Abstract
Mitochondrial complex I (NADH dehydrogenase) is a major contributor to neuronal energetics, and mutations in complex I lead to vision loss. Functional, neuroanatomical and transcriptional consequences of complex I deficiency were investigated in retinas of the Ndufs4 knockout mouse. Whole-eye ERGs and multielectrode arrays confirmed a major retinal ganglion cell functional loss at P32, and retinal ganglion cell loss at P42. RNAseq demonstrated a mild and then sharp increase in innate immune and inflammatory retinal transcripts at P22 and P33, respectively, which were confirmed with QRT-PCR. Intraperitoneal injection of the inflammogen lipopolysaccharide further reduced retinal ganglion cell function in Ndufs4 KO, supporting the connection between inflammatory activation and functional loss. Complex I deficiency in the retina clearly caused innate immune and inflammatory markers to increase coincident with loss of vision, and RGC functional loss. How complex I incites inflammation and functional loss is not clear, but could be the result of misfolded complex I generating a ‘non-self’ response, and induction of innate immune response transcripts was observed before functional loss at P22, including β-2 microglobulin and Cx3cr1, and during vision loss at P31 (B2m, Tlr 2, 3, 4, C1qa, Cx3cr1 and Fas). These data support the hypothesis that mitochondrial complex I dysfunction in the retina triggers an innate immune and inflammatory response that results in loss of retinal ganglion cell function and death, as in Leber's hereditary Optic Neuropathy and suggests novel therapeutic routes to counter mitochondrial defects that contribute to vision loss.
Introduction
Mitochondrial diseases are typically characterized by decreased energy production due to defects in the oxidative phosphorylation (OXPHOS) system. In humans, the most frequent deficiency is in complex I (1). Mitochondrial complex I activity is essential for neuronal homeostasis, and deficiency of complex I causes neurotoxicity. The most common symptoms linked to complex I deficiency include mental and motor retardation, blindness, heart rhythm disturbances, heart failure, exercise intolerance, and hypotonia—clinically presenting as Leigh's disease (1).
All mitochondrial complexes (except complex II) are under dual genetic control (mitochondrial and nuclear DNA), in which a mutation in either causes mitochondrial dysfunction (1). Mitochondrial complex I is by far the most complicated complex of the OXPHOS system. It comprises 45 subunits, 38 of which are encoded by nuclear DNA and 7 are encoded by mitochondrial DNA (2). Human genetic optic neuropathies such as, Leber's hereditary optic neuropathy has been linked to mtDNA point mutations at positions 11778/ND4, 3460/ND1 and 14484/ND6, which affect complex I and leads to a loss of retinal ganglion cells (RGCs) and degeneration of the optic nerve (3).
Other genetic mitochondrial defects that indirectly affect complex I is seen in autosomal dominant optic atrophy (DOA), which is caused by a mutation in the OPA1 gene that leads to a reduced rate of mitochondrial ATP synthesis at the level of complex I. The OPA1 gene also interacts with other mitochondrial proteins such as AIF. These interactions lead to apoptotic death of RGCs that further progresses to neurodegeneration and optic atrophy (4).
Rotenone, a known inhibitor of complex I, has been shown to induce neurotoxicity of rat brain areas through astroglial activation and apoptosis (5), and rotenone administered intravitreally causes RGC loss (6). In another study, subcutaneous injection of rotenone in rats caused a loss of photoreceptors in the outer retina and reduced synaptic connectivity between the remaining photoreceptors and their postsynaptic neurons (7).
These genetic and toxicity studies taken together clearly implicate the role of mitochondrial complex I in visual function. The precise mechanism for how complex I causes neuronal death is not known, however bioenergetic, excitotoxic and apoptotic mechanisms have been proposed (1,3). We have investigated the consequences of severe complex I deficiency in the retina, using the Ndufs4 KO mice originally developed as a model of Leigh's disease, an encephalomyelopathic disease resulting from complex I defects in the brain that results in infantile death. This mouse model presents with a loss of vision at postnatal 30 days (P30), relating complex I deficiency to visual function (8). However, the cellular and molecular underpinnings of this complex I dependent visual defect have not been identified.
Our goal for this study was to investigate the functional and neuroanatomical defects in retinas with complex I deficiency. A second goal of this study was to investigate the molecular differences in the retina between knockout and wild-type mice, to better understand how complex I deficiency could cause blindness. To accomplish this, we examined Ndufs4 mice before, during and after the onset of pathogenesis. We compared retinas of Ndufs4 KO mice and wild-type mice, and showed that the period of visual impairment is accompanied by a prominent innate immune and inflammatory response in the Ndufs4 KO retina that can be further enhanced by lipopolysaccharide (LPS) injection. Thus, complex I deficiency in the retina leads to increase in inflammatory signals and retinal ganglion cell functional decrements and death in the Ndufs4 KO mouse.
Results
Complex I defects result in decreased retinal ganglion cell function in Ndufs4 KO mice
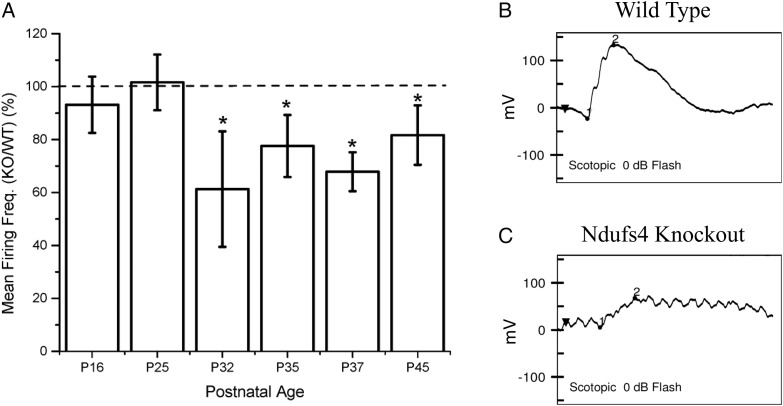
Microelectrode arrays (MEA) specifically measures the physiological activity of RGCs in living tissue. MEAs were conducted on Ndufs4 KO and wild-type littermate controls; at P16 (P = 0.3408) and P25 (P = 0.1750), no significant functional defects were observed in RGCs when comparing Ndufs4 KO to wild-type (Fig. 1A). In contrast, at P32 (P = 0.0325), P35 (P = 0.0445), P37 (P = 0.0009) and P45 (P = 0.0256), there was a significant decrease in RGC firing frequency when comparing Ndufs4 KO to wild-type. Retinal mitochondrial activity was measured by Seahorse; Ndufs4 mice had a significant deficit in glutamate/malate-driven mitochondrial oxygen consumption (data not shown). These data indicate that complex I defects in the KO mice specifically decrease RGC function. In order to measure overall retinal function in vivo, we also performed ERGs on P34 Ndufs4 KO mice. ERG testing of three Ndufs4 KO mice and three wild-type mice showed that all KO mice had a decreased and sometimes absent b-wave at the brightest light stimulus (0 dB) indicating a markedly reduced retinal response to light stimulus (Fig. 1B and C).
Figure 1.
(A) Decreased firing of RGC neurons from Ndufs4 KO mice begins at P32. MEA recording of Ndufs4 KO mice at P16, P25, P32, P35, P37, P45. Representative ERG of (B) wild-type control and (C) Ndufs4 KO mouse at P34. Statistical significance determined Kruskal–Wallis ANOVA test. *P < 0.05.
Complex I deficiency results in cell loss in the RGC layer after 30 days
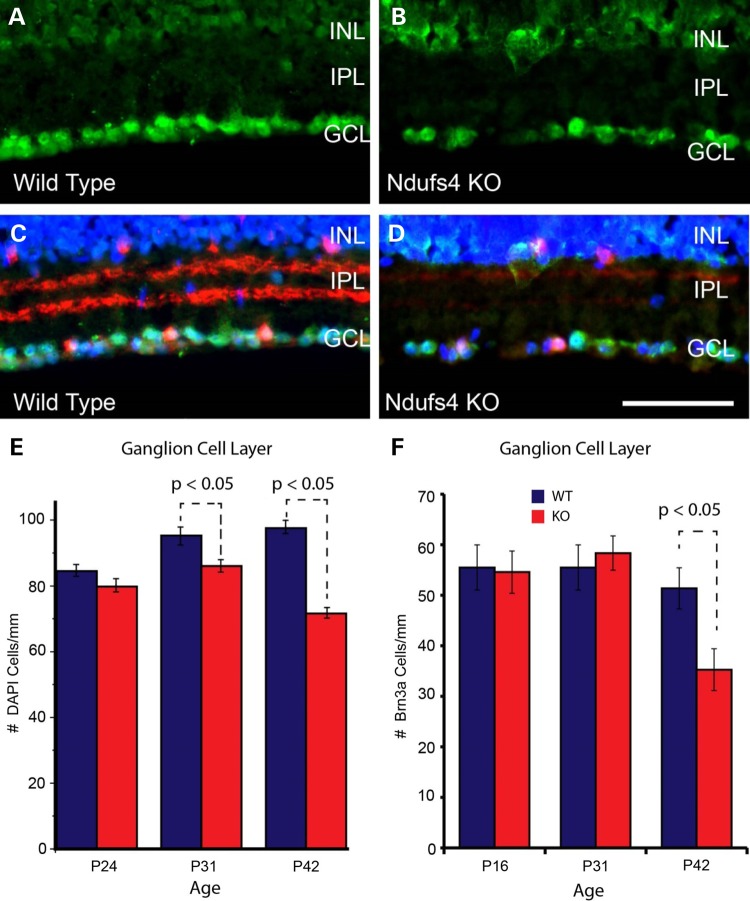
To determine if loss of Ndufs4 affected RGC number, we performed counts on DAPI-labeled cells within the RGC layer (Fig. 2). Compared with wild-type littermates, Ndufs4 KO mice had significantly reduced numbers of cells in the RGC layer at P31 and P42 (Fig. 2E). Decrease in RGC number was greater at P42 compared with P31. No difference in the number of DAPI-labeled cells was observed at P24 suggesting there was a progressive decline over time (Fig. 2E). Because DAPI labels all cells, we performed a similar analysis on cells immunolabeled for the POU domain, class 4, transcription factor 1 (Pou4f1 aka Brn3a), which is enriched in a subset of RGCs (9,10). There was a significant reduction in the number of Brn3a-positive cells at P42 in Ndufs4 KO compared with wild-type littermates (Fig. 2F). In contrast to the loss of DAPI-positive cells in the RGC layer, no differences in Brn3a cell number was observed at P16 or P31. Thus, in addition to the early loss of RGC function shown above at P32, there is a clear loss of RGCs in this complex I deficient mouse model at P42, consistent with pathological changes seen in multiple complex I deficient diseases that involve vision loss.
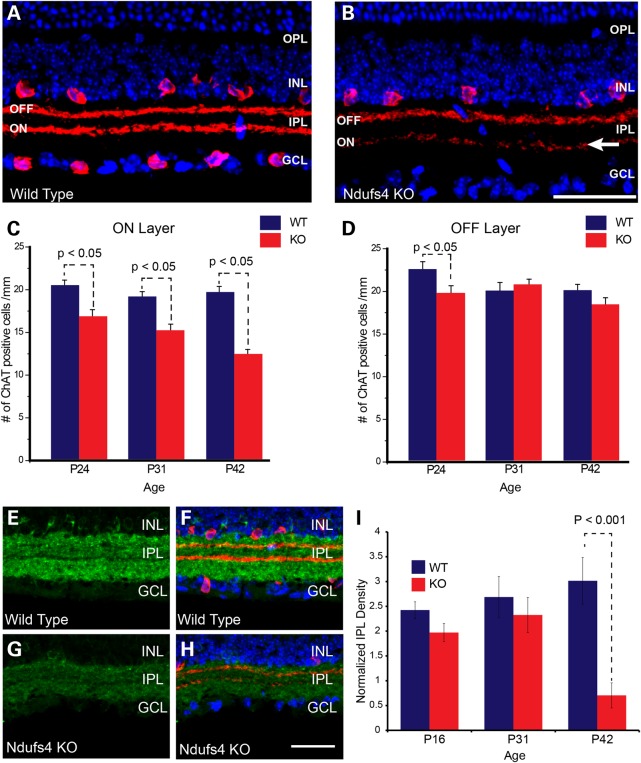
Figure 2.
Cell loss in the RGC layer of Ndufs4 mice measured by DAPI staining and Brn3a immunofluorescent labeling. (A–D) Representative images of immunofluorescent labeling for Brn3a (green; marker for retinal ganglion cells) in transverse retinal sections from P42 wild-type (A and C) and littermate Ndufs4 KO (B and D) animals. (C) and (D) show ChAT (red; marker for starburst amacrine cells) immunolabeling with DAPI cell staining (blue) overlaid on the images from (A) and (B), respectively. (E) Bar graph showing counts of DAPI positive, ChAT negative cells in the GCL of wild-type and Ndufs4 KO mice at P24, P31 and P42.(F) Bar graph showing counts of Brn3a positive, ChAT negative cells in the GCL of P16, P31 and P42 wild-type and Ndufs4 KO mice. Statistical comparisons were performed using 2-tailed Student’s t-test. Scale bar = 100 mm. Statistical significance determined by Student's t-test.
RNAseq indicates an increased inflammatory response
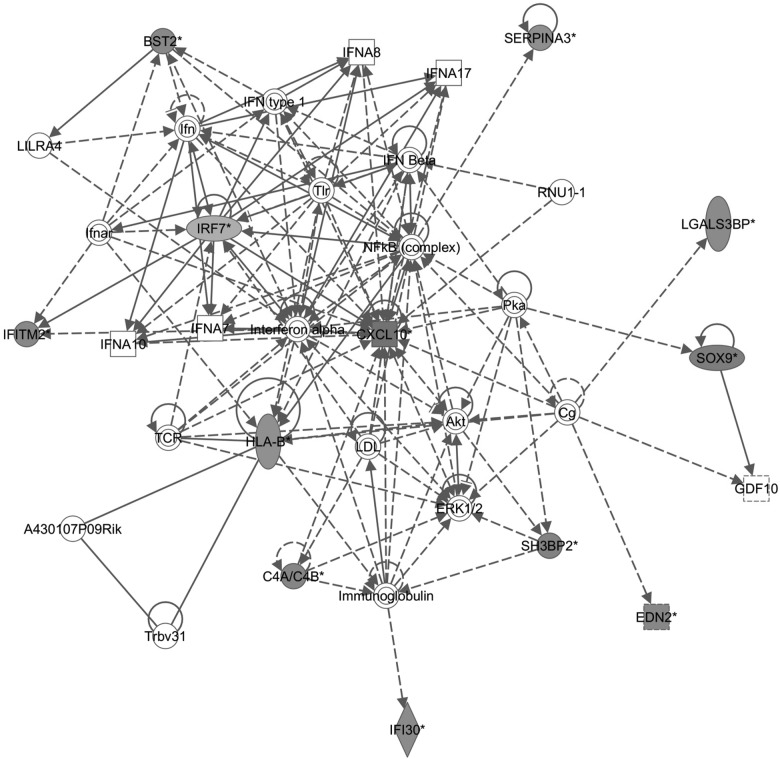
To gain insight into pathomechanism of RGC functional and cellular loss in Ndufs4 KO mice, we analyzed global gene expression in retina by RNAseq at P22 (data not shown) and P33, respectively. At P22, only a handful of significant genes showed differences, including Gfap and Nes, which are associated with activation of astrocytes. In contrast, at P33, several hundred genes were significantly induced, and innate immunity and inflammation genes were dominant. For example, in Table 1A (RNAseq of P33 KOs versus wild-types), the top five out of five most significant KEGG pathways were all related to inflammation/immunity. Similarly, in P33 retinas from KO versus wild-type mice that had been subjected to ERG, 9 out of 13 of the most significantly induced pathways are inflammatory/immune pathways, and the response is even stronger, presumably because the ERG procedure induced a stronger response (Table 1B). In addition, ingenuity pathway analysis provided support for a network of inflammatory/immune genes centered on Cxcl10 (Fig. 3). Thus, complex I deficiency in the retina induces immune and inflammatory transcripts that are further aggravated by the somewhat invasive technique of ERG.
Table 1.
KEGG pathways from DAVID analysis of RNAseq data from (A) P31 Ndufs4 KO retina with no prior testing or treatment and (B) P33 Ndufs4 KO retina after ERG testing
| RNAseq pathway analysis | ||
|---|---|---|
| KEGG pathway | Gene count | P-value |
| (A) | ||
| Systemic lupus erythematosus | 4 | 7.70E−03 |
| RIG-I-like receptor signaling pathway | 3 | 3.00E−02 |
| Complement and coagulation cascades | 3 | 3.60E−02 |
| Antigen processing and presentation | 3 | 5.10E−02 |
| Toll-like receptor signaling pathway | 3 | 5.90E−02 |
| (B) | ||
| Antigen processing and presentation | 8 | 1.50E−05 |
| Allograft rejection | 6 | 1.60E−04 |
| Graft-versus-host disease | 6 | 1.60E−04 |
| Viral myocarditis | 7 | 1.80E−04 |
| Toll-like receptor signaling pathway | 7 | 2.40E−04 |
| Type I diabetes mellitus | 6 | 2.40E−04 |
| RIG-I-like receptor signaling pathway | 6 | 3.40E−04 |
| Cell adhesion molecules (CAMs) | 8 | 4.20E−04 |
| Autoimmune thyroid disease | 6 | 4.40E−04 |
| Cytosolic DNA-sensing pathway | 5 | 1.50E−03 |
| Chemokine signaling pathway | 7 | 5.70E−03 |
| Fc gamma R-mediated phagocytosis | 5 | 1.20E−02 |
| Systemic lupus erythematosus | 5 | 1.40E−02 |
| Natural killer cell mediated cytotoxicity | 5 | 2.40E−02 |
Pathways consistent in both RNAseq experiments are bolded.
Figure 3.
Ingenuity pathway analysis of significantly upregulated genes that appeared in RNAseq analysis of both P33 Ndufs4 mice that underwent ERG and Ndufs4 mice that received no treatment.
Experimentally induced inflammation further decreases RGC function in mutant mice
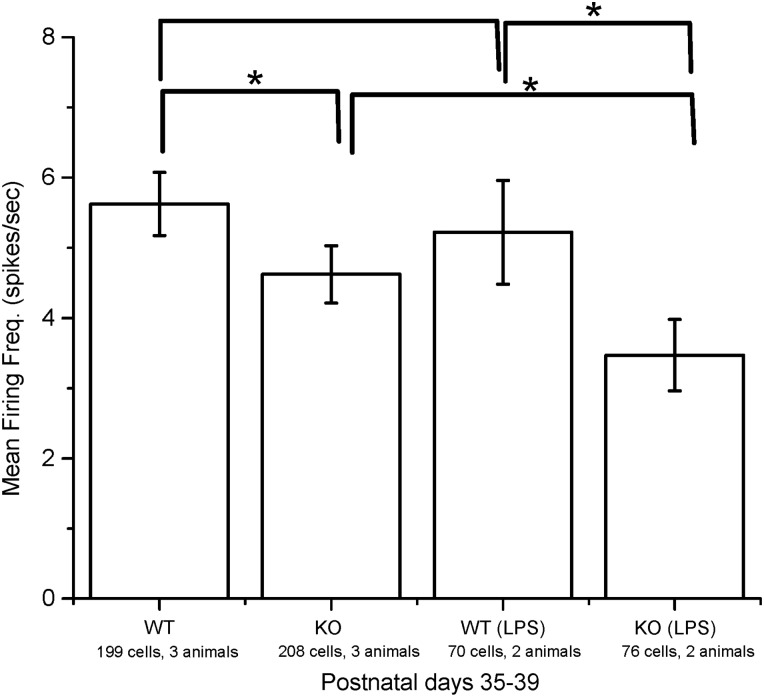
If increased inflammatory activity observed by RNAseq was the basis for decreased visual function observed by MEA, a further aggravation of inflammatory activity would be predicted to further decrease RGC function. LPS is a commonly used inflammogen known to induce microglial activation (11). Ndufs4 KO mice were treated with a 0.5 mg/kg intraperitoneal injection of LPS, and RGC function was measured by MEA the next day. MEA recordings demonstrated that LPS-treated mutants had worse RGC function than LPS-treated wild-type animals (P = 0.0399) as well as mutant animals not exposed to LPS (P = 0.0497), consistent with the idea that complex I defects induces inflammation which is responsible for RGC functional deficits (Fig. 4).
Figure 4.
MEA recording of Ndufs4 KO and wild-type mice with and without LPS treatment. Statistical significance determined by Kruskal–Wallis ANOVA test. *P < 0.05.
Induction of innate immunity, microglial and astroglial markers precedes visual loss in Ndufs4 mice
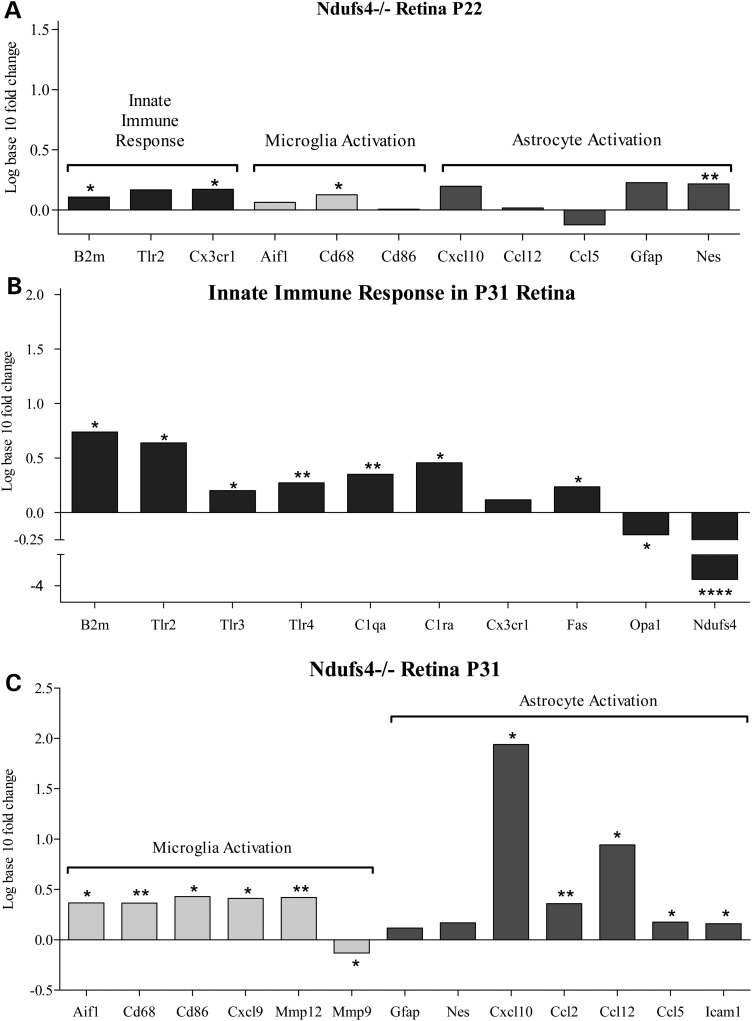
Since significant functional loss occurs between P20 and P30, these time points were investigated in more detail with respect to the prominent immune markers by QRT-PCR (Fig. 5). Just as in the RNAseq, there were very few significant differences between KO and wild-type animals at P22 (Fig. 5A). However, increases in the expression of genes associated with innate immunity (B2M; P = 0.0220 and Cx3cr1; P = 0.0334), microglial activation (Cd68; P = 0.0198) and astroglial activation (Nes; P = 0.0011) were observed (Fig. 5A). Thus, small increases in innate immunity, microglial and astroglial markers precede visual functional loss.
Figure 5.
(A) QRT-PCR of Ndufs4 KO retina at P22 suggests early onset of microglia activation, astrocyte activation and cell-mediated immune response genes. (B) QRT-PCR of Ndufs4 KO retina at P31 suggests further increased genetic markers for microglia and astrocyte activation along with (C) an increase in cell-mediated immune response genes. Statistical significance determined by Student's t-test. *P < 0.05, **P < 0.01 ***P < 0.001, ****P < 0.0001.
Strong induction of innate immunity genes, as well as microglial and astroglial markers coincides with visual functional loss in Ndufs4 mice
When retinas from P33 were examined, i.e. the time of visual functional loss, a much more robust set of gene expression changes associated with innate immunity, microglial and astroglial activation was observed between Ndufs4 KO and littermate wild-type controls (Fig. 5B). The innate immunity markers induced were B2m (fold change = 5.48, P = 0.0091), Tlr2 (fold change = 4.36, P = 0.0037), Tlr3 (P = 0.0161), C1qa (P = 0.0077), C1ra (P = 0.0431), Cx3cr1 (P = 0.1433) and Fas (P = 0.0426).
In terms of astroglial markers at P33, the chemokines, Cxcl10 (fold change = 87.02; P = 0.0055), Ccl2 (P = 0.0023) and Ccl5 (P = 0.0487) are significantly overexpressed and are associated with activated astrocytes, suggesting that the complex I-dependent immune response involves astrogliosis (12). Induction of the adhesion molecule, Icam1 (P = 0.0266), is also consistent with activation of astrocytes (12).
Increases in microglial markers at P33 included Cd68 (P = 0.0028), Cd86 (P = 0.0179), Aif1 (P = 0.0498) and Mmp12 (P = 0.0044), observed along with a significant decrease in Mmp9 (P = 0.0338) (Fig. 5B).
Rapamycin inhibits induction of inflammatory genes
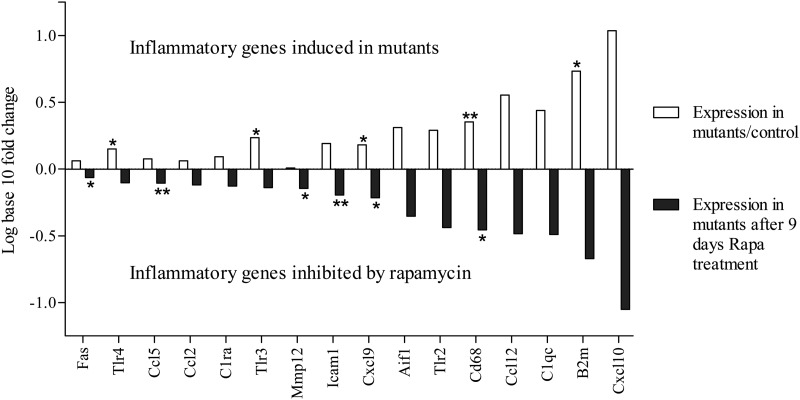
In the rapamycin treatment experiment, the Ndufs4 KO mice showed elevated inflammatory markers compared with wild-type control as expected. After 9 days of rapamycin treatment, Ndufs4 KO mice exhibited an inhibition of inflammatory gene induction with gene expression levels similar to wild-type levels (Fig. 6). These data suggest that complex I deficiency triggers the inflammatory response, that is repressible by a known immunosuppressant that has been shown to extend lifespan in this Ndufs4 KO model.
Figure 6.
Experimental reversal of inflammatory gene induction by rapamycin. White bars, a mean increase in transcript level of 16/16 inflammatory markers was observed by QRT-PCR in retinas of Ndufs4 KO mice compared with wild-type. Black bars, a uniform inhibition of mean amplitude of induction of inflammatory genes was observed in 16/16 transcripts in retinas of mice dosed intraperitoneally with rapamycin (8 mg/kg) for 9 days compared with vehicle-injected Ndufs4 KO mice. Statistical significance determined by Student's t-test. *P < 0.05, **P < 0.01.
Confirmation of inflammatory response at the protein level
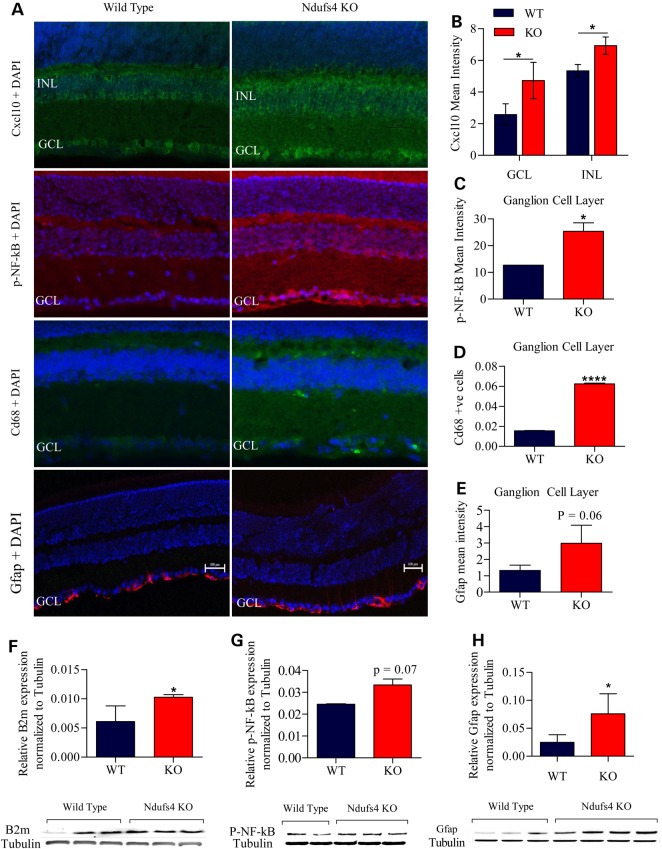
Immunofluorescent staining and western blot of Ndufs4 KO and wild-type retinas at P25–P30 provide supporting evidence for: activation of astrocytes through Cxcl10 and Gfap expression (Fig. 7B, E and H); microgliosis through Cd68 (Fig. 7D) and Iba 1 (Fig. 9) expression; along with innate immune and inflammatory proteins, such as B2m (Fig. 7F) and phosphorylated NF-kB (Fig. 7C and G), respectively. This lends further support to the idea that astrocytes and microglia are activated in the retinal ganglion cell layer at the time of reduced retinal function and RGC loss.
Figure 7.
Confirmation of astrocyte activation, microgliosis and inflammation through protein expression. (A) Immunofluorescent staining of Ndufs4 KO and wild-type retina at P30 of Cxcl10, p-NF-kB, Cd68 and Gfap along with DAPI. (B) Bar graph of Cxcl10 mean staining intensity of the GCL and INL minus background mean intensity. (C) Bar graph of p-NF-kB mean staining intensity of the GCL minus background mean intensity. (D) Bar graph of Cd68 positive cells normalized to total DAPI cell count. (E) Bar graph of Gfap mean staining intensity of the GCL minus background mean intensity. (F) Graphical western blot analysis of B2m normalized to tubulin with western blot image below. (G) Graphical western blot analysis of p-NF-kB normalized to tubulin with western blot image below. (H) Graphical western blot analysis of Gfap normalized to tubulin with western blot image below. Statistical significance determined by Student's t-test. *P < 0.05, ****P < 0.0001. Scale bar = 100 mm.
Figure 9.
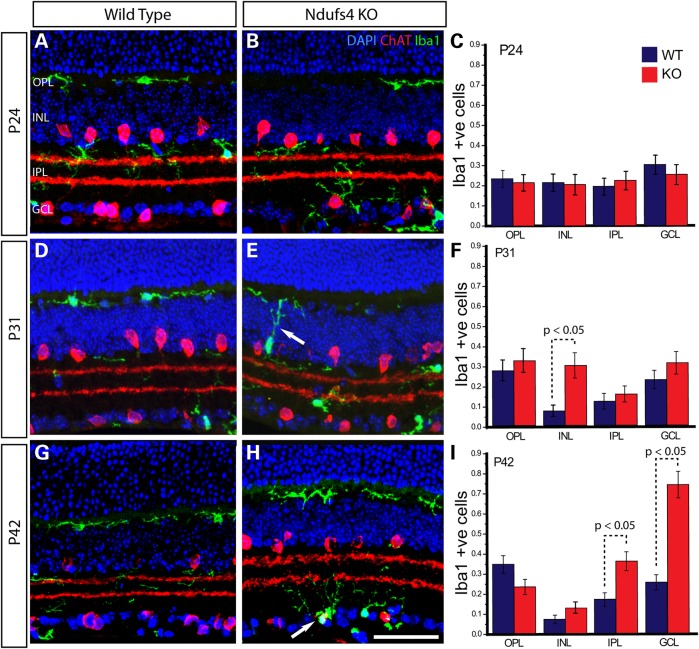
Representative images and graphical representation of counts within the retina from P24 (A–C), P31 (D–F) and P42 (G–I) depicting the migration of Iba1 positive microglia in Ndufs4 KO retina over time. Immunofluorescently labeled retina from wild-type (A, D and G) and littermate Ndufs4 KO (B, E and H) for Iba1 (green) a marker of microglia and ChAT (red) a starburst amacrine cell marker. DAPI (blue) stained nuclei identify lamina within the retina. Bar graphs (C, F and I) showing cell counts of Iba1 positive microglia with a nucleus in defined lamina of the retina at P24 (C), P31 (F) and P42 (I). Statistical comparisons were performed using 2-tailed Student’s t-test. Scale bar 50 mm.
Confirmation of microglia activation by immunofluorescent staining in KO mice at time of vision loss
The microglial activation suggested by increased Aif1 and Cd68 gene expression at P33 was assessed at the cellular level by immunofluorescent labeling of allograft inflammatory factor 1 (Aif1; aka Iba1) (Fig. 9), which is specifically localized in microglia throughout the brain and in retina (13). At P24, prior to onset of functional and cellular changes within the RGC layer, we found no difference between wild-type and Ndufs4 KO mice in the number of Iba1 positive cells in any of the retinal sublamina (Fig. 9A–C). Roughly equal numbers of cells were observed in all layers. At P31, however, the number of Iba1 positive cells was significantly increased within the INL of Ndufs4 KO animals (Fig. 9D–F). An increase in Iba1-positive cells was also observed at P42, but not within the INL. Instead, Iba1 was significantly upregulated in the IPL and the GCL (Fig. 9G–I). The progressive nature of increased Iba1 positive cells from the INL to the IPL and RGC layer over time suggests there could be a migration of microglia from outer to inner retina. Indeed, we observed Iba1 positive cells at P31 that seemed to have neurites branching across the INL in a potential migratory pattern (Fig. 9E, arrow). Additionally, at P42, Iba1 positive cells were observed to be colabeled with ChAT, a specific marker of SBACs (Fig. 9H, arrow) suggesting they were enveloping starburst amacrine cells. Taken together, the rise of Iba1 expression in Ndufs4 retinas is consistent with an inflammatory response initiated by damaged SBACs resulting in the migration of Iba1-positive activated microglia that remove dying/dead amacrine cells.
Starburst amacrine cell death also occurs as early as 24 days in KO mice
As noted earlier, there was a significant loss of cells in the RGC layer determined by DAPI staining that preceded cell loss measured with the more specific RGC associated factor Brn3a (Fig. 2). This suggests another cell type within the RGC layer that may be more vulnerable to complex I deficiency, the loss of which may induce microglial and astroglial activation and inflammation. After RGCs, displaced amacrine cells are the second most numerous cell type within the RGC layer (14). To address potential loss of amacrine cells, we examined immunoreactivity for ChAT, which is critical for synthesis of the transmitter acetylcholine and expressed exclusively by starburst amacrine cells (SBACs) (Fig. 8A and B). The number of SBACs stained by anti-ChAT antibody in the ON layer (RGC layer) was significantly decreased at P24, P31 and P42 in the Ndufs4 KO retina (Fig. 8C). In contrast, SBACs in the OFF layer (Inner nuclear layer) only showed a significant decrease in the P24 Ndufs4 KO retina (Fig. 8D). To determine whether this reduction in cell number was specific to SBACs, we examined immunoreactivity for Gad67, a general marker for all GABA-ergic amacrine cells of which SBACs are a subset (15) (Fig. 8E–H). A significant loss of Gad67 staining was observed in Ndufs4 retinas, but only at P42, (Fig. 8I). These data suggest that starburst amacrine cells are also sensitive to complex I depletion.
Figure 8.
Starburst Amacrine cell loss in Ndufs4 KO retinas. (A and B) Representative images of immunofluorescent labeling for ChAT (red), a marker of starburst Amacrine cells in transverse retinal sections from P42 wild-type (A) and Ndufs4 KO (B) mice. The two strong bands of ChAT immunolabeling within the IPL correspond to terminal arbors originating from OFF starburst amacrine cells residing in the innermost region of the INL or ON starburst amacrine cells residing in the GCL (so-called displaced amacrine cells). DAPI cell staining (blue) highlights cell lamina within the retina. Arrow depicts loss of synaptic processes in the ON layer of ChAT labeled amacrine cells. (C and D) Bar graphs showing cell counts of ChAT labeled neurons within the GCL (C, ON) or INL (D, OFF) in wild-type or Ndufs4 KO mice at P24, P31 and P42. (E–H) Representative images of immunolabeling for GAD67 (green) in transverse retinal sections from P42 wild-type and littermate Ndufs4 KO mice. (F and H) Overlaid images of immunolabeling for ChAT (red) and cell staining for DAPI (blue) on the same sections demarcate retinal lamina as well as the starburst amacrine cell terminal zones in the IPL. (I) bar graph showing normalized intensity levels of GAD67 immunoreactivity within the IPL of wild-type and Ndufs4 KO mice at P16, P24 and P42. Scale bar = 50 mm. Statistical significance determined by Student's t-test.
Discussion
Ndufs4 KOs as a model of mitochondrial complex I deficiency in the retina
We observed that Complex I deficiency in the Ndufs4 mouse induces decreased visual function by ERG and MEA functional tests, and analyzed transcriptomics, protein expression and neuroanatomy to clarify the mechanism of visual loss. We clearly observe specific RGC functional defects at P32 by MEA, cell loss in the ganglion cell layer at P31, and Brn3a-specific RGC cell loss at P42. These functional defects are preceded by an early (P21) increase in innate immunity, microglial and astroglial transcripts, which expands to a major increase in these transcripts and protein at P32, the time of visual functional loss. Further incitement of inflammation with LPS produces further decrement of RGC function, as measured by MEA, suggesting that an immune/inflammatory response is critical in the pathomechanism connecting complex I deficiency to vision loss.
Chemokine expression and astrogliosis
Chemokines mediate the recruitment and activation of leukocytes and other cells to the site of inflammation during an immune response. The chemokines Cxcl10 and Ccl2 have been reported to be secreted by activated astrocytes (12,16). Cxcl10 transcript was increased >80-fold in Ndufs4 KO retina (Fig. 5C), and Cxcl10 protein expression strongly increased in the ganglion cells layer and inner nuclear layer (Fig. 7B). The increase in Icam-1 is a further marker of astrogliosis, as well as the rise in astrocyte-specific GFAP (12). These data indicate that reactive astrocytes are playing a role in inflammation downstream of complex I deficiency.
Microglial activation in retinas of Ndufs4 KO mice
Microglial activation is also apparent at P33 in KO retinas as seen from genetic increases in Aif1, Cd68, Cd86, Cxcl9 and Mmp12 expression (Fig. 5C). Immunohistochemical data also support an increase in Iba1 (Fig. 9), which is the protein encoded by the gene Aif1, and Cd68, another marker of microglial activation (Fig. 7D). Over-activated microglia has been shown to have detrimental neurotoxic effects due to production of superoxide, nitric oxide and tumor necrosis factor-α (TNF-α), which suggests that microglia may have a causative role in neuroinflammation (17). LPS is only neurotoxic in the presence of microglia (17), thus the differentially larger defect in MEA in Ndufs4 KO mice (Fig. 4) further supports microglial involvement in their functional visual loss. Furthermore Tlr4 is the primary receptor for LPS (11), and Tlr4 is overexpressed in Ndufs4 KO retinas at P31 (Fig. 5B).
Innate immune response
Further, we observe a strong induction of an innate immune response as seen with increases in transcripts of MHC Class 1 molecule B2m, as well as Tlr2, Tlr3, Tlr4, Icam-1 and Fas in the Ndufs4 KO retina. Binding of Fas ligand or TNF-α to Fas receptor leads to apoptosis. Fas ligand is expressed on activated T cells and natural killer cells and is mainly expressed in immune privileged sites, such as the eye (18). The higher level of transcripts associated with innate immunity at P22 is consistent with the pathomechanism described in Figure 10.
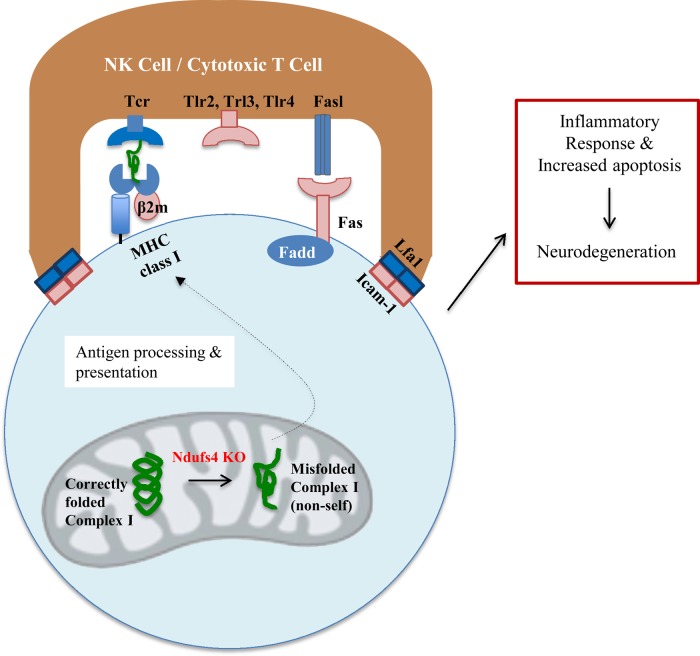
Figure 10.
A potential mechanism of Ndufs4-mediated neurodegeneration. Ndufs4 deficiency causes misfolding of complex I, uncovering novel antigens, which are presented at an increased rate through the immune system through increased β-2-microglobulin expression, and the consequences of NK cell activation are further amplified through increased Fas receptor, Tlr2, Tlr3 and Tlr4 expression. Gene products in pink/red are overexpressed in Ndufs4 retinas.
Potential mechanisms for complex I-dependent inflammation in Ndufs4 KO mice
The finding of a massive increase in immune and inflammatory genes coincident with vision loss in KO mice suggests that inflammation is an intermediate between complex I deficiency and vision loss. But how complex I deficiency increases inflammation is less clear. One possible mechanism (Mechanism 1) is that loss of Ndufs4, a major assembly-factor protein for complex I (19), leads to misfolding and/or aggregation of complex I, which is recognized by the host as ‘non-self’, activating an inflammatory response (Fig. 10). The misfolded complex I protein is processed and presented as an antigen by MHC class I to MHC class I receptors on T lymphocytes or natural killer cells (20). Binding of MHC class I receptors would initiate pathways for apoptosis of the target cell and cytokine signaling to other cells.
Consistent with this mechanism 1 we propose in the eye, Jin et al. (21) recently reported that complex I deficiency leads to a shift from fatty acid oxidation to glycolysis, which causes an induction of inflammation by the accumulation of fatty acids and lactate. They created a Ndufs4/TLR2/TLR4 triple KO mouse and found that TLR2/TLR4 deletion dampened palmitic acid-stimulated inflammatory gene expression and ROS production, implicating TLR2/TLR4 in complex I deficiency-mediated inflammation.
A second mechanism (Mechanism 2) is that complex I deficiency causes a severe bioenergetic defect selectively in the Starburst Amacrine Cells that start dying at P24 necrotically, inciting a microglial, astrocytic and innate immune response activation, which further damages retinal ganglion cells. This second mechanism is also a completely novel explanation of complex I-dependent vision loss and RGC death.
Both mechanisms invoke a strong innate immune response as an intermediate between complex I deficiency and cell death. Thus, drugs known to block the innate immune response we expect to be protective. Recently, it was shown that the lifespan and neurological damage of Ndufs4 KO mice could be rescued by rapamycin, a potent MHC/innate immunity response immunosuppressant that is used in transplantation (22), without alteration of mitochondrial function. We find similarly that rapamycin uniformly suppresses the overexpression of every inflammatory gene elevated in Ndufs4 KO retina (Fig. 6). We interpret that rapamycin rescues from neurodegeneration because it suppresses inflammation that is downstream of the mitochondrial dysfunction and upstream of visual loss.
Summary and prospects
The Ndufs4 KO mouse is a reasonable model of complex I-dependent decline in RGC function and RGC death, as occurs in Leber's hereditary Optic Neuropathy, LHON. Our data supports the mechanism complex I deficiency → immune/inflammatory response → RGC functional defects and death. Both mechanisms provide novel insight into the pathophysiology of complex I deficient vision loss. Our data suggest rational therapy, including rapamycin and other immunosuppressants in the context of LHON and other mitochondrial blindness.
Materials and Methods
Animals
All experiments were performed in accordance with the National Institutes of Health and institutional guidelines regarding animal use and were approved by the Animal Care and Use Committee of the University of California, Davis (IACUC). Animals were housed in IACUC approved animal facilities under controlled environmental conditions. Ndufs4 KO mice were provided by Richard Palmiter (University of Washington, Seattle, USA). Phenotypically, these mice appear smaller than wild-type and these mice were characterized as having visual deficits at P30 as measured by Morris water maze, visual cliff test and visual placing (23). Mice were sacrificed at P16, P22, P24, P25, P31, P32, P33, P34, P35, P37, P42 and P45. Eighty-four mice were used in the current study; a detailed description of the number of KO and wild-type mice, their age and sex is provided below when describing each experiment. Detailed methods of tissue fixation or storage can be found in Supplementary Material online.
LPS injection
Two Ndufs4 KO mice and two littermate wild-type mice received intraperitoneal injection of LPS (0.5 mg/kg). As a comparison, one Ndufs4 KO and one wild-type mouse received intraperitoneal PBS injection as vehicle controls. Mice were within the age of P35–P39. The retinas were removed 12 h after LPS injection and used for MEA as described in what follows.
Electroretinogram
Eight P33 (four Ndufs4 KO and four littermate control with an equal distribution of male and female mice in each group) mice underwent ERG testing in both eyes (UTAS-EPIC XL; LKC Technologies, Gaitersburg, MD, USA). The mice were dark-adapted overnight and were anesthetized with Nembutal (50 mg/kg dose). The body temperature of the mice was maintained at 37°C with a heating unit placed under the mouse. Drops of 1% tropicamide and 2.5% phenylephrine were administered in each eye for dilation. Proparacaine (0.5%) eye drops were applied for topical anesthesia. The eyes were lubricated with 1% methylcellulose. ERG recordings were performed with a mouse gold wire/contact electrode and 30 gauge needle reference electrodes (LKC Technologies, Gaitersburg, MD, USA). Needle electrodes were inserted under the skin on each side of the cheek as the references and at the base of the tail as the ground. ERGs were generated with the following program: scotopic blue filter (0 dB) at 20 μV/div single flash; scotopic white (0 dB) at 50 μV/div single flash; photopic white (0 dB) 10 μV/div single flash; and photopic white (0 dB) 20 μV/div flicker, average of 10. Five responses evoked by light were averaged in response to each luminance step. After recording, the animals were recovered and sacrificed the next day.
Multi-electrode array recordings
MEA recordings were performed as previously described (24,25) on Ndusfs4 KO and wild-type mice (18 mice total) at: P16 (two KO; two wild-type), P25 (two KO; two wild-type), P32 (one KO; one wild-type), P35 (one KO; one wild-type), P37 (two KO; two wild-type) and P45 (one KO; one wild-type). Mice were euthanized with a lethal dose of pentobarbital (0.1–0.2 ml) via intraperitoneal injection. The eyes were enucleated and the retinas were removed and stored in buffered and oxygenated media [Eagle's minimum essential medium (MEME), M7278; Sigma-Aldrich, St. Louis, MO, USA] at room temperature. A piece of retina was placed ganglion side down onto a 60-channel MEA (Multi-Channel Systems, Tubingen, Germany), and held in place with a piece of dialysis membrane (Spectrapore 132130; Spectrum, Los Angeles, CA, USA). The tissue was superfused with buffered MEME at 1–2 ml/min at 37°C. The array electrodes were 30 µm in diameter and arranged in an 8 × 8 rectilinear grid with an interelectrode spacing of 200 µm. At this distance, the signal for a given cell appeared on only one electrode, so each cell isolated was assigned the spatial coordinates of the electrode that recorded its signal. Analog data were acquired at 20 kHz per channel simultaneously from each electrode. After the retina was set up on the MEA, the tissue was allowed to acclimate for 5–20 min. Recordings were performed for 15–20 min during which time overall firing rates appeared stable. MEA spike identification is described in the Supplemental Material online. Data were analyzed by applying the Kruskal–Wallis ANOVA test with significance set at P < 0.05.
Rapamycin treatment
Rapamycin was dissolved in DMSO to 100 mg/ml. This was diluted in 5% PEG-400/5% Tween-20 (vehicle) to a concentration of 0.6 mg/ml, sterile filtered and stored at −80 for long-term storage. Rapamycin treated mice were injected with 132 ml/10 g body weight for a final dose of 8.0 mg/kg (22). Vehicle mice were injected with equal volume of PBS and DMSO lacking rapamycin. Injections were performed intraperitoneally using a 30 gauge needle. Two Ndufs4 KO and two wild-type mice received rapamycin treatment and two Ndufs4 KO and two wild-type mice received vehicle injections. Each animal was daily starting from P22 to P31. At P31, the mice were sacrificed and retinas were surgically removed and placed in RNALater and further processed for QRT-PCR as described in what follows.
Gene expression
Total RNA was extracted from whole retinas using affinity column purification (Qiagen, Valencia, CA, USA) and processed by the UC Davis DNA Technologies & Expression Analysis Core for RNAseq. Remaining total RNA was used to generate cDNA template (Bio-Rad, Hercules, CA, USA) for validation of candidate genes by QRT-PCR. RNAseq was done on three separate occasions (22 mice total), first with the eight P34 mice (four Ndufs4 KO and four wild-type with an equal distribution of male and female mice in each group) that underwent ERG testing, six P33 mice [three Ndufs4 KO (one male and two female) and three wild-type (one male and two female)] with no prior testing done and eight P22 mice [four Ndufs4 KO (three male and one female) and four wild-type (one male and three female)] with no prior testing. Detailed RNAseq run and analysis procedures can be found in the Supplemental Material online.
QRT-PCR was performed with the Roche LightCycler® 480 System (Roche, Indianapolis, IN, USA) using select gene primers that were chosen based on the results of the RNAseq data (Table 1). Tissues tested include the P22 retina used for RNAseq and eight additional P31 retinas (four KO and four wild-type; equal sex distribution in each group). Primers were designed using the NCBI database and created by Integrated DNA Technologies (IDT; Coralville, IA, USA). Primers were optimized by running QRT-PCR and validated by inspecting the length of the product using gel electrophoresis and inspecting the melt curve. The reference genes used in this experiment were Mapk1, Gapdh and Actb (26). A list of primers can be found in Supplementary Material, Table S1. The change in PCR products as the reaction proceeded was monitored by SYBR green dye (Invitrogen, Grand Island, NY, USA). Sample volume was 10 µl and samples were run in technical triplicates. Quantification cycle (Cq) values obtained by the Lightcycler were analyzed by normalizing the values to the reference genes and delta Cq values were calculated to determine relative gene expression values compared with wild-type. Delta delta Cq was also calculated to determine the fold change difference between Ndufs4 knockout and wild-type. Significance, P < 0.05 was determined using two sample independent Student's t-test statistics with equal sample size and assuming equal variance.
Multiple immunofluorescent labeling
All antibodies used in this current study are listed in Supplementary Material, Table S2. Immunohistochemical labeling was carried out using the indirect fluorescence method. Controls included no primary, and no primary or secondary for each antibody used. Sections were incubated in a blocking buffer containing 10% normal donkey serum, 2% Bovine serum albumen (BSA) and 0.5% Triton-X-100 in phosphate buffer saline (PBS, pH 7.2) for 1 h at room temperature. Sections were incubated overnight at 4°C in blocking buffer containing primary antibodies. After briefly washing in PBS, secondary antibodies diluted in PBS were applied for 1 h at room temperature. Secondary antibodies were conjugated to Alexa 488 or Alexa 594 (1:500; Molecular Probes, Eugene, OR, USA). Finally, sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1:500; Kirkegaard & Perry Laboratories, Gaithersburg, MD, USA) for 1 min. Sections were coverslipped using either Vectashield (Vector Laboratories, Burlingame, CA, USA) mounting media or Prolong Gold Antifade reagent (Life Technologies, Grand Island, NY, USA). Fluorescent images were captured using the Nikon Eclipse E1000 (Melville, NY, USA) and Olympus FV500 (Tokyo, Japan) confocal microscopes (found in Supplementary Material online).
Cell counts and immunofluorescence quantification
Quantification of cell number and/or immunofluorescence labeling intensity was obtained from four animals (two KO and two wild-type; 16 total) at the following ages; P16, P25, P31 and P42. A mix of both male and female (75% male, 25% female) mice was used for analyses and we observed no obvious differences in cell number or immunofluorescence between the sexes. Because thickness and cell number varies across the retina (14), sections were chosen for immunohistochemistry and cell counts from regions evenly spaced ∼150–200 µm apart so as to sample regions from central retina out to the periphery. Conventional inclusion/exclusion criteria were followed (27); any cell touching the left line of the grid was included in the count, any cell touching the right line of the grid was excluded. To be included in the counts immunoreactive cells had to have a nucleus as determined by DAPI staining. Equal numbers of images per age group and condition were analyzed. Values across each section from nasal to temporal portions of the retina were pooled together and included in the averages for each condition. All measures were obtained exclusively from the dorsal half of each retina and this process was identical for each eye examined. Observers were blind to genotype.
For ChAT immunolabeled sections, ∼450–850 cells, depending on the age and/or genotype, were counted for each condition. These counts were collected from 7 to 10 images collected from 7 to 10 evenly spaced sections. Using Neurolucida software, (MicroBrightfield, VT, USA), a grid of 150 µm2 was placed in the center of each image and cell counts were made within this region. The same procedure was followed for Iba1 cell counts (between 60 and 200 cells depending on age and/or genotype) using collapsed confocal stack images.
Quantification of Brn3a cell number and GAD67 immunofluorescence levels was done using ImageJ software (28,29). For Brn3a, ∼700–1000 cells, depending on age and/or genotype, were counted for each condition. These counts were collected from 3 to 4 images from 8 to 10 sections. Immunopositive cells were counted along a 150–200 µm region from the central portion of each image of the retinal ganglion cell layer. ChAT positive cells that colocalized Brn3a were removed from the counts. For GAD67 labeling, since no discernable somal labeling was detected, we measured the intensity of immunofluorescence within the IPL where the dendrites and terminals of amacrine cells reside (30). To account for potential variability in immunolabeling between samples not related to genotype, we normalized IPL values against non-specific background label overlying the outer segments of photoreceptor cells. Measures were made from a similar region and number of sections as those for Brn3a.
Statistical analyses of cell number and immunofluorescence were made in either Origin (OriginLab, Northampton, MA, USA) or Excel software (Microsoft Corp., Redmond, WA, USA). Significance, P < 0.05 was determined using two sample independent Student's t-test statistics with equal sample size and assuming equal variance.
Western blot analysis
Retinas were homogenized in ×2 lysis buffer (Cell Signaling Technology, Danvers, MA, USA) and 20 µg of protein per sample were analyzed in 4–12% NuPAGE Bis-Tris gel (Novex, Grand Island, NY, USA). After electrophoresis, proteins were transferred to nitrocellulose membranes using an Iblot dry blotting system (Invitrogen). The membranes were blocked with LI-COR Odyssey (Lincoln, NE, USA) block buffer, and subsequently incubated with anti-GFAP (Cell Signaling) and anti-Tubulin (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA) antibodies overnight. The LI-COR Odyssey scanner was used for western blot detection with goat-anti mouse IRDye 680RD and goat-anti rabbit IRDye 800CW secondary antibodies (LI-COR). Image studio (LI-COR) was used to analyze western blot results.
Intensity measurements and quantification of Gfap were made using three wild-type and three Ndufs4 KO mice. Using Nikon NIS-elements imaging software, three images of one section of each animal retina were analyzed. Gfap mean staining intensity of the GCL was subtracted from the background mean intensity.
Supplementary Material
Funding
This work was supported by the National Eye Institute (grant number P30 EY0 12576, R01 EY0 12245).
Supplementary Material
Acknowledgements
We thank Richard Palmiter for providing Ndufs4 mice, the UC Davis DNA Technologies Core for performing RNAseq and the UC Davis Bioinformatics Core for processing the RNAseq data.
Conflict of Interest statement. None declared.
References
- 1.Koene S., Willems P.H., Roestenbery P., Koopman W.J., Smeitink J.A. (2011) Mouse models for nuclear DNA-encoded mitochondrial complex I deficiency. J. Inherit. Metab. Dis., 34, 293–307. [DOI] [PubMed] [Google Scholar]
- 2.Tucker E.J., Compton A.G., Calvo S.E., Thorburn D.R. (2011) The molecular basis of human complex I deficiency. IUBMB Life, 63, 669–677. [DOI] [PubMed] [Google Scholar]
- 3.Carelli V., Ross-Cisneros F.N., Sadun A.A. (2002) Optic nerve degeneration and mitochondrial dysfunction: genetic and acquired optic neuropathies. Neurochem. Int., 40, 573–584. [DOI] [PubMed] [Google Scholar]
- 4.Zanna C., Ghelli A., Porcelli A.M., Karbowski M., Youle R.J., Schimpf S., Wissinger B., Pinti M., Cossarizza A., Vidoni S., et al. (2008) OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain, 131, 352–367. [DOI] [PubMed] [Google Scholar]
- 5.Swarnkar S., Goswami P., Kamat P.K., Patro I.K., Singh S., Nath C. (2013) Rotenone-induced neurotoxicity in rat brain areas: a study on neuronal and neuronal supportive cells. Neuroscience, 230, 172–183. [DOI] [PubMed] [Google Scholar]
- 6.Heitz F.D., Erb M., Anklin C., Robay D., Pernet V., Gueven N. (2012) Idebenone protects against retinal damage and loss of vision in a mouse model of Leber's hereditary optic neuropathy. PLoS One, 7, e45182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteve-Rudd J., Fernandez-Sanchez L., Lax P., De Juan E., Martin-Nieto J., Cuenca N. (2011) Rotenone induces degeneration of photoreceptors and impairs the dopaminergic system in the rat retina. Neurobiol. Dis., 44, 102–115. [DOI] [PubMed] [Google Scholar]
- 8.Kruse S.E., Watt W.C., Marcinek D.J., Kapur R.P., Schenkman K.A., Palmiter R.D. (2008) Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab., 7, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang M., Zhou L., Macke J.P., Yoshioka T., Hendry S.H., Eddy R.L., Shows T.B., Nathans J. (1995) The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J. Neurosci., 15, 4762–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quina L.A., Pak W., Lanier J., Banwait P., Gratwick K., Liu Y., Velasquez T., O'Leary D.D., Goulding M., Turner E.E. (2005) Brn3a-expressing retinal ganglion cells project specifically to thalamocortical and collicular visual pathways. J. Neurosci., 25, 11595–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin L., Li G., Qian X., Liu Y., Wu X., Liu B. (2005) Interactive role of the toll-like receptor 4 and reactive oxygen species in LPS-induced microglia activation. Glia, 52, 78–84. [DOI] [PubMed] [Google Scholar]
- 12.Oh L.W., Schwiebert L.M., Benveniste E.N. (1999) Cytokine regulation of CC and CXC chemokine expression by human astrocytes. J. Neurovirol., 5, 82–94. [DOI] [PubMed] [Google Scholar]
- 13.Ito D., Imai Y., Ohsawa K., Nakajima K., Fukuuchi Y., Kohsaka S. (1998) Microglia-specific localisation of a novel calcium binding protein, Iba1. Mol. Brain Res., 57, 1–9. [DOI] [PubMed] [Google Scholar]
- 14.Jeon C.J., Strettoi E., Masland R.H. (1998) The major cell populations of the mouse retina. J. Neurosci., 18, 8936–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voinescu P.E., Kay J.N., Sanes J.R. (2009) Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J. Comp. Neurol., 517, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhowmick S., Duseja R., Das S., Appaiahgiri M.B., Vrati S., Basu A. (2006) Induction of IP-10 (CXCL10) in astrocytes following Japanese encephalitis. Neurosci. Lett., 414, 45–50. [DOI] [PubMed] [Google Scholar]
- 17.Block M.L., Zecca L., Hong J.S. (2007) Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat. Rev. Neurosci., 8, 57–69. [DOI] [PubMed] [Google Scholar]
- 18.Brint E., O'Callaghan G., Houston A. (2013) Life in the Fas lane: differential outcomes of Fas signaling. Cell. Mol. Life Sci., 70, 4085–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvaruso M.A., Willems P., van den Brand M., Valsecchi F., Kruse S., Palmiter R., Smeitink J., Nijtmans L. (2012) Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet., 21, 115–120. [DOI] [PubMed] [Google Scholar]
- 20.Warren H.S., Smyth M.J. (1999) NK cells and apoptosis. Immunol. Cell Biol., 77, 64–75. [DOI] [PubMed] [Google Scholar]
- 21.Jin Z., Wei W., Yang M., Du Y., Wan Y. (2014) Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab., 20, 483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson S.C., Yanos M.E., Kayser E.B., Quintana A., Sangesland M., Castanza A., Uhde L., Hui J., Wall V.Z., Gagnidze A., et al. (2013) mTor inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science, 342, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintana A., Kruse S.E., Kapur R.P., Sanz E., Palmiter R.D. (2010) Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc. Natl Acad. Sci. USA, 107, 10996–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warland D.K., Huberman A.D., Chalupa L.M. (2006) Dynamics of spontaneous activity in the fetal macaque retina during development of retinogeniculate pathways. J. Neurosci., 26, 5190–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun C., Speer C.M., Wang G.Y., Chapman B., Chalupa L.M. (2008) Epibatidine application in vitro blocks retinal waves without silencing all retinal ganglion cell action potentials in developing retina of the mouse and ferret. J. Neurophysiol., 100, 3253–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha-Martins M., Njaine B., Silveira M.S. (2012) Avoiding pitfalls of internal controls: validation of reference genes for analysis by qRT-PCR and western blot throughout rat retinal development. PLoS One, 7, e43028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox D.A., Hamilton W.R., Johnson J.E., Xiao W., Chaney S., Mukherjee S., Miller D.B., O'Callaghan J.P. (2011) Gestational lead exposure selectively decreases retinal dopamine amacrine cells and dopamine content in adult mice . Toxicol. Appl. Pharmacol., 256, 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasband W.S. (1997) ImageJ, U.S. National Institutes of Health, Bethesda, MD, http://imagej.nih.gov/ij/, 1997–2012. [Google Scholar]
- 29.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masland R.H. (2012) The neuronal organization of the retina. Neuron, 76, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.