleoproteins. Anti-La human autoantibodies react with U1 RNA-protein complexes. J Biol Chem. 1984;259:1929–1933. [PubMed] [Google Scholar]
- 14.Wieben ED, Nenninger JM, Pederson T. Ribonucleoprotein organization of eukaryotic RNA. XXXII. U2 small nuclear RNA precursors and their accurate 3′ processing in vitro as ribonucleoprotein particles. J Mol Biol. 1985;183:69–78. doi: 10.1016/0022-2836(85)90281-5. [DOI] [PubMed] [Google Scholar]
- 15.Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 16.Ciliberto G, Dathan N, Frank R, Philipson L, Mattaj IW. Formation of the 3′ end on U snRNAs requires at least three sequence elements. Embo J. 1986;5:2931–2937. doi: 10.1002/j.1460-2075.1986.tb04589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vegvar HE, Lund E, Dahlberg JE. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986;47:259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. Embo J. 1985;4:1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- 20.Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. Embo J. 2003;22:925–934. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uguen P, Murphy S. The 3′ ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. Embo J. 2003;22:4544–4554. doi: 10.1093/emboj/cdg430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartkowiak B, Mackellar AL, Greenleaf AL. Updating the CTD Story: From Tail to Epic. Genet Res Int. 2011;2011:623718. doi: 10.4061/2011/623718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Ezzeddine N, Waltenspiel B, Albrecht TR, Warren WD, Marzluff WF, Wagner EJ. An RNAi screen identifies additional members of the Drosophila Integrator complex and a requirement for cyclin C/Cdk8 in snRNA 3′-end formation. RNA. 2012;18:2148–2156. doi: 10.1261/rna.035725.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O’Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell AG, Charette JM, Spencer DF, Gray MW. An early evolutionary origin for the minor spliceosome. Nature. 2006;443:863–866. doi: 10.1038/nature05228. [DOI] [PubMed] [Google Scholar]
- 30.Cazalla D, Xie M, Steitz JA. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol Cell. 2011;43:982–992. doi: 10.1016/j.molcel.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Wagner EJ. snRNA 3′ end formation: the dawn of the Integrator complex. Biochem Soc Trans. 2010;38:1082–1087. doi: 10.1042/BST0381082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albrecht TR, Wagner EJ. snRNA 3′ end formation requires heterodimeric association of integrator subunits. Mol Cell Biol. 2012;32:1112–1123. doi: 10.1128/MCB.06511-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- 35.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, Warren WD, Marzluff WF, Wagner EJ. A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3′-end formation. Mol Cell Biol. 2011;31:328–341. doi: 10.1128/MCB.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol Cell. 2012;45:111–122. doi: 10.1016/j.molcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. Embo J. 2005;24:4154–4165. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem. 2010;285:20564–20569. doi: 10.1074/jbc.M110.132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 44.Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang K, Manley JL, Tong L. The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat Commun. 2012;3:946. doi: 10.1038/ncomms1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ach RA, Weiner AM. The highly conserved U small nuclear RNA 3′-end formation signal is quite tolerant to mutation. Mol Cell Biol. 1987;7:2070–2079. doi: 10.1128/mcb.7.6.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuello P, Boyd DC, Dye MJ, Proudfoot NJ, Murphy S. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. Embo J. 1999;18:2867–2877. doi: 10.1093/emboj/18.10.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egloff S, Al-Rawaf H, O’Reilly D, Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol Cell Biol. 2009;29:4002–4013. doi: 10.1128/MCB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang XC, Sullivan KD, Marzluff WF, Dominski Z. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol Cell Biol. 2009;29:31–42. doi: 10.1128/MCB.00776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elela SA, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 52.Abou Elela S, Ares M., Jr Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. Embo J. 1998;17:3738–3746. doi: 10.1093/emboj/17.13.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. Embo J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chanfreau G, Elela SA, Ares M, Jr, Guthrie C. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 1997;11:2741–2751. doi: 10.1101/gad.11.20.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Hoof A, Lennertz P, Parker R. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5. 8S, U4, U5, RNase MRP and RNase P RNAs in yeast. Embo J. 2000;19:1357–1365. doi: 10.1093/emboj/19.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seipelt RL, Zheng B, Asuru A, Rymond BC. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 1999;27:587–595. doi: 10.1093/nar/27.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morlando M, Greco P, Dichtl B, Fatica A, Keller W, Bozzoni I. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol Cell Biol. 2002;22:1379–1389. doi: 10.1128/mcb.22.5.1379-1389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mischo HE, Proudfoot NJ. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta. 2013;1829:174–185. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeMarini DJ, Winey M, Ursic D, Webb F, Culbertson MR. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2154–2164. doi: 10.1128/mcb.12.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinmetz EJ, Brow DA. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci U S A. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 64.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 65.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 66.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154:557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hobor F, Pergoli R, Kubicek K, Hrossova D, Bacikova V, Zimmermann M, Pasulka J, Hofr C, Vanacova S, Stefl R. Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein. J Biol Chem. 2011;286:3645–3657. doi: 10.1074/jbc.M110.158774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porrua O, Hobor F, Boulay J, Kubicek K, D’Aubenton-Carafa Y, Gudipati RK, Stefl R, Libri D. In vivo SELEX reveals novel sequence and structural determinants of Nrd1-Nab3-Sen1-dependent transcription termination. Embo J. 2012;31:3935–3948. doi: 10.1038/emboj.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J Biol Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 73.Steinmetz EJ, Brow DA. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol Cell Biol. 2003;23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. Embo J. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 76.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 77.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- 79.Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P. A flexible template boundary element in the RNA subunit of fission yeast telomerase. J Biol Chem. 2008;283:24224–24233. doi: 10.1074/jbc.M802043200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 81.Kannan R, Hartnett S, Voelker RB, Berglund JA, Staley JP, Baumann P. Intronic sequence elements impede exon ligation and trigger a discard pathway that yields functional telomerase RNA in fission yeast. Genes & Development. 2013;27:627–638. doi: 10.1101/gad.212738.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell. 2010;39:385–395. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mayas RM, Maita H, Semlow DR, Staley JP. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proceedings of the National Academy of Sciences. 2010;107:10020–10025. doi: 10.1073/pnas.0906022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang W, Kannan R, Blanchette M, Baumann P. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature. 2012;484:260–264. doi: 10.1038/nature10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 93.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 94.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ford LP, Bagga PS, Wilusz J. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol Cell Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilusz JE, Jnbaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]